Abstract
Colorectal cancer (CRC) is the third most common malignancy and ranks as the second leading cause of cancer-related deaths worldwide. Despite the improvements in CRC diagnosis and treatment approaches, a considerable proportion of CRC patients still suffer from poor prognosis due to late disease detection and lack of personalized disease management. Recent evidences have not only provided important molecular insights into their mechanistic behavior but have also indicated that identification of cancer-specific long non-coding RNAs (LncRNAs) could benefit earlier disease detection and improve treatment outcomes in patients suffering from CRC. LncRNAs have raised extensive attention as they participate in various hallmarks of CRC. The mechanistic evidence gleaned in the recent decade clearly reveals that lncRNAs exert their oncogenic role by regulating autophagy, epigenetic modifications, enhancing stem phenotype and modifying tumor microenvironment. In view of their pleiotropic functional role in malignant progression, and their frequently dysregulated expression in CRC patients, they have great potential to be reliable diagnostic and prognostic biomarkers, as well as therapeutic targets for CRC. In the present review, we will focus on the oncogenic roles of lncRNAs and related mechanisms in CRC as well as discuss their clinical potential in the early diagnosis, prognostic prediction and therapeutic translation in patients with this malignancy.
Keywords: colorectal cancer, long non-coding RNAs, signaling pathways, biomarkers
1. Background
Colorectal cancer (CRC) remains the third most common malignancy, and ranks as the second leading cause of cancer-related deaths worldwide[1]. Surgical resection, chemotherapy and radiotherapy are currently the primary therapeutic strategies for the management of CRC patients, among which surgical resection is preferentially considered for patients with an earlier-stage cancer while chemoradiotherapy is a treatment modality often offered in later disease stages[2]. Despite encouraging advances in the earlier disease diagnosis and improved treatment options, approximately 40–50% of patients with a stage II and III CRC suffer from poor survival outcomes due to a variety of reasons including inadequacy of the currently-available treatments and lack of availability of adequate risk-assessment biomarkers[3]. Classic serological biomarkers, including CEA, CA19–9, CA724 and CA125, are often routinely used for the early diagnosis for CRC, however, their sensitivity and specificity is far from satisfactory[4]. Hence, there is an urgent need for the identification of novel and reliable biomarkers for the individualized management of CRC, which potentially would lead to an improved overall outcome from this disease.
Noncoding RNAs (ncRNAs) represent a class of RNAs that do not directly participate in the transcription of mRNAs, but indirectly influence gene regulation. With the routine application of sophisticated next generation sequencing approaches, the identification of novel classes of ncRNAs continue to evolve each day. Presently, the major categories of such RNA include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), microRNAs (miRNAs) and long noncoding RNAs (lncRNAs)[5]. The LncRNAs are characterized as a group of ncRNAs that are more than 200 nucleotides long and exert their gene regulatory function through a variety of cellular processes such as chromatin modification, transcriptional regulation and post-transcriptional alterations[6]. Increasing evidence suggests that lncRNAs play an important role in tumor progression[7], metastasis[8], autophagy[9] and chemoresistance[5] via multiple mechanisms. Furthermore, in the recent years, lncRNAs have garnered increasing attention as potential clinical biomarkers for the early cancer screening, determining patient prognosis, and predicting response to various drug treatments[6]. A recent review has provided various strategies to develop lncRNA-based targeted drugs in pancreatic ductal adenocarcinoma (PDAC) such as small interfering RNA, CRISPR-Cas9 and antisense oligonucleotides[10]. In breast cancer, emerging evidence has highlighted the crucial role of exosomal lncRNAs in drug resistance, firmly supporting their therapeutic potential[11]. Taken together, due to their vital role in cancer biology and therapy, a deep and comprehensive understanding of lncRNAs will undoubtedly benefit the biomarker-directed precision medicine in cancer patients.
In the present review, we firstly focus on the biological functions and related oncogenic mechanisms associated with lncRNAs during CRC development. We also comprehensively discuss the roles of lncRNAs in predicting therapeutic efficacy in CRC. Finally, we summarize the potential clinical significance of lncRNAs in the management of patients with CRC. Taken together, our review aims to provide novel insights into the critical role of lncRNAs in CRC, offers a bench to bedside perspective, and highlights their important clinical potential as disease biomarkers for directing precision medicine approaches in cancer patients.
