Abstract
We present a new methodology for generating a stepwise concentration gradient in a series of micro-droplets by using monolithic micro valves that act as ‘faucets’ in micrometer-scale. A distinct concentration gradient of a substrate was generated for the determination of the kinetic parameters of two different enzymes using only ten picoliter-scale droplets. With a single experiment on a chip, we obtained KM and kcat values of matrix metalloproteinase 2 (MMP-2) and matrix metalloproteinase 9 (MMP-9), and compared the catalytic competence of the two enzymes. The present system and method are highly suitable for applications where the reagents or samples are limited and precious.
Introduction
Chemical and biochemical reactions in pico/nanoliter-sized droplets have been studied intensively during the past decade1–4. The droplet-based reactions offer a number of advantages over conventional microliter or milliliter scale reactions. The reduction in droplet volume enables rapid reactions due to extremely small heat capacity and shorter diffusion distances5. The ability to conduct a large number of parallel reactions with droplets is another advantage6–8 in addition to confinement of reagents with distinct reaction conditions, and isolation of droplets to avoid nonspecific binding of biomolecules to the channel walls3, 9. To explore these advantages, different droplet formation methods have been reported to achieve control over size, shape, and mono-dispersity of droplets2, 3, 9.
The flow-control droplet generation method, also known as T-junction method, is one of the widely used methods4, 8–14. In the flow-control method, two fluidic channels intersect perpendicular to each other. Typically, a continuous phase flows through the horizontal channel while immiscible dispersed phase flows through the vertical channel. When the dispersed phase comes inside the continuous phase channel, it starts forming droplets due to competition between surface tension and shear forces13. Another popular approach3, 15, 16 of droplet generation is flow- focusing where the dispersed phase enters into a co-flowing continuous phase stream so as to produce droplets. Because both flow-control and flow-focusing methods are based on shearing of the dispersed phase by the continuous phase, the sizes of the droplets can be controlled by regulating flow speeds and the channel widths or by changing the viscosity of each phase8, 13, 17. Both of the shear-stress based methods of droplet generation, however, pose critical disadvantages of limiting the control of individual droplets in terms of the size and frequency of droplet formation18. Consequently, flexible control of the composition in each droplet reactor is challenging4. Lorenz et al19 and Damean et al20 independently reported concentration gradients in droplets with continuous supply of reagents in fluidic channels through a tree-like channel structures21, 22. In these cases, the extension of the concentration gradients is directly limited by the number of the fluidic branches19, 20. Another unique approach by Du et al23 used capillary tubes of 75 to 200 µm in inner diameter to generate concentration gradients. Because this capillary method needs a conventional external syringe pump to uptake different liquid samples from the end of the capillary, there are intrinsic limitations in achieving high accuracy of nanoliter or picoliter resolution. Moreover, this method is not free from preventing complete cross-contamination because of the direct contact of the tip of the capillary to the reagent reservoirs. With an ultrasonic actuator, several groups24, 25 showed ejection of nanoliter volumes of samples for printing arrays of biomaterials. However, none of the above referred methods demonstrated the creation of stepwise concentration gradient inside closed microchannels in picoliter resolution.
Here, we demonstrate a new methodology to realize a distinct concentration gradient in consecutive micro droplets by utilizing series of monolithic micro-valves26, 27. The present method could be implemented with a simple pneumatic control system and it can easily generate different droplet compositions by adjusting dispensing time of each reagent. To show the potential of the present droplet system, a concentration gradient of a substrate is generated for the evaluation of the catalytic competence of MMP-2 and MMP-9 in a single experiment.
MATERIALS AND METHODS
Materials
Matrix metalloproteinases-2 (MMP-2), matrix metalloproteinases-9 (MMP-9) and MMP substrate III (QXL™520-Pro-Leu-Gly-Cys-(Me)-His-Ala-D-Arg-Lys-(5-carboxyfluorescein)-NH2) were used as a model system. MMP-2 and MMP-9 are considered to play an important role in metastasis and are targets for drug development.28, 29 MMP-2 and MMP-9 were purchased from R&D Systems, Inc., and MMP substrate III was from AnaSpec Inc. The MMP substrate III used for the present work is a specially designed and commercially available peptide substrate. The substrate has an attached fluorophore, 5-carboxy-fluoresein-Pro- Leu-OH (FAM), on one end of the peptide substrate and a quencher, QXL520, on the other end. Upon enzymatic cleavage of the peptide substrate, the fluorescence of FAM is emitted, and the fluorescence signals are observed as an indicator of enzymatic reaction (Supplementary Figure S1). Stock solutions of 200 nM of MMP-2 and 9 in a reaction buffer (50 mM Tris, 10 mM CaCl2, 150 mM NaCl, 0.05% Brij-35, and 0.1% BSA at pH 7.5) were prepared. Immediately before use, the thawed stock solutions were diluted with the reaction buffer solution. All the experiments were conducted at room temperature (25 ± 1.5 °C). During the on-chip experiments, mineral oil (Sigma Inc., product number M5904) was used as a continuous phase and no surfactant was used in the microchannels.
Chip fabrication
The mask was designed using AutoCAD software (AutoDesk Inc., San Rafael, CA) and printed on a transparent film at 20,000 dpi (CAD/Art Services, Inc., Bandon, OR). The molds for the two layers, fluidic and control, were fabricated by a photolithographic technique. At first, the positive photoresist (AZ P4620, AZ electronic materials) was spin-coated onto a 4- inch silicon wafer. This was followed by UV exposure and development. For reliable opening and closing of the valves, the photoresists for fluidic layer was rounded by heating the mold at 130°C for 2 min.
