Abstract
This meta-analysis aims to synthesize global evidence on the risk of reinfection among people previously infected with SARS-CoV-2. We systematically searched PubMed, Scopus, Embase and Web of Science as of April 5, 2021. We conducted: (1) meta-analysis of cohort studies containing data sufficient for calculating the incidence rate of SARS-CoV-2 reinfection; (2) systematic review of case reports with confirmed SARS-CoV-2 reinfection cases. The reinfection incidence was pooled by zero-inflated beta distribution. The hazard ratio (HR) between reinfection incidence among previously infected individuals and new infection incidence among infection-naïve individuals was calculated using random-effects models. Of 906 records retrieved and reviewed, 11 studies and 11 case reports were included in the meta-analysis and the systematic review, respectively. The pooled SARS-CoV-2 reinfection incidence rate was 0.70 (standard deviation [SD] 0.33) per 10,000 person-days. The incidence of reinfection was lower than the incidence of new infection (HR = 0.12, 95% confidence interval 0.09–0.17). Our meta-analysis of studies conducted prior to the emergency of the more transmissible Omicron variant showed that people with a prior SARS-CoV-2 infection could be re-infected, and they have a lower risk of infection than those without prior infection. Continuing reviews are needed as the reinfection risk may change due to the rapid evolution of SARS-CoV-2 variants.
Subject terms: Infectious diseases, Diseases
Introduction
Since the first case of COVID-19 was reported in the early December 20191, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19, has infected 420 million people and has been associated with over 5 million deaths worldwide. The rapid spread of this disease is mainly due to the high efficiency of respiratory transmission and universal susceptibility to the virus in the general population2,3. The pandemic may lose its increasing momentum only after a high proportion of population become immune to the virus or develop herd immunity. Individuals can obtain immunity through infection or vaccination4. Since the first COVID-19 vaccine was available in December 2020, over 60% of the world population has received at least one dose of a COVID-19 vaccine by middle February of 2022. However, there is significant disparity in access to the vaccine by nation, such as 75% in European Union countries and 17% in African countries5. SARS-CoV-2 incidence rates may vary by geographic region, but population infection rates have been continuously increasing globally6–9. Breakthrough infections were also reported among vaccinated individuals and reinfections were increasing common10–12. Then, an urgent public health question is how likely people are to be reinfected.
Studies have shown that SARS-CoV-2 infection-induced immunity may last at least 5–6 months after infection13,14, while some small case studies have shown that repeat infections could occur even within 1–3 months after first infection15–17. Little is known about the risk of repeat infection among previously infected individuals14. Some studies found that the incidence rate of repeat infection was below one percent18,19, while other studies showed a higher reinfection rate20,21. Other factors may also contribute to the difference of reinfection rates in the studies among general population: (1) diagnostic criteria. Some studies defined a possible reinfection based on an interval of > 30 days between two positive PCR tests21 while other studies used an interval ≥ 90 days 19; some studies used non-PCR diagnosis approaches22,23; (2) different SARS-CoV-2 prevalence and incidence and dominant circulating variants in the study population4,20. The demographic characteristics of study population may also account for reinfection rates to some extent, such as proportions of immunocompromised and elderly participants19,24–26 and special groups at high risks such as health care workers4,27. To better estimate the risk of reinfection and describe the characteristics of reinfection cases, we conducted a systematic review and meta-analysis of the global literature.
Methods
Search strategy and selection criteria
This systematic review and meta-analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines28. We proposed two research questions: 1) What is the incidence rate of reinfection among people who have been previously infected with SARS-CoV-2? 2) Is the infection risk of SARS-CoV-2 lower among individuals who have been previously infected than among infection-naïve individuals?
We systematically searched 4 electronic databases PubMed, Scopus, Embase, and Web of Science for publications between January 1, 2020 and April 5, 2021. The search terms included: (reinfection*) AND ("COVID-19" OR "Covid-19" OR "SARS-CoV-2" OR "novel coronavirus" OR "2019-nCov" OR "severe acute respiratory syndrome coronavirus 2") AND (cohort OR follow-up OR "followed up" OR longitudinal). We also manually checked the bibliography of each selected paper for additional studies and searched Google Scholar for additional articles. For studies that did not have enough data, we contacted authors to request data, if appropriate.
