Abstract
The chlamydiae are obligate intracellular pathogens that occupy a nonacidified vacuole, termed an inclusion, throughout their developmenal cycle. When an epithelial cell is infected with multiple Chlamydia trachomatis elementary bodies, they are internalized by endocytosis into individual phagosomal vacuoles that eventually fuse to form a single inclusion. In the course of large-scale serotyping studies in which fluorescent antibody staining of infected cells was used, a minority of strains that had an alternate inclusion morphology were identified. These variants formed multiple nonfusogenic inclusions in infected cells, with the number of independent inclusions per cell varying directly with the multiplicity of infection. Overall the nonfusogenic phenotype was found in 1.5% (176 of 11,440) of independent isolates. Nonfusing variants were seen in C. trachomatis serovars B, D, D−, E, F, G, H, Ia, J, and K. The nonfusing phenotype persisted through repeated serial passage, and the phenotype was consistent in four mammalian host cell lines. Fluorescence microscopy and immunoblotting with antisera directed at proteins in the C. trachomatis inclusion membrane revealed that one such protein, IncA, was not detected in the inclusion membrane in each tested nonfusogenic strain. The distributions of other chlamydial proteins, including one additional Inc protein, were similar in wild-type and variant strains. The incA coding and upstream regions were amplified and sequenced from the prototype serovar D and two nonfusing serovar D(s) strains. Three nucleotide changes were discovered in the D(s) incA gene, leading to two amino acid changes within the predicted D(s) IncA sequence. These studies demonstrate a subgroup of variant C. trachomatis isolates that form nonfusing inclusions; the variant phenotype is associated with the absence of detectable IncA and with an altered incA sequence that modifies the characteristic hydrophobic domain of the IncA protein.
Many intracellular pathogens develop within distinct vacuoles that do not fuse with lysosomes. Examples include Mycobacterium tuberculosis, a species that resides in a vacuole with similarity to an early endosomal compartment (18), and Legionella pneumophilia, a species that localizes to a vacuole with similarity to the endoplasmic reticulum (24). The obligatory intracellular chlamydiae also occupy a nonacidified vacuole (termed an inclusion) throughout their intracellular growth, but little is known about inclusion structure and development. A single fluorescent lipid marker, NBD-ceramide, transiently resides in the inclusion membrane and stably is found in chlamydiae within the inclusion. This localization led to the proposal that the inclusion is placed within the host cell exocytic pathway, receiving material from the Golgi apparatus in a vesicle-mediated process (5–7).
With the exception of NBD-ceramide, no host cell markers that might be useful in the characterization of the origins of the inclusion have been identified (9). There is, however, a family of chlamydial proteins (termed Inc proteins) that are localized to the inclusion membrane (1, 2, 15). These proteins have been identified in Chlamydia trachomatis and C. psittaci, and an apparent homolog is also encoded within the C. pneumoniae genome sequence (11). It is anticipated that an elucidation of Inc protein function will greatly enhance our understanding of the chlamydial inclusion and its interaction with the host cell. However, this effort is complicated by the lack of amino acid sequence identity of known Incs with other proteins in the global sequence databases. This fact, in combination with the absence of a workable genetic system for directed mutagenesis or deletion of chlamydial genes, makes the functional analysis of these proteins difficult. Mutant strains that do not produce one or more of the Inc proteins would thus be valuable to further our understanding of their role in inclusion development.
One distinctive trait that varies among chlamydial species and strains is the fusogenicity of the developing inclusion. With prototypic C. trachomatis strains, infection of single cells with multiple elementary bodies (EB) results in multiple inclusions, but these inclusions eventually fuse to form a single vacuole (10, 12). This fusion does not occur at 32°C in HeLa cells, indicating that the processes involved are temperature dependent (23). Ridderhof and Barnes (13) demonstrated that the inclusions harboring different serovars of C. trachomatis could fuse during the infectious process, leading to the possibility for genetic exchange between reticulate bodies. In contrast, Matsumoto et al. (12) showed that C. trachomatis serovar L2 and C. psittaci Cal 10 inclusions within the same cell will not fuse with one another. Additionally, many strains of C. psittaci form inclusions that not only are not fusogenic but appear to actively divide during the infectious process (16). It is likely that the collective distinctions among these different inclusion structures is a result of selective differences between protein interactions at the surface of the inclusion, possibly between distinct Inc proteins and host cell mediators of vesicle fusion. The first inclusion membrane protein identified, IncA of C. psittaci, is a serine/threonine phosphoprotein that has contact with the cytoplasm of infected cells and is likely phosphorylated by host cell protein kinases (17). This is a candidate molecule for affecting host cell vesicular trafficking. Genes encoding IncA homologs have been identified in C. trachomatis (1) and C. pneumoniae (2a), and the protein products have been shown to be localized to the inclusion membrane in each species. The overall identity shared by these proteins is relatively low (20 to 22%), but each possesses a characteristic 50- to 70-amino-acid hydrophobic domain. The function of IncA or any other candidate inclusion membrane protein remains unknown.