The top thick fluidic layer of the chip was prepared by pouring uncured polydimethylsiloxane (PDMS, GE RTV615; elastomer:crosslinker = 10:1) onto the fluidic layer mold to achieve a thickness of 5 mm. The bottom control layer of the chip was fabricated by spin-coating uncured PDMS (elastomer:crosslinker = 20:1) onto the control layer mold at 3000 rpm for 1 min. The two layers were cured for 1 h (fluidic layer) and 45 min (control layer) at 80°C, respectively. The fluidic layer was peeled off from the mold, and holes were punched for inlet/outlet ports to the flow channels through the thick layer with a 19-gauge punch (Technical Innovations Inc., Brazoria, TX). The fluidic layer was aligned over the control layer. The two layers were bonded by baking at 80 °C for 45 min. The bonded layers were peeled off from the control layer mold, and holes were punched for inlet ports to the control channels. Finally, the PDMS chip was placed on a pre-cleaned glass slide (Fisher Scientific Pittsburgh, PA) and kept it in an oven at 80 °C for 18 h to advance curing.
Labview based pneumatic control
The pneumatic control of each valve was carried out with eight-channel manifolds (Fluidigm Corp.). The source of driving pressure was pressurized nitrogen gas. We used multiple two-way splitters to connect each dispensed channels to the manifolds. The gas pressure was finely attenuated by using a regulator (Alicat Scientific, Inc., Model- LSPM). We mounted a digital I/O card (NI PCI-6533 (DIO-32HS), National Instruments) in the computer to digitally control the switching of each channel of the manifold. For the accurate control of the droplet generation process, a custom-built LabVIEW (National Instruments) program was used for sending the appropriate signal with predetermined dispensing time, and the corresponding droplet volume was calculated from our predictive model published before27. Preliminary experiments were conducted to establish the relation between the dispensing time and the corresponding droplet volumes at the start of each experiment.
Image acquisition and processing
A series of fluorescence images of the enzyme reactions were acquired with a modified biochip reader (arrayWoRx®, Applied Precision, WA). All fluorescence images shown in Figure 3a and b were digitized and analyzed by using the time series analyzer of ImageJ software (NIH, USA). 5-FAM (5-carboxyfluorescein) droplets were generated of concentration range, 0.3 µM to 1.5 µM with the increment of 0.3 µM, and measured the fluorescent intensities by collecting fluorescence images. During the measurement, it was observed that 0.7% of the fluorescent intensity was decreased due to photobleaching of the fluorescent molecule during each fluorescence scan (Figure S7 of the supplementary materials). The loss of fluorescent intensities was compensated30.
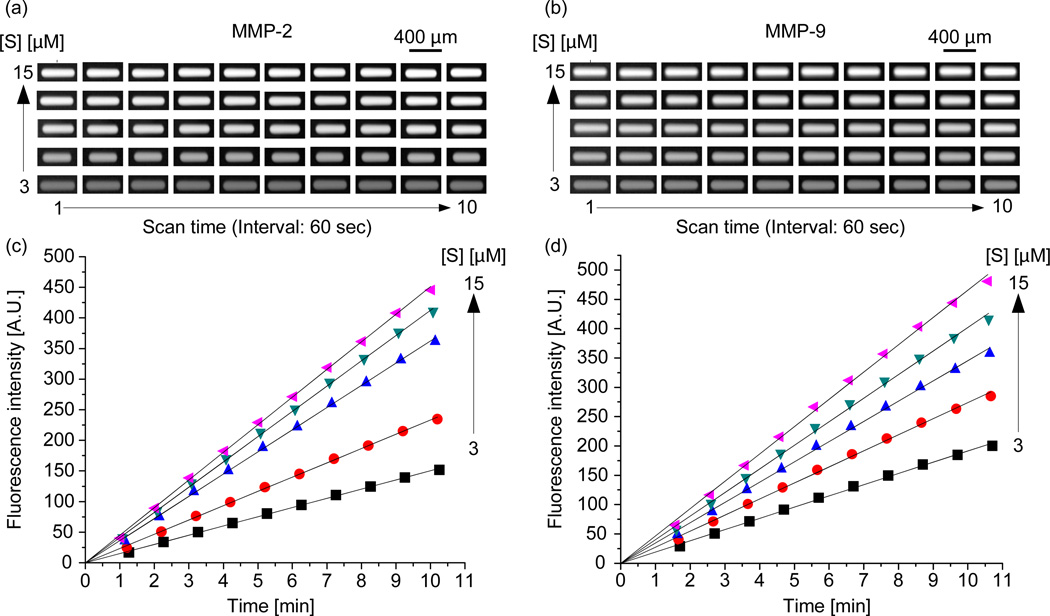
Figure 3.
(a) and (b) Series of scanned images of the droplet microreactors for the enzymatic reactions of MMP-2 and MMP-9 with MMP substrate III. (c) and (d) Fluorescent intensity of droplet reactors during enzymatic reactions of MMP-2 and MMP-9 with MMP substrate III.
