Abstract
A coinfection assay was developed to examine Mycobacterium tuberculosis genes suspected to be involved in resistance to killing by human macrophages. THP-1 macrophages were infected with a mixture of equal numbers of recombinant Mycobacterium smegmatis LR222 bacteria expressing an M. tuberculosis gene and wild-type M. smegmatis LR222 bacteria expressing the xylE gene. At various times after infection, the infected macrophages were lysed and the bacteria were plated. The resulting colonies were sprayed with catechol to determine the number of recombinant colonies and the number of xylE-expressing colonies. M. smegmatis bacteria expressing the M. tuberculosis glutamine synthetase A (glnA) gene or open reading frame Rv2962c or Rv2958c demonstrated significantly increased survival rates in THP-1 macrophages relative to those of xylE-expressing bacteria. M. smegmatis bacteria expressing M. tuberculosis genes for phospholipase C (plcA and plcB) or for high temperature requirement A (htrA) did not.
It is estimated that Mycobacterium tuberculosis, the causative agent of tuberculosis, infects about one-third of the world's population, and about three million people die of tuberculosis each year (24). M. tuberculosis is an intracellular pathogen which survives and replicates within cells of the host immune system, primarily macrophages. Following phagocytosis into the macrophage, M. tuberculosis prevents acidification of the phagosome and fusion with lysosomes by altering the maturation of the phagosome (1, 5, 8, 17, 29). The precise survival strategies used by M. tuberculosis and the genes required for intracellular survival remain to be elucidated, although several candidate genes and activities, such as superoxide dismutase and catalase/peroxidase, have been proposed to be involved (6, 21, 23, 32).
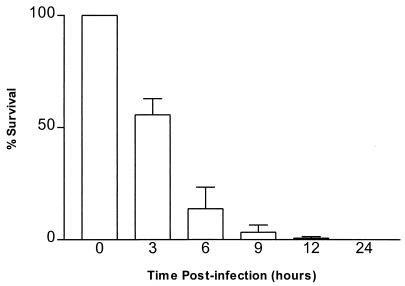
To examine the potential involvement of M. tuberculosis genes in survival in macrophages, a coinfection assay was developed in which the survival of a recombinant Mycobacterium smegmatis strain relative to that of a wild-type strain could be directly measured. For these assays, cells of THP-1, a human monocyte-derived macrophage line (2), were used. The THP-1 cells were maintained in RPMI 1640 medium (Gibco BRL, Gaithersburg, Md.) containing 10% fetal calf serum (FCS) (Gibco BRL) at 37°C in 5% CO2 and differentiated into macrophage-like cells by treatment with 10 μM phorbol myristate acetate (Sigma Chemical Company, St. Louis, Mo.) as previously described (2). A coinfection assay was necessary because wild-type M. smegmatis bacteria are rapidly killed in the first few hours after phagocytosis by differentiated THP-1 macrophages (Fig. 1). By 24 h postinfection, less than 0.01% of the phagocytized bacteria are viable (<100 CFU per well). The determination of the precise kinetics of survival of the bacteria is confounded by well-to-well variability in the numbers of bacteria phagocytized and the small numbers of CFU recovered. This results in large standard deviations which might mask relatively small differences in survival rates.
FIG. 1.
Survival of M. smegmatis in THP-1 macrophages. THP-1 macrophages were infected with wild-type M. smegmatis bacteria as described in the text. Time zero is defined as immediately after the 2-h phagocytosis period. Percent survival at a given time was calculated by dividing the number of CFU recovered at that time by the number of CFU recovered at time zero (1.35 × 106 CFU) and multiplying by 100. Error bars represent the standard deviations for three replicate cultures. Percent survival at 12 h was 0.67 ± 0.32, and that at 24 h was 0.015 ± 0.005.
In order to distinguish the two strains in the coinfection, the wild-type strain was engineered to express catechol 2,3-dioxygenase. The xylE gene of Pseudomonas putida was isolated from the plasmid pTKmx (14) and cloned into the expression vector pHIP, and the construct was electroporated into wild-type M. smegmatis LR222 (18). Colonies of mycobacteria expressing the xylE gene turn yellow when sprayed with catechol due to the conversion of catechol to 2-hydroxymuconic semialdehyde by catechol 2,3-dioxygenase (33). By measuring the ratio of white to yellow colonies over time in macrophages infected with a mixture of nonexpressing bacteria (white) and xylE-expressing bacteria (yellow), the survival rates of the two strains can be directly compared.
