Abstract
Legionella pneumophila has been shown to possess multiple genetic loci that play roles in its ability to survive within host cells. The mil (macrophage-specific infectivity loci) mutants of L. pneumophila exhibit a spectrum of defects in intracellular survival in and cytopathogenicity to macrophages and alveolar epithelial cells. This study characterizes one of the mil mutants (GB111). Intracellular growth of GB111 in macrophages was approximately 100- to 1,000-fold less than that of AA100, the parental strain, at 24 and 48 h postinfection. This defect in turn corresponded to a defect in cytopathogenicity. Sequence analysis of the affected GB111 open reading frame (ORF) revealed it to encode a putative transport protein, and the ORF was designated milA. The phenotypic defect of the milA mutant was complemented with a PCR fragment containing only milA, indicating that the defect in GB111 was due to the disruption of milA. Intracellular trafficking of the mutant was examined by laser scanning confocal microscopy. The data showed that 50% of the GB111 phagosomes colocalized with the late endosomal/lysosomal marker LAMP-2 (2 and 4 h postinfection), while less than 10% of the AA100 phagosomes colocalized with this marker. On the other hand, over 80% of the GB111 phagosomes were similar to the AA100 phagosome in that they were devoid of LAMP-1 and cathepsin D, and they were colocalized with the endoplasmic reticulum (ER) marker BiP. However, the number of GB111 phagosomes that colocalized with BiP decreased to 50% 6 h postinfection compared to that of AA100, which remained constant (80% colocalization). Thus, compared to AA100, the milA mutation caused a defect in intracellular replication, which was associated with colocalization of the phagosome with LAMP-2 and BiP, while colocalization with LAMP-1 and cathepsin D was not affected.
Legionella pneumophila, the causative agent of Legionnaires' disease, is recognized as a major cause of community and nosocomial epidemic pneumonia (8, 13, 26). The ability of L. pneumophila to cause disease is dependent on its ability to invade and replicate within human alveolar cells. Furthermore, the capacity of L. pneumophila to survive in host cells is closely related to the nature of its subcellular localization (32, 35–37). At the ultrastructural level, the phagosome inhabited by L. pneumophila is surrounded by the endoplasmic reticulum (ER) and the mitochondria (1, 5, 23). In addition, this phagosome does not fuse to lysosomes and is devoid of the late endosomal/lysosomal markers LAMP-1, LAMP-2, cathepsin D, and CD-63 (11, 24, 35, 37). Furthermore, the L. pneumophila phagosome is devoid of the transferrin receptor and major histocompatibility complex classes I and II, suggesting that L. pneumophila excludes these molecules from its phagosome (11). Thus, the phagosome inhabited by L. pneumophila has been described as an endosome maturation-blocked phagosome (1, 5), at least during early stages of the infection.
In the environment L. pneumophila replicates within protozoa, a feature linked to its ability to cause infection in humans (6, 14). Interestingly, the subcellular characteristics of the L. pneumophila phagosome appear to be similar in protozoan and mammalian cells (2, 7). In addition, necrosis-mediated killing of the host cell upon termination of intracellular replication is similar for both evolutionarily distant host cells (17; L.-Y. Gao, B. J. Stone, O. S. Harb, and Y. Abu Kwaik, submitted for publication). Taken together, these findings indicate that besides the similarities at the subcellular level, L. pneumophila utilizes similar molecular mechanisms to exploit mammalian and protozoan cells. Conversely, we have identified several macrophage-specific infectivity loci (mil) that are not required by L. pneumophila for intracellular growth in Acanthamoeba polyphaga but are required for growth in human macrophages (18, 20). In addition, L. pneumophila induces caspase-3-mediated apoptosis in mammalian cells, but no apoptosis is induced in protozoa (16, 17). These observations suggest a complex adaptation of L. pneumophila to mammalian cells and protozoa. This adaptation may enable L. pneumophila to differentially utilize genes for the infection of its different host cells.
The mil mutants of L. pneumophila exhibit a range of defects in intracellular growth within and cytopathogenicity to U937 macrophage-like cells and WI-26 alveolar epithelial cells (18, 20). None of the mil mutants contain insertions in the dot and icm genes or are resistant to NaCl (18). Current work in our laboratory is focused on the genetic characterization of the mil mutants and determining effects that the mutated genes may have on the subcellular features of the L. pneumophila phagosome. In this study, one of the mil mutants, mutant GB111, is characterized. Genetic analysis of the affected gene (milA) and data pertaining to the role of milA in intracellular survival and proper subcellular localization of L. pneumophila are presented.
MATERIALS AND METHODS
Bacterial strains and vectors.
The virulent strain of L. pneumophila (AA100) is a clinical isolate and has been described previously (6). Plasmid PBC-SK+ (Stratagene, La Jolla, Calif.) was used to subclone L. pneumophila DNA. Cloning experiments were performed with Escherichia coli DH5α (Gibco BRL, Gaithersburg, Md.) as a host strain. The L. pneumophila chromosomal cosmid DNA library has been previously described (22).
DNA manipulations and sequence analysis.
L. pneumophila chromosomal DNA was prepared by using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). Transfections, restriction enzyme digestions, and DNA ligations were performed as described elsewhere (3) unless specified otherwise. Restriction enzymes were purchased from Promega (Madison, Wis.), and T4 DNA ligase was obtained from Gibco BRL. Plasmid and cosmid DNA preparations were performed with Qiagen mini and midi plasmid kits, respectively (Qiagen Inc., Chatsworth, Calif.). Transformations were done with a Gene Pulser as recommended by the manufacturer (Bio-Rad, Hercules, Calif.). Purification of DNA fragments from agarose gels for subcloning or labeling for Southern hybridization was carried out with a QIAquick gel purification kit (Qiagen). Fluorescein labeling of DNA probes for Southern hybridization was done with the Amersham ECL random prime labeling system, version II (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Oligonucleotide synthesis for PCR was done by Integrated DNA Technologies Inc. (Coralville, Calif.). Sequencing was carried out by Genemed Synthesis Inc. (South San Francisco, Calif.). Sequence comparisons and alignments were performed with the BlastX and Blast2 programs, respectively. Hydropathy profiles were performed by using the Kyte-Doolittle algorithm and MacVector sequence analysis program (Oxford Molecular Group, Inc., Campbell, Calif.).