RESULTS AND DISCUSSION
Droplet generation and concentration gradient
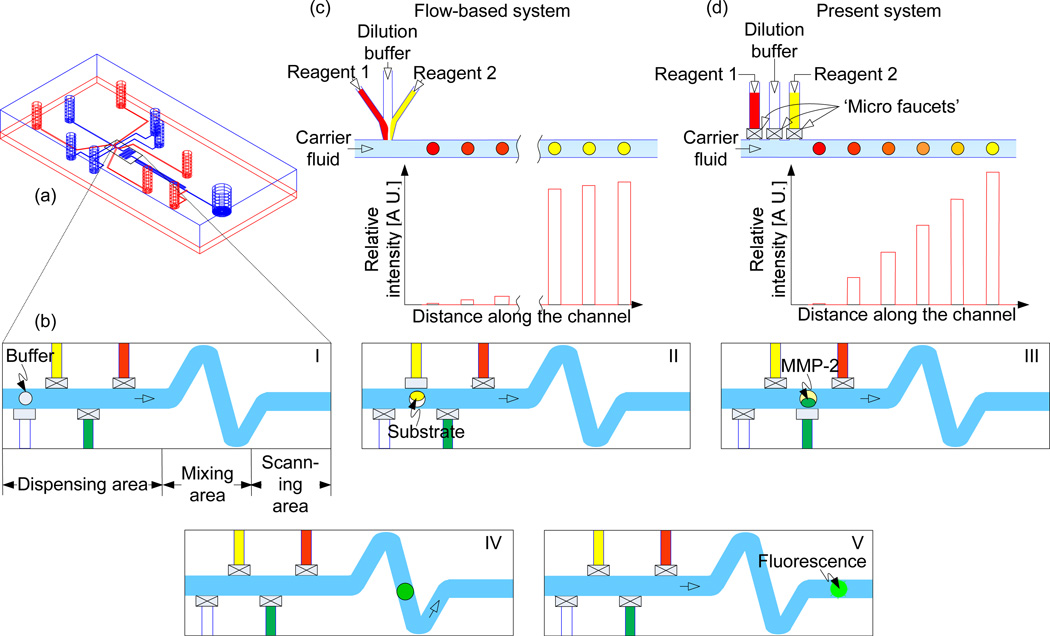
The system to generate picoliter-scale droplets consists of a continuous phase channel that is running left to right and dispensing channels which are perpendicular to the continuous phase channel (Figure 1a, b). The distinct difference between the present system and other analog or low concentration gradient droplet systems4 is the inclusion of a mechanical valve as shown in Figure 1c, d. We positioned a set of microvalves at the end of the four dispensing channels as shown in Figure 1b. Based on the functionality, the continuous phase channel can be divided into three parts: dispensing area, mixing area and scanning area. In the dispensing area, the picoliter-scale droplets were generated by the opening and closing of the valves and the generated droplets were moved through the continuous phase channel. For instance, by the operation of the valve of the first dispensing channel, we can generate a droplet of buffer. This droplet moves forward due to the flow of continuous phase. When the buffer droplet approaches the second dispensing channel, the second droplet of a substrate is ejected and merged to the buffer droplet. A schematic explanation of the processes of conducting enzymatic reactions with generating and merging picoliter-scale droplets is depicted in Figures 1d-I to V. The enzymatic reactions consist of MMP-2 and MMP-9 enzymes, and MMP substrate III. For the sake of simplicity, we have explained only one reaction with MMP-2 and MMP substrate III. Firstly, the buffer droplet was ejected from the buffer dispensing channel (Figure 1b-I). When the buffer droplet approached the substrate dispensing channel, as a second step, the substrate droplet was ejected to form a merged buffer-substrate droplet (Figure 1b-II). In third step, the merged buffer-substrate droplet was combined with the third droplet of enzyme after its ejection from the MMP-2 dispensing channel (Figure 1b-III). The reagents inside the individual merged droplets were mixed when flowing through the winding channel (Figure 1b-IV), known as the mixing area, through the process of chaotic advection11. Due to mixing of enzyme and substrate in the merged droplet, fluorescence from the droplet was intensified continuously (Figure 1b-V). Finally, this increasing fluorescence intensity was detected by using the biochip reader at a 60-second interval. In parallel, the reaction for the MMP-9 enzyme and MMP substrate III was conducted by using the same procedure of five steps on the same chip.
Figure 1.
Concentration gradients in droplet reactors. (a) Design of the microfluidic system. Blue and red colors indicate fluidic channels and control channels, respectively. (b) Step-by-step generation of droplets, I to V, with different compositions (See text for explanation). (c) Gradient generation with other flow-based systems. (d) Gradient generation with the present system. Reagent 1 (MMP substrate III), dilution buffer, and reagent 2 (MMP-2 or MMP-9) were fed in the form of droplets through the vertical dispensing channels that were controlled by micro valves.
