Abstract
Background
Medication adherence is the main determinant of effective management of type 2 diabetes, yet there is no gold standard method available to screen patients with high-risk non-adherence. Developing machine learning models to predict high-risk non-adherence in patients with T2D could optimize management.
Methods
This cross-sectional study was carried out on patients with T2D at the Sichuan Provincial People's Hospital from April 2018 to December 2019 who were examined for HbA1c on the day of the survey. Demographic and clinical characteristics were extracted from the questionnaire and electronic medical records. The sample was randomly divided into a training dataset and a test dataset with a radio of 8:2 after data preprocessing. Four imputing methods, five sampling methods, three screening methods, and 18 machine learning algorithms were used to groom data and develop and validate models. Bootstrapping was performed to generate the validation set for external validation and univariate analysis. Models were compared on the basis of predictive performance metrics. Finally, we validated the sample size on the best model.
Results
This study included 980 patients with T2D, of whom 184 (18.8%) were defined as medication non-adherence. The results indicated that the model used modified random forest as the imputation method, random under sampler as the sampling method, Boruta as the feature screening method and the ensemble algorithms and had the best performance. The area under the receiver operating characteristic curve (AUC), F1 score, and area under the precision-recall curve (AUPRC) of the best model, among a total of 1,080 trained models, were 0.8369, 0.7912, and 0.9574, respectively. Age, present fasting blood glucose (FBG) values, present HbA1c values, present random blood glucose (RBG) values, and body mass index (BMI) were the most significant contributors associated with risks of medication adherence.
Conclusion
We found that machine learning methods could be used to predict the risk of non-adherence in patients with T2D. The proposed model was well performed to identify patients with T2D with non-adherence and could help improve individualized T2D management.
Keywords: medication adherence, T2D, machine learning, prediction model, ensemble model
Introduction
Diabetes mellitus (DM) is a common chronic disease with disordered metabolism and hyperglycemia. Type 2 diabetes (T2D) accounts for more than 90% of diabetic cases (1, 2). As morbidity and prevalence continue to rise worldwide, T2D greatly increases healthcare costs and imposes a tremendous economic burden on society and public health systems (3, 4). Total healthcare costs for diabetics are estimated ~$2.1 trillion by 2030 (5).
Pharmacotherapy is one of the most commonly used treatment modalities for controlling the progression of chronic diseases, especially diabetes. In most cases, the benefits of high adherence to medications have been well-determined in diabetes (6–8). The extent to which patients follow prescribed treatments determines the outcome. However, poor adherence to oral hypoglycemic drugs is common in patients with T2D (9). As reported, between a third and a half of drugs prescribed for patients with T2D were not taken as recommended, and estimates varied widely depending on the population studied (10–13). Evidence suggested that non-adherence was an important contributor associated with poor glycemic control and other negative health outcomes, such as the increased risk of hospitalization and complications (14, 15). In a decade, studies indicated that telephone calls, text messages, and educational interventions played an important role in improving adherence to medication (16–18). However, for patients with good compliance, additional interventions are a waste of healthcare resources that are already limited. Thus, the early detection of patients with a high risk of poor adherence to medication is the premise of these effective interventions.
So, we considered whether it is possible to identify patients with a high risk of poor medication adherence early and provide individualized methods to improve their compliance. In our previous study, we reported predictive models of the risks of medication adherence in patients with T2D (19), and the area under the receiver operating characteristic curve (AUC) of the ensemble model was 0.866. The results confirmed that machine learning could be used to predict the risk of drug non-adherence in patients with T2D. Thus, in this study, we used a larger sample size, more variables, more data preprocessing algorithms, and machine learning algorithms to develop models that could more accurately predict medication adherence in patients with T2D.
Methods
Data sources and participants
The cross-sectional study was conducted at the Sichuan Provincial People's Hospital from 1 April 2018 to 31 December 2019. We performed a face-to-face questionnaire interview and filled out questionnaires according to the responses of the patients who participated in the survey. Participants were selected according to the following criteria: (1) diagnosed as patients with T2D; (2) examined HbA1c on the day of the questionnaire; (3) interested to take part in the survey and provide information to the investigators, as well as signed the informed consent forms; (4) received hypoglycemic agency treatment; and (5) over 18 years of age. Ethics approval was obtained through the Ethics Committee of the Sichuan Provincial People's Hospital (approval # 2018-53).
Data collection and outcome definition
The data in this study were collected from electronic medical records (EMRs) and face-to-face questionnaires. Clinical laboratory results, such as HbA1c value and fasting blood glucose (FBG) value, were collected according to EMRs. Body mass index (BMI) was calculated using the following formula: BMI = weight (kg)/height2 (m2). Information on self-glycemic monitoring, diet, exercise, and mental state were provided by patients in face-to-face questionnaires. The questionnaire consists of four parts. The first part is about basic characteristics, including age, nationalities, waistline, occupation, marital status, and so on. The second part is related to self-glycemic monitoring, containing regular measurements frequency of FBG, measurement interval between previous and present, etc. The third part was about exercise, diet, and mental state. The last part was treatment regimen and medication adherence, in which we recorded the duration of the treatment regimen, type and dose of insulin used, etc. The adherence status, which was determined as the outcome variable, was defined according to the proportion of days covered (PDC). PDC higher than 80% was regarded as good medication compliance (20, 21).
Data preprocessing
Data were preprocessed by removing (1) the variables with missing values >90%, (2) the variables with a single value occupying >90%, and (3) the variables with coefficients of variation < 0.01. After the above steps, the data were further processed.
Data partition and dataset building
The data were randomly divided into two subsets (namely, training set and test set) at a ratio of 8:2, which would be used to train and test models, respectively.
Missing data were inevitable in practice. In case of questionable data or missing data in the part of the questionnaire, patients were contacted via telephone for certainty or addition. However, the clinical characteristics of the patients comprised several missing values, such as FBG and postprandial blood glucose (PBG). Missing data were filled in using four imputing methods, including not imputing (marked as Not), simple imputing, random forest, and modified random forest.
Due to the imbalanced data of medication adherence, five sampling methods were applied, including not sampling (marked as Not), Synthetic Minority Oversampling Technique (SMOTE), Borderline SMOTE, Random Over Sampler, and Random Under Sampler.
Three variable selection methods were considered in this study, including no screening (marked as Not), Boruta, and LassoCV. The importance of variables was evaluated according to the output of Boruta and LassoCV (variable importance scores). A high score suggested that the variable could improve predictive accuracy.
Thus, a total of 60 datasets were derived from the training set and set up by using four imputing methods, five sampling methods, and three feature screening methods.
Model development
In this process, several machine learning algorithms were trained for binary classification and applied to develop predictive models, including AdaBoost, Extreme Gradient Boosting (XGBoost), gradient boosting, Bagging, Bernoulli Naive Bayes, Gaussian Naive Bayes, Multinomial Naive Bayes, decision tree, extra tree, K-nearest neighbor (KNN), linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), logistic regression, passive-aggressive, random forest, Stochastic Gradient Descent (SGD), support vector machine (SVM), and ensemble algorithm. The ensemble algorithm summarized the output of the five best models [assessed by area under the receiver operating characteristic curve (AUC)] among the trained models and generated output according to the voting principle.
Model evaluation
Internal validation was conducted with 10-fold cross-validation in 60 datasets, and 10 independent repeated values among indices were collected. Then, the test set was used for external validation. The predictive performances of those models were assessed by the AUC, accuracy, precision, recall, F1-score, and area under the precision-recall curve (AUPRC). AUPRC was calculated by taking the average of precision across all recall values corresponding to different thresholds, and a high value represented both high recall and precision (22, 23).
To elucidate the contribution of different imputing methods, sampling methods, screening methods, machine learning algorithms, and variables, univariate analysis was performed. The whole process could be described as follows: (1) before analysis, the test set was expanded using the Bootstrap method with 2,000 times resampling from the test set. (2) Additionally, the average performance metrics of each method were calculated, respectively. (3) Univariate analysis was used for statistical analysis. The highest values of performance metrics meant that the method was the best than others. If the average performance metrics of models when the variable was included were significantly higher than the average performance indicators when the variable was excluded (P < 0.05), the variable would be judged as a positive contribution to the prediction improvement.
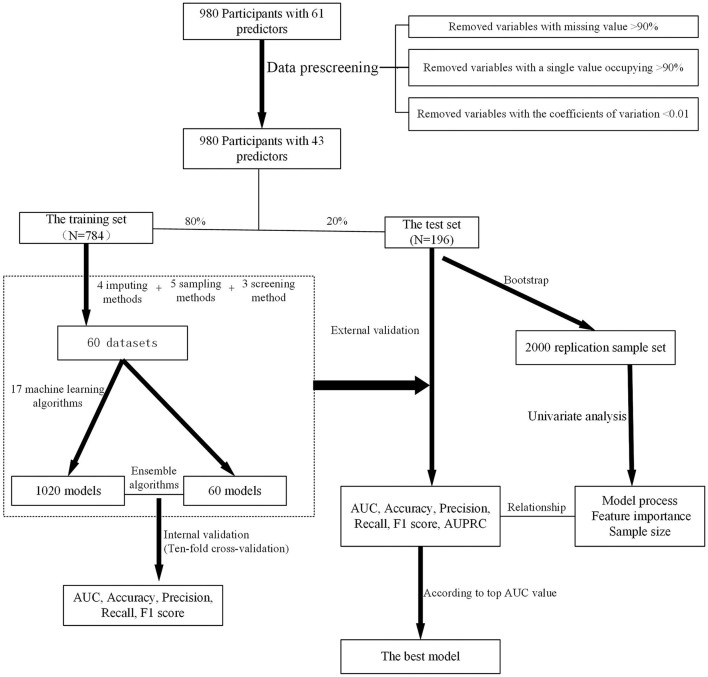
Above all, the overall process of model development and validation is shown in Figure 1.