Studies were eligible for our meta-analysis if they met the following eligibility criteria:
The study included a group of participants who have previously been infected with SARS-CoV-2;
The study was published in English;
The sample size was no less than 100;
The study reported quantitative data that allowed for the calculation of the incidence rate of SARS-CoV-2 infection or reinfection;
If more than one study was based on the same cohort, only the study with the largest sample size was included;
The study included the outcome of SARS-CoV-2 reinfection which was defined as suspected case or confirmed case based on the definitions by PAHO/WHO29, and those reported recurrent or re-positive or reactivation of SARS-CoV-2 infection were excluded. In addition, we also included case reports and case series in our systematic review, but not the meta-analysis. The language was limited to English.
Data screening
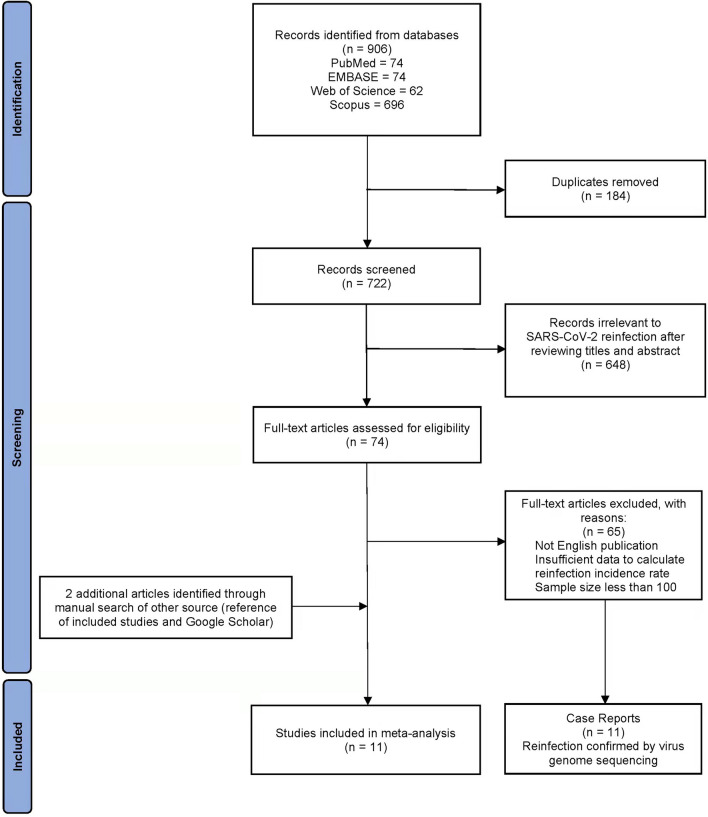
Four authors (LD, PL, QJ, and YG) performed initial screening independently. Duplicate records were removed using EndNote X9 software30. The titles and abstracts were screened to assess whether articles met eligibility criteria. Full texts were assessed if the title and abstract did not provide sufficient information for assessing eligibility. Disagreements were resolved through discussion with the senior investigator (HZQ). Figure 1 describes the literature search and study selection procedures.
Figure 1.
Flow diagram of literature search and study selection.
Data extraction and quality assessment
Four reviewers (LD, XZ, PL, and QJ) extracted the data from individual studies independently. Two standardized data extraction forms (Table 1 and Table 2) were used to extract information from the included epidemiological studies and case reports. Data extracted from epidemiological studies included study location, population (general population or health care workers), start and end dates of participant accrual and follow-up, age, sex, cohort follow-up time, laboratory testing of SARS-CoV-2 infection, sample size, and number of new infections. The data on cohort follow-up person-days were either extracted directly from the studies or calculated by multiplying the mean/median follow-up days with sample size. Data extracted from case reports included study country, age, sex, health status other than COVID-19, time interval of two infections, severity and duration of infection and reinfection.
Table 1.
Characteristics of included epidemiological studies.