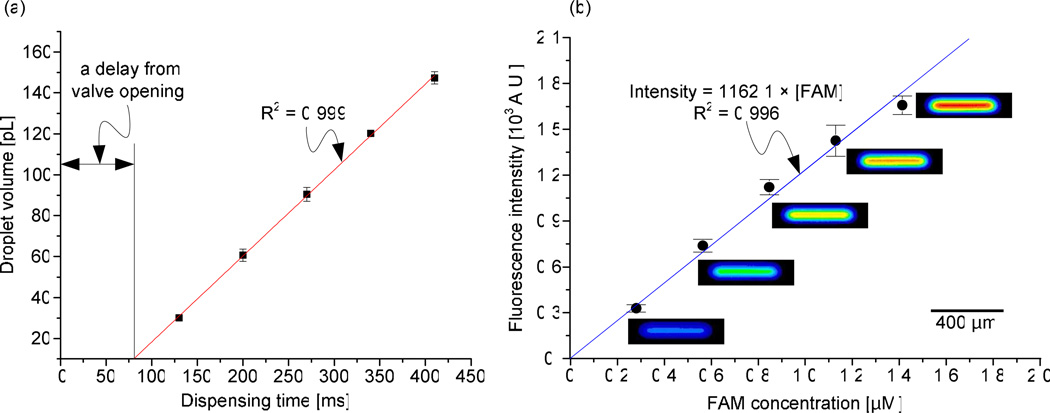
We charted the relationship between droplet volumes from 30 pL to 150 pL and their corresponding dispensing time that is equivalent to the valve opening time as shown in Figure 2a. This linear relationship between the valve opening time and the droplet volume was confirmed for three sets of experiments with a standard deviation of 1.6% in the droplet volume. These results confirm the accuracy and reproducibility of the present droplet system.
Figure 2.
(a) Dispensing time vs. volume of droplets. (b) Standard curve of the fluorescent intensity vs. FAM concentration. Scanned images show fluorescence of the micro-droplets for corresponding concentrations of FAM.
Standard curve
To calibrate the velocity characterization of enzymatic reactions of MMP-2 and MMP-9 with MMP substrate III, we generated a series of droplets having different concentrations of fluorescent molecules, 5-carboxy-fluoresein-Pro-Leu-OH (FAM) on a chip and plotted a standard curve (Figure 2b). The FAM emits fluorescent light at 520 nm, which is identical to the wavelength of fluorescence emitted by the product of MMP-2 and 9 when MMP substrate III is used. Internally quenched MMP substrate III does not emit any fluorescent signal without enzymatic reaction. Thus, the increase in the fluorescence intensity can be used to monitor the rate of catalytic reactions of MMP-2 and 9 when MMP substrate III is used as a substrate. Figure 2b shows the scanned images of the five droplet microreactors containing different concentrations of FAM. The concentration was varied linearly from 0 to 1.5 µM with an increment of 0.3 µM. The changes in colors of the droplets indicate the increasing fluorescence intensity of the droplets. We measured the intensity of the whole area of each droplet to calculate the velocity of the enzyme reactions. We confirmed that the intensities at the center of the droplets were higher compared to those at the edge of droplets. We attribute this intensity variation in the droplet to the rounded shape of the fluidic channel (Supplementary Figure S2). To prevent the non-specific binding of proteins and chemicals on the channel walls31, we added 0.1% of bovine serum albumin (BSA) which is widely used as a blocking reagent32, 33 to the buffer. We also generated a standard curve of FAM for conventional enzyme experiments by using a spectrophotometer (Supplementary Figure S3).
Determination of kinetic parameters
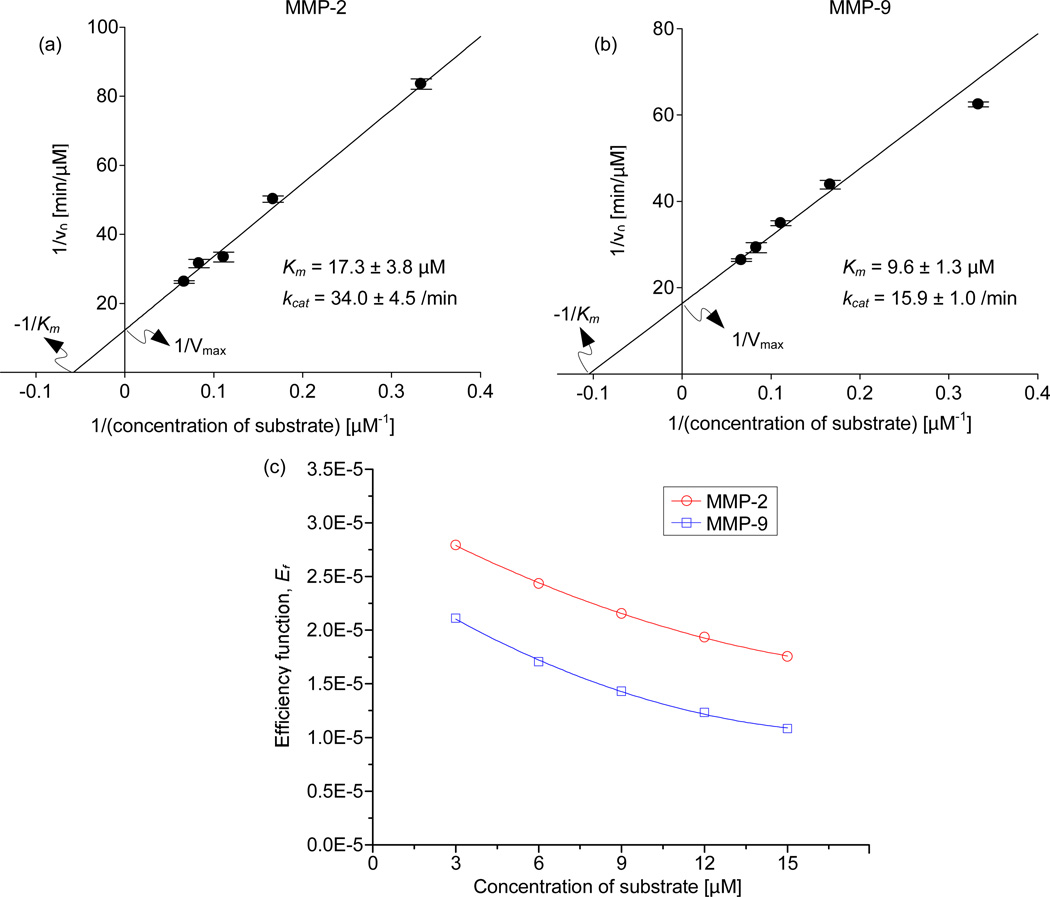
The accuracy of kinetic parameters such as KM and kcat is affected by the enzyme concentration34, 35 and its linear response to a range of substrate concentrations35. To determine the enzyme concentration suitable for the experiments, the substrate concentration needs to be maintained high in order to avoid substrate depletion36 and to help an enzyme to reach its saturation limit without falling short of substrate molecules35. We varied the enzyme concentrations using conventional methods from 0.5 to 35 nM for both MMP-2 and MMP-9 by keeping the MMP substrate III concentration high, 30 µM. We traced the reaction velocities for 120 min. For MMP-2, with the substrate concentration at 30 µM, we observed linear velocity responses up to 11 min with 0.5 nM and 2.5 nM concentrations of the enzymes. At higher concentrations, above 2.5 nM of the enzyme, we witnessed nonlinear