PCR.
Amplification of the GB111 open reading frame (ORF) was done with primers Bbam, complementary to a region 260 nucleotides upstream of the GB111 start codon (5′-gcgggatcctgagacggc-3′), and Bxho, complementary to a region 135 nucleotides downstream of the GB111 stop codon (5′-gcgctcgagctgacacaac-3′). Bbam and Bxho were designed to generate BamHI and XhoI sites, respectively, to facilitate cloning as we described previously (22). A 1.5-kb fragment was amplified by PCR with a Gene Amp PCR System 2400 (Perkin-Elmer, Norwalk, Conn.). This fragment was subcloned into PBC-SK+, and the recombinant clone was designated V1B.
Bacterial cultures.
For all experiments described, L. pneumophila AA100 and GB111 were grown to the postexponential phase in the following manner. Bacteria were grown from frozen stocks on buffered charcoal-yeast extract agar (supplemented with 20 μg of kanamycin per ml for strain GB111) for 48 h at 37°C. The bacteria were resuspended to an optical density at 550 nm (OD550) of 0.1 to 0.4 in buffered yeast extract broth and grown at 37°C in a shaking incubator for 15 to 18 h to an OD550 of 2.1 to 2.2, when bacterial replication stopped (postexponential growth phase).
Tissue culture and infections.
U937 macrophage-like cells were maintained at 37°C and 5% CO2 in RPMI 1640 tissue culture medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum (Gibco BRL). For infections, cells (105/well in 96- or 24-well plates) were differentiated for 48 h, using phorbol 12-myristate 13-acetate as described previously (3).
WI-26 human type I alveolar epithelial cells have been previously used for L. pneumophila infections (10, 20). These cells were maintained in minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C and 5% CO2. For infections, 104 cells/well were seeded in 96-well plates and incubated at 37°C and 5% CO2 overnight.
Infection protocol.
A multiplicity of infection (MOI) of 10 was used to determine bacterial growth kinetics in U937 macrophage-like cells or WI-26 alveolar epithelial cells, and an MOI of 1 was used to determine cytopathogenicity. An MOI of 1 was also used in laser scanning confocal microscopy experiments (see below).
Infections were performed as described previously (4, 18, 19, 22). Briefly, bacteria were diluted to the appropriate concentrations in tissue culture medium and added to monolayers in 96-well plates to determine intracellular growth kinetics and cytopathogenicity and in 24-well plates for microscopy experiments. Plates were then spun at 1,000 × g for 5 min to synchronize the infection. For cytopathogenicity and microscopy experiments, plates were then incubated at 37°C for several time intervals. To examine intracellular growth and survival, extracellular bacteria were killed by exposure to gentamicin (50 mg/ml) for 1 h following an initial 1-h infection at 37°C. Infected cultures were then incubated at 37°C for several time intervals, and the intracellular bacteria were enumerated following growth on buffered charcoal-yeast extract plates.
Cytopathogenicity was determined by treatment of infected or noninfected monolayers with 10% Alamar Blue dye (Alamar Bioscience Inc., Sacramento, Calif.). Viability of monolayers was measured optically at a wavelength of 570 nm and corrected for background at 600 nm, using a VMAX kinetic microplate reader (Molecular Devices, Menlo Park, Calif.). The relative degree of cytopathogenicity was expressed as percent cytopathogenicity compared to noninfected cells, calculated as (mean OD of infected cells/mean OD of noninfected cells) × 100 as we described previously (19). Noninfected cells were considered 100% viable.
Antibodies, stains, and laser scanning confocal microscopy.
U937 cells were differentiated on 12-mm-diameter (0.13- to 0.17-mm-thick) circular glass coverslips (Fisher Scientific, Pittsburgh, Pa.) in 24-well culture plates. Following infections, cells were washed three times with culture medium and fixed in 4% paraformaldehyde for 1 h. Paraformaldehyde was removed by washing wells three times with phosphate-buffered saline, pH 7.5. All subsequent steps were performed at room temperature, and each step was followed by three washes in phosphate-buffered saline. Blocking was performed with 3% bovine serum albumin for 1 h.
To differentiate between intracellular and extracellular bacteria, a polyclonal anti-L. pneumophila antibody (16) at a dilution of 1:1,000 was used to label extracellular bacteria for 1 h. Cells were then permeabilized with 0.5% Triton X-100 for 10 min. Intracellular and extracellular bacteria were stained with 2 μM nucleic acid stain Toto-3 (excitation at 642 nm, emission at 660 nm; visualized as blue pseudocolor) (Molecular Probes Inc., Eugene, Oreg.) for 1 h during the secondary antibody incubation step (see below).
Both primary and secondary antibodies were diluted in blocking solution, and incubations were performed for 1 h at room temperature. The antibodies used to label lysosomal and late endosomal compartments were anti-LAMP-1 (H3B3) at a dilution of 1:100 and anti-LAMP-2 (H4B4) at a dilution of 1:300. These monoclonal antibodies (developed by J. T. August and J. E. K. Hildreth) were obtained from the Developmental Studies Hybridoma Bank maintained by The University of Iowa. Anti-cathepsin D monoclonal antibody was purchased from Transduction Laboratories (Lexington, Ky.) and used at a dilution of 1:100. Mouse monoclonal antibody specific for Grp-78 (BiP) (StressGen Biotechnologies Corp., Victoria, British Columbia, Canada) was used at a dilution of 1:300 to label the ER. Secondary antibodies, anti-rabbit conjugated to Alexa Red (excitation at 590 nm, emission at 617 nm; visualized as red pseudocolor) and anti-mouse conjugated to Oregon Green (excitation at 511 nm, emission at 530 nm; visualized as green pseudocolor) (Molecular Probes, Eugene, Oreg.), were used at a concentration of 1:300. Coverslips were mounted on glass slides by using a ProLong antifade kit (Molecular Probes).