| Study | Study country & city | Study population | Start and end dates of participant accrual and follow-up | Accumulative cohort follow-up time, person-days | Laboratory testing of SARS-CoV-2 infection | Age mean (SD) or median [IQR] | Sample size | Male sex, n (%) | Number of infections | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previously-infected group | Previously-noninfected group | Previously-infected group | Previously-noninfected group | Previously-infected group | Previously-noninfected group | Previously-infected group | Previously-noninfected group | Previously-infected group | Previously-noninfected group | |||||
| Mumoli 202041 | Legnano & Milan, Italy | general | 72,360* | 804 | 0 | |||||||||
| Xu 202042 | Guangzhou, China | general | 2020.1.20–2020.4.10 | 8545.9* | PCR, IgG, IgM | IgG positive 49.1 (14.4) IgG negative 43.2 (12.8) | IgG positive 154 IgG negative 33 | IgG positive 68 (44.2) IgG negative14 (42.4) | 10 | |||||
| Abu-Raddad 202140 | Qatar | all SARS-CoV-2 cases | 2020.2.28–2020.8.12 | 3,466,461 | Nasopharyngeal and/or oropharyngeal swabs PCR | 133,266 | 4 | |||||||
| Hall 20214 | England, UK | health care workers | 2020.6.18–2020.12.31 | 2,047,113 | 2,971,436 | Anterior nasal swabs or combined nose and oropharyngeal swabs PCR, sero-Ab | 45.6 [34.6–53.8] | 45.7[35.8–53.9] | 8278 | 17,383 | 1425 (17.2) | 2585 (14.9) | 155 | 1704 |
| Hanrath 202122 | Newcastle upon Tyne, UK | health care workers | 2020.3.10- 2020.11.20 | 179,574* | 1,753,701* | PCR, Ab (IgG) | 39.5 [30–49] | 40 [30–50] | 1038 | 10,137 | 17.50% | 19.50% | 0 | 290 |
| Hansen 202123 | Denmark | general | 2020.2.26–2020.12.31 | 1,346,920 | 62,151,056 | Throat swabs PCR | 11,068 | 514,271 | 72 | 16,819 | ||||
| Leidi 202145 | Geneva, Switzerland | general | 2020.4–2021.1.25 | 178* | 5343.8* | RT-PCR naso- or oropharyngeal | 46.6 (16.6) | 47.3 (16.3) | 498 | 996 | 242 (48.6) | 486 (48.8) | 5 | 154 |
| Lumley 202114 | Oxford, UK | health care workers | 2020.4.23–2020.11.30 | 152,983 | 2,036,358 | nasal and oropharyngeal swab PCR, sero-IgG | 38 [29–49] | 11,276: 38 [29–49] 88: 41 [28–49] | 1177 | 11,364 | 339 (28.8) | 2920 (25.7) | 2 | 26 |
| Masia 202126 | Spain | general | 26,280* | PCR, Ab (IgG), genome sequencing | Median: 64 | 146 | 60.30% | 1 | ||||||
| Murillo-Zamora 202143 | Mexico | general | PCR | 100,432 | 258 | |||||||||
| Pilz 202144 | Austria | general | 3,116,400* | 1,865,984,400* | PCR | 14,840 | 8,885,640 | 40 | 253,581 | |||||
SD standard deviation; IQR interquartile range; PCR polymerase chain reaction; IgG immunoglobulin G; IgM immunoglobulin M; Ab antibody.
*The duration of follow-up was calculated through multiplying sample size by median or mean follow-up time.
Table 2.
Characteristics of included case reports.
| Article | Country | Age, sex | Health status other than COVID-19 | Time interval (Date of first laboratory PCR positive-date of first laboratory PCR positive during reinfection) | Severity of reinfection compared with prime infection | Note (e.g., different variants, severity, vaccination history, …) | Duration of reinfection compared with prime infection (duration of prime infection, duration of reinfection) (Diagnostic criterion ‡) |
|---|---|---|---|---|---|---|---|
| Larson 202046 | USA | 42, M# | Healthy | 65 (2020.03.20–2020.05.24) | More severe | Several potential variations, including one high confidence variation | Longer (10, 14) (Clinical) |
| Goldman 202051 | USA | 60 ~ 69, ND | Severe emphysema (FEV1 34% predicted) on home oxygen, and hypertension | 140 (ND) | Less severe | Revealed 10 high confidence intra-host single nucleotide variants (iSNVs) of which 5 type the March sequence to clade 19B, and 5 type the July sequence to 20A | ND |
| Lee 202015 | South Korea | 23, F | Healthy | 26 (2020.03.11–2020.04.06) | Similar | Different SARS-CoV-2 subtype (pike protein D614G substitution, mutations characterizing the clade “V” (ie, nsp6 L37F and ORF3a G251V) | Shorter (15,13) (Laboratory) |
| To 202052 | China | 33, M | Healthy | 142 (2020.03.26–2020.08.15) | Less severe | The first viral genome belongs to GISAID clade V, Nextstrain clade 19A, and Pangolin lineage B.2 with a probability of 0.99. The second viral genome belongs to GISAID clade G, Nextstrain clade 20A, and Pangolin lineage B.1.79 with a probability of 0.70 | Shorter (3,0) (Clinical) |
| Gousseff 202049 | Switzerland | 36, F# | Healthy | 204 (2020.4.10–2020.10.31) | Similar | Two different SARS-CoV-2 genomes both belonging to clade 20A | Shorter (14,10) (Clinical) |