Chlamydial serotyping has been a powerful tool in the epidemiologic study of chlamydial sexually transmitted infections. The techniques that have been used for serotypic analysis of chlamydial isolates include microimmunofluorescence (25), a solid-phase enzyme-linked immunosorbent assay (3), and a microtiter plate format (22). Our research group uses the latter method to routinely analyze clinical isolates. This method involves culturing chlamydiae in monolayers grown in 96-well microtiter plates and serotyping the developing organisms by fluorescence microscopy with subspecies- and serovar-specific monoclonal antibodies (MAbs). This low-passage technique is sensitive and specific, and it allows the rapid determination of serotype soon after initial isolation of chlamydiae from infected patients.
During our analysis of these clinical isolates, strains with an unusual inclusion morphology were identified. The phenotype manifested as multiple inclusions within infected cells and was observed in approximately 1.5% of all isolates examined. The nonfusing phenotype, designated by the subscript “(s)”, was observed in isolates of each chlamydial serovar, was consistent regardless of the host cell line used, and was stable over many passages. Additionally, these strains produced inclusions that lacked detectable IncA on the membrane while retaining other inclusion membrane proteins, and they demonstrated altered incA nucleotide sequences that modified the characteristic hydrophobic domain of the protein.
(Results of this investigation were presented at the American Society for Microbiology conference “A Cell Biology Approach to Microbial Pathogenesis,” Portland, Oreg., April 1999.)
MATERIALS AND METHODS
Chlamydial culture.
The nonfusing chlamydial phenotype was detected visually by observing inclusion morphology during large-scale serotyping studies in a low-passage microtiter plate format (22). The patient population studied consisted of 7,096 women (cervix) and 4,344 men (urethra) who had a culture-documented C. trachomatis genital infection at any of the Seattle King County Health Department Sexually Transmitted Disease clinics between 1988 and 1996. The collection and isolation techniques have previously been described in detail (26). Briefly, patient swabs were collected and stored in chlamydia transport medium at 4°C and transported within 24 h to the laboratory. Each specimen was inoculated onto McCoy cells, centrifuged at 1,200 × g, aspirated, and overlaid with the growth medium, minimal essential medium with 10% fetal bovine serum and cycloheximide (1.0 μg/ml) added. Cells were incubated at 37°C in 4% CO2 for 48 h and fixed with methanol. Chlamydial inclusions were detected by fluorescence microscopy using genus-specific MAb CF-2 to the lipopolysaccharide determinant of Chlamydia (Washington Research Foundation, Seattle). Specimens producing inclusions were stored at −70°C before serotyping.
For subsequent studies, we used three C. trachomatis strains with the normal inclusion phenotype (serovars J/UW-36/cx, K/UW-31/cx, and D/UW-3/cx) and four strains manifesting the respective nonfusing phenotype [serovars J(s)/MT-893/ur, K(s)/MT-2481/cx, D(s)/MT-2923/cx, and D(s)/MT-8039/ur]. We also used C. psittaci GPIC (guinea pig inclusion conjunctivitis).
For incubation temperature experiments, wild-type and nonfusing serovars were infected in HeLa cells on coverslips at a multiplicity of infection (MOI) of 10 and centrifuged; the inoculum was removed, growth medium was added, and the cultures were incubated at various temperatures for 40 h. Cells were then fixed in methanol, blocked with 2% bovine serum albumin in phosphate-buffered saline (PBS), and incubated for 30 min with anti-major outer membrane protein (MOMP) MAb 2C1 (MicroTrak; SYVA Co., Palo Alto, Calif.). Duplicate coverslips were incubated with a 1:100 dilution of anti-IncA antiserum. Cells were rinsed with PBS and incubated for 1 h with fluorescein isothiocyanate (FITC)-labeled anti-rabbit immunoglobulin G (IgG).
Growth characteristics of wild-type and nonfusing phenotype strains were observed in HeLa 229 (human), McCoy (mouse), HaK (hamster), and Vero (simian) cell lines obtained from the American Type Culture Collection (Manassas, Va.). All strains used for fluorescent antibody double-labeling experiments and for ultrastructural studies were grown in HeLa 229 cells.
Antibodies.
Serovar J/J(s)-specific MAb CC-1 (IgM) and serovar K/K(s) specific MAb KK-1 (IgG) were used as primary antibodies (25). FITC-labeled anti-mouse IgM and aminomethylcoumarin acetate (AMCA)-labeled anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, Pa.) were used as second antibodies in the fluorescent antibody double-labeling experiments. The genus-specific MAb CF-2 was used to stain C. psittaci GPIC. Anti-IncA monospecific antibody was produced in rabbits by using purified maltose-binding protein–IncA fusion protein as previously described (2). Antiserum to an additional Inc protein (anti-p229), encoded by open reading frame (ORF) D229 of the C. trachomatis genome, was produced in BALB/C mice by using a similar maltose-binding protein fusion. These sera reacted specifically with recombinant p229 in Western blot experiments. This ORF encodes a previously unidentified Inc protein that will be described elsewhere (2a).
Double-label fluorescence microscopy.