Samples were analyzed with a Leica TCS SP laser scanning confocal microscope (Leica Microscopy and Scientific Instruments Group, Heerburg, Switzerland) equipped with three lasers: argon (488-nm excitation line), krypton (568-nm excitation line), and helium neon (633-nm excitation line). On average, fifteen 0.2-μm sections of each image were captured and stored for further analyses, using Photoshop 5.0 (Adobe Inc.). Duplicate samples from at least three independent experiments were analyzed. Colocalization was assessed by counting 100 to 150 intracellular bacteria per experiment. Fluorescence intensity was measured with Leica TCS SP software (Leica Microscopy and Scientific Instruments Group).
Hemolysis assays.
Hemolysis of sheep erythrocytes (sRBCs) was performed as previously described (27). Bacteria were mixed with sRBCs at an MOI of 25, pelleted for 2 min at 13,000 × g, and incubated at 37°C for 60 min. Hemolysis of sRBCs was assessed spectrophotometrically at OD415.
Nucleotide sequence accession number.
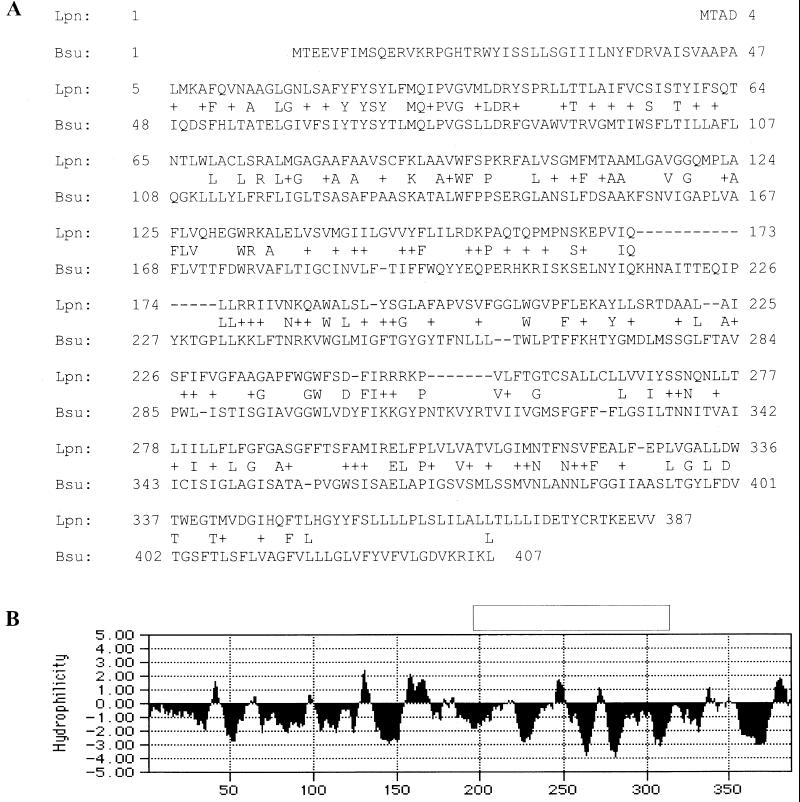
The GB111 sequence shown in Fig. 3A has been assigned GenBank accession no. AF153695.
FIG. 3.
Analysis of the GB111 sequence. (A) Alignment of the predicted amino acid sequence of the GB111 ORF to a predicted permease of B. subtilis, using the Blast2 sequence analysis program. (B) Hydropathy profile of the predicted GB111 amino acid sequence based on the Kyte-Doolittle algorithm with a window size of seven amino acids. Negative scores indicate relative hydrophobicity.
RESULTS
Growth kinetics of GB111 in U937 macrophage-like cells.
Recently, the growth phase of L. pneumophila has been shown to have detrimental effects on the outcome of an infection such that postexponential-phase bacteria exhibit enhanced virulence (9). We have recently described the isolation of 121 transposon mutants of L. pneumophila (18, 19). The initial characterization of these mutants was performed with broth-grown L. pneumophila at an OD550 of 1 (mid to late exponential phase). The phenotypes of a large number of these mutants have been reconfirmed in assays using postexponential-phase bacteria; only one (GB111) deviated from its original phenotype for intracellular growth and cytopathogenicity, although this mutant was still defective for these phenotypes.
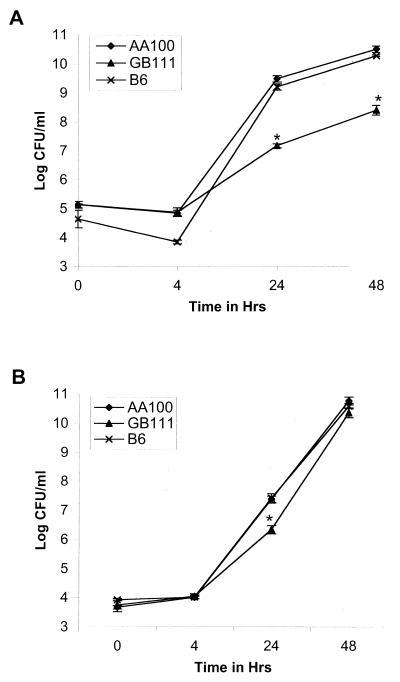
During the initial 4 h postinfection, the survival of GB111 in U937 macrophages was similar to that of the parental strain (AA100) of L. pneumophila (Fig. 1A). However, the number of GB111 bacteria within macrophages was 100- to 1,000-fold less than that of the parental strain (AA100) at 24 and 48 h postinfection (Fig. 1A). Interestingly, the numbers of GB111 and AA100 bacteria recovered were similar at 72 h postinfection (data not shown). These results indicated that mutant GB111 exhibited a delay in intracellular replication within macrophages.