| Gupta 202047 | India | 25, M# | ND | 108 (2020.05.05–2020.08.21) | Similar | A genetic variant 22882 T > G (S: N440K) found during reinfection in I2 | Longer (8,14) (Laboratory) |
| 28, F# | ND | 111 (2020.05.17–2020.09.05) | Similar | Shorter (12,6) (Laboratory) | |||
| Prado-Vivar 202116 | Ecuador | 46, M | ND | 63 (2020.05.20–2020.07.22) | More severe | The first infection variant belonged to clade 20A and lineage B1.p9, whereas the second infection variant belonged to clade 19B and lineage A.1.1 | shorter(22, 15)(Clinical and laboratory ) |
| Klein 202118 | USA | 60 ~ 70, M | Renal transplantation 2 years prior end-stage renal disease | 233(ND) | Less severe | The virus genome sequenced from the reinfection had 12 mutations not observed in the virus sequenced from the primary infection | Shorter (27,15) (Clinical) |
| Van Elslande 202117 | Belgium | 51, F | Asthma | 90 (2020.03.09–2020.06.10) | Less severe | Distinct: the initial infection was caused by a lineage B.1.1 SARS-CoV-2 virus and the relapsing infection by a lineage A. Eleven mutations were identified | Shorter (49,7) (Clinical) |
| Salehi-Vaziri 202148 | Iran | 32, F | Healthy | 63 (2020.04.20-ND) | More severe | D614G mutation | Longer (28,30) (Laboratory) |
| 42, M | Healthy | 111 (2020.3.10-ND) | Less severe | D614G mutation | Longer (39,5) (Laboratory) | ||
| Tillett 202150 | USA | 25, M | Healthy | 48 (2020.03.25–2020.06.05) | More severe | Both specimens were members of clade 20C, but have different mutations | ND |
#Health care workers; ‡Clinical diagnosis such as presence of duration or hospital discharge, or laboratory results of polymerase chain reaction (PCR) testing.
M—Male.
F—Female.
ND–no data.
The Newcastle–Ottawa Quality Assessment Form for Cohort Studies31 was used to appraise the methodological quality (supplementary Table 1). The total score is 9, and the higher score means better quality. An additional criterion was added to assess the quality of laboratory testing of reinfection—whether whole-genome sequencing of SARS-CoV-2 was used to assess reinfection.
During data extraction and quality assessment, discrepancies were resolved through discussion with the senior investigator (HZQ).
Data analysis
Data analysis was performed using R software (version 4.0.2)32 and gamlss (version 5.3.4)33, gamlss.dist (version 5.3.2)34, meta (version 4.19.0)35, metafor (version 3.0.2)36, dmetar (version 0.0.9)37 packages.
Our main outcome of interest was SARS-CoV-2 reinfection. Two primary analyses were performed to calculate the pooled incidence rate of SARS-CoV-2 reinfection and to compare the pooled incidence rates among those who were previously infected and among those who were never infected with SARS-CoV-2.
The incidence rates were calculated by dividing number of cases by total person-time followed up among these participants. The total person-time for each study was either directly provided or calculated based on the time span and number of participants. To include studies with zero event into analysis, the two-part zero-inflated beta (ZIB) distribution was used to calculate the pooled incidence rate33,34. The pooled incidence rate was defined as the marginal mean of ZIB38. We also simulated the incidence rate for 1000 times using ZIB distribution with estimated parameters and used the standard errors of these simulated samples to calculate 95% confidence intervals (CI) of the pooled incidence rate.
To compare the incidence rates between previously infected individuals and uninfected individuals, we calculated hazard ratio (HR) by computing the incidence ratios between previously infected and uninfected groups. Heterogeneity was assessed by Cochran’s Q test, and the degree of heterogeneity was assessed by I2 statistics. Due to the high heterogeneity of included studies, we used random-effects models. HRs were pooled with DerSimonian-Laird method39 Significance was considered to have a two-sided P value < 0.05.
Subgroup analyses were also conducted among health care workers and the general population.
Registration
The study protocol was registered with the International Prospective Register of Systematic Reviews (PROSPERO) (https://www.crd.york.ac.uk/PROSPERO/, ID: CRD42021265784).
Results
A total of 906 articles were retrieved from electronic database searches. After removing duplicates, 722 records were retained. After 648 records were excluded through reviewing the titles or abstracts, 74 full texts were reviewed, and 2 additional articles were identified through manual searches of the references and Google Scholar. After review, 11 epidemiological studies and 11 case reports were eligible (Fig. 1).