To determine the effects of cohabitation of different combinations of wild-type and nonfusing mutants within the same inclusion, fluorescence double-labeling experiments were conducted. HeLa cells were grown in glass vials on sterile coverslips and infected either with both wild-type strains J and K, with both nonfusing variants J(s) and K(s), with wild-type K and the nonfusing variant J(s), or with both wild-type strains J and K plus the nonfusing variant J(s). Strains were used to infect HeLa cells simultaneously or staggered by 1 h. MOIs were approximately 10 to 20. After each inoculation, cells were centrifuged at 1,200 × g for 1 h. Monolayers were then rinsed with PBS, and the growth medium was added. Infected cells were incubated at 37°C in 4% CO2 for 24 to 30 h, fixed with methanol, rinsed with PBS, and blocked with 2% bovine serum albumin in PBS. Cell layers were first incubated with MAb CC-1 for 1 h at room temperature (RT), rinsed three times with PBS, and then incubated with the FITC-labeled anti-mouse IgM second antibody for 30 min at RT. These cell layers were rinsed again three times with PBS and then labeled with MAb KK-1 for 1 h at RT, rinsed three times with PBS, and reacted with the AMCA-labeled anti-mouse IgG second antibody for 30 min at RT. Finally, cells were rinsed three times with PBS and counterstained for 5 min with Evans blue (0.005%) in distilled H2O. Coverslips were removed and inverted onto a microscope slide in a drop of mounting fluid. Fluorescence was observed with a 63× objective of a Zeiss epifluorescence microscope, and photomicrographs were taken with a Nikon UFX-11A camera.
Electron microscopy.
HeLa cells used for transmission electron microscopy were grown on four-well glass chamber slides (Nunc, Inc., Naperville, Ill.) prior to infection with serovar J or J(s) at an MOI of 10. Cultures were incubated for 30 h before being fixed with Karnovsky's fixative (8) and refrigerated at 4°C for 24 h. Cells were then postfixed in 1% osmium tetroxide in distilled H2O for 1 h, followed by two washes for 5 min each in 0.1 M sodium cacodylate buffer, pH 7.4. Cells were dehydrated in a graduated ethanol series, embedded in Eponate resin, and allowed to polymerize for 24 h at 60°C. Thin sections were cut on a Riechert ultramicrotome, placed on 200-mesh hexagonal copper grids, stained with 1% uranyl acetate and Reynolds lead citrate, and observed on a Philips CM-10 transmission electron microscope (Philips, Eindhoven, The Netherlands).
Temporal analysis of inclusion growth.
HeLa cells were grown on sterile glass 12-mm-diameter coverslips and infected with a wild-type C. trachomatis strain (J/UW-36), a nonfusing phenotype C. trachomatis strain [J(s)/MT-893], or C. psittaci GPIC at a low MOI of 0.01 or a high MOI of 10. Coverslips were fixed at 12, 24, 48, and 60 h postinfection (p.i.) and stained with appropriate antibodies. Morphology and number of inclusions were observed by fluorescent antibody microscopy.
Immunoblotting.
Polyacrylamide gel electrophoresis and immunoblotting of the wild-type strain D and the corresponding nonfusing strain D(s) were performed as previously described by Rockey and Rosquist (14), using antiserum against IncA. Briefly, lysates of infected cells were collected 18 and 30 h p.i. with electrophoresis sample dye (1% sodium dodecyl sulfate, 50 mM Tris [pH 6.8], 1% 2-mercaptoethanol, 10% glycerol), boiled for 5 min, and frozen at −20°C. Mock-infected control cells were prepared by identical methods. These lysates were electrophoresed, blotted, and developed with 35S-labeled staphylococcal protein A followed by exposure to autoradiography film.
Sequence analysis of IncA from prototype and nonfusing isolates.
Thermal cycling was used to amplify gene sequences from prototypic and the corresponding nonfusing isolates. All oligonucleotides were derived from sequence information provided in the C. trachomatis genome database. Two sets of primers were generated for cloning and sequencing. The first set (5′-GATCTGCTATGATTTCTTGCG-3′; 5′-GTTTTTTCATGGCCTCTTCTCT-3′) amplified a region 400 nucleotides upstream of incA through position 532 of the incA coding sequence. The second set (5′-AGCCATAGGATCTGGTTTCAGCGA-3′; 5′-GCGCGGATCCTAGGAGCTTTTTGTAGAGGGTGA-3′) amplified the complete incA coding sequence. Both products were used in sequence analysis of the prototypic and nonfusing isolates from each tested serovar. All sequencing was conducted on an automated sequencer, and data were analyzed with MacVector software (Oxford Molecular, Oxford, England).
Nucleotide sequence accession number.
The D(s) incA gene sequence has been assigned GenBank accession no. AF163773.
RESULTS
Isolation and preliminary characterization of the nonfusing strains.