In this study, we investigated six genes that have been suggested to contribute to the ability of M. tuberculosis to survive in the macrophage: glnA, plcA, plcB, htrA, Rv2962c, and Rv2958c. In pathogenic mycobacteria, glutamine synthetase A catalyzes the extracellular synthesis of l-glutamine, an important cell wall component (10). Phospholipase C might enable the tubercle bacillus to escape from the phagosome into the cytoplasm, as has been observed in some studies (17, 20). In Listeria monocytogenes, two phospholipase C proteins are produced, one of which is involved in escaping the phagosome while the other is involved in cell-to-cell spread (16, 27). The M. tuberculosis high temperature requirement A gene (htrA) encodes a protein that has homology with the HtrA family of serine proteases (30). Disruption of the htrA genes of Yersinia enterocolitica and Salmonella enterica serovar Typhimurium results in decreased rates of survival of the bacteria in cultured murine macrophages relative to those of wild-type bacteria (13, 22, 31). The precise role of HtrA in intracellular survival of these bacteria is not known. M. tuberculosis open reading frames (ORF) Rv2962c and Rv2958c display homology to a Mycobacterium leprae ORF that was shown to confer increased survival in J774 macrophages on both Escherichia coli and M. smegmatis recipients (19).
To construct M. smegmatis bacteria expressing the M. tuberculosis glutamine synthetase A (glnA), phospholipase C A (plcA), phospholipase C B (plcB), or high temperature requirement A (htrA) gene or Rv2962c or Rv2958c, each ORF was PCR amplified from M. tuberculosis H37Rv (28) genomic DNA and cloned individually downstream of the M. tuberculosis hsp65 promoter in the pHIP vector. The vector pHIP was constructed by cloning the hsp65 promoter into pBHIN (25). The resulting plasmids were electroporated into M. smegmatis, and hygromycin-resistant bacteria were selected. The pHIP constructs are stably maintained in mycobacteria by virtue of integration of a single copy of the plasmid into the mycobacteriophage L5 attachment site in the mycobacterial genome. The hsp65 promoter should provide a high level of transcription during macrophage infection (4).
For a coinfection assay, the recombinant bacteria and the xylE-expressing wild-type bacteria were each grown separately to mid-log phase (optical density at 600 nm = ∼0.3) in Middlebrook 7H9 medium (Difco Laboratories, Detroit, Mich.) containing 0.05% (vol/vol) Tween 80 (Sigma). The bacteria were harvested by centrifugation for 1 min at 16,000 × g, washed twice with RPMI 1640–10% FCS, and resuspended in RPMI 1640–10% FCS at a concentration of 1.5 × 108 bacteria/ml. Equal volumes of the two bacterial suspensions were mixed, and a portion of the combined suspension was plated onto tryptic soy agar (TSA) plates (Difco Laboratories) to determine the number of viable bacteria of each strain in the initial inoculum (−2-h time point). The combined suspension was diluted to approximately 5 × 107 bacteria/ml and 3 ml was added to each well of THP-1 macrophages (∼1 × 106 cells per well), giving a multiplicity of infection of 50 bacteria per macrophage. Phagocytosis of the bacteria was allowed to proceed for 2 h at 37°C, after which each well was washed twice with RPMI 1640–10% FCS. This results in approximately one phagocytized bacterium per THP-1 macrophage. To kill remaining extracellular bacteria, 3 ml of fresh medium containing 200 μg of amikacin (Sigma) per ml was added to each well. Cultures were incubated at 37°C in 5% CO2. At various times after phagocytosis, the medium was removed from each of three wells and 1 ml of 0.1% (vol/vol) Triton X-100 in H2O was added to each well to lyse the macrophages. Each lysate was diluted as necessary and portions were plated onto TSA plates. The wells which were assayed immediately after the addition of amikacin served as the standard for measuring the number and ratio of phagocytized bacteria; the time at which these wells were assayed was considered time zero. The TSA plates from each time point were incubated at 37°C for 3 days and then stored overnight at 4°C. The following day, the plates were sprayed with 0.5 M catechol in 50 mM potassium phosphate (pH 7.5) to distinguish the xylE-expressing colonies (yellow) from the recombinant colonies (white). Storing the plates overnight at 4°C results in a stronger yellow color.