Figure 1.
The schematic flow of the main steps in this study.
Sample size validation
The best model (assessed by AUC) was employed to estimate the impact of sample sizes on predictive performance (19). The total samples were randomly separated into 80% training set and 20% test set. First, 10% of the samples were randomly extracted from the training set to train the model, and AUC was evaluated in the test set. The training samples increased from 10 to 100% in increments of 10%. These steps were repeated 10 times so that ten independent repeated values of AUC were generated. The contribution of a sample size to improve the prediction performance of models was assessed according to the inflection point change of the line graph.
Statistical analysis
Continuous variables were described by mean and standard deviation, whereas categorical variables were expressed in terms of frequencies and percentages. Analysis of variance (ANOVA) and rank sum test were used for univariate analysis.
Statistical analysis was implemented using the stats package, and model development was performed using the sklearn package in Python (Python Software Foundation, Python Language Reference, version 3.6.8) on PyCharm (developed by JetBrains.r.o., version 11.0.4). The results of variable valuation assessed using univariate analysis were summarized and presented by box plots using R (R software, version 4.0.2).
Results
Participant characteristics
Overall, 980 patients completed the survey, among which 571 were male and 409 were female. The mean age was 59.2 ± 11.9 years. In total, 184 patients were defined as having poor medication adherence (18.8%). Detailed characteristics of participants are shown in Table 1.
Table 1.
The detailed information of participants.
| Variable | Identifier | Parameter |
Value
(N = 980) |
|---|---|---|---|
| Basic characteristics | |||
| Age | X1 | N | 980 |
| Mean ± SD | 59.2 ± 11.9 | ||
| Median | 59 | ||
| Minimum, maximum | 21, 90 | ||
| Nationalities | X2 | N | 979 |
| Han | 945 (96.5%) | ||
| Tibetan | 31 (3.2%) | ||
| Qiang | 3 (0.3%) | ||
| Gender | X3 | N | 980 |
| Male | 571 (58.3%) | ||
| Female | 409 (41.7%) | ||
| Height (m) | X4 | N | 978 |
| Mean ± SD | 1.6 ± 8.0 | ||
| Median | 1.6 | ||
| Minimum, maximum | 1.4, 1.9.0 | ||
| Waistline (cm) | X5 | N | 913 |
| Mean ± SD | 85.3 ± 9.5 | ||
| Median | 83.3 | ||
| Minimum, maximum | 66.6, 128.0 | ||
| Weight (kg) | X6 | N | 976 |
| Mean ± SD | 64.2 ± 10.5 | ||
| Median | 64 | ||
| Minimum, maximum | 40.0, 110.0 | ||
| Marital status | X7 | N | 976 |
| Unmarried | 9 (0.9%) | ||
| Married | 940 (96.3%) | ||
| Divorced | 4 (0.4%) | ||
| Widowed | 23 (2.4%) | ||
| Occupational status | X8 | N | 978 |
| Unemployed | 133 (13.6%) | ||
| Empolyed | 358 (36.6%) | ||
| Retirement | 482 (49.3%) | ||
| Others | 5 (0.5%) | ||
| Education level | X9 | N | 978 |
| Illiteracy | 92 (9.4%) | ||
| Junior middle school | 366 (37.4%) | ||
| High school or special secondary school | 264 (27.0%) | ||
| College and above educational level | 256 (26.2%) | ||
| Family history of diabetes mellitus | X10 | N | 970 |
| No | 629 (64.8%) | ||
| Yes | 341 (35.2%) | ||
| BMI (kg/m2) | X11 | N | 975 |
| Mean ± SD | 24.3 ± 3.3 | ||
| Median | 24.0 | ||
| Minimum, maximum | 16.2, 45.2 | ||
| Health status scores (%) | X12 | N | 980 |
| Mean ± SD | 77.3 ± 10.8 | ||
| Median | 80 | ||
| Minimum, maximum | 30, 100 | ||
| Clinical information | |||
| Course of diabetes (in months) | X13 | N | 980 |
| Mean ± SD | 90.3 ± 76.5 | ||
| Median | 72 | ||
| Minimum, maximum | 1, 540 | ||
| Medicare status | X14 | N | 518 |
| unreimbursement | 233 (45.0%) | ||
| reimbursement | 285 (55.0%) | ||
| Frequency of FBG measurements | X15 | N | 980 |
| Irregular measurements | 139 (14.2%) | ||
| Two to three times a week | 323 (33.0%) | ||
| Three to four times a month | 400 (40.8%) | ||
| Two to three times per three months | 118 (12.0%) | ||
| Interval of measurement (in days) | X16 | N | 613 |
| Mean ± SD | 212.5 ± 213.7 | ||
| Median | 150 | ||
| Minimum, maximum | 2.0, 2920.0 | ||
| Previous HbA1c values (%) | X17 | N | 676 |
| ≤ 7% | 269 (39.8%) | ||
| 7%-9% | 328 (48.5%) | ||
| >9% | 79 (11.7%) | ||
| Present HbA1c values (%) | X18 | N | 980 |
| Mean ± SD | 7.5 ± 1.6 | ||
| Median | 7.1 | ||
| Minimum, maximum | 4.6, 15.0 | ||
| Present FBG level | X19 | N | 838 |
| 3.8–6.1 | 54 (6.4%) | ||
| 6.1–7 | 257 (30.7%) | ||
| ≥7 | 527 (62.9%) | ||
| Present FBG values (mmoL/L) | X20 | N | 197 |
| Mean ± SD | 9.3 ± 3.56 | ||
| Median | 8.1 | ||
| Minimum, maximum | 3.3, 22.0 | ||
| Present RBG values (mmoL/L) | X21 | N | 517 |
| Mean ± SD | 11.6 ± 5.1 | ||
| Median | 10.4 | ||
| Minimum, maximum | 3.1, 34.1 | ||
| Present PBG values (mmoL/L) | X22 | N | 16 |
| Mean ± SD | 9.8 ± 2.4 | ||
| Median | 9.3 | ||
| Minimum, maximum | 6.9, 13.8 | ||
| Type of operation or other communicable diseases | X23 | N | 979 |
| No | 775 (79.2%) | ||
| Abdominal surgery | 114 (11.6%) | ||
| Thoracic surgery | 31 (3.2%) | ||
| Others | 59 (6.0%) | ||
| Number of comorbid diseases | X24 | N | 979 |
| 0 | 500 (51.1%) | ||
| 1 | 299 (30.5%) | ||
| 2 | 143 (14.6%) | ||
| 3 | 34 (3.5%) | ||
| 4 | 3 (0.3%) | ||
| Hypertension | X25 | N | 980 |
| No | 663 (67.7%) | ||
| Yes | 317 (32.3%) | ||
| Hyperlipidemia | X26 | N | 979 |
| No | 768 (78.4%) | ||
| Yes | 211 (21.6%) | ||
| With or without complications | X27 | N | 980 |
| No | 884 (90.2%) | ||
| Yes | 96 (9.8%) | ||
| Vascular complications | X28 | N | 980 |
| No | 977 (99.7%) | ||
| Yes | 3 (0.3%) | ||
| Neurological complication | X29 | N | 980 |
| No | 926 (94.5%) | ||
| Yes | 54 (5.5%) | ||
| Complications with lesions of the extremities | X30 | N | 980 |
| No | 975 (99.5%) | ||
| Yes | 5 (0.5%) | ||
| Ocular complications | X31 | N | 980 |
| No | 973 (99.3%) | ||
| Yes | 7 (0.7%) | ||
| Nephropathy complications | X32 | N | 980 |
| No | 972 (99.2%) | ||
| Yes | 8 (0.8%) | ||
| Complications(other diseases) | X33 | N | 980 |
| No | 957 (97.7%) | ||
| Yes | 23 (2.3%) | ||
| Exercise, diet and mental state | |||
| Intensity of exercise | X34 | N | 980 |
| None | 153 (15.6%) | ||
| Low intensity | 664 (67.8%) | ||
| Moderate intensity | 124 (12.7%) | ||
| High intensity | 39 (3.9%) | ||
| Exercise session (mins/day) | X35 | N | 980 |
| Mean ± SD | 53.4 ± 55.4 | ||
| Median | 45 | ||
| Minimum, maximum | 0, 600 | ||
| Had a ration and reasonable eating | X36 | N | 980 |
| No | 256 (26.1%) | ||
| Yes | 724 (73.9%) | ||
| Sleep duration | X37 | N | 980 |
| Good | 453 (46.2%) | ||
| Ordinary | 333 (34.0%) | ||
| Lose sleep | 194 (19.8%) | ||
| Psychological status | X38 | N | 980 |
| Well | 459 (46.8%) | ||
| General | 493 (50.3%) | ||
| Depression | 28 (2.9%) | ||
| EQ-5D scores | X39 | N | 980 |
| Mean ± SD | 0.9 ± 0.1 | ||
| Median | 1 | ||
| Minimum, maximum | 0.5, 1.0 | ||
| Treatment regimen and medication adherence | |||
| Compliance | X40 | N | 980 |
| No | 183 (18.6%) | ||
| Yes | 797 (83.4%) | ||
| Duration of treatment regimen (in months) | X41 | N | 979 |
| Mean ± SD | 24.8 ± 34.0 | ||
| Median | 12 | ||
| Minimum, maximum | 1.0, 240.0 | ||
| Type of insulin used | X42 | N | 980 |
| 0 | 731 (74.6%) | ||
| 1 | 228 (23.3%) | ||
| 2 | 21 (2.1%) | ||
| Use of insulin | X43 | N | 980 |
| No | 744 (75.9%) | ||
| Yes | 236 (24.1%) | ||
| Times of insulin use | X44 | N | 980 |