In all, 176 (1.5%) of 11,440 C. trachomatis clinical isolates serotyped in the course of epidemiological studies were observed to form nonfusing inclusions. The nonfusing variants were seen in equal proportions in male and female patients and were represented by most of the major genital serovars, B, D, D−, E, F, G, H, Ia, J, and K. The heritability of the nonfusogenic phenotype was confirmed by serial passage of at least one isolate from each serotype. Two nonfusing isolates were cultured in four different mammalian cell lines (HeLa, McCoy, HaK, and Vero) to determine if the phenotype was a function of the host cell or host cell species. Both isolates [D(s)/MT-2923 and J(s)/MT-893] formed nonfusogenic inclusions in each tested cell line.
Fluorescence microscopy and ultrastructural analysis.
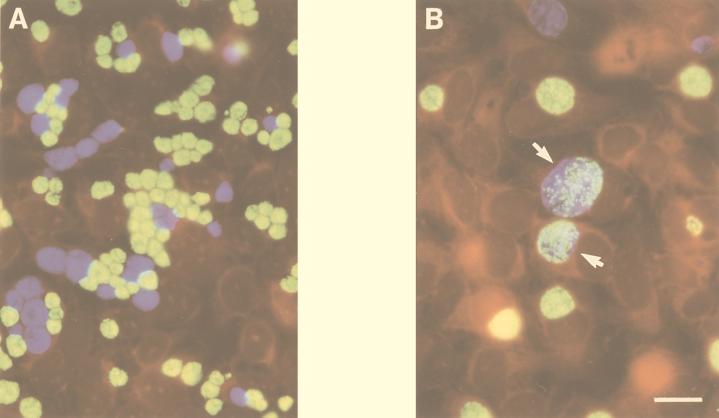
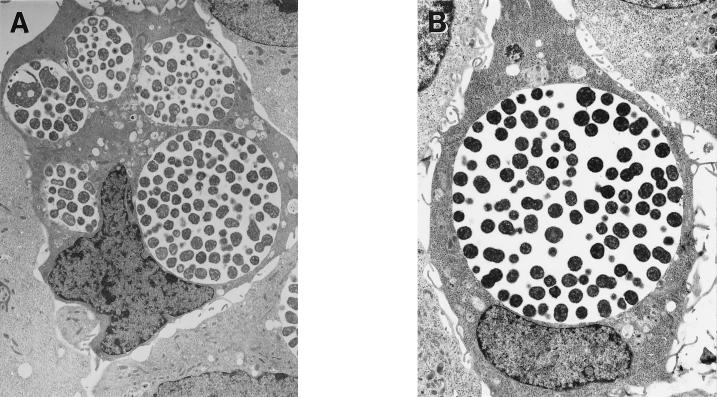
Sequential infection of a single monolayer with two different nonfusing serovariants followed by double-label fluorescent antibody staining using serovar-specific anti-MOMP MAbs demonstrated complete segregation of each nonfusing serovar (Fig. 1A). Similar experiments with wild-type strains demonstrated fusion as evidenced by mixed fluorescent antibody staining of chlamydia within inclusions (Fig. 1B). In very rare instances, the two nonfusing variants or a nonfusing variant and a fusing wild-type serovar could be found in a single vacuole, in cells infected with both strains in the same inocula (not shown). However, if the infection of each distinct serovar was staggered by 1 h, this was never observed. These observations imply that the rare occurrence of distinct nonfusing strains within a single vacuole is a result of multiple chlamydiae entering the cell coincidentally within a single phagosome during chlamydial uptake. Finally, fusion was observed between two wild-type strains infected sequentially with a nonfusing strain (not shown). Collectively, these data suggest that when wild-type strains and those with the nonfusing phenotype cohabit within the same cell, the strains have no effect on the fusion of each other's inclusion membrane. Electron microscopy was also used to characterize the inclusions formed by wild-type and nonfusogenic strains. These analyses were conducted at an MOI of 10, and infected monolayers were fixed 30 h p.i. Consistent with a previously published report (13), the wild-type inclusions fused into a single large vacuole in each cell (Fig. 2B). However, multiple distinct inclusions were observed in cells infected by the nonfusogenic variants (Fig. 2A).
FIG. 1.
Dual infection of nonfusing (A) and fusion-competent (B) C. trachomatis strains cultured in HeLa cells and fixed for microscopy 30 h p.i. Within the nonfusing inclusions, serovar J(s) (green developmental forms) and serovar K(s) (blue developmental forms) remain segregated in individual vacuoles. Consistent with results of Ridderhof and Barnes (13), developmental forms of wild-type J (green) and K (blue) cells can be found within a single inclusion (arrows). The bar in panel B indicates 10 μm for both panels.
FIG. 2.
(A) Electron micrographs of HeLa cells infected with serovar J(s) EB at an MOI of 10 and fixed for electron microscopy 30 h p.i. Notice multiple vacuoles within single cells (magnification, ×6,200). (B) Parallel micrograph of HeLa cells infected with wild-type serovar J-infected HeLa cells (magnification, ×8,200).
Immunostaining of inclusion membrane proteins in wild-type or nonfusing C. trachomatis strains.