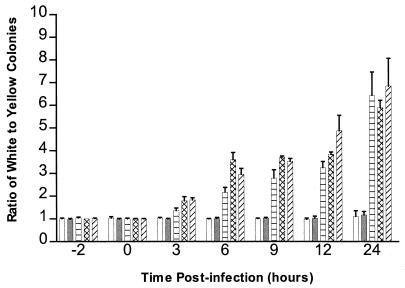
To determine if the expression of the xylE gene or pHIP vector genes affected survival, THP-1 macrophages were infected with a mixture of M. smegmatis bacteria expressing xylE and wild-type M. smegmatis bacteria by using a multiplicity of infection that results in uptake of about one bacterium per macrophage. The ratio of xylE-expressing colonies (yellow) to wild-type colonies (white) was 1:1 at 0 h and remained 1:1 throughout the experiment (Fig. 2), indicating that the two strains are phagocytized and survive equally well and that the xylE-expressing strain is a suitable reference or internal control for comparison of the survival rates of other M. smegmatis strains.
FIG. 2.
Survival of M. smegmatis bacteria expressing xylE relative to that of recombinant M. smegmatis bacteria expressing an M. tuberculosis gene. THP-1 macrophages were infected with a mixture of equal numbers of recombinant bacteria (white) and bacteria expressing the xylE gene (yellow). The ratio of white to yellow colonies is shown for −2 (initial inoculum), 0 (immediately after phagocytosis), 3, 6, 9, 12, and 24 h after phagocytosis. The mixtures tested contained control bacteria expressing the xylE gene and wild-type M. smegmatis bacteria (open bars) or recombinant bacteria expressing the M. tuberculosis htrA (shaded bars), glnA (horizontally striped bars), Rv2962c (cross-hatched bars), or Rv2958c (hatched bars) genes. The results are the averages of three independent experiments. Error bars represent the standard deviations.
During the course of an infection of THP-1 macrophages with a mixture of M. smegmatis bacteria expressing the M. tuberculosis glnA gene and wild-type M. smegmatis bacteria expressing the xylE gene, both strains of mycobacteria were rapidly killed but the ratio of glnA-expressing colonies to wild-type colonies increased from 1:1 at 0 h (9.6 × 105 white to 9.8 × 105 yellow colonies) to 3.2:1 at 12 h (4.6 × 104 white to 1.4 × 104 yellow colonies) to 6:1 at 24 h (150 white to 25 yellow colonies) after phagocytosis (Fig. 2). The differences between the ratios at time zero and the subsequent time points are statistically significant (P < 0.005, two-sample t test) for all time points.
Previous studies (10, 11) demonstrated that expression of the M. tuberculosis glnA gene in M. smegmatis results in a recombinant protein that is identical to the M. tuberculosis protein and that both the M. smegmatis recombinant and M. tuberculosis bacilli secrete the M. tuberculosis GlnA protein. The M. smegmatis GlnA protein is not secreted. One possible explanation for the ability of the secreted M. tuberculosis GlnA protein to enhance the survival of M. smegmatis is that the GlnA protein may modulate the pH of the phagosome by altering the levels of phagosomal ammonia (9, 10). So, by secreting the M. tuberculosis GlnA protein, the recombinant M. smegmatis bacteria may interfere with the acidification of the phagosome and thereby delay fusion with lysosomes and exposure to the antimicrobial activities in the lysosome. Expression of GlnA is not sufficient, however, to prevent acidification and fusion, because the recombinant M. smegmatis bacteria are still efficiently killed by the macrophages. Indeed, no viable mycobacteria were recovered from samples harvested 48 h postinfection.
The M. tuberculosis ORFs Rv2962c and Rv2958c were identified through their homology to an ORF of M. leprae suspected to be involved in survival in murine J774 macrophages (19). The M. leprae ORF has 72% identity with Rv2962c and 79% identity with Rv2958c (7). The two M. tuberculosis ORFs have 77% identity in their DNA sequences and 74% identity in their amino acid sequences (7). THP-1 macrophages were coinfected with bacteria expressing either Rv2962c or Rv2958c and wild-type bacteria expressing xylE (Fig. 2). For Rv2962c, the ratio of white to yellow colonies increased from 1:1 at 0 h to ∼4:1 at 12 h to ∼6:1 at 24 h. The ratio of white to yellow colonies for Rv2958c was 1:1 at 0 h, ∼5:1 at 12 h, and ∼8:1 at 24 h. The differences between the ratios at time zero and at the subsequent time points are statistically significant (P < 0.005) for all time points.