FIG. 1.
Intracellular growth kinetics of L. pneumophila GB111 and AA100 (wild type) in U937 macrophage-like cells (A) and WI-26 alveolar epithelial cells (B) as determined by a gentamicin protection assay. Strain B6 is a cosmid-complemented clone of GB111. ∗, significantly fewer intracellular bacteria than in the wild-type (AA100)-infected cells based on Student's t test (P < 0.05).
Intracellular growth kinetics of GB111 in WI-26 alveolar epithelial cells.
It has been hypothesized that alveolar epithelial cells may constitute a potential site of L. pneumophila replication (1, 5, 20). Interestingly, the ability of GB111 to survive within lungs of A/J mice was similar to that of the AA100 strain 48 h postinfection (20). It is possible that replication of GB111 within alveolar epithelial cells in lungs of infected mice compensates for the growth defect in macrophages. To test this possibility, we examined the intracellular growth of postexponential-phase GB111 within WI-26 alveolar epithelial cells. In general, the intracellular growth of GB111 was similar to that of AA100 in WI-26 cells throughout the infection (Fig. 1B). However, a slight difference in replication between GB111 and AA100 was consistently evident at 24 h postinfection (Fig. 1B).
Cytopathogenicity of GB111 to U937 and WI-26 cells.
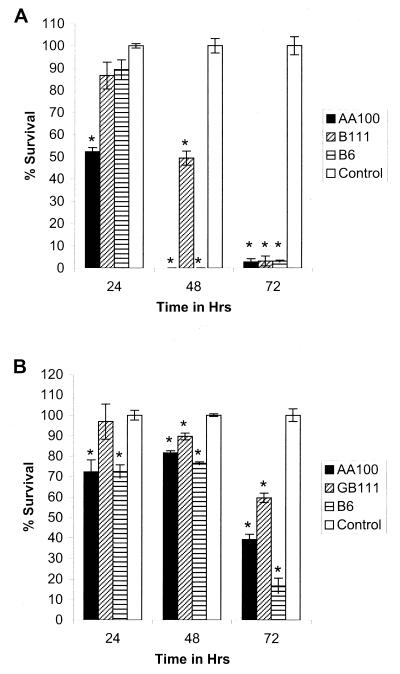
In general, the extent of intracellular replication of L. pneumophila correlates with the extent of its cytopathogenicity to its host cells. For all previously described mutants of L. pneumophila, there is a correlation between the defect in intracellular replication and cytopathogenicity (18, 19, 31, 33, 38). While AA100 was 50% cytopathogenic at 24 h, GB111 was completely noncytopathogenic (Fig. 2A). Cytopathogenicity of GB111 to U937 cells increased to 50% at 48 h, compared to 100% for AA100 (Fig. 2A). Following 72 h of infection, both GB111 and AA100 were completely cytopathogenic to macrophages (Fig. 2A).
FIG. 2.
Cytopathogenicity of L. pneumophila GB111 and AA100 (wild type) to U937 macrophage-like cells (A) and WI-26 alveolar epithelial cells (B) as determined by using Alamar Blue dye. Strain B6 is a cosmid-complemented clone of GB111. Percent killing of cells was normalized to that for uninfected cells, which were considered 100% viable. ∗, significantly less survival than for uninfected control cells based on Student's t test (P < 0.05).
Cytopathogenicity of GB111 to WI-26 cells was similar to that of AA100 at all time points tested (Fig. 1B). Cytopathogenicity to WI-26 alveolar epithelial cells never reached 100% for both strains, most likely because WI-26 cells continue to divide throughout the course of the infection. Thus, cytopathogenicity of GB111 to both macrophages and epithelial cells correlated with its intracellular growth.
In vitro growth of mutant GB111.
To determine whether the defective phenotype of GB111 in macrophages and epithelial cells was due to a general defect in growth, we compared its growth to that of the wild-type strain in two types of media (buffered yeast extract rich medium and semidefined Casamino Acids medium). The growth of GB111 was similar to that of AA100 in both types of media (data not shown). Thus, the defect in GB111 did not result in a defect in in vitro growth. However, the similarity in growth of GB111 and AA100 in culture media does not rule out the possibility that the intracellular defect in GB111 is due to the deficiency in another component that is present in the culture media but absent from the intracellular environment.
Hemolysis of sRBCs.
We have recently proposed biphasic killing of the host cell by L. pneumophila (1, 15–17). Cytopathogenicity of L. pneumophila to mammalian cells is mediated through induction of caspase-3-mediated apoptosis during early stages of the infection (15, 16), followed by pore formation-mediated necrosis upon termination of intracellular replication (9; Gao et al., submitted). We have previously shown that GB111 is capable of inducing apoptosis at a level similar to that for the wild-type strain (16). Therefore, we tested the ability of GB111 to form pores upon contact with cells. Pore formation can be tested by determining hemolysis of sRBCs upon contact with the bacteria (27). Since GB111 exhibited a defect in cytopathogenicity to U937 macrophage-like cells, it is possible that this defect is due to a decrease in the pore-forming activity. In this regard, we determined whether postexponential-phase GB111 expressed pore-forming capability similar to that of AA100. The results showed that GB111 and AA100 exhibited similar levels of hemolysis of sRBCs (data not shown). Thus, GB111 was not defective in pore formation.