2. LncRNAs and hallmarks of CRC
Data gathered in the last two decades have clearly highlighted the biological roles of various lncRNAs in a variety of diseases, and particularly in cancer. In this context, a few specific examples of lncRNAs that associate with various hallmarks of CRC are illustrated in Figure 1. The subsequently text will delineate these features for their biological and clinical significance in further detail.
Figure 1:
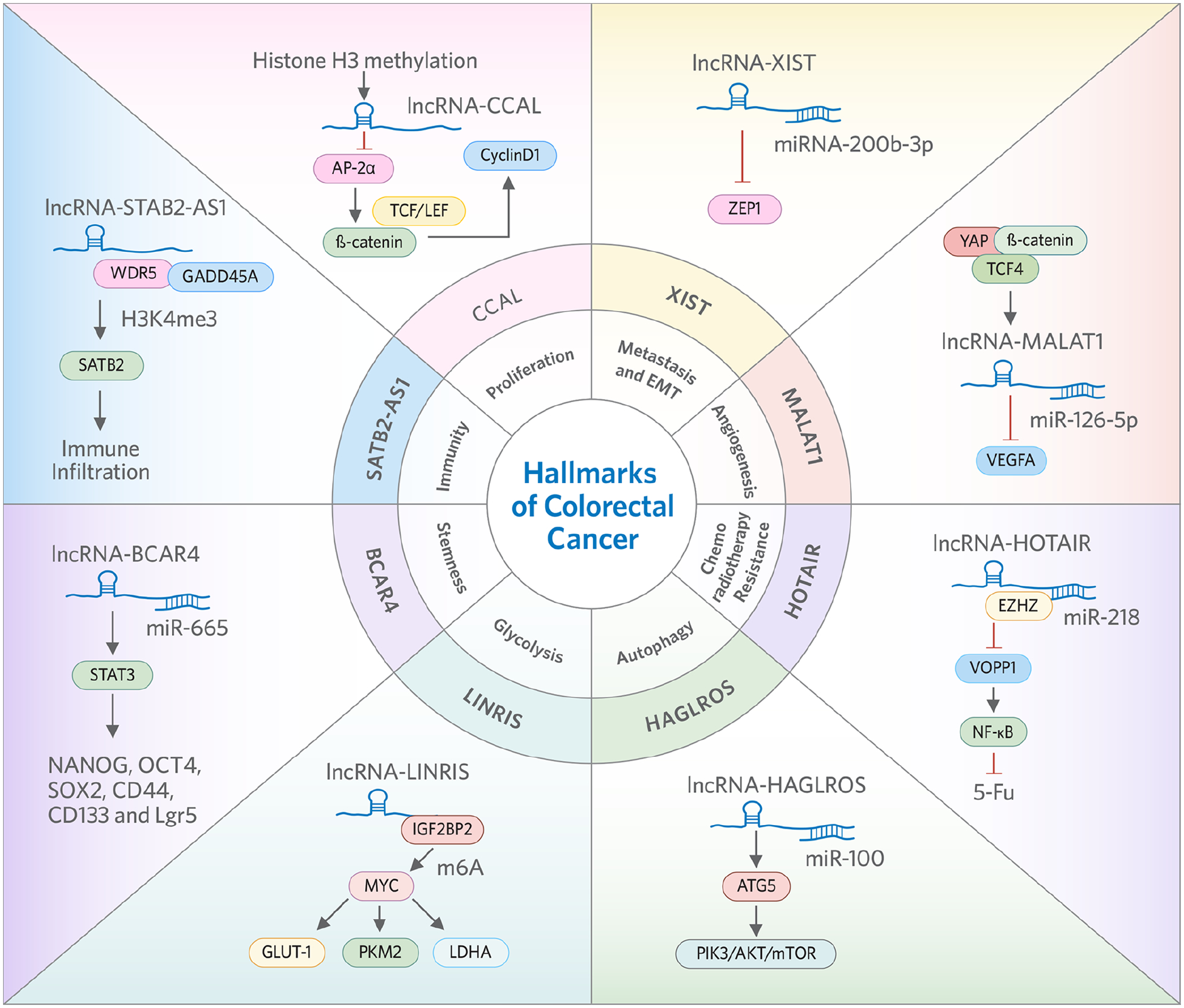
Representative LncRNAs and related oncogenic mechanisms in the proliferation, metastasis, angiogenesis, chemoradiotherapy, autophagy, glycolysis, stemness and immunity of colorectal cancer.