ORFs Rv2962c and Rv2958c encode proteins which display homology to glycosylases and glycosyltransferases. For example, the putative glycosyltransferase of Streptomyces capreolus has 31.7% identity with that of Rv2962c in a 375-amino-acid overlap (7). The mycobacterial glycosylases might affect intracellular survival through the glycosylation of surface or cell wall moieties, detoxification activities, or modification of host phagosomal proteins.
The M. tuberculosis genes plcA and plcB encode two phospholipase C proteins that have 68.9% identity (12, 15). In a semiquantitative assay for phospholipase C activity (3), lysates of M. smegmatis bacteria expressing either the M. tuberculosis plcA or plcB gene were able to cleave p-nitrophenol from p-nitrophenylphosphorylcholine, which turned the lysate yellow, whereas lysates of wild-type M. smegmatis bacteria did not cleave the substrate and remained colorless. During infection of THP-1 macrophage cultures with mixtures of bacteria expressing either plcA or plcB and wild-type bacteria expressing xylE, the ratio of white to yellow colonies was 1:1 at 0 h and remained 1:1 throughout the experiment for both pairs of strains (data not shown).
Expression of M. tuberculosis htrA mRNA in the M. smegmatis strain containing the M. tuberculosis htrA gene was confirmed by detecting a reverse transcription-PCR amplicon corresponding to htrA mRNA in the recombinant strain but not in wild-type M. smegmatis (data not shown). In coinfections of THP-1 macrophages, the ratio of white to yellow colonies at 0 h was 1:1 and remained 1:1 throughout the experiment (Fig. 2).
The failure of the M. tuberculosis genes for phospholipase C and high temperature requirement A to confer increased rates of intracellular survival on M. smegmatis recipients does not rule out an involvement of these genes in the ability of M. tuberculosis to survive in the human macrophage, because the assay measures primarily resistance to killing, which is a subset of intracellular survival. That is, using the approach described in this report, only certain gene products with a significant positive effect on preventing the killing of M. smegmatis bacteria, such as those that directly inhibit or inactivate one of the killing mechanisms of the macrophage, are likely to be identified (19). Genes that are not likely to be identified include those encoding proteins that require interaction with other proteins to prevent killing, that are part of a multienzyme biosynthetic pathway whose end product prevents killing, that regulate genes involved in intracellular survival, or that are essential for intracellular survival or replication but which do not directly affect the killing processes (19, 26). For example, a phospholipase C enzyme may be only one of several enzymes needed to modify the phagosomal membrane to block phagosome-lysosome fusion or to allow escape from the host phagosome. For the htrA gene, an additional limitation is that the presence of htrA mRNA does not show that functional HtrA protein is produced. For example, the recombinant expressed RNA or protein may not be posttranscriptionally modified in M. smegmatis in the same manner as in M. tuberculosis.
Further work is needed to elucidate possible roles of the glnA, Rv2962c, and Rv2958c genes in the ability of M. tuberculosis to survive in the human macrophage. The analysis of M. tuberculosis strains with disruptions in the glnA, Rv2962c, or Rv2958c gene is needed to determine these genes' roles in intracellular survival. Finally, the ability to preferentially recover recombinant strains with a greater resistance to killing by macrophages suggests that the culture system could be modified to enrich for recombinants with increased survival rates from a library of M. smegmatis bacteria carrying large fragments of M. tuberculosis DNA, perhaps thereby allowing identification of additional M. tuberculosis genes involved in resistance to killing by macrophages.
Acknowledgments
We thank the Biological Products Branch, National Center for Infectious Disease (NCID), CDC, for providing THP-1 cells, the Biotechnology Core Facility, NCID, CDC, for providing oligonucleotide primers, and Jack Crawford, Tuberculosis/Mycobacteriology Branch, NCID, CDC, for providing M. smegmatis LR222.