| 0 | 730 (74.5%) | ||
| 1 | 104 (10.6%) | ||
| 2 | 112 (11.4%) | ||
| 3 | 15 (1.5%) | ||
| 4 | 19 (2.0%) | ||
| Dose of basal insulin (U) | X45 | N | 980 |
| Mean ± SD | 2.0 ± 5.7 | ||
| Median | 0 | ||
| Minimum, maximum | 0, 35 | ||
| Dose of non-basal insulin in morning (U) | X46 | N | 980 |
| Mean ± SD | 2.2 ± 5.8 | ||
| Median | 0 | ||
| Minimum, maximum | 0, 33 | ||
| Dose of non-basal insulin in noon (U) | X47 | N | 980 |
| Mean ± SD | 0.4 ± 2.5 | ||
| Median | 0 | ||
| Minimum, maximum | 0, 32 | ||
| Dose of non-basal insulin in afternoon (U) | X48 | N | 980 |
| Mean ± SD | 2.2 ± 5.7 | ||
| Median | 0 | ||
| Minimum, maximum | 0, 32 | ||
| Number of oral drugs | X49 | N | 980 |
| 0 | 71 (7.2%) | ||
| 1 | 328 (33.5%) | ||
| 2 | 419 (42.8%) | ||
| 3 | 153 (15.6%) | ||
| 4 | 8 (0.8%) | ||
| 5 | 1 (0.1%) | ||
| Use of other types of drugs | X50 | N | 979 |
| None | 804 (82.1%) | ||
| National medicine | 11 (1.1%) | ||
| Chinese medicine | 88 (9.0%) | ||
| Health care products | 71 (7.3%) | ||
| Others | 5 (0.5%) | ||
| Use of metformin | X51 | N | 979 |
| None | 313 (32.0%) | ||
| Once a day | 175 (17.9%) | ||
| Twice a day | 399 (40.8%) | ||
| Three times a day | 92 (9.3%) | ||
| Dose of metformin | X52 | N | 976 |
| None | 313 (32.1%) | ||
| 0.25 g | 50 (5.1%) | ||
| 0.425 g | 2 (0.2%) | ||
| 0.5 g | 154 (15.8%) | ||
| 0.75 g | 1 (0.1%) | ||
| 0.85 g | 447 (45.8%) | ||
| 1.0 g | 9 (0.9%) | ||
| Type of manufacturers of metformin | X53 | N | 976 |
| Unknown | 313 (32.1%) | ||
| Generic drugs | 205 (21.0%) | ||
| Guthentic drugs | 458 (46.9%) | ||
| α-Glucosidase inhibitors | X54 | N | 980 |
| No | 616 (62.9%) | ||
| Yes | 364 (37.1%) | ||
| Sulfonylureas | X55 | N | 980 |
| No | 637 (65.0%) | ||
| Yes | 343 (35.0%) | ||
| Glinides | X56 | N | 980 |
| No | 911 (93.0%) | ||
| Yes | 69 (7.0%) | ||
| DPP-4 inhibitors | X57 | N | 980 |
| No | 845 (86.2%) | ||
| Yes | 135 (13.8%) | ||
| Thiazolidinediones | X58 | N | 980 |
| No | 928 (94.7%) | ||
| Yes | 52 (5.3%) | ||
| GLP-1 RAs | X59 | N | 980 |
| No | 979 (99.9%) | ||
| Yes | 1 (0.1%) | ||
| SGLT2 inhibitors | X60 | N | 980 |
| No | 976 (99.6%) | ||
| Yes | 4 (0.4%) | ||
| Use of Chinese medicine | X61 | N | 980 |
| No | 974 (99.4%) | ||
| Yes | 6 (0.6%) |
BMI, body mass index; HbA1c, glycated hemoglobin; FBG, fasting blood glucose; RBG, random blood glucose; EQ-5D, EuroQol five dimensions questionnaire; DPP-4 inhibitors, dipeptidylpeptidase-4 inhibitors; GLP-1 Ras, glucagon-like peptide-1 receptor agonists; SGLT2 inhibitors, sodium-dependent glucose transporters 2 inhibitors.
Dataset building
After data preprocessing, 43 variables were retained, and 18 variables were deleted. Sixty datasets were set up by applying different imputing methods, sampling methods, and screening methods with 43 variables. Additionally, the different number of variables and samples in each dataset is listed in Table 2.
Table 2.
The detailed information of 60 datasets.
| Number | Imputing methods | Sampling methods | Screening methods | Number of variables | Number of train samples |
|---|---|---|---|---|---|
| 1 | Not | Not | Not | 33 | 773 |
| 2 | Not | Not | Boruta | 21 | 773 |
| 3 | Not | Not | LassoCV | 12 | 773 |
| 4 | Not | SMOTE | Not | 33 | 1,260 |
| 5 | Not | SMOTE | Boruta | 16 | 1,260 |
| 6 | Not | SMOTE | LassoCV | 22 | 1,260 |
| 7 | Not | Borderline SMOTE | Not | 33 | 1,260 |
| 8 | Not | Borderline SMOTE | Boruta | 17 | 1,260 |
| 9 | Not | Borderline SMOTE | LassoCV | 23 | 1,260 |
| 10 | Not | Random OverSampler | Not | 33 | 1,260 |
| 11 | Not | Random OverSampler | Boruta | 16 | 1,260 |
| 12 | Not | Random OverSampler | LassoCV | 20 | 1,260 |
| 13 | Not | Random UnderSampler | Not | 33 | 286 |
| 14 | Not | Random UnderSampler | Boruta | 21 | 286 |
| 15 | Not | Random UnderSampler | LassoCV | 8 | 286 |
| 16 | Simple | Not | Not | 43 | 784 |
| 17 | Simple | Not | Boruta | 21 | 784 |
| 18 | Simple | Not | LassoCV | 11 | 784 |
| 19 | Simple | SMOTE | Not | 43 | 1,274 |
| 20 | Simple | SMOTE | Boruta | 21 | 1,274 |
| 21 | Simple | SMOTE | LassoCV | 30 | 1,274 |
| 22 | Simple | Borderline SMOTE | Not | 43 | 1,274 |
| 23 | Simple | Borderline SMOTE | Boruta | 20 | 1,274 |
| 24 | Simple | Borderline SMOTE | LassoCV | 30 | 1,274 |
| 25 | Simple | Random OverSampler | Not | 43 | 1,274 |
| 26 | Simple | Random OverSampler | Boruta | 19 | 1,274 |
| 27 | Simple | Random OverSampler | LassoCV | 25 | 1,274 |
| 28 | Simple | Random UnderSampler | Not | 43 | 294 |
| 29 | Simple | Random UnderSampler | Boruta | 26 | 294 |
| 30 | Simple | Random UnderSampler | LassoCV | 9 | 294 |
| 31 | Random forest | Not | Not | 43 | 784 |
| 32 | Random forest | Not | Boruta | 23 | 784 |
| 33 | Random forest | Not | LassoCV | 12 | 784 |
| 34 | Random forest | SMOTE | Not | 43 | 1,274 |
| 35 | Random forest | SMOTE | Boruta | 22 | 1,274 |
| 36 | Random forest | SMOTE | LassoCV | 30 | 1,274 |
| 37 | Random forest | Borderline SMOTE | Not | 43 | 1,274 |
| 38 | Random forest | Borderline SMOTE | Boruta | 21 | 1,274 |
| 39 | Random forest | Borderline SMOTE | LassoCV | 31 | 1,274 |
| 40 | Random forest | Random OverSampler | Not | 43 | 1,274 |
| 41 | Random forest | Random OverSampler | Boruta | 18 | 1,274 |
| 42 | Random forest | Random OverSampler | LassoCV | 25 | 1,274 |
| 43 | Random forest | Random UnderSampler | Not | 43 | 294 |
| 44 | Random forest | Random UnderSampler | Boruta | 24 | 294 |
| 45 | Random forest | Random UnderSampler | LassoCV | 19 | 294 |
| 46 | Modified random forest | Not | Not | 43 | 784 |
| 47 | Modified random forest | Not | Boruta | 22 | 784 |
| 48 | Modified random forest | Not | LassoCV | 13 | 784 |
| 49 | Modified random forest | SMOTE | Not | 43 | 1,274 |
| 50 | Modified Random Forest | SMOTE | Boruta | 21 | 1,274 |
| 51 | Modified Random Forest | SMOTE | LassoCV | 31 | 1,274 |
| 52 | Modified Random Forest | Borderline SMOTE | Not | 43 | 1,274 |
| 53 | Modified Random Forest | Borderline SMOTE | Boruta | 22 | 1,274 |
| 54 | Modified Random Forest | Borderline SMOTE | LassoCV | 30 | 1,274 |
| 55 | Modified Random Forest | Random OverSampler | Not | 43 | 1,274 |
| 56 | Modified Random Forest | Random OverSampler | Boruta | 18 | 1,274 |
| 57 | Modified Random Forest | Random OverSampler | LassoCV | 24 | 1,274 |
| 58 | Modified Random Forest | Random UnderSampler | Not | 43 | 294 |
| 59 | Modified Random Forest | Random UnderSampler | Boruta | 24 | 294 |
| 60 | Modified Random Forest | Random UnderSampler | LassoCV | 18 | 294 |
Model validation
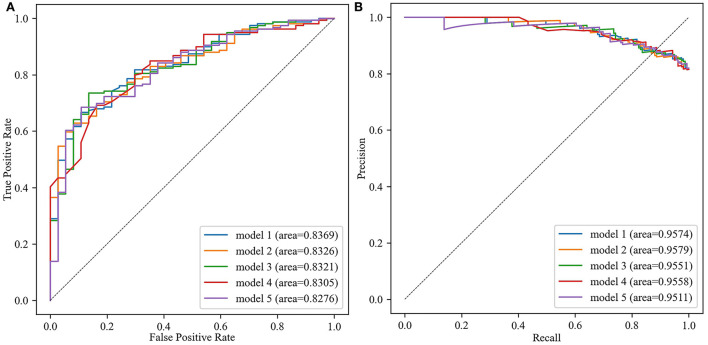
A total of 1,080 models were validated in the test set, considered as external validation, and the performance metrics were output. As shown in Table 3, the best five models were listed in sequence according to the AUC value. The best model (model 1) was applied the ensemble algorithm and trained in the No. 59 dataset (applied modified random forest as imputing method, random under sampler as sampling method, and Boruta as screening method). AUC, accuracy, precision, recall, F1 score, and AUPRC of the best model (model 1) were 0.8369, 0.9474, 0.6792, 0.7912, and 0.9574, respectively (Table 3; Figure 2). Especially in unbalanced data, the high value of AUPRC indicated that the best model (model 1) performed well to identify patients at risk for non-adherence.