2.1. Proliferative signaling pathways
There is unequivocal evidence that specific lncRNAs play a central role in mediating the oncogenic cellular signaling in CRC, by impacting key signaling pathways including the Wnt/b-catenin, PI3K/AKT, JAK2/STAT3 and p53 pathways shown in more detail in Table 1.
Table 1.
Summary of the oncogenic roles of long non-coding RNAs in colorectal cancer.
| LncRNA | Effector | Functions | Targets | Signaling pathway | Cell lines | Reference |
|---|---|---|---|---|---|---|
| CCAL | oncogene | proliferation, invasion and migration | AP-2α | Wnt/β-catenin | LOVO | [18] |
| CCAT2 | oncogene | proliferation and metastasis | TCF7L2 | Wnt/β-catenin | HCT116,COLO320 | [19] |
| CCAS11 | oncogene | proliferation and metastasis | hnRNP-K | Wnt/β-catenin | SW480, SW620 | [20] |
| CRNDE | oncogene | proliferation and chemoresistance | miR-181a-5p | Wnt/β-catenin | HCT116, SW480 | [21] |
| SNHG1 | oncogene | proliferation and metastasis | β-catenin/TCF-4 | Wnt/β-catenin | SW480, LOVO | [28] |
| SNHG1 | oncogene | proliferation | miR-154-5p, EZH2 | miR-154-5p/CCND2 | HCT116, HCT8 | [46] |
| SNHG1 | oncogene | proliferation and migration | miR-137 | miR-137/RICTOR | LOVO, HT29 | [47] |
| ZEB1-AS1 | oncogene | proliferation and apoptosis | miR-181a-5p | Wnt/β-catenin | RKO, HCT116, LOVO | [22] |
| H19 | oncogene | proliferation | miR-200a | Wnt/β-catenin | HCT116, SW480 | [23] |
| H19 | oncogene | migration and invasion | RAS | RAS/MAPK | SW480, HCT116 | [48] |
| H19 | oncogene | proliferation, invasion and EMT | miR-29b-3p | Wnt/β-catenin | HT29, SW480 | [64] |
| H19 | oncogene | proliferation and EMT | miR-138, miR-200a | miR-138/ Vimentin, miR-200a/ZEB1,ZEB2 | SW620, HT29 | [56] |
| HCG18 | oncogene | proliferation and invasion | miR-1271 | Wnt/β-catenin | HCT116, SW480 | [24] |
| PlncRNA-1 | oncogene | proliferation, migration and invasion | PI3K | PI3K/AKT | SW480, HCT116 | [36] |
| PlncRNA-1 | oncogene | proliferation, migration, invasion and EMT | miR-204 | Wnt/β-catenin | SW480, SW620, HCT116, HT29 | [25] |
| HOTAIR | oncogene | proliferation and chemoresistance | miR-203a-3p | Wnt/β-catenin | CoLo205, SW620 | [26] |
| HOTAIR | oncogene | proliferation, metastasis and chemoresistance | miR-214 | JAK2/STAT3 | SW620, SW480, HCT-8 | [41] |
| XIST | oncogene | proliferation and invasion | miR-34a | Wnt/β-catenin | SW480, HCT116 | [27] |
| XIST | oncogene | proliferation, EMT and stem cell formation | miR-200b-3p | miR-200b-3p/ZEB 1 | HCT116, SW620 | [55] |
| HNF1A-AS1 | oncogene | proliferation, migration and invasion | β-catenin | Wnt/β-catenin | HCT116, SW620 | [29] |
| HIF1A-AS2 | oncogene | proliferation, invasion and EMT | miR-129-5p | miR-129-5p/DNMT3A | SW620, HCT116 | [75] |
| BCAT1 | suppressor | proliferation and invasion | β-catenin | Wnt/β-catenin | HCT116, SW480, | [32] |
| LINC00365 | oncogene | proliferation, migration, and invasion | CDK1 | Wnt/β-catenin | SW480, HT29 | [30] |