Sequence analysis.
A chromosomal fragment containing the kanamycin insertion region (Kan cassette) and flanking Legionella DNA was isolated from GB111, subcloned, and sequenced. Analysis of the sequence revealed that the Kan cassette was inserted in a 1,164-bp ORF which was designated milA. Comparison of the predicted amino acid sequence of this ORF with sequences of other proteins by using the BlastX program revealed small degrees of similarity to several transporter proteins. The highest similarities were to an ORF from Bacillus subtilis (24% identity and 43% similarity) predicted to be similar to ABC permeases and to UhpC from E. coli (26% identity, 44% similarity) and Salmonella enterica serovar Typhimurium (21% identity and 41% similarity) (Fig. 3A). Hydropathy analyses of the predicted MilA protein revealed it to be extremely hydrophobic (Fig. 3B). Topology analysis using the TopPred2 program (www.biokemi.su.se) indicated that the predicted MilA protein might contain up to 11 transmembrane domains (data not shown). Based on these results, it is most likely that the MilA protein is an integral membrane protein that may function as part of a transport system.
Complementation of GB111.
To subclone the wild-type gene of GB111 and determine if the Kan insertion resulted in polar effects on other genes, an L. pneumophila cosmid DNA library was screened with a probe derived from a DNA fragment containing regions flanking the Kan cassette in GB111. Two cosmid clones were confirmed by Southern hybridization (data not shown) to contain the corresponding DNA fragment containing milA. Both clones were electrotransformed into GB111, and intracellular growth kinetics and cytopathogenicity were determined. One of the clones (B6) complemented GB111 for growth and cytopathogenicity (Fig. 1 and 2). To exclude the possibility that the Kan insertion resulted in polar effects that may account for the defective phenotype of GB111, a 1.5-kb PCR-generated fragment containing milA was subcloned and designated V1B. V1B was transformed into GB111 and tested for its ability to complement intracellular growth and cytopathogenicity. The results showed that milA was sufficient to completely complement the growth and cytopathogenicity defects of GB111 (data not shown). Thus, the phenotypic defect in GB111 is due to the disruption of milA.
GB111 exhibited an enhanced colocalization with LAMP-2.
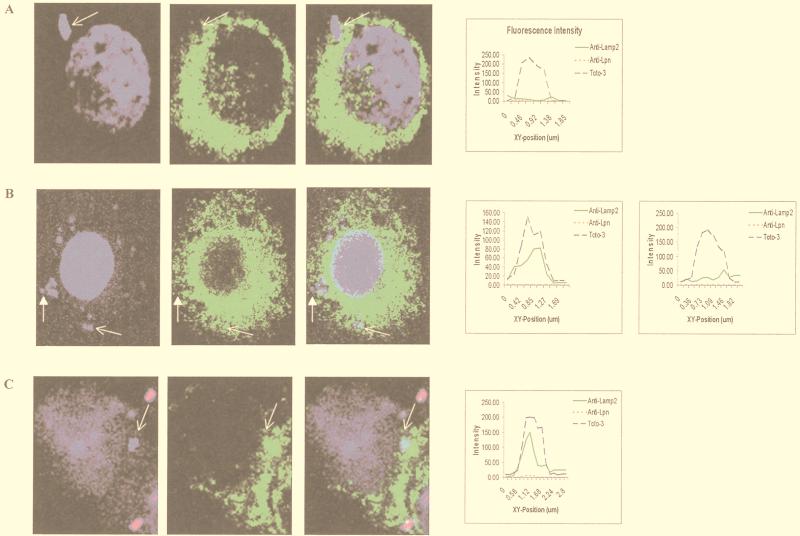
To determine whether GB111 differed from AA100 in its intracellular trafficking, we examined colocalization of their phagosomes with the late endosomal/lysosomal marker LAMP-2 at 2 and 4 h after infection of U937 cells. The visual assessment of colocalization was corroborated by measurement of fluorescence intensity across the phagosome. Colocalization was determined by the presence of two fluorescence peaks in the same spot (one green for late endosomal/lysosomal markers and one blue for the bacteria) (e.g., Fig. 4).
FIG. 4.
Colocalization of phagosomes with LAMP-2. (A) Wild-type L. pneumophila (AA100); (B) GB111; (C) paraformaldehyde-killed AA100. LAMP-2 is visualized by using secondary antibodies conjugated to Oregon Green (green pseudo-color). Extracellular bacteria were visualized with secondary antibody conjugated to Alexa Red (red pseudocolor). Nucleic acids were stained with Toto-3 (blue pseudocolor), which labels both intracellular and extracellular bacteria and the cell nucleus. Intensities of fluorescence across the indicated phagosomes (arrows) are shown at the right. Darkened arrows in panel B point to a GB111 phagosome that does not colocalize with LAMP-2, and fluorescence intensity across this phagosome is shown on the far right.
The GB111 phagosomes were assessed for the presence of LAMP-2 as described above. Colocalization coincided with two fluorescent peaks across the phagosome (Fig. 4). Forty to 50% of the GB111 phagosomes colocalized with LAMP-2 at 2 and 4 h postinfection (Fig. 4B and Table 1). In contrast, only 5 to 10% of the AA100 phagosomes colocalized with LAMP-2 (Fig. 4A and Table 1). As expected, 80 to 90% of the paraformaldehyde-killed bacterial phagosomes were positive for LAMP-2 at 2 and 4 h postinfection (Fig. 4C and Table 1). Thus, the defect in GB111 was associated with a defect in LAMP-2 sorting to the L. pneumophila phagosome.
TABLE 1.