Table 3.
The summary of the performance of five best models.
| ID | Algorithms | AUC | Accuracy | Precision | Recall | F1 Score | AUPRC |
|---|---|---|---|---|---|---|---|
| Model 1 | Ensemble | 0.8369 | 0.7092 | 0.9474 | 0.6792 | 0.7912 | 0.9574 |
| Model 2 | Ensemble | 0.8326 | 0.7041 | 0.9469 | 0.6730 | 0.7868 | 0.9579 |
| Model 3 | Bernoulli Naive Bayes | 0.8321 | 0.7500 | 0.9435 | 0.7358 | 0.8269 | 0.9551 |
| Model 4 | Ensemble | 0.8305 | 0.8010 | 0.9000 | 0.8491 | 0.8738 | 0.9558 |
| Model 5 | SGD | 0.8276 | 0.6786 | 0.9615 | 0.6289 | 0.7605 | 0.9511 |
AUC, area under the receiver operating characteristic curve; AUPRC, area under the precision-recall curve.
Figure 2.
The area under the receiver operating characteristic curve (AUC) and area under the precision-recall curve (AUPRC) of the best five models. (A) The receiver operator characteristic curve. (B) The precision-recall curves.
As shown in Table 4, the effects of various factors on model performance were compared using univariate analysis. With a decrease in the number of samples (AUC=-0.071, P < 0.0001) and an increase in the number of variables (AUC=0.047, P < 0.0001), the prediction model would achieve a high AUC value. Among the three imputing methods, modified random forest (AUC = 0.726 ± 0.076, vs. not 0.657 ± 0.075, simple 0.702 ± 0.087, and random forest 0.723 ± 0.081, P < 0.0001) was performed to improve performance of models, as well as random under sampler (AUC = 0.724 ± 0.076, vs. not 0.723 ± 0.080, random over sampler 0.698 ± 0.090, SMOTE 0.683 ± 0.086, and Border line SMOTE 0.682 ± 0.081, P < 0.0001) in five sampling methods, and Boruna (AUC = 0.709 ± 0.083, vs. not 0.700 ± 0.084, and LassoCV 0.698 ± 0.087, P < 0.0001) in three screening methods. In addition, the ensemble algorithm also performed well compared with other 17 algorithms (AUC = 0.790 ± 0.053, P < 0.0001). It should be mentioned that the above results were the same as the methods applied in the best model (model 1).
Table 4.
The results of univariate analysis ( ± SD).
| Classification | AUC | Accuracy | Precision | Recall | F1 score |
|---|---|---|---|---|---|
| Number of samples | –0.071 | 0.251 | –0.134 | 0.236 | 0.255 |
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
| Number of variables | 0.047 | 0.063 | 0.024 | 0.040 | 0.056 |
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
| Imputing methods | |||||
| Not | 0.657 ± 0.075 | 0.701 ± 0.088 | 0.859 ± 0.039 | 0.762 ± 0.143 | 0.799 ± 0.078 |
| Simple | 0.702 ± 0.087 | 0.723 ± 0.094 | 0.863 ± 0.047 | 0.791 ± 0.157 | 0.813 ± 0.087 |
| Random Forest | 0.723 ± 0.081 | 0.733 ± 0.079 | 0.871 ± 0.046 | 0.795 ± 0.136 | 0.822 ± 0.070 |
| Modified Random Forest | 0.726 ± 0.076 | 0.735 ± 0.079 | 0.871 ± 0.045 | 0.797 ± 0.136 | 0.824 ± 0.070 |
| P values | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| Sampling methods | |||||
| Not | 0.723 ± 0.080 | 0.802 ± 0.039 | 0.832 ± 0.035 | 0.951 ± 0.062 | 0.885 ± 0.028 |
| Random over sampler | 0.698 ± 0.090 | 0.711 ± 0.071 | 0.873 ± 0.041 | 0.757 ± 0.112 | 0.805 ± 0.062 |
| Random under sampler | 0.724 ± 0.076 | 0.623 ± 0.068 | 0.907 ± 0.042 | 0.598 ± 0.086 | 0.716 ± 0.068 |
| SMOTE | 0.683 ± 0.086 | 0.741 ± 0.068 | 0.859 ± 0.033 | 0.815 ± 0.089 | 0.834 ± 0.052 |
| Borderline SMOTE | 0.682 ± 0.081 | 0.738 ± 0.064 | 0.859 ± 0.032 | 0.811 ± 0.084 | 0.832 ± 0.050 |
| P values | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| Screening methods | |||||
| Not | 0.700 ± 0.084 | 0.722 ± 0.091 | 0.865 ± 0.044 | 0.786 ± 0.151 | 0.813 ± 0.082 |
| Lasso | 0.698 ± 0.087 | 0.724 ± 0.086 | 0.865 ± 0.044 | 0.789 ± 0.144 | 0.816 ± 0.077 |
| Boruta | 0.709 ± 0.083 | 0.722 ± 0.080 | 0.868 ± 0.045 | 0.783 ± 0.136 | 0.814 ± 0.073 |
| P values | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| Algorithms | |||||
| Logistic regression | 0.716 ± 0.064 | 0.732 ± 0.068 | 0.869 ± 0.043 | 0.797 ± 0.127 | 0.823 ± 0.060 |
| SGD | 0.693 ± 0.095 | 0.727 ± 0.078 | 0.874 ± 0.054 | 0.788 ± 0.150 | 0.816 ± 0.075 |
| KNN | 0.667 ± 0.085 | 0.711 ± 0.073 | 0.854 ± 0.043 | 0.784 ± 0.135 | 0.809 ± 0.063 |
| Decision tree | 0.672 ± 0.065 | 0.682 ± 0.106 | 0.870 ± 0.051 | 0.726 ± 0.182 | 0.774 ± 0.110 |
| Gaussian Naive Bayes | 0.673 ± 0.086 | 0.689 ± 0.075 | 0.874 ± 0.038 | 0.722 ± 0.106 | 0.786 ± 0.064 |
| Bernoulli Naive Bayes | 0.753 ± 0.069 | 0.731 ± 0.060 | 0.881 ± 0.041 | 0.777 ± 0.099 | 0.821 ± 0.051 |
| Multinomial Naive Bayes | 0.661 ± 0.084 | 0.678 ± 0.088 | 0.853 ± 0.043 | 0.736 ± 0.157 | 0.779 ± 0.080 |
| SVM | 0.698 ± 0.057 | 0.752 ± 0.070 | 0.850 ± 0.042 | 0.849 ± 0.127 | 0.842 ± 0.061 |
| QDA | 0.689 ± 0.091 | 0.727 ± 0.069 | 0.869 ± 0.041 | 0.786 ± 0.109 | 0.819 ± 0.058 |
| Random forest | 0.743 ± 0.057 | 0.769 ± 0.093 | 0.862 ± 0.043 | 0.861 ± 0.159 | 0.850 ± 0.084 |
| Extra tree | 0.624 ± 0.080 | 0.679 ± 0.090 | 0.853 ± 0.045 | 0.739 ± 0.157 | 0.780 ± 0.087 |
| LDA | 0.735 ± 0.070 | 0.738 ± 0.063 | 0.880 ± 0.040 | 0.789 ± 0.109 | 0.826 ± 0.054 |
| Passive aggressive | 0.620 ± 0.090 | 0.657 ± 0.073 | 0.854 ± 0.042 | 0.699 ± 0.105 | 0.764 ± 0.066 |
| AdaBoost | 0.736 ± 0.061 | 0.725 ± 0.078 | 0.873 ± 0.044 | 0.782 ± 0.138 | 0.815 ± 0.071 |
| Bagging | 0.724 ± 0.059 | 0.746 ± 0.099 | 0.860 ± 0.038 | 0.827 ± 0.158 | 0.832 ± 0.091 |
| Gradient boosting | 0.730 ± 0.056 | 0.738 ± 0.084 | 0.866 ± 0.042 | 0.808 ± 0.145 | 0.826 ± 0.075 |
| XGBoost | 0.717 ± 0.068 | 0.756 ± 0.081 | 0.859 ± 0.039 | 0.842 ± 0.136 | 0.842 ± 0.073 |
| Ensemble | 0.790 ± 0.053 | 0.776 ± 0.067 | 0.886 ± 0.045 | 0.838 ± 0.122 | 0.854 ± 0.058 |
| P values | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
Feature selection and validation
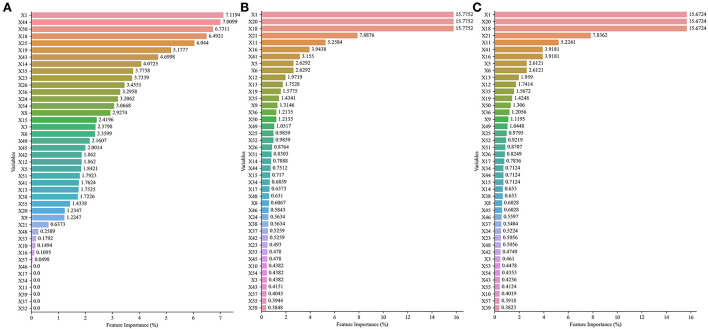
The best five models involved the following three datasets: No. 27, No. 44, and No. 59. In those datasets, the variable importance scores are ranked in Figure 3. Age, times of insulin use, use of other types of drugs, present HbA1c values, and hypertension were top 5 highest variable importance in No. 27 dataset (Figure 3A). The top 5 variables with the highest importance score in No. 44 dataset and No. 59 dataset were age, present FBG values, present HbA1c values, present random blood glucose (RBG) values, and BMI (Figures 3B,C).