| NEAT1 | oncogene | proliferation, migration and invasion | DDX5 | Wnt/β-catenin | HCT116, SW1116, HT29 | [31] |
| AB073614 | oncogene | proliferation, migration and invasion | 740Y-P | PI3K/AKT | SW480 | [34] |
| AB073614 | oncogene | migration, invasion and EMT | JAK | JAK/STAT3 | SW480, HCT116 | [76] |
| LncRNA-422 | suppressor | proliferation, migration and invasion | PI3K, AKT, mTOR | PI3K/AKT | SW480, SW620 | [35] |
| TTN-AS1 | oncogene | proliferation, migration, invasion and EMT | miR-497 | PI3K/AKT | SW480, SW620 | [37] |
| CASC2 | oncogene | proliferation | miR-18a | JAK2/STAT3 | CACO2, HT29 | [42] |
| PURPL | oncogene | proliferation | MYBBP1A | p53 | HCT116, RKO, SW48, DLD1 | [44] |
| ROR | oncogene | proliferation and viability | p53 | p53 | M5, HT29 | [45] |
| EPB41L4A-AS1 | oncogene | proliferation, migration, invasion and EMT | RhoA, Rac1 | Rho/ROCK pathway | HCT116, SW620 | [49] |
| B3GALT5-AS1 | suppressor | proliferation and EMT | miR-203 | miR-203/ZEB2, SNAI2 | HCT116, SW620 | [59] |
| GNAT1-1 | suppressor | proliferation and metastasis | RKIP | RKIP/NF-κB/Snail | SW480, LOVO | [50] |
| UICLM | oncogene | proliferation, EMT and stem cell formation | miR-215 | miR-215/ZEB2 | SW620, DLD-1 | [57] |
| SNHG6 | oncogene | proliferation, migration, invasion | UPF1 | TGF-β/Smad | RKO, HCT116 | [58] |
| SNHG6 | oncogene | EMT | miR-101-3p | miR-101-3p/ZEB 1 | RKO, HCT116 | [58] |
| LINC01413 | oncogene | proliferation, migration, invasion and EMT | hnRNP-K | hnRNP-K/ZEB 1 | HT29, LOVO | [60] |
| CHRF | oncogene | migration, invasion and EMT | miR-489 | miR-489/ TWIST1 | HCT116, SW480 | [61] |
| SNHG15 | oncogene | proliferation and migration | Slug | NA | SW1116, HCT116, SW480,SW620 | [62] |
| TUG1 | oncogene | migration and invasion and EMT | miR-600 | miR-600/KIAA1199 | HCT116, LOVO, SW620 | [63] |
| SLCO4A1-AS1 | oncogene | proliferation, migration, invasion and EMT | β-catenin | Wnt/β-catenin | HCT116, SW480 | [65] |
| CYTOR | oncogene | migration, invasion and EMT | β-catenin | Wnt/β-catenin | HCT116, SW620, HCT-8 | [66] |
| CTD903 | suppressor | invasion, migration and EMT | NA | Wnt/β-catenin | RKO, SW480, SW620, HCT116, DLD-1 | [67] |
| TCF7 | oncogene | proliferation, migration and invasion | NA | Wnt/β-catenin | DLD-1, LOVO | [68] |
| MALAT1 | oncogene | EMT and chemoresistance | E-cadhrin | EZH2 | HT29, SW480, SW620 | [70] |
| HOXD-AS1 | oncogene | proliferation, invasion, EMT and stem cell formation | miR-217 | EZH2 | HCT116, LOVO | [73] |
| BDNF-AS | suppressor | proliferation, migration and invasion | EZH2-mediated H3K27me3 | GSK-3β | HCT116, LOVO | [74] |
| BC200 | oncogene | proliferation, invasion and EMT | STAT3, β-catenin | JAK/STAT3, Wnt/β-catenin | HCT116, HT29 | [77] |
| UCA1 | oncogene | proliferation, invasion, migration and EMT | CAFs | mTOR | SW480 | [78] |
| ZFAS1 | oncogene | proliferation, EMT and angiogenesis | miR-150-5p | miR-150-5p/ VEGFA, Akt/mTOR | HCT116, HCT8 | [79] |