Intracellular trafficking of AA100, GB111, and paraformaldehyde-killed AA100 phagosomes in U937 macrophage-like cells
| Marker | Strain | % Colocalization (mean ± SE) aftera:
|
||
|---|---|---|---|---|
| 2 h | 4 h | 6 h | ||
| LAMP-1 | AA100 | 17 ± 2.6 | 4.3 ± 4 | ND |
| GB111 | 21.3 ± 2.5 | 10.9 ± 5.5 | ND | |
| Pf-AA100b | 80 ± 5 | 83.9 ± 7.6 | ND | |
| LAMP-2 | AA100 | 5.5 ± 2.3 | 10.7 ± 4 | ND |
| GB111 | 53.5 ± 7.7 | 46 ± 6.6 | ND | |
| Pf-AA100 | 88.3 ± 7 | 90.6 ± 9.3 | ND | |
| Cathepsin D | AA100 | 8 ± 1 | 13.5 ± 4.3 | ND |
| GB111 | 8.3 ± 2.5 | 26 ± 5.6 | ND | |
| Pf-AA100 | 84 ± 7.9 | 82.2 ± 6.6 | ND | |
| BiP | AA100 | ND | 85 ± 5.1 | 88.7 ± 5.5 |
| GB111 | ND | 77.7 ± 7.5 | 50.7 ± 4 | |
| Pf-AA100 | ND | 4.2 ± 2.6 | 3.7 ± 1.5 | |
Mean percentage of colocalization of bacterial phagosomes with LAMP-1, LAMP-2, cathepsin D, or BiP at the indicated time points from a representative experiment. ND, not determined.
Pf-AA100, paraformaldehyde-killed AA100.
GB111 does not colocalize with the late endosomal/lysosomal markers LAMP-1 and cathepsin D.
We also examined colocalization of the phagosomes of GB111 and AA100 with the late endosomal/lysosomal markers LAMP-1 and cathepsin D at 2 and 4 h after infection of U937 cells. The visual assessment of colocalization was corroborated by measurement of fluorescence intensity across the phagosome. Colocalization was determined by the presence of two fluorescence peaks in the same spot (one green for late endosomal/lysosomal markers and one blue for the bacteria) (Fig. 4).
At 2 h postinfection, approximately 80% of the GB111 and AA100 phagosomes did not colocalize with LAMP-1, whereas 80% of the paraformaldehyde-killed bacteria colocalized with LAMP-1 (Table 1 and data not shown). Colocalization with LAMP-1 remained relatively unchanged at 4 h postinfection (Table 1 and data not shown).
The proportions of GB111 and AA100 phagosomes that colocalized with cathepsin D ranged from 5 to 20% at 2 and 4 h postinfection, respectively (Table 1 and data not shown). Cathepsin D colocalized with 80% of the paraformaldehyde-killed AA100 phagosomes at both time points (Table 1 and data not shown). Thus, the defect in GB111 did not affect sorting of LAMP-1 and cathepsin D to the phagosome.
Colocalization of GB111 with the ER.
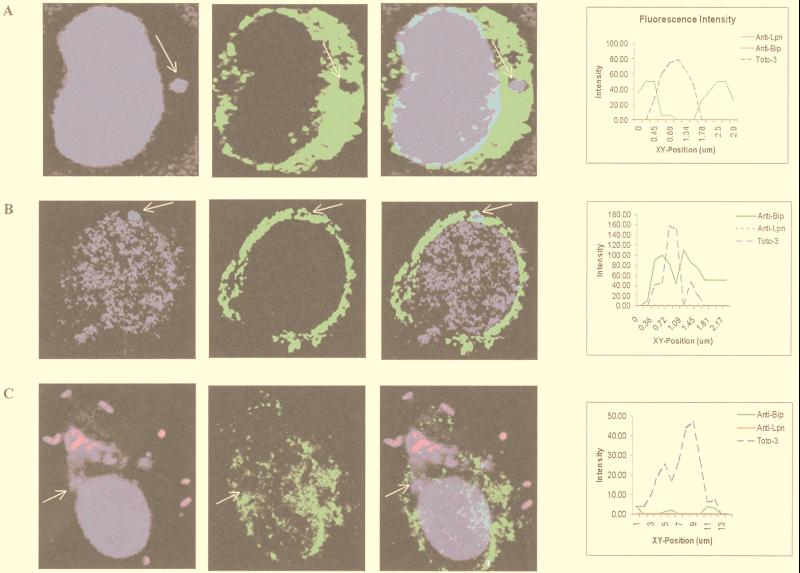
Since L. pneumophila is known to recruit the ER (23, 36), we tested whether the defect in intracellular survival of GB111 was reflected in a defect in recruitment of this organelle. Using anti-BiP antibody (luminal ER protein) and laser scanning confocal microscopy, recruitment of the ER was determined by an accumulation of BiP around the L. pneumophila phagosome (Fig. 5). Intensity of fluorescence around the bacterial phagosome was measured and corresponded to the visual assessment of ER recruitment (Fig. 5A and Table 1). Green fluorescence (used to visualize BiP) peaked on either end of a line drawn across the phagosome, while blue fluorescence (used to visualize bacteria) peaked only in the center of the phagosome (Fig. 5). Lack of colocalization was determined by the absence of a green fluorescence peak around the bacterial phagosome. Recruitment of the ER was scored in AA100, GB111, and paraformaldehyde-killed AA100-infected U937 macrophages. At 4 and 6 h postinfection, 85% of the AA100 phagosomes were colocalized with BiP (Table 1); 78% of the GB111 phagosomes colocalized with BiP at 4 h postinfection (Table 1). Interestingly, the number of GB111 phagosomes that colocalized with BiP decreased to 50% 6 h postinfection (Table 1). As expected, 95% of the phagosomes containing paraformaldehyde-killed AA100 were not colocalized with BiP (Table 1).
FIG. 5.