responses. Consequently, we used 2.5 nM of MMP-2 for the rest of the experiments. For MMP-9, linear velocity was observed up to 11 min at 0.5 nM and 4 nM of the enzyme concentration. Hence, we used 4 nM of MMP-9 for the rest of the experiments. (Supplementary Figure S4). We used these enzyme concentrations hereafter. The difference in the enzyme concentrations for MMP-2 and MMP-9 is due to different upper limit of linear velocity responses37–40. For enzymatic reactions inside the droplets on a chip, we generated two sets of substrate concentration gradient from 3 to 15 µM by adjusting volume of the buffer droplet and the substrate droplet. Then, 2.5 nM of MMP-2 was introduced for the first set of the five droplets and 4.0 nM of MMP-9 for the next five droplets. Detailed information on volume of each component of the merged droplets is found in Table 1. The fluorescent signals resulting from the enzymatic reaction of MMP-2 and MMP-9 inside the droplet reactors were scanned for a period of 11 min using 1 min intervals (Figure 3a and 3b). These scanned images were digitized to obtain the fluorescence values of each droplet. The digitized intensity values that represent the progress of the reactions are plotted in Figure 3c and 3d. Although each droplet is scanned at shifted time points, the scanning interval for each droplet is same (Supplementary Figure S5). Hence the initial velocity of reaction in each droplet reactor was calculated from respective reaction progress plots. In order to determine the kinetic parameters, the initial velocities and substrate concentrations were plotted with Lineweaver-Burk plots (Figure 4a and 4b). In the curve fitting, the nonlinear regression analysis was used to find the Y- intercept (1/Vmax) and the X-intercept (−1/KM). The half-maximal activity, KM of MMP-2 and 9 determined from three independent experiments were 17.3 ± 3.8 µM and 9.6 ± 1.3 µM, respectively. The calculated turnover number, kcat, of MMP-2 and 9 were 34.0 ± 4.5 /min and 15.9 ± 1.0 /min, respectively. To validate our on-chip results, we also conducted conventional experiments (Supplementary Figure S6). The KM values of MMP-2 and 9 were 14.8 ± 1.8 µM and 8.1 ± 1.1 µM, respectively. The turnover number kcat, from the conventional methods, of MMP-2 and 9 were 32.3 ± 2.3 /min and 14.1 ± 0.9 /min. The deviations of KM and kcat from our droplet and conventional experiments were 16.9% and 5.3% for MMP-2 and 8.5% and 12.8% for MMP-9. The deviations are acceptable for enzymatic reactions because enzymes are biological samples with variable properties and the experimental errors involved in the kinetic measurements of enzymes could never be completely eliminated41.
Table 1.
Compositional changes of droplet reactors.
| Substrate dispensing volume [pL] |
Dilution buffer dispensing volume [pL] |
Enzyme dispensing volume [pL] |
Concentration of substrate [µM] |
Concentration of MMP-2 enzyme [nM] |
Concentration of MMP-9 enzyme [nM] |
|---|---|---|---|---|---|
| 30 ± 1.4 | 147.3 ± 3.0 | 180 ± 3.2 | 3.0 | 2.5 | 4.0 |
| 60.7 ± 3.0 | 120.1 ± 1.3 | 180 ± 3.0 | 6.0 | ||
| 90.5 ± 3.4 | 90.5 ± 3.4 | 180 ± 2.7 | 9.0 | ||
| 120.1 ± 1.3 | 60.7 ± 3.0 | 180 ± 2.4 | 12.0 | ||
| 147.3 ± 3.0 | 30 ± 1.4 | 180 ± 2.1 | 15.0 |
Figure 4.
(a) and (b) Lineweaver-Burk plots of MMP-2 and MMP-9. (c) Variation of the efficiency function (Ef) for MMP-2 and MMP-9.