2.1.1. Wnt/β-catenin signaling
The Wnt/β-catenin signaling cascade is one of the most important pathways that controls a variety of biological processes in CRC, including cell proliferation and apoptosis[12][13]. The Wnt/β-catenin signaling cascade is regulated by the β-catenin complex, which consists of APC, Axin2, CK1 and GSK-β[14]. More specifically, the β-catenin interacts with various transcription factors such as the TCF/LEF family members to activate the downstream gene targets[15]. Previous studies have reported Wnt/β-catenin pathway members were frequently activated in CRC[16][17]. Several lncRNAs, such as CCAL[18], CCAT2[19] and CASC11[20] have been validated to affect Wnt/β-catenin signaling pathway through their indirect binding to key proteins. The CCAT2 lncRNA enhances Wnt pathway activity through TCF7L2 and facilitates overactivation of oncogenic MYC[19], while CASC11 activates Wnt/β-catenin signaling to promote tumor growth by targeting hnRNP-K in CRC cells[20]. Some lncRNAs can exert effects on Wnt/β-catenin pathway by sponging or binding to specific miRNAs, and this list of lncRNAs include CRNDE[21], ZEB1-AS1[22], H19[23], HCG18[24], PlncRNA-1[25], HOTAIR[26] and XIST[27].
Furthermore, other lncRNAs can directly regulate the core effectors of this signaling pathway. For instance, lncRNA SNHG1 can upregulate β-catenin, TCF-4, cyclin D1 and MMP-9 expression to influence growth of CRC cells[28]. Likewise, the lncRNA HNF1A-AS1 can directly regulate the expression of β-catenin, cyclinD1, and c-myc – all of which play a vital role in Wnt/β-catenin signaling pathway[29]. Upregulation of the LINC00365 can lead to increased CDK1 protein expression and resultant activation of the Wnt/β-catenin signaling pathway[30]. On similar lines, NEAT1 indirectly activates the Wnt/β-catenin signaling pathway by regulating protein DDX5[31], while BCAT1 acts as a tumor suppressor by downregulating β-catenin expression in CRC cells[32].
2.1.2. PI3K/AKT signaling pathway
It is well-established that PI3K/AKT signaling plays a crucial role in the proliferative ability of CRC cells[33]. Recent studies have demonstrated numerous lncRNAs promote CRC proliferation via PI3K/AKT signaling pathway. The lncRNA AB073614[34], lncRNA-422[35] and PlncRNA-1[36] regulate proliferation in CRC cells through PI3K/AKT signaling pathway. The siRNA mediated knockdown of AB073614 expression significantly inhibited the proliferation, migration, and invasion of SW480 cells by targeting the PI3K/AKT-mediated signaling pathway[34]. LncRNA-422 overexpression inhibited cell proliferation, migration and invasion, and can also be regarded as a tumor suppressor through its ability to regulate PI3K/AKT/mTOR pathway in CRC[35]. In addition, PlncRNA-1 exerts its proliferative effects by targeting the PI3K/Akt signaling pathway in CRC[36] and TTN-AS1 enhances the proliferative ability of CRC cells by activating miR-497-mediated PI3K/Akt/mTOR signaling[37].