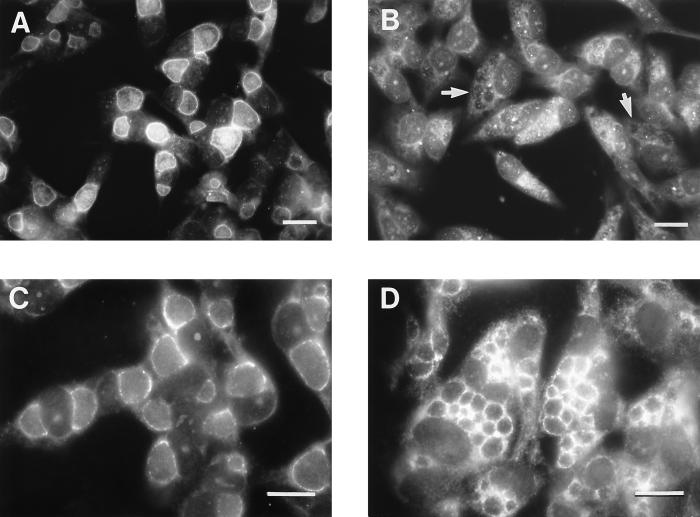
Rabbit antiserum to the C. trachomatis inclusion protein IncA was used to immunostain methanol-fixed monolayers of C. trachomatis-infected HeLa cells. The anti-IncA antibody labeled the inclusion membrane of the wild-type J strain but not the inclusion membrane of the corresponding nonfusing J(s) strain (Fig. 3A and B). In contrast, inclusion membranes of both wild-type J and nonfusing J(s) strains were labeled with an antiserum directed at a novel C. trachomatis inclusion membrane protein, p229, that will be described elsewhere (Bannantine and Rockey, unpublished data) (Fig. 3C and D). In addition to the J and J(s) strains, the same experiment was conducted with D/D(s) and K/K(s). Normal strains showed bright fluorescent staining of the inclusion membrane. With each serovar, bacteria with the nonfusing phenotypes showed bright staining of the inclusion membrane with anti-p229 but showed no anti-IncA staining of the inclusion membrane. An additional novel Inc protein, p223, was also localized to the inclusion membranes of tested fusing and nonfusing isolates (not shown). Therefore, while the nonfusing strains lack IncA on their inclusion membrane, they do produce other Inc proteins that localized there.
FIG. 3.
Fluorescence microscopic analysis of HeLa cells infected with serovar J (A and C) or serovar J(s) (B and D) fixed for microscopy 24 h p.i. (A and B) Cells labeled with an antiserum directed at C. trachomatis IncA; (C and D) cells labeled with an antiserum directed at another protein localized to the inclusion membrane, p229. Notice that IncA is apparently not localized to the inclusion membrane in serovar J(s)-infected cells (arrows in panel B). The bar in each panel indicates 10 μm.
Immunoblot analysis.
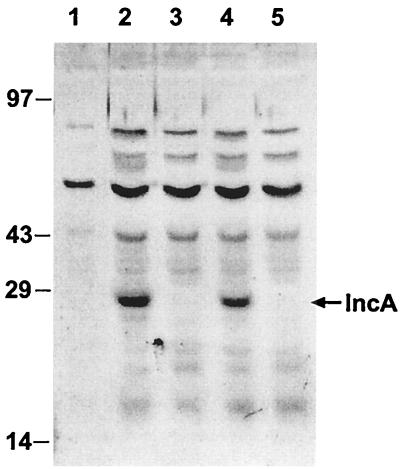
Immunoblot analysis of HeLa cells infected with either wild-type strain D or that with the nonfusing phenotype, D(s), at both 18 and 30 h p.i. and uninfected HeLa cells was performed with polyclonal IncA rabbit antiserum. A 27-kDa band was present in the D-infected cells but not in the lysates of cell infected with strain D(s) (Fig. 4).
FIG. 4.
Immunoblot, probed with anti-C. trachomatis IncA serum, of HeLa cells infected with either serovar D or serovar D(s) collected at 18 or 30 h p.i. Lanes: 1, uninfected HeLa cells; 2, serovar D-infected HeLa cells (30 h); 3, nonfusing D(s)-infected HeLa cells (30 h); 4, serovar D-infected HeLa cells (18 h); 5, nonfusing D(s)-infected HeLa cells (18 h).
Temporal and incubation temperature analysis of inclusion morphology.
Quantitative analysis of the inclusions produced by C. trachomatis prototype strain J, by the corresponding nonfusing J(s) strain and by C. psittaci GPIC at high and low MOIs is shown in Table 1. Except at very early time points, monolayers infected singly and multiply with the prototype J strain resulted in a single inclusion. C. psittaci GPIC always formed many inclusions independent of the MOI as a result of its characteristic lobing nature (16). The nonfusing J(s) strain, however, formed a single inclusion at a low MOI and multiple inclusions at a high MOI that remained segregated at even 60 h p.i. Therefore, the number of inclusions present within the nonfusing strains varied directly with the number of entering EB. In contrast, C. psittaci GPIC always forms multiple or lobed inclusions regardless of the MOI.
TABLE 1.