Figure 3.
The importance scores and ranking of each variable in No. 27 dataset, No. 44 dataset, and No. 59 dataset with different variable selection methods. (A) Details of No. 27 dataset. (B) Details of No. 44 dataset. (C) Details of No. 59 dataset.
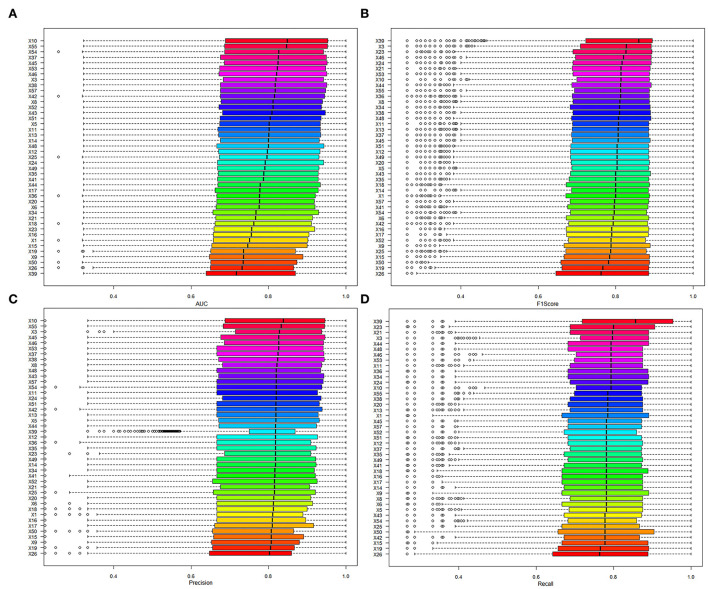
In addition, the contribution of variables was evaluated by comparing the AUC of models to identify whether the variable was included or excluded. In addition, the mean AUC of variables was from 0.689 to 0.724 in the included cohort and between 0.669 and 0.762 in the excluded cohort (details in Table 5; Figure 4). The variable that had higher AUC when the variable was included would be considered as a positive contribution to the prediction model. Those variables provided positive contributions and were in line with variables that had high variable importance scores, which was output in No. 59 dataset (the best model applied).
Table 5.
The influence of model performances whether the variable was included or excluded.
| Variables | Code of | Included or | AUC | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|---|---|---|
| variables | excluded | Mean ±SD | Mean ±SD | Mean ±SD | Mean ±SD | Mean ±SD | |
| Age | X1 | No | 0.672 ± 0.085 | 0.671-0.672 | 0.733 ± 0.070 | 0.732-0.733 | 0.859 ± 0.032 |
| Yes | 0.707 ± 0.084 | 0.707-0.707 | 0.721 ± 0.088 | 0.721-0.721 | 0.867 ± 0.046 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Gender | X3 | No | 0.712 ± 0.128 | 0.698 ± 0.122 | 0.717 ± 0.142 | 0.716 ± 0.180 | 0.709 ± 0.148 |
| Yes | 0.807 ± 0.142 | 0.780 ± 0.116 | 0.815 ± 0.142 | 0.791 ± 0.135 | 0.795 ± 0.118 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Waistline (cm) | X5 | No | 0.760 ± 0.141 | 0.747 ± 0.122 | 0.775 ± 0.145 | 0.773 ± 0.167 | 0.767 ± 0.140 |
| Yes | 0.797 ± 0.145 | 0.766 ± 0.122 | 0.800 ± 0.150 | 0.771 ± 0.141 | 0.778 ± 0.127 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0478 | P = 0.0117 | |||
| Weight (Kg) | X6 | No | 0.780 ± 0.142 | 0.765 ± 0.115 | 0.796 ± 0.140 | 0.796 ± 0.150 | 0.788 ± 0.125 |
| Yes | 0.784 ± 0.146 | 0.758 ± 0.125 | 0.789 ± 0.151 | 0.765 ± 0.151 | 0.769 ± 0.133 | ||
| P = 0.2130 | P = 0.0284 | P = 0.2102 | P < 0.0001 | P < 0.0001 | |||
| Occupational status | X8 | No | 0.688 ± 0.102 | 0.701 ± 0.110 | 0.718 ± 0.124 | 0.755 ± 0.202 | 0.729 ± 0.154 |
| Yes | 0.802 ± 0.145 | 0.771 ± 0.121 | 0.805 ± 0.149 | 0.776 ± 0.139 | 0.783 ± 0.125 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.1838 | P < 0.0001 | |||
| Education level | X9 | No | 0.815 ± 0.138 | 0.776 ± 0.117 | 0.810 ± 0.145 | 0.778 ± 0.135 | 0.786 ± 0.120 |
| Yes | 0.759 ± 0.146 | 0.747 ± 0.125 | 0.776 ± 0.149 | 0.768 ± 0.162 | 0.764 ± 0.139 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0686 | P < 0.0001 | |||
| Family history of diabetes mellitus | X10 | No | 0.760 ± 0.141 | 0.747 ± 0.121 | 0.775 ± 0.145 | 0.769 ± 0.162 | 0.765 ± 0.138 |
| Yes | 0.819 ± 0.144 | 0.777 ± 0.122 | 0.814 ± 0.151 | 0.778 ± 0.133 | 0.787 ± 0.121 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0558 | P < 0.0001 | |||
| BMI (kg/m2) | X11 | No | 0.767 ± 0.141 | 0.750 ± 0.122 | 0.778 ± 0.146 | 0.768 ± 0.163 | 0.766 ± 0.138 |
| Yes | 0.795 ± 0.147 | 0.766 ± 0.122 | 0.800 ± 0.150 | 0.775 ± 0.142 | 0.779 ± 0.126 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0002 | |||
| Health status scores (%) | X12 | No | 0.733 ± 0.130 | 0.736 ± 0.116 | 0.761 ± 0.135 | 0.777 ± 0.183 | 0.761 ± 0.146 |
| Yes | 0.795 ± 0.146 | 0.764 ± 0.123 | 0.797 ± 0.150 | 0.771 ± 0.143 | 0.777 ± 0.128 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0001 | P = 0.0136 | |||
| Course of diabetes (in months) | X13 | No | 0.736 ± 0.128 | 0.735 ± 0.117 | 0.761 ± 0.136 | 0.766 ± 0.182 | 0.756 ± 0.146 |
| Yes | 0.795 ± 0.146 | 0.765 ± 0.123 | 0.798 ± 0.150 | 0.774 ± 0.143 | 0.778 ± 0.127 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.9355 | P < 0.0001 | |||
| Medicare status | X14 | No | 0.771 ± 0.146 | 0.755 ± 0.123 | 0.785 ± 0.147 | 0.774 ± 0.158 | 0.772 ± 0.136 |
| Yes | 0.796 ± 0.143 | 0.763 ± 0.122 | 0.796 ± 0.149 | 0.771 ± 0.144 | 0.775 ± 0.127 | ||
| P < 0.0001 | P = 0.0008 | P < 0.0001 | P = 0.0906 | P = 0.9577 | |||
| Frequency of FBG measurements | X15 | No | 0.810 ± 0.140 | 0.776 ± 0.116 | 0.810 ± 0.144 | 0.785 ± 0.137 | 0.790 ± 0.119 |
| Yes | 0.768 ± 0.145 | 0.750 ± 0.125 | 0.779 ± 0.150 | 0.765 ± 0.159 | 0.765 ± 0.137 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Interval of measurement (in days) | X16 | No | 0.794 ± 0.143 | 0.766 ± 0.121 | 0.798 ± 0.148 | 0.776 ± 0.148 | 0.779 ± 0.129 |
| Yes | 0.772 ± 0.146 | 0.752 ± 0.123 | 0.783 ± 0.149 | 0.768 ± 0.155 | 0.768 ± 0.134 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0736 | P < 0.0001 | |||
| Previous HbA1c value (%) | X17 | No | 0.783 ± 0.143 | 0.762 ± 0.120 | 0.793 ± 0.145 | 0.776 ± 0.151 | 0.777 ± 0.131 |
| Yes | 0.785 ± 0.148 | 0.755 ± 0.125 | 0.787 ± 0.153 | 0.767 ± 0.151 | 0.768 ± 0.132 | ||
| P = 0.5126 | P = 0.0201 | P = 0.0623 | P = 0.0117 | P = 0.0009 | |||
| Present HbA1c values (%) | X18 | No | 0.881 ± 0.116 | 0.805 ± 0.115 | 0.849 ± 0.150 | 0.787 ± 0.086 | 0.808 ± 0.095 |
| Yes | 0.776 ± 0.144 | 0.756 ± 0.122 | 0.787 ± 0.147 | 0.771 ± 0.155 | 0.771 ± 0.134 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.2544 | P < 0.0001 | |||
| Present FBG level | X19 | No | 0.812 ± 0.142 | 0.777 ± 0.118 | 0.812 ± 0.146 | 0.786 ± 0.135 | 0.791 ± 0.120 |
| Yes | 0.757 ± 0.142 | 0.742 ± 0.124 | 0.771 ± 0.148 | 0.760 ± 0.164 | 0.758 ± 0.140 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Present FBG values (mmoL/L) | X20 | No | 0.781 ± 0.146 | 0.757 ± 0.125 | 0.787 ± 0.150 | 0.767 ± 0.159 | 0.769 ± 0.137 |
| Yes | 0.784 ± 0.144 | 0.761 ± 0.121 | 0.792 ± 0.147 | 0.775 ± 0.147 | 0.776 ± 0.129 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Present RBG values (mmoL/L) | X21 | No | 0.678 ± 0.083 | 0.678-0.678 | 0.708 ± 0.089 | 0.708-0.708 | 0.864 ± 0.042 |
| Yes | 0.719 ± 0.082 | 0.719-0.719 | 0.734 ± 0.083 | 0.733-0.734 | 0.868 ± 0.046 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Type of operation or other communicable diseases | X23 | No | 0.803 ± 0.141 | 0.742 ± 0.131 | 0.768 ± 0.161 | 0.725 ± 0.132 | 0.739 ± 0.131 |
| Yes | 0.777 ± 0.146 | 0.765 ± 0.119 | 0.798 ± 0.143 | 0.788 ± 0.154 | 0.785 ± 0.130 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Number of comorbid diseases | X24 | No | 0.767 ± 0.139 | 0.744 ± 0.121 | 0.771 ± 0.146 | 0.760 ± 0.159 | 0.759 ± 0.136 |