Of the 11 included epidemiological studies for meta-analysis (Table 1), three were conducted in the United Kingdom4,14,22, and one in each of 8 countries Denmark23, Qatar40, Italy41, Spain26, China42, Mexico43, Austria44, and Switzerland45. All 11 studies were either prospective or retrospective cohort studies published between 2020 and 2021. Length of follow-up for assessing new SARS-CoV-2 infections among participants with exposure to index cases ranged from 3 to 9 months. No study reported vaccination status of study participants. All 11 studies were included in calculating the pooled incidence rate, and 6 were also included in the analysis of comparing previously-infected and uninfected individuals4,14,22,23,44,45. Of 11 epidemiological studies, 27% (3/11) were conducted among health care workers and 73% among the general population. A total of 9,711,525 participants in these 11 epidemiological studies were included in meta-analysis, including 271,734 with a history of SARS-CoV-2 infection and 9,439,791 without. The mean age of all participants was 42.6 years.
The pooled incidence rate of SARS-CoV-2 reinfection was 0.70 (standard deviation [SD] 0.33) per 10,000 person-days or 2.5% (1.2%) person-years (Table 3). Subgroup analyses showed the incidence rate was 0.30 (SD 0.18) per 10,000 person-days or 1.1% (SD 0.6%) person-years among health care workers and 0.85 (SD 0.49) per 10,000 person-days or 3.1% (SD 1.8%) person-years among the general population (Table 3). The difference between the two groups was statistically significant (P = 0.02) (Table 3).
Table 3.
Pooled incidence rate of reinfection and subgroup analysis.
| Number of included studies | Number of studies with zero reinfection | Total number of participants | Total number of reinfections | Pooled incidence rate per 10,000 person-days (SD)* | Group difference P value | |
|---|---|---|---|---|---|---|
| Total | 11 | 3 | 271,734 | 547 | 0.70 (0.33) | |
| Subgroups | ||||||
| Health care workers | 3 | 1 | 10,493 | 157 | 0.30 (0.18) | 0.02 |
| General population | 8 | 2 | 261,241 | 390 | 0.85 (0.49) | |
SD standard deviation.
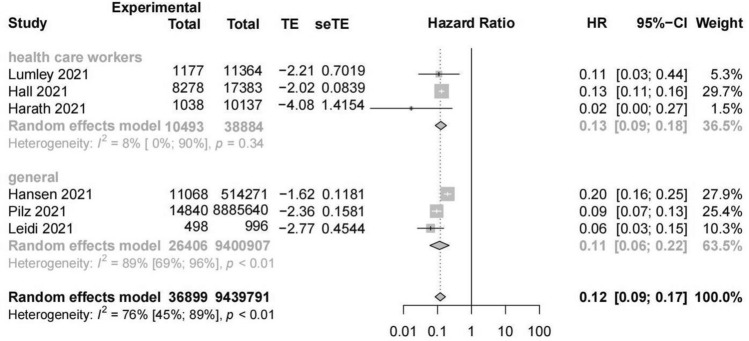
A random-effects meta-analysis of six studies reporting new SARS-CoV-2 infection among the previously-infected group and noninfected group showed a significantly lower risk of infection in the previously-infected group (HR = 0.12, 95% CI = 0.09–0.17, number of estimates (k) = 6, I2 = 76%, 95% CI = 45%-89%). The HR was slightly lower among the general population (HR = 0.11, 95% CI = 0.06–0.22, k = 3, I2 = 89%, 95% CI = 69%-96%) than health care workers (HR = 0.13, 95% CI = 0.09–0.18, k = 3, I2 = 8%) (Fig. 2). Influence analysis indicated the robustness of the results (Supplementary Fig. 1). Egger’s test showed no publication bias in these studies (P = 0.34).
Figure 2.
Forest plot of association between previous infection and SARS-CoV-2 reinfection.
Nine studies4,14,22,23,40,42–45 were ranked as good or fair quality31. Two studies26,41 were considered as poor quality.
A total of 13 SARS-CoV-2 reinfection cases were reported in the 11 case reports or case series15–18,46–52 and all were confirmed via whole-genome sequencing. More than half of cases (7/13) were reported in 2020. Participant age ranged from 25 to 70 years, and about half (7/13) were men. Four cases were health care workers, and three had comorbid diseases (Table 2).
The time interval between two episodes of infection ranged from 26 to 233 days. Five cases had less severe symptoms of reinfection than first infection, four had more severe symptoms, and four had similar severity. Eight cases had a shorter duration of disease in the second infection than the first, while three patients had longer duration in their second infection (Table 2).