Temporal analysis of number of inclusions formed in infected HeLa cells at different MOIs by a prototype C. trachomatis strain, a nonfusing C. trachomatis strain, and C. psittaci GPIC, a strain that forms multiple inclusions regardless of MOI (16)
| MOI | Time (h p.i.) | No. of inclusions/cell
|
||
|---|---|---|---|---|
|
C. trachomatis
|
C. psittaci GPIC | |||
| Prototype J | Nonfusing J(s) | |||
| <0.01 | 12 | 1 | 1 | 5–10 |
| 24 | 1 | 1 | 10–20 | |
| 48 | 1 | 1 | >20 | |
| 60 | 1 | 1 | >20 | |
| 10 | 12 | 1–3 | 5–10 | >20 |
| 24 | 1 | 5–10 | >20 | |
| 48 | 1 | 5–10 | >20 | |
| 60 | 1 | 5–10 | >20 | |
To assess the effect of incubation temperature on inclusion morphology, duplicate coverslips of a wild-type J strain and a nonfusing J(s) strain were incubated at 32, 37, and 40°C for 40 h p.i., fixed, and incubated with appropriate antibodies. Anti-MOMP staining of the J strain incubated at 32°C showed the previously reported result (23) of multiple inclusions. Anti-MOMP staining of the same J strain incubated at 37 and 40°C showed one large inclusion as a result of the fusion of the early phagosomes. Staining of the corresponding nonfusing J(s) strain with anti-MOMP revealed multiple inclusions at all three incubation temperatures. Anti-IncA staining of the J strain revealed the presence of IncA in the inclusion membrane at each incubation temperature. These results imply that production of IncA is not temperature dependent and therefore not associated with the lack of fusion at 32°C. Staining of the nonfusing strain J(s) with anti-IncA revealed no localization of IncA to the inclusion membrane at each temperature tested.
Nucleotide sequence analysis.
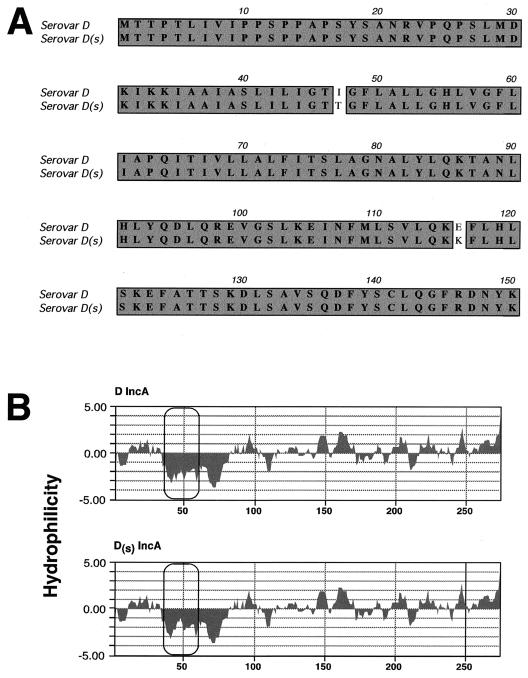
Sequence analysis was conducted on the incA gene from isolates of serotypes D and D(s) (Fig. 5). The incA gene sequence of the serovar D isolate was identical to the sequence found in the C. trachomatis genome database. Two independent D(s) incA sequences differed from the prototype at three nucleotides, leading to two amino acid changes (Fig. 5A) and predicted proteins with virtually identical weights. The amino acid change at position 47 resulted in a significant modification of the characteristic hydrophobicity profile identified in all Inc proteins tested to date (Fig. 5B).
FIG. 5.
Amino acid sequence (A) and hydrophobicity (B) comparisons between mutant and wild-type IncA. (A) Amino acid sequence comparison between prototypic serovar D IncA and serovar D(s) strain MT-2923. Only the first 150 amino acids are shown; there are no differences between the wild type and mutant over the remaining section of the protein. Both isolates of serovar D(s) incA contain three identical nucleotide changes, resulting in two amino acid differences between wild-type and mutant IncA. Each of these mutations results in a changed but apparently intact IncA, but each variant shows a modification in the first hydrophobic domain of the protein (B).
DISCUSSION
Because of their importance to intracellular survival, the composition and function of the chlamydial inclusion membrane are of great interest. It has recently been shown that the inclusion membrane does not acquire lysosomal markers of early or late endosomes from the endosomal/lysosomal pathway and thus avoids phagosome-lysosome fusion (9). Conversely, inclusions are likely fusogenic with intercepted exocytic vesicles containing sphingomyelin, derived endogenously from host cell lipids in the trans-Golgi network, originally routed to the host cell plasma membrane (5, 6). Recently, three C. psittaci-specific proteins, designated IncA, IncB, and IncC, have been identified and characterized (1). Analogous C. trachomatis-specific genes encoding IncA, IncB, and IncC have also been identified by search of the C. trachomatis serovar D genome sequence (11). C. trachomatis IncA is localized to the inclusion membrane and has been demonstrated in all serovars of C. trachomatis tested, including members of each C. trachomatis biovar (2). Recently, a new set of C. trachomatis inclusion membrane proteins, designated IncD, -E, -F, and -G, have been identified (serovar D genome sequence ORFs D116 to D119) (19), and there may be several more (2a). To date, however, no function has been attributed to any Inc protein.