| Yes | 0.794 ± 0.148 | 0.769 ± 0.122 | 0.804 ± 0.149 | 0.780 ± 0.145 | 0.784 ± 0.128 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Hypertension | X25 | No | 0.727 ± 0.130 | 0.776 ± 0.094 | 0.808 ± 0.110 | 0.850 ± 0.140 | 0.824 ± 0.109 |
| Yes | 0.793 ± 0.145 | 0.756 ± 0.127 | 0.788 ± 0.154 | 0.758 ± 0.149 | 0.765 ± 0.133 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Hyperlipidemia | X26 | No | 0.827 ± 0.138 | 0.793 ± 0.108 | 0.830 ± 0.137 | 0.801 ± 0.109 | 0.808 ± 0.100 |
| Yes | 0.754 ± 0.142 | 0.737 ± 0.126 | 0.764 ± 0.150 | 0.753 ± 0.171 | 0.751 ± 0.145 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Intensity of exercise | X34 | No | 0.787 ± 0.139 | 0.759 ± 0.120 | 0.789 ± 0.145 | 0.768 ± 0.147 | 0.772 ± 0.129 |
| Yes | 0.780 ± 0.150 | 0.759 ± 0.125 | 0.792 ± 0.151 | 0.776 ± 0.155 | 0.776 ± 0.134 | ||
| P = 0.0188 | P = 0.4927 | P = 0.1874 | P = 0.0002 | P = 0.0145 | |||
| Exercise session (mins/day) | X35 | No | 0.751 ± 0.139 | 0.745 ± 0.118 | 0.774 ± 0.142 | 0.784 ± 0.174 | 0.770 ± 0.141 |
| Yes | 0.791 ± 0.145 | 0.763 ± 0.123 | 0.795 ± 0.150 | 0.769 ± 0.145 | 0.775 ± 0.129 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.8408 | |||
| Had a ration and reasonable eating | X36 | No | 0.769 ± 0.141 | 0.722 ± 0.139 | 0.748 ± 0.164 | 0.687 ± 0.154 | 0.710 ± 0.150 |
| Yes | 0.784 ± 0.145 | 0.762 ± 0.121 | 0.794 ± 0.147 | 0.778 ± 0.149 | 0.778 ± 0.129 | ||
| P = 0.0545 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Sleep duration | X37 | No | 0.758 ± 0.137 | 0.749 ± 0.117 | 0.777 ± 0.140 | 0.776 ± 0.161 | 0.770 ± 0.135 |
| Yes | 0.807 ± 0.148 | 0.768 ± 0.126 | 0.803 ± 0.155 | 0.769 ± 0.141 | 0.778 ± 0.129 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0008 | P = 0.0224 | |||
| Psychological status | X38 | No | 0.765 ± 0.140 | 0.750 ± 0.120 | 0.778 ± 0.143 | 0.771 ± 0.158 | 0.768 ± 0.134 |
| Yes | 0.806 ± 0.148 | 0.770 ± 0.125 | 0.806 ± 0.153 | 0.773 ± 0.143 | 0.781 ± 0.128 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.9048 | P < 0.0001 | |||
| EQ-5D scores | X39 | No | 0.813 ± 0.140 | 0.751 ± 0.132 | 0.781 ± 0.162 | 0.729 ± 0.131 | 0.747 ± 0.131 |
| Yes | 0.750 ± 0.143 | 0.769 ± 0.109 | 0.802 ± 0.130 | 0.822 ± 0.157 | 0.804 ± 0.125 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Duration of treatment regimen (in months) | X41 | No | 0.709 ± 0.116 | 0.759 ± 0.098 | 0.786 ± 0.112 | 0.838 ± 0.172 | 0.806 ± 0.132 |
| Yes | 0.790 ± 0.145 | 0.759 ± 0.124 | 0.791 ± 0.151 | 0.766 ± 0.148 | 0.771 ± 0.131 | ||
| P < 0.0001 | P = 0.1224 | P = 0.0054 | P < 0.0001 | P < 0.0001 | |||
| Type of insulin used | X42 | No | 0.760 ± 0.138 | 0.755 ± 0.116 | 0.785 ± 0.138 | 0.785 ± 0.158 | 0.778 ± 0.132 |
| Yes | 0.803 ± 0.147 | 0.762 ± 0.128 | 0.796 ± 0.156 | 0.762 ± 0.145 | 0.770 ± 0.131 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0008 | |||
| Use of insulin | X43 | No | 0.764 ± 0.141 | 0.750 ± 0.119 | 0.779 ± 0.143 | 0.776 ± 0.159 | 0.770 ± 0.134 |
| Yes | 0.804 ± 0.146 | 0.769 ± 0.125 | 0.804 ± 0.153 | 0.768 ± 0.143 | 0.778 ± 0.129 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0095 | |||
| Times of insulin use | X44 | No | 0.774 ± 0.142 | 0.750 ± 0.121 | 0.779 ± 0.147 | 0.762 ± 0.153 | 0.763 ± 0.133 |
| Yes | 0.788 ± 0.146 | 0.764 ± 0.123 | 0.797 ± 0.149 | 0.778 ± 0.150 | 0.780 ± 0.130 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Dose of basal insulin (U) | X45 | No | 0.751 ± 0.138 | 0.744 ± 0.118 | 0.771 ± 0.141 | 0.775 ± 0.164 | 0.766 ± 0.137 |
| Yes | 0.812 ± 0.145 | 0.772 ± 0.125 | 0.808 ± 0.153 | 0.770 ± 0.139 | 0.781 ± 0.127 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0001 | P < 0.0001 | |||
| Dose of non-basal insulin in morning (U) | X46 | No | 0.763 ± 0.138 | 0.746 ± 0.121 | 0.773 ± 0.144 | 0.763 ± 0.160 | 0.761 ± 0.136 |
| Yes | 0.805 ± 0.149 | 0.774 ± 0.123 | 0.809 ± 0.151 | 0.782 ± 0.141 | 0.787 ± 0.125 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Dose of non-basal insulin in afternoon (U) | X48 | No | 0.776 ± 0.142 | 0.755 ± 0.120 | 0.785 ± 0.145 | 0.771 ± 0.153 | 0.771 ± 0.132 |
| Yes | 0.795 ± 0.149 | 0.766 ± 0.125 | 0.800 ± 0.153 | 0.774 ± 0.149 | 0.779 ± 0.131 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.3904 | P = 0.0016 | |||
| Number of oral drugs | X49 | No | 0.731 ± 0.127 | 0.742 ± 0.115 | 0.770 ± 0.133 | 0.773 ± 0.187 | 0.765 ± 0.149 |
| Yes | 0.791 ± 0.146 | 0.762 ± 0.123 | 0.794 ± 0.150 | 0.772 ± 0.145 | 0.775 ± 0.129 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0158 | P = 0.5867 | |||
| Use of other types of drugs | X50 | No | 0.692 ± 0.084 | 0.692-0.692 | 0.738 ± 0.065 | 0.738-0.739 | 0.860 ± 0.036 |
| Yes | 0.707 ± 0.084 | 0.707-0.707 | 0.716 ± 0.093 | 0.716-0.716 | 0.869 ± 0.048 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |||
| Use of metformin | X51 | No | 0.687 ± 0.098 | 0.715 ± 0.108 | 0.736 ± 0.120 | 0.782 ± 0.205 | 0.751 ± 0.156 |
| Yes | 0.798 ± 0.145 | 0.766 ± 0.123 | 0.799 ± 0.150 | 0.771 ± 0.141 | 0.777 ± 0.127 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0002 | |||
| Dose of metformin | X52 | No | 0.767 ± 0.141 | 0.761 ± 0.118 | 0.792 ± 0.141 | 0.784 ± 0.159 | 0.781 ± 0.133 |
| Yes | 0.798 ± 0.147 | 0.758 ± 0.126 | 0.790 ± 0.155 | 0.762 ± 0.144 | 0.767 ± 0.130 | ||
| P < 0.0001 | P = 0.5662 | P = 0.5470 | P < 0.0001 | P < 0.0001 | |||
| Type of manufacturers of metformin | X53 | No | 0.759 ± 0.138 | 0.747 ± 0.119 | 0.775 ± 0.142 | 0.767 ± 0.162 | 0.764 ± 0.136 |
| Yes | 0.805 ± 0.148 | 0.770 ± 0.124 | 0.805 ± 0.152 | 0.776 ± 0.141 | 0.782 ± 0.127 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0377 | P < 0.0001 | |||
| α-Glucosidase inhibitors | X54 | No | 0.708 ± 0.120 | 0.739 ± 0.112 | 0.764 ± 0.128 | 0.799 ± 0.180 | 0.776 ± 0.143 |
| Yes | 0.809 ± 0.144 | 0.766 ± 0.125 | 0.800 ± 0.154 | 0.763 ± 0.139 | 0.773 ± 0.128 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.0007 | |||
| Sulfonylureas | X55 | No | 0.746 ± 0.135 | 0.741 ± 0.119 | 0.768 ± 0.140 | 0.769 ± 0.168 | 0.762 ± 0.139 |
| Yes | 0.816 ± 0.145 | 0.775 ± 0.123 | 0.811 ± 0.152 | 0.775 ± 0.135 | 0.784 ± 0.123 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.8960 | P < 0.0001 | |||
| DPP-4 inhibitors | X57 | No | 0.761 ± 0.139 | 0.753 ± 0.117 | 0.783 ± 0.140 | 0.779 ± 0.159 | 0.774 ± 0.134 |
| Yes | 0.804 ± 0.147 | 0.765 ± 0.127 | 0.798 ± 0.155 | 0.766 ± 0.143 | 0.774 ± 0.130 | ||
| P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P = 0.5225 |
Figure 4.