Colocalization of BiP (ER) with phagosomes of wild-type L. pneumophila (AA100) (A), GB111 (B), and paraformaldehyde-killed AA100 (C). BiP is visualized by using secondary antibodies conjugated to Oregon Green (green pseudocolor). Extracellular bacteria were visualized with secondary antibody conjugated to Alexa Red (red pseudocolor). Nucleic acids were stained with Toto-3 (blue pseudocolor), which labels both intracellular and extracellular bacteria and the cell nucleus. Intensities of fluorescence across the indicated (arrows) phagosomes are shown at the right.
DISCUSSION
Recently, Gao and colleagues have identified 32 mutants (mil mutants) of L. pneumophila that exhibited defects ranging from mild to deleterious in cytopathogenicity to and intracellular survival in macrophages and alveolar epithelial cells (18, 20). None of the mil mutants are defective in the dot and icm genes (18).
The mil mutant GB111 exhibited a defect in intracellular replication in both U937 macrophages and WI-26 alveolar epithelial cells, albeit the defect was more severe in macrophages than in epithelial cells and the bacteria recovered sooner (48 h postinfection) in epithelial cells than in macrophages (72 h postinfection). This defect was not due to a defect in attachment or entry since GB111 was previously shown to attach as well as AA100 to its host cells (18). Similar numbers of GB111 and AA100 bacteria were recovered from macrophages and epithelial cells at the early time points of infection (0 and 4 h). Hence, the defect in GB111 is exhibited as slower intracellular replication, which recovered at 72 h postinfection. It is also possible that the defect is due to concomitant killing and replication of GB111.
We have recently proposed a biphasic model by which L. pneumophila exhibits cytopathogenicity to the host cell (15, 16). The first phase is mediated by induction of apoptosis during early stages of the infection (16, 29) and occurs through the activation of caspase-3 (15). The second phase is manifested by pore formation-mediated rapid necrosis (27) and occurs upon termination of intracellular bacterial replication (9; Gao et al., submitted). Compared to AA100, GB111 exhibited a decrease in cytopathogenicity that correlated with the decrease in its survival within macrophages. In WI-26 epithelial cells, cytopathogenicity also correlated with intracellular growth and was similar to that of AA100. This reduction in cytopathogenicity is most likely due to the defect in intracellular replication, since GB111 is not defective in pore formation (see above) or apoptosis (16).
Analysis of the predicted amino acid sequence indicated that the milA gene may encode a putative transport protein, based on a low degree of similarity to UhpC (uptake of hexose phosphates) from E. coli and Salmonella serovar Typhimurium and a predicted ABC permease from B. subtilis. UhpC has been localized to the inner membrane and most likely interacts with UhpB to transmit signals presented by external glucose-6-phosphate (25). However, uhpC is part of the uhpABC operon, and milA did not appear to be in an operon (25). Hydropathy and membrane topology analysis of MilA revealed it to be a largely hydrophobic protein containing 11 potential transmembrane domains. Although the above results are not sufficient to assign a function to MilA, it is most likely a transmembrane protein.
Since the mutation in GB111 was generated by using a mini-Tn10::Kan transposon insertion, polar effects may ensue. However, the defect in GB111 was complemented with a PCR-generated fragment containing only the milA gene. Both intracellular replication and cytopathogenicity were similar to that of wild-type L. pneumophila. Thus, disruption of this ORF alone was sufficient to result in the defective phenotype of GB111 in macrophages.
The L. pneumophila phagosome has been shown to avoid classical maturation along the endosomal/lysosomal pathway (11, 35, 37). Since GB111 was defective in intracellular replication, we determined whether the characteristics of its phagosome differed from those of wild-type L. pneumophila. The presence of four different markers in the GB111 phagosome was investigated. GB111 was competent in ER recruitment as determined by BiP accumulation around the phagosome, although a decrease was observed following 4 h of infection. GB111 was able to exclude the late endosomal/lysosomal markers LAMP-1 and cathepsin D from its phagosome. Surprisingly, however, GB111 had a decreased ability to exclude the late endosomal/lysosomal marker LAMP-2 from its phagosome. Whether this is sufficient to cause a reduction in the intracellular replication of GB111 is not known.
Classically, the maturation of an endosome into a phagolysosome is described as a sequential series of steps involving the gradual and regulated acquisition and clearance of endosomal/lysosomal markers (12, 28). However, the trafficking of a pathogen-containing phagosome is likely to be dictated by the pathogen itself and may not necessarily occur through this classical pathway. For example, the phagosome containing Mycobacterium avium has been shown to resemble an aberrant early endosome or recycling vesicle that is accessible to sorting endosomes and also acquires certain markers that are characteristic of late endosomes (34). This phagosome contains an immature form of the late endosomal marker cathepsin D that is prevented from becoming active by inhibition of phagosomal acidification (34). The mechanisms utilized by L. pneumophila to inhibit the maturation of its phagosome into a phagolysosome are not known. However, the inability of GB111 to exclude LAMP-2 from its phagosome may suggest that the L. pneumophila phagosome may transiently acquire late endosomal/lysosomal markers. Alternatively, GB111 may be defective in exclusion of alternatively spliced variants of LAMP-2 that have been shown to be predominantly distributed on the cell surface (21).