Several studies have compared the differences and similarities of the developing inclusion membranes among the various chlamydial species (12, 20, 28). These studies have focused on inclusion morphology, growth dynamics, and interaction of the inclusion membrane with multiple homotypic or heterotypic inclusions. When epithelial cells are multiply infected with strains of either C. trachomatis or C. pneumoniae or some strains of C. psittaci, multiple inclusions develop that eventually fuse to form a single vacuole (12). However, some strains of C. psittaci, including GPIC and Cal 10, form multiple inclusions (12, 16, 28). Another study showed no evidence of fusion between inclusions in which epithelial cells were infected with different chlamydial species, suggesting that fundamental properties of the heterotypic inclusion membranes must differ (12). Certain environmental conditions may also influence inclusion membrane biology. A recent study demonstrated that low incubation temperature (32°C) or chloramphenicol-induced inhibition of protein synthesis prevents fusion of C. trachomatis inclusions (23). Further investigation of the biogenesis and composition of the inclusion membrane and its interaction with host cells is key to the understanding of chlamydial pathogenesis and survival.
Our results describe a stable and novel phenotype of C. trachomatis manifested by multiple nonfusing inclusions. The nonfusing phenomenon is cell line and temperature independent, MOI dependent, and associated with the absence of immunostaining or the protein IncA in the inclusion membrane while retaining other inclusion membrane proteins. The formation of nonfusing inclusions and the apparent absence of detectable IncA in the inclusion membrane appear to be fundamental biological differences from wild-type C. trachomatis strains. It seems likely that the nonfusing variants represent stable mutants of wild-type strains, but whether the apparent lack of IncA in the inclusion membrane is related to the nonfusion of multiple inclusions is not yet clear. A paradox of this research is evident in the comparison between C. psittaci nonfusing strains, such as GPIC, and the C. trachomatis nonfusing variants described in this work. As highlighted in Table 1, the C. psittaci inclusions not only do not fuse but also appear to divide during growth (16). The nonfusogenic C. trachomatis variants form inclusions that do not divide but also do not fuse. While there is no direct evidence suggesting that the lack of IncA leads to the nonfusing phenotype, every isolate thus far examined does not localize IncA to the inclusion membrane. In contrast, the nonfusogenic C. psittaci strains do have IncA localized to the inclusion membrane. This apparent inconsistency may point to distinct differences in IncA function within these species. In fact, there is only 22% identity between the IncA proteins from each species, and as wild-type inclusions of C. trachomatis and GPIC do not fuse and are not similar in structure, it is likely the proteins do not have identical functions within each species.
One area of interest surrounds the identification of any selective advantage provided by these mutants by virtue of maintaining nonfusogenic inclusions. The phenotype likely did not arise from a single event, as it occurs in each of the 12 examined serovars at roughly similar rates (1.5%) that are higher than expected by random mutation. These variants have also never been identified as a product of passage in tissue culture, suggesting that the benefit may be a function of survival in vivo. Work is in progress to review clinical features of patients whose C. trachomatis isolates produced nonfusing inclusions to examine possible associations of clinical manifestations with the phenotype. It has been suggested that the multiple inclusions of C. psittaci GPIC offer an advantage by generating a greater surface area than would be possible with one large inclusion (16). Having more inclusion membrane in contact with the cytoplasm may provide a greater opportunity for the acquisition of host cell nutrients as well as more docking sites for Golgi apparatus-derived sphingomyelin vesicles and thus potentially more production of progeny. However, selection for nonfusion lessens the opportunity for genetic exchange between dividing reticulate bodies in cells that are infected with multiple serovars, thus acting as a physical barrier reducing the possibility of OMP1 antigenic variation or other recombinational events. Comparison of the protein compositions of the initial phagosome membrane, the association of annexin proteins with early phagosomes, and the presence or absence of recently discovered Inc proteins between wild-type and nonfusing strains may lead to further understanding of the nonfusing phenotype.
Inclusions from every tested nonfusing isolate lack IncA on their inclusion surface, and immunoblotting with anti-IncA antiserum shows no evidence of IncA within tissue culture cells infected with nonfusing strains. However, the sequence of incA from two D(s) strains revealed that the coding sequence of incA has only three nucleotide changes, leading to two amino acid substitutions. This should encode a structurally intact protein. Therefore, the mechanism leading to the nonfusing phenotype remains unclear. It is possible that IncA is produced, but due to secondary structural changes the resulting polypeptide is unstable and may be degraded. There is precedent for rapid turnover of unstable or improperly trafficked proteins in other systems (4, 27), and the genes associated with some these processes are present in the chlamydia genome (21). Another possibility is that an unidentified gene is altered, leading to transcriptional, translational, or secretory blocks in IncA production and localization. It is our opinion that this seems less likely, as other inclusion membrane proteins are produced and localized to the inclusion membrane of cells infected with the nonfusing phenotype. Finally, although the lack of IncA is a trait of all currently known nonfusing strains, the relationship clearly may be unrelated to the actual mechanism leading to the nonfusing phenotype. Further research is in progress to clarify the association of IncA with this phenotype and to identify the processes involved in the establishment of nonfusogenic inclusions. In turn this may facilitate our understanding of inclusion development in wild-type C. trachomatis.