The model performance of models when the variables were included. (A) The results of AUC. (B) The results of the F1 score. (C) The results of precision. (D) The results of recall.
Sample size assessment
As shown in Figure 5, with the size of sample data incorporated into the model from small to large, the values of AUC continued to increase. When the sample size was extremely small ( ≤ 30%), compared with the 100% sample size, the SDs of AUC were dispersed, and the AUCs were statistically significant (P < 0.05). As the sample size increased, the above situation was alleviated (P>0.05). In addition, the growth rate of AUC slowed down when the sample size was more than or equal to 40%. These results indicated that the performance of the proposed model might be affected less when expanding the sample size. The sample size was suitable for the prediction model construction.
Figure 5.
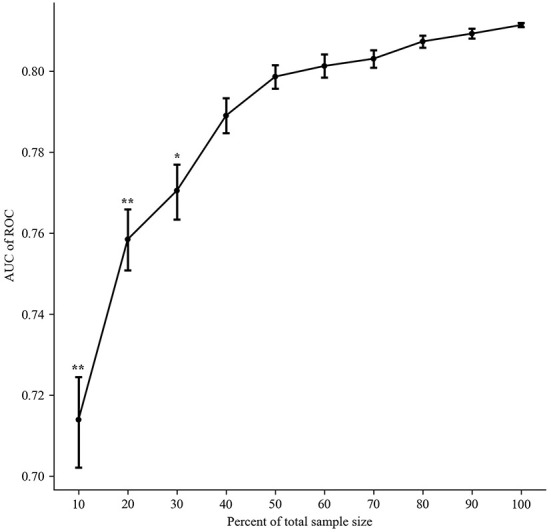
The impact of sample data size on model performances (mean ± SD).
Discussion
Early detection of non-adherence to medication in patients with T2D will help devise strategies for personalized treatment. In this research, we developed a total of 1,080 models for the prediction of adherence in patients with T2D. The AUC, accuracy, precision, recall, F1 score, and AUPRC of the best model were 0.8369, 0.9474, 0.6792, 0.7912, and 0.9574, respectively. Meanwhile, various methods in model development and variables were validated by univariate analyses. Interestingly, the imputing method, the sampling method, the variable selection method, and the machine learning algorithm applied in the best model were the same as the results of univariate analysis. Additionally, variables with high importance scores in the best model were similar to the results of variable validation, which provided a positive contribution to the model prediction.
The adherence to the medication of patients with T2D has received great attention worldwide (24, 25). Nonadherence is associated with bad outcomes, including increased mortality and avoidable healthcare costs. Previous studies reported models to predict drug non-adherence in Crohn's disease maintenance therapy (26), patients with hypertension (27), and patients with heart failure (28). However, few studies reported on prediction models of non-adherence to medication in patients with T2D. Intelligence technology is becoming more prevalent in healthcare as a tool to improve practice patterns and patient outcomes (29–31). With technology development, ensemble models have been commonly used to explore disease progression in the field of molecular biology (32–36). Recently, the ensemble algorithm has been frequently applied to develop prediction models (37, 38). In our prior study, we reported that the ensemble algorithm was superior to the Bayesian network, KNN, SVM, C&R Tree, and CHAID (19). In this study, we added more machine learning classifications, including XGBoost, Bernoulli Naive Bayes, SGD, etc. Additionally, the ensemble algorithm was still the best.
Many variables have previously been reported to associate with drug adherence, such as age, population, level of education, etc. For example, according to the data from the National Health Insurance Service-National Sample Cohort (NHIS-NSC) of Korea, adherence consistently increased as the age increased until 69 years and started to decrease from the age of 70 years. When the same number of drugs was taken, the proportion of adherent patients according to age featured an inverted U-shape with a peak at 60–69 years (39). Additionally, Aditama et al. (25) stated that the factors influencing non-adherence included complex instructions for taking medication, the absence of a reminder, the unwanted side effects of the drug, feeling of repetition, feeling that the drugs were ineffective, and concern for the effects of the drug on the kidney. Therefore, more patient-related and drug-related variables were considered in this study, including the number of comorbid diseases, EQ-5D scores, number of oral drugs, use of other types of drugs, and so on.
The results of the univariate analysis suggested that more variables can improve the accuracy of the prediction model (AUC = 0.063, P < 0.001). In clinical research, more variables mean collecting more data and increasing the missing data. Thus, feature selection plays an important role in the field of machine learning. In this study, no screening (marked as Not), Boruta, and LassoCV were performed. Boruta is a feature selection algorithm to identify the minimal set of relevant variables, which was applied in the best model. According to the variable importance score, the ten most important variables were age, present FBG values, present HbA1c values, present RBG values, BMI, duration of the treatment regimen, interval of measurement, waistline, weight, and course of diabetes. Glycemic control in patients with T2D can be accessed via the following three key parameters: glycated hemoglobin (HbA1c), FBG, and RBG. The results of variable importance demonstrated that patients with non-adherence should strongly encourage to monitor their blood glucose and receive reinforced education.
Limitation
First, this was a single-center study, and the patient profile might be biased and not representative of the Chinese as a whole. People from Sichuan Province may have different distributions of risk factors than patients in other areas of China. A large multicenter sample study is desired, which can verify the applicability of the model. However, for some variables, recall bias still exists, such as the intensity of exercise and exercise sessions.
Conclusion
In summary, the present research introduced 1,080 machine-learning models to predict non-adherence in patients with T2D and proposed an ensemble model with better classifier performance. This study also reconfirmed that variables including age, BMI, and interval of measurement were risk factors for non-adherence. We are in the process of developing a mobile App or a web server for caregivers and patients in an effort to integrate the adherence enhancement intervention into daily T2D management.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Sichuan Provincial People's Hospital (Approval # 2018-53). The patients/participants provided their written informed consent to participate in this study.
Author contributions
ML and XL contributed to data analysis and writing and approval of the final manuscript. HY and RY assisted in the face-to-face questionnaire. RT and YY were responsible for designing and coordinating the research. XW was involved in the questionnaire design, data analysis, model design, and contributed to revision of the manuscript. All authors agree to be accountable for the content of this study.