Evaluation of catalytic competence
The evaluation of catalytic competence of multiple enzymes consists of reactions with substrate concentration gradients under uniform experimental conditions42. Ideally, for keeping the experimental conditions uniform, the reactions needs to be conducted in parallel under the same experimental conditions. The ability of the present system to generate substrate concentration gradient in parallel architecture makes it possible for the direct evaluation of catalytic competence. For this evaluation, the catalytic efficiency, kcat/KM is one of the commonly used second order rate constants43. From the on-chip experiments, the calculated values of kcat/KM for MMP-2 and 9 were 2.0 ± 0.5 and 1.7 ± 0.2 µM−1min−1, respectively. The values of kcat/KM using conventional methods were found to be 2.2 ± 0.3 µM−1min−1 for MMP-2 and 1.7 ± 0.3 µM−1min−1 for MMP-9 (Table 2). The difference between on-chip values and conventional values of kcat/KM for MMP-2 and MMP-9 were 9.5 and 0.0 %, respectively. Because the catalytic efficiency is the ratio of kinetic parameters, its variation depends on the variation of individual values of KM and kcat 35, 43. Therefore, the deviation in kcat/KM of MMP-2 could be attributed to the aforementioned large variation in KM.
Table 2.
Comparison of kinetic parameters of MMP-2 and MMP-9 for enzymatic reaction with MMP substrate III.
| Kinetic parameters | MMP-2 | MMP-9 | ||
|---|---|---|---|---|
| On-chip | Off-chip | On-chip | Off-chip | |
| KM(µM) | 17.3 ± 3.8 | 14.8 ± 1.8 | 9.6 ± 1.3 | 8.1 ± 1.1 |
| kcat (min−1) | 34.0 ± 4.5 | 32.3 ± 2.3 | 15.9 ± 1.0 | 14.1 ± 0.9 |
| kcat/KM(µM−1 min−1) | 2.0 ± 0.5 | 2.2 ± 0.3 | 1.7 ± 0.2 | 1.7 ± 0.3 |
Throughout the literature of enzyme kinetics44–46 scientists use kcat/KM for reporting the catalytic competence of an enzyme. However, it has been proven that the kcat/KM alone is an incomplete measure for the evaluation of catalytic competence because it is valid only for substrate concentration, [S], approaching zero47, 48. To circumvent the limitation of kcat/KM, we have used another parameter known as efficiency function, Ef. The efficiency function is also known as practical catalytic efficiency as it takes into consideration the variation of catalytic efficiency with the change in substrate concentration47, 48. However, to use Ef as the parameter for the evaluation of catalytic competence, similar physiological experimental conditions, such as precise concentration gradient for multiple reagents, need to be maintained for all the reactions42, 47. This is quite difficult in conventional experiments. However, with the present system, parallel reactions under identical conditions can be realized by only changing the target enzymes. We determined the values of Ef for both MMP-2 and MMP-9 with variable substrate concentration. As shown in Figure 4c, for the substrate concentrations ranging from 3 to 15 µM, higher values of Ef for MMP-2 were found in comparison to MMP-9. This indicates that, MMP-2 is more effective enzyme than MMP-9 when reacting with MMP substrate III under identical experimental conditions. The results were confirmed by performing off-chip experiments (See Figure S6). Due to the capability of the simultaneous evaluation of competing enzymes, our microfluidic system would be a unique enzyme screening tool for pharmaceutical49, 50 and biotechnology51, 52 field.
Stepwise concentration gradient
The uniqueness of our system lies in its ability to generate multiple droplet reactors having different concentrations of reagents without increasing the complexity of the system. The general formulation of concentration of a reagent in ith droplet could be given as
Here, i and k are identification numbers for the droplet and the required concentration gradient number, respectively; I, K indicate the corresponding total number of droplets and total number of concentration gradients required. In the present case, I=5 to accommodate five droplets with different concentration of substrate and K=1 because only concentration gradient of substrate was created. In the present system, the independent droplets of enzyme, substrate and buffer are designated as sub-droplets. Here, the term signifies that, we have a control over the volume of individual sub-droplet and, hence, can create concentration gradient in the consecutive droplets. Thus, we can conduct multiple, parallel, and combinatorial reactions that are difficult to conduct with existing channel-based32, 53 or other droplet-based systems4, 54. The concentration gradient can be expanded beyond 1:10,000 though we showed the gradient of 1:5 with MMP substrate III in the present manuscript. The smallest volume of a droplet we generated with the current method was 0.014 pL27 and the largest volume was 150 pL. By combining these two droplets of 0.014 pL and 150 pL volumes, the dilution ratio of 1 to 10,714 could be achieved.
One exciting potential application opened up by the presented method is the possibility of determining target potency and selectivity55, using only a few picoliter-scale droplets in a single experiment. In addition, it could also open the window for generating the concentration-response plots of multiple proteins or drug candidates when the reactions under the same conditions are performed in parallel. In practice, the concentration-response experiments are commonly performed by inhibitor titration experiments using microwell plates56 with millilitre scale drug samples. These experiments could be realized by adding dispensing channels to the present droplet system and by generating ‘n’ picoliter-scale droplets for each drug candidate besides the two droplets for positive and negative control of the experiment. The ‘n’ droplets for each drug are used for varying the concentration of inhibitor and to measure the velocity of the inhibited reaction. The droplet for positive control is without inhibitor while the droplet for negative control is with saturating concentration of inhibitor. The velocities obtained from ‘n’ droplets are used for constructing concentration-response plots and for determining the IC50 values of the target drug candidate.