ACKNOWLEDGMENTS
We thank Peter Cummings for the transmission electron microscopy work. We also thank Wasna Viratyosin and Ryan Griffiths for valuable technical support.
A portion of this work was supported by Public Health Service grants U1931448 (W.E.S.) and R29AI42869 (D.D.R.).
REFERENCES
- 1.Bannantine J P, Rockey D D, Hackstadt T. Tandem genes of Chlamydia trachomatis that encode proteins localized to the inclusion membrane. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine J P, Stamm W E, Suchland R J, Rockey D D. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect Immun. 1998;66:6017–6021. doi: 10.1128/iai.66.12.6017-6021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol., in press. [DOI] [PubMed]
- 3.Barnes R C, Wang S-P, Kuo C-C, Stamm W E. Rapid immunotyping of Chlamydia trachomatis with monoclonal antibodies in a solid-phase enzyme immunoassay. J Clin Microbiol. 1985;22:609–613. doi: 10.1128/jcm.22.4.609-613.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackstadt T, Scidmore M A, Rockey D D. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 7.Hackstadt T, Fischer E R, Scidmore M A, Rockey D D, Heinzen R A. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 8.Hayat M A. Basic electron microscopy techniques. New York, N.Y: Van Nostrand Reinhold Company; 1972. p. 53. [Google Scholar]
- 9.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Differential interaction [sic] with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetti and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodinka R L, Davis C H, Choong J, Wyrick P B. Ultrastructural study of endocytosis of Chlamydia trachomatis by McCoy cells. Infect Immun. 1988;56:1456–1463. doi: 10.1128/iai.56.6.1456-1463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalman S, Mitchell W, Marathe R, Lamme C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto A, Bessho H, Uehira K, Suda T. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J Electron Microsc. 1991;40:356–363. [PubMed] [Google Scholar]
- 13.Ridderhof J C, Barnes R C. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect Immun. 1989;57:3189–3193. doi: 10.1128/iai.57.10.3189-3193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockey D D, Rosquist J L. Protein antigens of Chlamydia psittaci present in infected cells but not detected in the infectious elementary body. Infect Immun. 1994;62:106–112. doi: 10.1128/iai.62.1.106-112.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockey D D, Heinzen R A, Hackstadt T. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 16.Rockey D D, Fischer E R, Hackstadt T. Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect Immun. 1996;64:4269–4278. doi: 10.1128/iai.64.10.4269-4278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockey D D, Grosenbach D, Hruby D E, Peacock M G, Heinzen R A, Hackstadt T. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol Microbiol. 1997;24:217–228. doi: 10.1046/j.1365-2958.1997.3371700.x. [DOI] [PubMed] [Google Scholar]
- 18.Russell D G, Dant J, Sturgill-Koszycki S. Mycobacterium avium- and Mycobacterium tuberculosis-containing vacuoles are dynamic, fusion-competent vesicles that are accessible to glycosphingolipids from host cell plasmalemma. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- 19.Scidmore-Carlson M A, Shaw E I, Dooley C A, Fischer E R, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 20.Spears P, Storz J. Biotyping of Chlamydia psittaci based on inclusion morphology and response to diethylaminoethyl-dextran and cycloheximide. Infect Immun. 1979;24:224–232. doi: 10.1128/iai.24.1.224-232.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aarvind L, Mitchell W, Olinger I, Tatusov R L, Zhao O, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 22.Suchland R J, Stamm W E. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J Clin Microbiol. 1991;29:1333–1338. doi: 10.1128/jcm.29.7.1333-1338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Ooij C V, Homola E, Kincaid E, Engel J. Fusion of Chlamydia trachomatis-containing inclusions is inhibited at low temperatures and requires bacterial protein synthesis. Infect Immun. 1998;66:5364–5371. doi: 10.1128/iai.66.11.5364-5371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel J P, Isberg R R. Cell biology of Legionella pneumophilia. Curr Opin Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 25.Wang S-P, Kuo C-C, Barnes R C, Stephens R S, Grayston J T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 1985;152:791–800. doi: 10.1093/infdis/152.4.791. [DOI] [PubMed] [Google Scholar]
- 26.Workowski K, Suchland R J, Pettinger M, Stamm W E. Association of genital infection with specific Chlamydia trachomatis serovars and race. J Infect Dis. 1992;166:1445–1449. doi: 10.1093/infdis/166.6.1445. [DOI] [PubMed] [Google Scholar]
- 27.Wu W F, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HsIUV) protease. J Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyrick P B, Richmond S J. Biology of chlamydiae. J Am Vet Med Assoc. 1989;195:1507–1512. [PubMed] [Google Scholar]