REFERENCES
- 1.Armstrong J A, Hart P D. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asseffa A, Dickson L A, Mohla S, Bremner T A. Phorbol myristate acetate-differentiated THP-1 cells display increased levels of MHC class I and class II mRNA and interferon-gamma-inducible tumoricidal activity. Oncol Res. 1993;5:11–18. [PubMed] [Google Scholar]
- 3.Berka R M, Gray G L, Vasil M L. Studies of phospholipase C (heat-labile hemolysin) in Pseudomonas aeruginosa. Infect Immun. 1981;34:1071–1074. doi: 10.1128/iai.34.3.1071-1074.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burlein J E, Stover C K, Offutt S, Hanson M S. Expression of foreign genes in mycobacteria. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 239–252. [Google Scholar]
- 5.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohn M L, Kovitz C, Oda U, Middlebrook G. Studies on isoniazid and tubercle bacilli. II. The growth requirements, catalase activities, and pathogenic properties of isoniazid-resistant mutants. Am Rev Tuberc. 1954;70:641–664. doi: 10.1164/art.1954.70.4.641. [DOI] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Crowle A J, Dahl R, Ross E, May M H. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun. 1991;59:1823–1831. doi: 10.1128/iai.59.5.1823-1831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart P D, Young M R. Ammonium chloride, an inhibitor of phagosome-lysosome fusion in macrophages, concurrently induces phagosome-endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J Exp Med. 1991;174:881–889. doi: 10.1084/jem.174.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harth G, Clemens D L, Horwitz M A. Glutamine synthetase of Mycobacterium tuberculosis: extracellular release and characterization of its enzymatic activity. Proc Natl Acad Sci USA. 1994;91:9342–9346. doi: 10.1073/pnas.91.20.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harth G, Horwitz M A. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J Biol Chem. 1997;272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 12.Johansen K A, Gill R E, Vasil M L. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect Immun. 1996;64:3259–3266. doi: 10.1128/iai.64.8.3259-3266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson K, Charles I, Dougan G, Pickard D, O'Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 14.Kenney T J, Churchward G. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol. 1996;178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leão S C, Rocha C L, Murillo L A, Parra C A, Patarroyo M E. A species-specific nucleotide sequence of Mycobacterium tuberculosis encodes a protein that exhibits hemolytic activity when expressed in Escherichia coli. Infect Immun. 1995;63:4301–4306. doi: 10.1128/iai.63.11.4301-4306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquis H, Doshi V, Portnoy D A. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonough K A, Kress Y, Bloom B R. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect Immun. 1993;61:2763–2773. doi: 10.1128/iai.61.7.2763-2773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mundayoor S, Shinnick T M. Identification of genes involved in the resistance of mycobacteria to killing by macrophages. Ann N Y Acad Sci. 1994;730:26–36. doi: 10.1111/j.1749-6632.1994.tb44236.x. [DOI] [PubMed] [Google Scholar]
- 20.Myrvik Q N, Leake E S, Wright M J. Disruption of phagosomal membranes of normal alveolar macrophages by the H37Rv strain of Mycobacterium tuberculosis. A correlate of virulence. Am Rev Respir Dis. 1984;129:322–328. [PubMed] [Google Scholar]
- 21.O'Brien L, Carmichael J, Lowrie D B, Andrew P W. Strains of Mycobacterium tuberculosis differ in susceptibility to reactive nitrogen intermediates in vitro. Infect Immun. 1994;62:5187–5190. doi: 10.1128/iai.62.11.5187-5190.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 23.Pascopella L, Collins F M, Martin J M, Lee M H, Hatfull G F, Stover C K, Bloom B R, Jacobs W R., Jr Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect Immun. 1994;62:1313–1319. doi: 10.1128/iai.62.4.1313-1319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelicic V, Reyrat J M, Gicquel B. Genetic advances for studying Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 1998;28:413–420. doi: 10.1046/j.1365-2958.1998.00807.x. [DOI] [PubMed] [Google Scholar]
- 25.Plikaytis B B, Crawford J T, Shinnick T M. IS1549 from Mycobacterium smegmatis forms long direct repeats upon insertion. J Bacteriol. 1998;180:1037–1043. doi: 10.1128/jb.180.5.1037-1043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinnick T M, King C H, Quinn F D. Molecular biology, virulence, and pathogenicity of mycobacteria. Am J Med Sci. 1995;309:92–98. doi: 10.1097/00000441-199502000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steenken W, Gardner L U. History of H37 strain of tubercle bacillus. Am Rev Tuberc. 1946;54:62–66. doi: 10.1164/art.1946.54.1.62. [DOI] [PubMed] [Google Scholar]
- 29.Sturgill-Koszycki S, Schlesinger P H, Chakraborty P, Haddix P L, Collins H L, Fok A K, Allen R D, Gluck S L, Heuser J, Russell D G. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q-L, Kong D, Lam K, Husson R N. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto T, Hanawa T, Ogata S, Kamiya S. The Yersinia enterocolitica GsrA stress protein, involved in intracellular survival, is induced by macrophage phagocytosis. Infect Immun. 1997;65:2190–2196. doi: 10.1128/iai.65.6.2190-2196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Lathigra R, Garbe T, Catty D, Young D. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol Microbiol. 1991;5:381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x. [DOI] [PubMed] [Google Scholar]
- 33.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]