Discussion
This systematic review and meta-analysis summarized the evidence on likelihood of SARS-CoV-2 reinfection. The pooled incidence rate was 0.70 per 10,000 person-days or 2.5 per 100 person-years. People who were previously infected were 87% less likely to get reinfection than those who were never infected (HR = 0.12). Although the risk of reinfection may be low for individuals, the global number of reinfections could be several millions in one year, considering over 440 million people had been infected worldwide by February of 2022. It is suggested that people who have been infected should also receive vaccinations and use personal protections to reduce the risk of reinfection. Given the global number of reinfections, this guidance is warranted.
Our meta-analysis showed that health care workers had lower incidence of reinfection than the general population (0.30 vs 0.85). Health care workers have more COVID-19 exposure than the general population, but they may have higher risk awareness and better use of personal protections than the general population, leading to a lower likelihood of reinfection.
In the results of our analysis, the I2 statistics and Cochran’s Q test result indicated relatively strong heterogeneity among included studies. The reinfection rates could vary across different study geographic locations and time periods. The extent of local virus spread could also affect the reinfection rates. Such differences may underlie the heterogeneity and cause a significant Cochran’s Q test result in the general population sub-group. Due to lack of data, we cannot control such factors in our meta-analysis.
The risk of reinfection could be affected by numerous factors. For example, the likelihood for a person to get an infectious disease depends on the chance of exposure and use of personal protection, and reinfection is also associated with decline in immunity and virus mutation and circulating SARS-CoV-2 variants53,54. Our analysis has limitations. First, the median follow-up time of participants was less than 6 months in most studies included in this meta-analysis. The infection-induced immunity may wane over time, and the risk of reinfection may increase. Studies have found that the vaccine-induced neutralizing antibody response against the spike protein of five major SARS-CoV-2 variants declined over time55 , and infection-induced humoral immunity against SARS-CoV-2 (IgG level) might not be long lasting in persons with mild illness56. Real-world research is needed to assess and the duration of infection- and vaccine-induced immunity against SARS-CoV-2 reinfection. The duration of immune response may also be moderated by other factors such as age. A study on immunogenicity of an mRNA vaccine showed that serum neutralization and levels of binding IgG or IgA after the first vaccine dose were lower in older individuals, but neutralization against SARS-CoV-2 variants was detectable regardless of age57. Second, our literature search was limited to publications before April 5, 2021, when the Omicron variant has not emerged. Studies have shown both immune evasion by Omicron variant contributed to a higher transmission rate than other variants58–60 Third, As the vaccination status was not reported in the included studies, vaccination status was not considered in assessing the risk of reinfection. Before April 2021, most countries had not started to vaccinate their populations, or if had started, might still have low vaccination rates. Therefore, the estimated risk of reinfection is unlikely to be significantly confounded by vaccination. Updated meta-analysis is needed to estimate the risk of reinfection in the circumstance of Omicron as the dominant variant. As the data related to SARS-CoV-2 reinfection becomes more available, sub-analyses could be explored to examine the rates of reinfection by a variety of covariates, such as age, sex, comorbidities and history of vaccination. The results from this meta-analysis may serve as a comparison to future research on the risk of reinfection of Omicron and new emerging variants among the widely vaccinated population61,62.
Conclusion
In conclusion, our meta-analysis suggests that there is a risk of reinfection among people who have been previously diagnosed with COVID-19. Vaccination may produce higher neutralizing antibody titers compared to SARS-CoV-2 infection63, and people who are infected with SARS-CoV-2 can still benefit from vaccination, particularly for the purposes of preventing more transmissible variants.
Supplementary Information
Author contributions
L.D., P.L., X.Z., Q.J. and H.Z.Q. designed the study. L.D., P.L., X.Z., Q.J. and Y.G. collected the data. L.D., X.Z. and C.h.Z. analyzed the data. L.D., P.L., Q.J., D.T. and H.Z.Q. drafted the manuscript. All authors interpreted the results and revised the manuscript. All authors read and approved the final version of the manuscript. The corresponding author H.Z.Q. has final responsibility for the decision to submit for publication.
Funding
This research did not receive grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
The data analyzed in this meta-analysis are from previously published studies, which have been cited.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Luojia Deng, Peiqi Li, Xuezhixing Zhang and Qianxue Jiang.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-24220-7.
References
- 1.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B, et al. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rostami A, et al. Update on SARS-CoV-2 seroprevalence: Regional and worldwide. Clin. Microbiol. Infect. 2021;27(12):1762–1771. doi: 10.1016/j.cmi.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall VJ, et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: A large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397(10283):1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Our World in Data. https://ourworldindata.org/covid-vaccinations?country=OWID_WRL Accessed 18 Feb 2021.