2.1.3. JAK2/STAT3 pathway
Similar to the PI3K/AKT pathway, the JAK2/STAT3 signaling pathway exerts a significant growth regulatory influence in driving neoplastic progression in CRC[38][39]. Moreover, this signaling pathway is suggested to serve as a crucial regulatory mechanism for the persistent growth of cancer stem cells following radiotherapy treatment in CRC[40]. Accumulating evidence suggests that certain lncRNAs control CRC cell proliferation via JAK2/STAT3 pathway by influencing the expression of phosphorylated STAT3 or by acting as competing endogenous RNAs (ceRNAs). For instance, ST6GAL1 and HOTAIR have been demonstrated to serve as direct targets of miR-214, wherein the expression of ST6GAL1 is modulated by HOTAIR by acting as a sponge for miR-214. Furthermore, ST6GAL1 regulates the elevated metabolic sialylation of c-Met, which finally impacts the CRC cell proliferation through JAK2/STAT3 pathway. The HOTAIR/miR-214/ST6GAL1 axis controls the malignant behavior of CRC cells by activating JAK2/STAT3 pathway[41]. In addition, the lncRNA CASC2 can directly upregulate PIAS3 expression by functioning as a competing endogenous RNA (ceRNA) for miR-18a and leads to the suppression of expression of genes downstream of STAT3, consequentially inhibiting the CRC tumor growth - both in-vitro and in in-vivo experimental models[42].
2.1.4. p53 pathway
The p53 pathway is a well-established pathway that regulates cellular proliferation and apoptosis in cancer cells[43]. In the recent years, several studies have indicated that specific lncRNAs can modulate the growth of CRC cells through p53 pathway. In this context, PURPL was shown to suppress basal p53 levels by interacting with MYBBP1A, a protein that binds to and stabilizes p53, and promotes tumorigenicity in CRC[44]. The inhibition of lncRNA-ROR significantly suppresses cell proliferation and viability, but enhances cell apoptosis, partially by modulating p53[45].
2.1.5. Other mechanisms
In addition to the specific CRC-associated growth pathways described above, several lncRNAs have been reported to affect other mechanisms in CRC. For instance, SNHG1 was shown to be involved in CRC growth by interacting with EZH2 and miR-153–5p[46] or through miR-137/RICTOR axis[47]. Likewise, H19 has been shown to promote the proliferation of colon cancer cells via MAPK signaling pathway[48]. Furthermore, PB41L4A-AS1 may participate in the growth of CRC by activating the Rho/Rho-associated protein kinase signaling pathway[49], while GNAT1–1 acts as a tumor suppressor through regulation of the RKIP-NF-κB-Snail cascade[50].
2.2. Epithelial-mesenchymal transition (EMT) mediated invasion and migration
The EMT is a biological process where epithelial cells acquire a mesenchymal phenotype with enhanced invasive and migratory potential[51]. It is now well-established that EMT acts as a critical mechanism that accounts for tumor invasion, metastasis and chemotherapeutic resistance[52][53]. Previous studies have demonstrated that lncRNAs regulate EMT-mediated invasion and migration mainly through transcription factors, Wnt/β-catenin signaling and epigenetic modifiers in CRC.
2.2.1. Transcription factors related pathway
The EMT process is regulated by several transcription factors, such as the zinc finger E-box-binding homeobox-(ZEB)1, snail family transcriptional repressor 2 (SLUG), snail family transcriptional repressor 1 (SNAI1), TWIST-related protein 1 (TWIST) and ZEB2[54]. LncRNAs can interact with these transcription factors to induce EMT in CRC, leading to the invasion and migration of tumor cells. Some of the lncRNAs (XIST, H19, UICLM, SNHG6, B3GALT5-AS1 and LINC01413) also act as regulators of ZEB1/2 related pathway and most of them mediate their effects by acting as competing endogenous RNAs (ceRNAs). For instance, XIST regulates CRC metastasis by competing with miR-200b-3p to modulate the expression of ZEB1[55]. Likewise, H19 acts as a ceRNA for miR-138 and miR-200a, and inhibits the expression of Vimentin, ZEB1, and ZEB2[56]. On the other hand, UICLM promotes liver metastasis in CRC by regulating the expression of ZEB2 by serving as a ceRNA for miR-215[57], and SNHG6 plays an oncogenic role in CRC cells by inducing EMT through the regulation of ZEB1[58]. A recent research article revealed that B3GALT5-AS1 directly binds to the promoter region of miR-203, downregulated its expression, targeted ZEB2 and SNAI2 and induced EMT and liver metastasis in CRC[59]. In contrast, LINC01413/hnRNP-K/ZEB1 axis upregulates cell proliferation and EMT in CRC by inducing YAP1/TAZ1 translocation[60]. Furthermore, lncRNA CHRF-induced miR-489 loss facilitates metastasis and EMT process in CRC cells probably via TWIST1 signaling pathway[61]. Finally, SNHG15 maintains Slug stability and promotes invasion and migration of CRC through ubiquitin-proteasome signaling pathway[62].