Another potential application of the present system could be envisioned for the screening of whole cell assays57, 58. During the compartmentalization process, a single cell could be entrapped inside the droplet and later a number of cells could be grown from a single cell for the subsequent drug screening. Next, the cell-containing droplet is merged with other droplets for generating stepwise concentration gradients of a drug candidate, for example. The fate of the cells, live or dead, or the growth of cells could be tracked by direct observations under a microscope or other optical or electrochemical detection methods. The unique advantage of our system includes its ability to conduct an almost unlimited number of reactions with many chemicals or proteins in parallel with a single cell resolution. To adopt this method for the realization of such high throughput screening for comparative assays, the number of parallel dispensing channels could be increased depending on the required number of reagents. Therefore, the present system could be an ideal platform for the drug screening research as mentioned above.
Two practical issues need to be tackled for improving the performance and the application range of the present system. First is the improvement of scan rate and resolution of continuous fluorescence scanning of the droplets. This issue could be addressed by integrating the present device with light emitting diode (LED) based illumination systems and complementary metal oxide semiconductor (CMOS) based diode array for detection. In addition, recently developed wide field lens-free-microscopes59, 60 could also be incorporated with the present droplet system for broadening the application range. The second issue is the generation of femto or atto-liter scale droplets. Although we have reported a picoliter-scale concentration gradient, the present droplet generation method could be used to generate volumes in femto-liter scale through redesign of the chip.
CONCLUSION
In conclusion, the stepwise concentration gradient of reagents was created in ten picoliter- scale droplets for conducting two parallel reactions of enzymes. The simultaneous enzymatic reactions of MMP-2 and MMP-9 were performed in droplet microreactors with MMP substrate III. Five droplets having different compositions of the buffer and the substrate were precisely generated with different ratios of 1:5, 2:4, 3:3, 4:2, 5:1, and 6:0 of the buffer and the substrate. Although the volume ratios of the buffer and the substrate in the droplets were varied, the volumes of the two enzymes were kept constant in the droplet reactors. Two parallel reactions with only five picoliter-scale droplets for each enzyme were performed. Increase in the fluorescence of each droplet reactors after merging the three components, the buffer, the substrate, and the enzymes was observed. From the scanned images of the droplets, the kinetic parameters of KM and kcat of MMP-2 and MMP-9 were determined. The kinetic parameters were used to calculate efficiency function, Ef, and based on Ef values the catalytic competence of the two enzymes was successfully compared. Our system could be used for conducting kinetic analysis and catalytic competence evaluation of a vast number of other enzymes for laboratory research and industrial applications with its flexibility in generating different concentration gradients beyond 1:10,000 with different compositions of reagents.
Supplementary Material
ACKNOWLEDGEMENTS
This research was partially supported by a grant from Marine Bioprocess Research Center of the Marine Bio 21 Project funded by the Ministry of Land, Transport and Maritime, Republic of Korea. We also acknowledge partial support from the National Institutes of Health (NIH R01 008392). The authors would like to thank Kirn Cramer for her help during the preparation of the manuscript.
References
- 1.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proceedings of the National Academy of Sciences. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huebner A, Sharma S, Srisa-Art M, Hollfelder F, Edel JB, demello AJ. Lab on a Chip. 2008;8:1244–1254. doi: 10.1039/b806405a. [DOI] [PubMed] [Google Scholar]
- 3.Teh SY, Lin R, Hung LH, Lee AP. Lab on a Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 4.Song H, Ismagilov RJ. Am. Chem. Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabeling P. Introduction to microfluidics. USA: Oxford University Press; 2005. [Google Scholar]
- 6.Prakash M, Gershenfeld N. Science. 2007;315:832. doi: 10.1126/science.1136907. [DOI] [PubMed] [Google Scholar]
- 7.Hong J, Edel J, deMello A. Drug discovery today. 2009;14:134–146. doi: 10.1016/j.drudis.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Teh S, Lin R, Hung L, Lee A. Lab on a Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 9.Song H, Chen DL, Ismagilov RF. Angewandte Chemie (International ed. in English) 2006;45:7336. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, Tice JD, Ismagilov RF. Angewandte Chemie (International ed. in English) 2003;42:768. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 11.Bringer MR, Gerdts CJ, Song H, Tice JD, Ismagilov RF. Philos Transact A Math Phys Eng Sci. 2004;362:1087–1104. doi: 10.1098/rsta.2003.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srisa-Art M, Dyson EC, deMello AJ, Edel JB. Analytical Chemistry. 2008;80:7063–7067. doi: 10.1021/ac801199k. [DOI] [PubMed] [Google Scholar]
- 13.Thorsen T, Roberts R, Arnold F, Quake S. Physical Review Letters. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 14.Huebner A, Sharma S, Srisa-Art M, Hollfelder F, Edel J, demello A. Lab on a Chip. 2008;8:1244–1254. doi: 10.1039/b806405a. [DOI] [PubMed] [Google Scholar]