- 6.Anand S, et al. Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: A cross-sectional study. Lancet. 2020;396(10259):1335–1344. doi: 10.1016/S0140-6736(20)32009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JM, et al. Estimated US infection- and vaccine-induced SARS-CoV-2 seroprevalence based on blood donations, July 2020-May 2021. JAMA. 2021;326(14):1400–1409. doi: 10.1001/jama.2021.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar D, Burma A, Kumar Mandal A. A seroprevalence study of Covid 19 antibody after 1st wave of the pandemic in South Andaman district India. Clin. Epidemiol. Glob. Health. 2021;12:100901. doi: 10.1016/j.cegh.2021.100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallal PC, et al. Slow spread of SARS-CoV-2 in Southern Brazil over a 6-month period: Report on 8 sequential statewide serological surveys including 35 611 participants. Am. J. Public Health. 2021;111(8):1542–1550. doi: 10.2105/AJPH.2021.306351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson KB, et al. Postvaccination SARS-CoV-2 infections and incidence of the B.1.427/B.1.429 variant among healthcare personnel at a northern California academic medical center. MedRxiv. 2021;182(5):1284. doi: 10.1093/cid/ciab554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, et al. Mild breakthrough infection in a healthcare professional working in the isolation area of a hospital designated for treating COVID-19 patients—Shaanxi Province, China, March 2021. China CDC Wkly. 2021;3(19):397–400. doi: 10.46234/ccdcw2021.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyagi K, et al. Breakthrough COVID19 infections after vaccinations in healthcare and other workers in a chronic care medical facility in New Delhi India. Diabetes Metab. Syndr. 2021;15(3):1007–1008. doi: 10.1016/j.dsx.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lumley SF, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N. Engl. J. Med. 2021;384(6):533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JS, et al. Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clin. Infect. Dis. 2021;73(9):e3002–e3008. doi: 10.1093/cid/ciaa1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado-Vivar B, et al. A case of SARS-CoV-2 reinfection in Ecuador. Lancet Infect. Dis. 2021;21(6):e142. doi: 10.1016/S1473-3099(20)30910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Elslande J, et al. Symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2021;73(2):354–356. doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, J. et al. Case Study: Longitudinal immune profiling of a SARS-CoV-2 reinfection in a solid organ transplant recipient. medRxiv (2021).
- 19.Slezak J, et al. Rate and severity of suspected SARS-Cov-2 reinfection in a cohort of PCR-positive COVID-19 patients. Clin. Microbiol. Infect. 2021;27(12):1860.e7–1860.e10. doi: 10.1016/j.cmi.2021.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhry Z, et al. Short durations of corticosteroids for hospitalised COVID-19 patients are associated with a high readmission rate. J. Infect. 2021;82(6):276–316. doi: 10.1016/j.jinf.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finch E, et al. SARS-CoV-2 antibodies protect against reinfection for at least 6 months in a multicentre seroepidemiological workplace cohort. Plos Biol. 2022;20(2):e3001531. doi: 10.1371/journal.pbio.3001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanrath AT, Payne BAI, Duncan CJA. Prior SARS-CoV-2 infection is associated with protection against symptomatic reinfection. J. Infect. 2021;82(4):e29–e30. doi: 10.1016/j.jinf.2020.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen CH, et al. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: A population-level observational study. Lancet. 2021;397(10280):1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brawner CA, et al. Inverse relationship of maximal exercise capacity to hospitalization secondary to coronavirus disease 2019. Mayo Clin. Proc. 2021;96(1):32–39. doi: 10.1016/j.mayocp.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffery-Smith A, et al. Antibodies to SARS-CoV-2 protect against re-infection during outbreaks in care homes, September and October 2020. Eurosurveill. 2021 doi: 10.2807/1560-7917.ES.2021.26.5.2100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masiá M, et al. Incidence of delayed asymptomatic COVID-19 recurrences in a 6-month longitudinal study. J Infect. 2021;82(6):276–316. doi: 10.1016/j.jinf.2021.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adrielle Dos Santos L, et al. Recurrent COVID-19 including evidence of reinfection and enhanced severity in thirty Brazilian healthcare workers. J. Infect. 2021;82(3):399–406. doi: 10.1016/j.jinf.2021.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan American Health Organization/World Health Organization. Interim guidelines for detecting cases of reinfection by SARS-CoV-2. https://www.paho.org/en/documents/interim-guidelines-detecting-cases-reinfection-sars-cov-2 Accessed 29 Oct 2020.