Funding
This study was funded by the National Natural Science Foundation of China (Grant No. 72004020), the Key Research and Development Program of Science and Technology Department of Sichuan Province (Grant No. 2019YFS0514), the Postgraduate Research and Teaching Reform Project of the University of Electronic Science and Technology of China (Grant No. JYJG201919), the Research Subject of Health Commission of Sichuan Province (Grant No. 19PJ262), Sichuan Science and Technology Program (Grant No. 2021YJ0427), and Scientific Research Foundation of Sichuan Provincial People's Hospital (Grant No. 2022BH10).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.American Diabetes Association . 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2020. Diab Care. (2020). 43(Suppl 1):S14–31. 10.2337/dc20-S002 [DOI] [PubMed] [Google Scholar]
- 2.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. (2001) 414:782–7. 10.1038/414782a [DOI] [PubMed] [Google Scholar]
- 3.Lam DW, LeRoith D. The worldwide diabetes epidemic. Curr Opin Endocrinol Diab Obes. (2012) 19:93–6. 10.1097/MED.0b013e328350583a [DOI] [PubMed] [Google Scholar]
- 4.Arredondo A. Diabetes: a global challenge with high economic burden for public health systems and society. Am J Public Health. (2013) 103:e1–2. 10.2105/AJPH.2012.301106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bommer C, Sagalova V, Heesemann E, Manne-Goehler J, Atun R, Bärnighausen T, et al. Global economic burden of diabetes in adults: projections from 2015 to 2030. Diabetes Care. (2018) 41:963–70. 10.2337/dc17-1962 [DOI] [PubMed] [Google Scholar]
- 6.Simpson SH, Eurich DT, Majumdar SR, Padwal RS, Tsuyuki RT, Varney J, et al. A meta-analysis of the association between adherence to drug therapy and mortality. BMJ. (2006) 333:15. 10.1136/bmj.38875.675486.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterberg L, Blaschke T. Adherence to medication. N Engl J Med. (2005) 353:487–97. 10.1056/NEJMra050100 [DOI] [PubMed] [Google Scholar]
- 8.Lerman I. Adherence to treatment: the key for avoiding long-term complications of diabetes. Arch Med Res. (2005) 36:300–6. 10.1016/j.arcmed.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 9.Quisel T, Foschini L, Zbikowski SM, Juusola JL. The association between medication adherence for chronic conditions and digital health activity tracking: retrospective analysis. J Med Internet Res. (2019) 21:e11486. 10.2196/11486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryson CL, Au DH, Maciejewski ML, Piette JD, Fihn SD, Jackson GL, et al. Wide clinic-level variation in adherence to oral diabetes medications in the VA. J Gen Intern Med. (2013) 28:698–705. 10.1007/s11606-012-2331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farmer AJ, Rodgers LR, Lonergan M, Shields B, Weedon MN, Donnelly L, et al. Adherence to oral glucose-lowering therapies and associations with 1-year HbA1c: a retrospective cohort analysis in a large primary care database. Diabetes Care. (2016) 39:258–63. 10.2337/dc15-1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park LG, Howie-Esquivel J, Chung ML, Dracup K. A text messaging intervention to promote medication adherence for patients with coronary heart disease: a randomized controlled trial. Patient Educ Couns. (2014) 94:261–8. 10.1016/j.pec.2013.10.027 [DOI] [PubMed] [Google Scholar]
- 13.Kurtyka K, Nishikino R, Ito C, Brodovicz K, Chen Y, Tunceli K. Adherence to dipeptidyl peptidase-4 inhibitor therapy among type 2 diabetes patients with employer-sponsored health insurance in Japan. J Diabetes Investig. (2016) 7:737–43. 10.1111/jdi.12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentil L, Vasiliadis HM, Berbiche D, Préville M. Impact of depression and anxiety disorders on adherence to oral hypoglycemics in older adults with diabetes mellitus in Canada. Eur J Ageing. (2017) 14:111–21. 10.1007/s10433-016-0390-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor PJ, Schmittdiel JA, Pathak RD, Harris RI, Newton KM, Ohnsorg KA, et al. Randomized trial of telephone outreach to improve medication adherence and metabolic control in adults with diabetes. Diabetes Care. (2014) 37:3317–24. 10.2337/dc14-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dulmen S, Sluijs E, Van Dijk L, de Ridder D, Heerdink R, Bensing J. Patient adherence to medical treatment: a review of reviews. BMC Health Serv Res. (2007) 7:55. 10.1186/1472-6963-7-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azulay R, Valinsky L, Hershkowitz F, Magnezi R. Repeated automated mobile text messaging reminders for follow-up of positive fecal occult blood tests: randomized controlled trial. JMIR Mhealth Uhealth. (2019) 7:e11114. 10.2196/11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khachadourian V, Truzyan N, Harutyunyan A, Thompson ME, Harutyunyan T, Petrosyan V. People-centered tuberculosis care versus standard directly observed therapy: study protocol for a cluster randomized controlled trial. Trials. (2015) 16:281. 10.1186/s13063-015-0802-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu XW, Yang HB, Yuan R, Long EW, Tong RS. Predictive models of medication non-adherence risks of patients with T2D based on multiple machine learning algorithms. BMJ Open Diabetes Res Care. (2020) 8:e001055. 10.1136/bmjdrc-2019-001055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury R, Khan H, Heydon E, Shroufi A, Fahimi S, Moore C, et al. Adherence to cardiovascular therapy: a meta-analysis of prevalence and clinical consequences. Eur Heart J. (2013) 34:2940–8. 10.1093/eurheartj/eht295 [DOI] [PubMed] [Google Scholar]
- 21.Cramer JA, Benedict A, Muszbek N, Keskinaslan A, Khan ZM. The significance of compliance and persistence in the treatment of diabetes, hypertension and dyslipidaemia: a review. Int J Clin Pract. (2008) 62:76–87. 10.1111/j.1742-1241.2007.01630.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuwani R, Wadhwa S, Bagler G. BitterSweet: building machine learning models for predicting the bitter and sweet taste of small molecules. Sci Rep. (2019) 9:7155. 10.1038/s41598-019-43664-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes M, Mendes R, Vieira SM, Leite F, Palos C, Johnson A, et al. Predicting intensive care unit admission among patients presenting to the emergency department using machine learning and natural language processing. PLoS ONE. (2020) 15:e0229331. 10.1371/journal.pone.0229331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CS, Tan JH, Sankari U, Koh YL, Tan NC. Assessing oral medication adherence among patients with type 2 diabetes mellitus treated with polytherapy in a developed Asian community: a cross-sectional study. BMJ Open. (2017) 7:e016317. 10.1136/bmjopen-2017-016317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aditama L, Athiyah U, Utami W, Rahem A. Adherence behavior assessment of oral antidiabetic medication use: a study of patient decisions in long-term disease management in primary health care centers in Surabaya. J Basic Clin Physiol Pharmacol. (2020) 30:20190257. 10.1515/jbcpp-2019-0257 [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Fan R, Zhang C, Hong L, Zhang T, Chen Y, et al. Applying machine learning models to predict medication nonadherence in Crohn's disease maintenance therapy. Patient Prefer Adherence. (2020) 14:917–26. 10.2147/PPA.S253732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aziz F, Malek S, Ali AM, Wong MS, Mosleh M, Milow P. Determining hypertensive patients' beliefs towards medication and associations with medication adherence using machine learning methods. PeerJ. (2020) 8:e8286. 10.7717/peerj.8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karanasiou GS, Tripoliti EE, Papadopoulos TG, Kalatzis FG, Goletsis Y, Naka KK, et al. Predicting adherence of patients with HF through machine learning techniques. Healthc Technol Lett. (2016) 3:165–70. 10.1049/htl.2016.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lind ML, Mooney SJ, Carone M, Althouse BM, Liu C, Evans LE, et al. Development and validation of a machine learning model to estimate bacterial sepsis among immunocompromised recipients of stem cell transplant. JAMA Netw Open. (2021) 4:e214514. 10.1001/jamanetworkopen.2021.4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie F, Ong ME, Liew JN, Tan KB, Ho AF, Nadarajan GD, et al. Development and assessment of an interpretable machine learning triage tool for estimating mortality after emergency admissions. JAMA Netw Open. (2021) 4:e2118467. 10.1001/jamanetworkopen.2021.18467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue B, Li D, Lu C, King CR, Wildes T, Avidan MS, et al. Use of machine learning to develop and evaluate models using preoperative and intraoperative data to identify risks of postoperative complications. JAMA Netw Open. (2021) 4:e212240. 10.1001/jamanetworkopen.2021.2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Liu J, Nanga P, Liu Y, Cicek AE, Knoblauch N, et al. Detailed modeling of positive selection improves detection of cancer driver genes. Nat Commun. (2019) 10:3399. 10.1038/s41467-019-11284-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggerth A, Hayn D, Schreier G. Medication management needs information and communications technology-based approaches, including telehealth and artificial intelligence. Br J Clin Pharmacol. (2020) 86:2000–7. 10.1111/bcp.14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. (2006) 34:e112. 10.1093/nar/gkl480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacks MS, Zhang W, Wognum S. A novel fibre-ensemble level constitutive model for exogenous cross-linked collagenous tissues. Interface Focus. (2016) 6:20150090. 10.1098/rsfs.2015.0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JH, Laurent AH, Hilser VJ, Ostermeier M. Design of protein switches based on an ensemble model of allostery. Nat Commun. (2015) 6:6968. 10.1038/ncomms7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Wang H, Wang J, Liu H, Lu R, Duan T, et al. A novel model for malaria prediction based on ensemble algorithms. PLoS ONE. (2019) 14:e0226910. 10.1371/journal.pone.0226910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewin-Epstein O, Baruch S, Hadany L, Stein GY, Obolski U. Predicting antibiotic resistance in hospitalized patients by applying machine learning to electronic medical records. Clin Infect Dis. (2020) 72:e848–55. 10.1093/cid/ciaa1576 [DOI] [PubMed] [Google Scholar]
- 39.Kim SJ, Kwon OD, Han EB, Lee CM, Oh SW, Joh HK, et al. Impact of number of medications and age on adherence to antihypertensive medications: a nationwide population-based study. Medicine (Baltimore). (2019) 98:e17825. 10.1097/MD.0000000000017825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.