- 15.Lee C, Hsiung S, Lee G. IEEE. 2007:167–171. [Google Scholar]
- 16.Hsiung S, Chen C, Lee G. Journal of Micromechanics and Microengineering. 2006;16:2403. [Google Scholar]
- 17.Song H, Chen D, Ismagilov R. Angewandte Chemie (International ed. in English) 2006;45:7336. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joanicot M, Ajdari A. Science. 2005;309:887–888. doi: 10.1126/science.1112615. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz RM, Fiorini GS, Jeffries GDM, Lim DSW, He M, Chiu DT. Analytica Chimica Acta. 2008;630:124–130. doi: 10.1016/j.aca.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damean N, Olguin LF, Hollfelder F, Abell C, Huck WTS. Lab on a Chip. 2009;9:1707–1713. doi: 10.1039/b821021g. [DOI] [PubMed] [Google Scholar]
- 21.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Analytical Chemistry. 2001;73:1240–1246. [Google Scholar]
- 22.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 23.Du WB, Sun M, Gu SQ, Zhu Y, Fang Q. Analytical Chemistry. 2010:7336–7356. doi: 10.1021/ac1020479. [DOI] [PubMed] [Google Scholar]
- 24.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Trends in Biotechnology. 2003;21:157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 25.Moon SJ, Hasan SK, Song YS, Xu F, Keles HO, Manzur F, Mikkilineni S, Hong JW, Nagatomi J, Haeggstrom E. Tissue Engineering Part C: Methods. 2009;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 27.Lee W, Jambovane S, Kim D, Hong J. Microfluidics and Nanofluidics. 2009;7:431–438. [Google Scholar]
- 28.Egeblad M, Werb Z. Nature Reviews Cancer. 2002;2:163–176. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 29.Yoneda T. European Journal of Cancer. 1998;34:240–245. doi: 10.1016/s0959-8049(97)10132-0. [DOI] [PubMed] [Google Scholar]
- 30.McGrath J, Tardy Y, Dewey C, Jr, Meister J, Hartwig J. Biophysical journal. 1998;75:2070–2078. doi: 10.1016/S0006-3495(98)77649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toepke M, Beebe D. Lab on a Chip. 2006;6:1484. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 32.Jambovane S, Duin E, Kim S, Hong J. Analytical Chemistry. 2009;81:3239–3245. doi: 10.1021/ac8020938. [DOI] [PubMed] [Google Scholar]
- 33.Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A. Analytical Biochemistry. 2000;278:123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- 34.Segel I, Fisher J. Enzyme kinetics. New York: Wiley; 1993. [Google Scholar]
- 35.Copeland R. Enzymes: a practical introduction to structure, mechanism, and data analysis. Wiley-Vch; 2000. [Google Scholar]
- 36.Bisswanger H. Practical enzymology. Wiley Online Library; 2004. [Google Scholar]
- 37.Hutzler J, Dancis J. Biochimica et Biophysica Acta (BBA)-Enzymology. 1975;377:42–51. doi: 10.1016/0005-2744(75)90284-3. [DOI] [PubMed] [Google Scholar]
- 38.O'Sullivan M, Tipton K, McDevitt W. Archives of oral biology. 2002;47:399. doi: 10.1016/s0003-9969(02)00009-2. [DOI] [PubMed] [Google Scholar]
- 39.Eisenthal R, Danson M. Enzyme assays: a practical approach. USA: Oxford University Press; 2002. [Google Scholar]
- 40.Diamondstone T. Analytical Biochemistry. 1966;16:395–401. [Google Scholar]
- 41.Cornish-Bowden A. Analysis of enzyme kinetic data. USA: Oxford University Press; 1995. [Google Scholar]
- 42.Benner S. Chemical Reviews. 1989;89:789–806. [Google Scholar]
- 43.Eisenthal R, Danson M, Hough D. Trends in Biotechnology. 2007;25:247–249. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Oyeyemi O, Sours K, Lee T, Resing K, Ahn N, Klinman J. Proceedings of the National Academy of Sciences. 2010;107:10074. doi: 10.1073/pnas.1003678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox R, ElsonShing-Hui N, Griffin M. Journal of molecular biology. 1971;58:197–204. doi: 10.1016/0022-2836(71)90241-5. [DOI] [PubMed] [Google Scholar]
- 46.Hubatsch I, Ridderström M, Mannervik B. Biochemical Journal. 1998;330:175. doi: 10.1042/bj3300175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albery W, Knowles J. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 48.Ceccarelli E, Carrillo N, Roveri O. Trends in Biotechnology. 2008 doi: 10.1016/j.tibtech.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Frase H, Shi Q, Testero S, Mobashery S, Vakulenko S. Journal of Biological Chemistry. 2009;284:29509. doi: 10.1074/jbc.M109.011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith D, Jones B. Biochemical pharmacology. 1992;44:2089. doi: 10.1016/0006-2952(92)90333-e. [DOI] [PubMed] [Google Scholar]
- 51.Wu P, Hanlon M, Eddins M, Tsui C, Rogers R, Jensen J, Matunis M, Weissman A, Wolberger C, Pickart C. The EMBO Journal. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamiya N, Okazaki S, Goto M. Biotechnology Techniques. 1997;11:375–378. [Google Scholar]
- 53.Hadd A, Raymond D, Halliwell J, Jacobson S, Ramsey J. Anal. Chem. 1997;69:3407–3412. doi: 10.1021/ac970192p. [DOI] [PubMed] [Google Scholar]
- 54.Miller E, Wheeler A. Anal. Chem. 2008;80:1614–1619. doi: 10.1021/ac702269d. [DOI] [PubMed] [Google Scholar]
- 55.Terrett N. Drug Discovery Today: Technologies. 2010 [Google Scholar]
- 56.Copeland R. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. John Wiley and Sons; 2005. [PubMed] [Google Scholar]
- 57.Thomas G. Medicinal chemistry. Wiley-Interscience; 2007. [Google Scholar]
- 58.Doris C, Jun W, Lorraine H, Sheo S, Srinivas K. 2006 [Google Scholar]
- 59.Ozcan A, Demirci U. Lab on a Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 60.Coskun A, Su T, Ozcan A. Lab on a Chip. 2010;10:824. doi: 10.1039/b926561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.