2.2.2. Wnt/β-catenin signaling pathway
The neoplastic progression of cancer cells through EMT is significantly associated with the activation of intracellular stem-associated pathways including Wnt/β-catenin, Notch, TGF-β, and Hedgehog pathways. As one of the major signaling pathways involved in EMT, the Wnt/β-catenin signaling contributes to the activation of transcription factors such as ZEB and Snail, upregulation of the expression of mesenchymal genes and concurrent downregulation of the E-cadherin expression[12][13]. In this regard, several lncRNAs can regulate EMT by targeting Wnt/β-catenin signaling pathway. For instance, TUG1[63], H19[64] and PlncRNA-1[25] has been shown to act as ceRNAs and accelerate metastasis and EMT in CRC through β-catenin signaling. Similarly, TUG1 promotes KIAA1199 expression leading to the induction of EMT and subsequent metastasis in CRC cells through inhibition of miR-600 expression[63]. H19/miR-29b-3p/PGRN axis promotes EMT in CRC through the Wnt signaling pathway as well[64]. PlncRNA-1/miR-204/Wnt/β-catenin regulatory network promotes cell proliferation, EMT and hepatic metastasis in CRC[25]. Other lncRNAs, such as SLCO4A1-AS1[65], CYTOR[66], CTD903[67] and TCF7[68] can modulate β-catenin expression to exert their oncogenic functions. On similar lines, SLCO4A1-AS1 promotes cellular proliferation, migration, invasion and EMT in CRC cells through activation of Wnt/β-catenin signaling pathway[65]. The positive feed-forward circuit of CYTOR-β-catenin promotes EMT and metastasis in CRC[66]; however, the underlying mechanism of CTD903 and TCF7 still require further exploration.
2.2.3. Epigenetic regulatory pathway
Increasing studies have demonstrated that epigenetic regulation plays a pivotal role during EMT, which is also manifested through methylation and acetylation changes[69]. The EZH2 protein has been proved as a critical factor in histone methylation[70], and DNMT3A methyl transferase is responsible for transferring methyl groups to CpG islands, leading to gene repression[71]. Specific lncRNAs, such as MALAT1[70], XIST[72], HOXD-AS1[73], BDNF-AS[74] and BCAT1[75] can exert regulatory function of EMT by targeting EZH2 and DNMT3A. In addition, lncRNA XIST promotes tumor metastasis and EMT in CRC possibly through the regulation of miR-137-EZH2 axis[72]. The HOXD-AS1 lncRNA promotes cancer progression and EMT by modulating the protein expression levels of AEG-1 and EZH2 in CRC cell by competing for miR-217[73]. LncRNA BDNF-AS functions as a tumor suppressor in CRC progression by suppressing GSK-3β expression through its binding to the EZH2 and H3K27me3 within the GSK-3β promoter, which leads to the inhibition of EMT-induced migration[74]. Finally, HIF1A-AS2 can directly bind to miR-129–5p, and positively affect EMT-mediated cell invasion by regulating the expression of miR-129–5p and DNMT3A[75].
2.2.4. Other mechanisms
There are various lncRNAs that regulate invasion, migration and EMT through other mechanisms. For instance, LncRNA AB073614 and BC200 induce invasion and EMT in CRC cells by regulating the JAK/STAT3 pathway[76][77]. Likewise, UCA1 and ZFAS1 can lead to migration and invasion of CRC through EMT through mTOR signaling pathway[78][79], while EPB41L4A-AS1 inhibits the migration, invasion, and EMT in CRC cells through Rho/ROCK pathway[49].