The intracellular bacterial pathogen Brucella abortus has been shown to transit early in an infection through a LAMP-positive, cathepsin D-negative compartment (30). Thus, acquisition of LAMPs into the bacterial phagosome may not be fatal for the pathogen. On the other hand, Roy and colleagues have shown that L. pneumophila excludes LAMP-1 from its phagosome as early as 5 min postinfection (32). This study also revealed the existence of a small population of phagosomes that are LAMP-1 positive (32). Whether these phagosomes were also positive for LAMP-2 was not reported. Half of the GB111 phagosomes were LAMP-1 and cathepsin D negative but LAMP-2 positive, indicating that GB111 may be partially defective in blocking the endosomal/lysosomal pathway. This altered trafficking of GB111 may account for the reduced intracellular replication and cytopathogenicity. It is possible that the inability of GB111 to exclude LAMP-2 inhibits its intracellular replication but does not kill it, since this phagosome excludes cathepsin D, making it unlikely that it matures into a phagolysosome. This possibility is supported by the finding that equal wild-type and GB111 bacterial numbers were recovered at 4 h postinfection prior to bacterial replication. However, the possibility that these phagosomes mature into phagolysosomes at a later time point cannot be excluded. Conversely, exclusion of LAMP-2 from these phagosomes at a later time point also cannot be excluded. Examining the nature of the Legionella phagosome at later time points may prove to be important for understanding the nature of its maturation.
Thus, the defect in GB111 seems to result in aberrant phagosomal maturation. Whether the milA gene is directly involved in dictating the nature of the L. pneumophila phagosome cannot be determined based on the above observations. In addition, based on the intracellular growth and sequence results, it is possible that the defect in GB111 affects the ability of the bacteria to replicate within macrophages. This defect in intracellular replication may be due to the inability of GB111 to transport a required metabolite or interact with an external signal. The decreased replication within macrophages in turn results in a delay in cytopathogenicity. Therefore, the altered intracellular trafficking of GB111 is a direct result of the milA mutation or is a secondary effect due to a defect in intracellular replication.
ACKNOWLEDGMENTS
Y.A.K. was supported by Public Health Service grant R29AI38410. O.S.H. was supported by NIH training grant 5T32CA09509.
We thank Bruce Maley, Richard Watson, and Mary Gail Engle at the University of Kentucky microscopy imaging suite for technical assistance with laser scanning confocal microscopy. We thank Oyebode Terry Alli for assistance with some of the experiments.
REFERENCES
- 1.Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–696. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Gao L-Y, Stone B J, Harb O S. Invasion of mammalian and protozoan cells by Legionella pneumophila. Bull Inst Pasteur. 1998;96:237–247. [Google Scholar]
- 6.Abu Kwaik Y, Gao L-Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba catellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozzoni M, Radice L, Frosi A, Vezzoli S, Cuboni A, Vezzoli F. Prevalence of pneumonia due to Legionella pneumophila and Mycoplasma pneumoniae in a population admitted to a department of internal medicine. Respiration. 1995;62:331–335. doi: 10.1159/000196475. [DOI] [PubMed] [Google Scholar]
- 9.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianciotto N P, Stamos J K, Kamp D W. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr Microbiol. 1995;30:247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- 11.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagolysosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjardins M, Huber L, Parton R, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein P H. Legionnaires' disease. Clin Infect Dis. 1993;16:741–747. doi: 10.1093/clind/16.6.741. [DOI] [PubMed] [Google Scholar]
- 14.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 15.Gao L-Y, Abu Kwaik Y. Activation of caspase 3 during Legionella pneumophila-induced apoptosis. Infect Immun. 1999;67:4886–4894. doi: 10.1128/iai.67.9.4886-4894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao L-Y, Abu Kwaik Y. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect Immun. 1999;67:862–870. doi: 10.1128/iai.67.2.862-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, L.-Y., and Y. Abu Kwaik. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ. Microbiol., in press. [DOI] [PubMed]
- 18.Gao L-Y, Harb O S, Abu Kwaik Y. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect Immun. 1998;66:883–892. doi: 10.1128/iai.66.3.883-892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L-Y, Stone B J, Brieland J K, Abu Kwaik Y. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb Pathog. 1998;25:291–306. doi: 10.1006/mpat.1998.0237. [DOI] [PubMed] [Google Scholar]
- 21.Gough N R, Fambrough D M. Different steady state subcellular distributions of the three splice variants of lysosome-associated membrane protein LAMP-2 are determined largely by the COOH-terminal amino acid residue. J Cell Biol. 1997;137:1161–1169. doi: 10.1083/jcb.137.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harb O S, Abu Kwaik Y. Identification of the aspartate-β-semialdehyde dehydrogenase gene of Legionella pneumophila and characterization of a null mutant. Infect Immun. 1998;66:1898–1903. doi: 10.1128/iai.66.5.1898-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz M A. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz M A. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Island M D, Wei B Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller D W, Hajjeh R, DeMaria A, Fields B S, Pruckler J M, Benson R S, Kludt P E, Lett S M, Mermel L A, Giorgio C, Breiman R F. Community outbreak of legionnaires' disease: an investigation confirming the potential for cooling towers to transmit Legionella species. Clin Infect Dis. 1995;22:257–261. doi: 10.1093/clinids/22.2.257. [DOI] [PubMed] [Google Scholar]
- 27.Kirby J E, Vogel J P, Andrews H L, Isberg R R. Evidence for pore-forming ability by Legionella pneumophila. Mol Microbiol. 1998;27:323–336. doi: 10.1046/j.1365-2958.1998.00680.x. [DOI] [PubMed] [Google Scholar]
- 28.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 29.Muller A, Hacker J, Brand B C. Evidence for apoptosis of human macrophage-like HL-60 cells by Legionella pneumophila infection. Infect Immun. 1996;64:4900–4906. doi: 10.1128/iai.64.12.4900-4906.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizarro-Cerda J, Meresse S, Parton R G, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel J P. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell M, Shuman H A. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy C R, Berger K H, Isberg R R. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 33.Sadosky A B, Wiater L A, Shuman H A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturgill-Koszycki S, Schaible U E, Russell D G. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996;15:6960–6968. [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson M S, Isberg R R. Analysis of the intracellular fate of Legionella pneumophila mutants. Ann NY Acad Sci. 1996;797:8–18. doi: 10.1111/j.1749-6632.1996.tb52944.x. [DOI] [PubMed] [Google Scholar]
- 36.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]