- 30.Hupe M. EndNote X9. J. Electron. Resour. Med. Libr. 2019;16(3–4):117–119. doi: 10.1080/15424065.2019.1691963. [DOI] [Google Scholar]
- 31.Wells G, et al. Newcastle-Ottawa quality assessment scale cohort studies. Ottawa: University of Ottawa; 2014. [Google Scholar]
- 32.R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 33.Rigby RA, Stasinopoulos DM. Generalized additive models for location, scale and shape. J. R. Stat. Soc. Ser. C Appl. Stat. 2005;54:507–554. doi: 10.1111/j.1467-9876.2005.00510.x. [DOI] [Google Scholar]
- 34.Mikis, S., Robert, R. Gamlss.dist: Distributions for generalized additive models for location scale and shape. R package version 5.3–2. https://CRAN.R-project.org/package=gamlss.dist.
- 35.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: A practical tutorial. Evid. Based Ment. Health. 2019;22(4):153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36(3):1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 37.Harrer, M., Cuijpers, P., Furukawa, T. & Ebert, D. D. Dmetar: Companion R package for the guide 'doing meta-analysis in R'. R package version 0.0.9000. URL http://dmetar.protectlab.org/ (2010).
- 38.Chai H, et al. A marginalized two-part beta regression model for microbiome compositional data. Plos Comput. Biol. 2018;14(7):e1006329. doi: 10.1371/journal.pcbi.1006329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin. Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 40.Abu-Raddad LJ, et al. Assessment of the risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection in an intense reexposure setting. Clin. Infect. Dis. 2021;73(7):e1830–e1840. doi: 10.1093/cid/ciaa1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mumoli N, Vitale J, Mazzone A. Clinical immunity in discharged medical patients with COVID-19. Int. J. Infect. Dis. 2020;99:229–230. doi: 10.1016/j.ijid.2020.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu G, et al. No evidence of re-infection or person-to-person transmission in cured COVID-19 patients in Guangzhou, a retrospective observational study. Front. Med. 2020;7:593133. doi: 10.3389/fmed.2020.593133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murillo-Zamora E, et al. Predictors of severe symptomatic laboratory-confirmed SARS-CoV-2 reinfection. Public Health. 2021;193:113–115. doi: 10.1016/j.puhe.2021.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilz S, et al. SARS-CoV-2 re-infection risk in Austria. Eur. J. Clin. Invest. 2021;51(4):e13520. doi: 10.1111/eci.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leidi A, et al. Risk of reinfection after seroconversion to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A population-based propensity-score matched cohort study. Clin. Infect. Dis. 2022;74(4):622–629. doi: 10.1093/cid/ciab495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson D, et al. A case of early reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2021;73(9):e2827–e2828. doi: 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta V, et al. Asymptomatic reinfection in 2 healthcare workers from India with genetically distinct severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2021;73(9):e2823–e2825. doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salehi-Vaziri M, et al. Clinical characteristics of SARS-CoV-2 by re-infection vs. reactivation: A case series from Iran. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:1713–1719. doi: 10.1007/s10096-021-04221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gousseff M, et al. Clinical recurrences of COVID-19 symptoms after recovery: Viral relapse, reinfection or inflammatory rebound? J. Infect. 2020;81(5):816–846. doi: 10.1016/j.jinf.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tillett RL, et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman JD, et al. Reinfection with SARS-CoV-2 and failure of humoral Immunity a case report. MedRxiv. 2020 doi: 10.1101/2020.09.22.20192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.To KK, et al. Coronavirus disease 2019 (COVID-19) re-infection by a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2021;73(9):e2946–e2951. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mlcochova P, et al. SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hui KPY, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603(7902):715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 55.Evans JP, et al. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022;14(637):eabn8057. doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ibarrondo FJ, et al. Rapid decay of Anti-SARS-CoV-2 antibodies in persons with mild covid-19. N. Engl. J. Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collier DA, et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596(7872):417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Y, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cameroni E, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews N, et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 2022;386(16):1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marinov GK, et al. SARS-CoV-2 reinfections during the first three major COVID-19 waves in Bulgaria. Plos One. 2022;17(9):e0274509. doi: 10.1371/journal.pone.0274509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang SL, et al. SARS-CoV-2 in Malaysia: A surge of reinfection during the predominantly Omicron period. Lancet Reg. Health West. Pac. 2022;26:100572. doi: 10.1016/j.lanwpc.2022.100572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Favresse J, et al. Neutralizing antibodies in COVID-19 patients and vaccine recipients after two doses of BNT162b2. Viruses. 2021;13:1369. doi: 10.3390/v13071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this meta-analysis are from previously published studies, which have been cited.