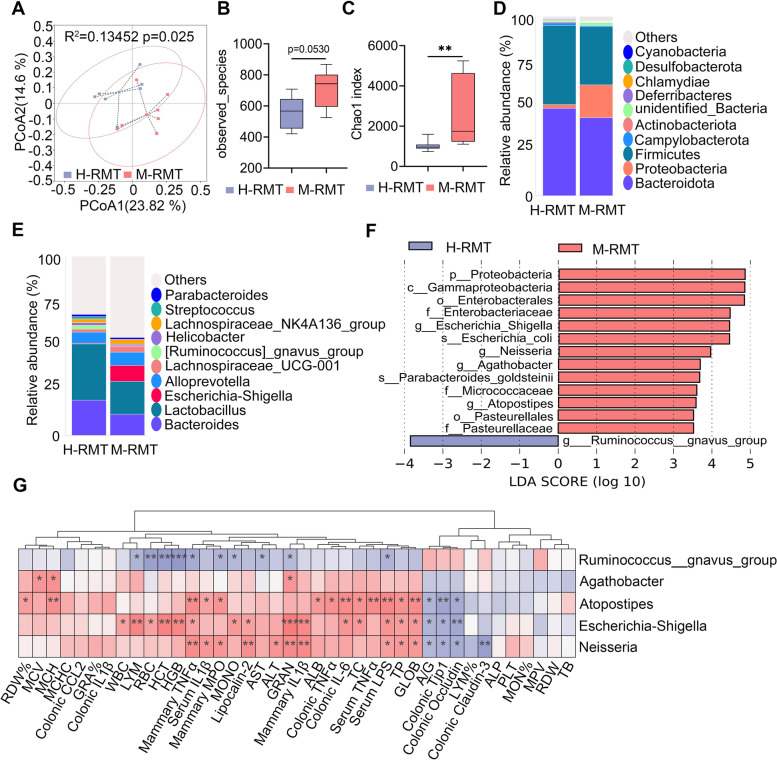
Fig. 6.
M-RMT induces gut dysbiosis in recipient mice. Mice were subjected to RMT as mentioned above, and the composition of the gut microbiota was determined using 16S rRNA sequencing. A PCoA score plots indicating the separation of the H-RMT and M-RMT gut microbiota structure (R2 = 0.13452, P = 0.025) based on the unweighted UniFrac distance (n = 7). B The M-RMT group had increased observed species compared with the H-RMT group (n = 7). C The Chao1 index showed that the M-RMT group had increased alpha diversity compared with the H-RMT group (n = 7). D, E Bacterial composition at the phylum (D) and genus (E) levels from the indicated groups. F LEfSe analysis was performed to determine the different bacterial taxa enriched in the H-RMT and M-RMT groups (log10LDA score > 3.5). J Spearman correlation between the intestinal microbiota and inflammatory parameters from the H-RMT and M-RMT groups. Red denotes a positive correlation, while blue color denotes a negative correlation. The intensity of the color is proportional to the strength of the Spearman correlation. Data are expressed using boxplots (B, C), and the Mann-Whitney U test was performed. **p < 0.01 indicates significance