2.3. Glycolysis metabolism
Tumor growth depends on a low-efficiency ATP production process which utilizes glucose uptake for aerobic glycolysis[80]. In this context, the lncRNA PKM2 catalyzes the last step during glycolysis by forming pyruvate and ATP from phosphoenolpyruvate (PEP) and ADP[81]. The glycolytic process can also be influenced by other lncRNAs in CRC. For instance, FEZF1-AS1 and MAFG-AS1 promote pyruvate kinase activity and aerobic glycolysis by regulating PKM2 signaling[82][83]. Furthermore, lncRNAs are able to influence glycolysis in CRC through MYC-mediated pathway, such as LINRIS[84] and GLCC1[83]. The lncRNA KCNQ1OT1 promotes CRC by enhancing aerobic glycolysis via hexokinase-2[85], while HNF1A-AS1 regulates glycolysis by modulating miR-124/MYO6 expression in CRC cells[86].
2.4. Tumor angiogenesis
Angiogenesis is a complex and multistep process driving tumor proliferation and metastasis. There are various regulators involved in tumor angiogenesis, such as vascular endothelial growth factor (VEGF), epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), platelet-derived growth factor (PDGF), angiopoietin 1 and 2 (Ang-1 and Ang-2), and transforming growth factor-α and -β (TGF-α and -β)[87]. Recent studies have found several lncRNAs promoted or inhibited CRC angiogenesis by various mechanisms. ZFAS1 and TPT1-AS1 induce CRC angiogenesis by upregulating VEGFA related pathway[79][88]. MALAT1 mediates angiogenesis by sponging miR-126–5p in CRC[89]. LncRNA AK001058 regulates tumor growth and angiogenesis in CRC via methylation of ADAMTS12[90]. LncRNA GAS5 inhibits angiogenesis and metastasis of CRC through the Wnt/β-catenin signaling pathway[91].
2.5. Chemoradiotherapy resistance
Chemotherapy and targeted drugs, including 5-fluorouracil (5-FU), platinum, hydroxycamptothecin, vincristine, methotrexate, irinotecan, paclitaxel and cetuximab have been widely used for treating CRC[92]. However, a considerable proportion of patients eventually acquire therapeutic resistance to these treatment modalities. The underlying mechanisms for chemoradioresistance are complex and may be related to DNA repair capacity, DNA damage response, apoptotic factors, loss of cell cycle checkpoint control and drug metabolism[5]. Recent studies have highlighted the crucial role of lncRNAs in the chemoradiotherapy resistance in CRC (Table 2).
Table 2.
Summary of the role of long non-coding RNAs in determining therapeutic efficacy in colorectal cancer.
| LncRNA | Effector | Therapy | Signaling pathway | Cell lines | Reference |
|---|---|---|---|---|---|
| HOTAIR | Resistance | 5-FU | NF-κB/TS | HT29, SW480 | [93] |
| HOTAIR | Resistance | Radiotherapy | miR-93/ATG12 | HCT116, SW480 | [127] |
| UCA1 | Resistance | 5-FU | miR-204-5p/CREB1/BCL2/RAB22A | HCT116, HT29, LOVO, SW480 | [94] |
| UCA1 | Resistance | Radiotherapy | NA | CCL244 | [126] |
| SNHG6 | Resistance | 5-FU | miR-26a-5p/ULK1 | RKO, HT29, HCT116 | [95] |
| TUG1 | Resistance | 5-FU | miR-197-3p/TYMS | HCT116, SW1116, HCT8 | [96] |
| TUG1 | Resistance | MTX | miR-186/CPEB2 | HT29, HCT8 | [117] |
| HOTAIRM1 | Sensitivity | 5-FU | miR-17-5p/BTG3 | HCT116, SW480 | [97] |
| HAND2-AS1 | Sensitivity | 5-FU | miR-20a/PDCD4 | HCT116, SW480 | [98] |
| LINC00152 | Resistance | 5-FU | miR-139-5p/NOTCH1 | HT29, LOVO, HCT116, SW480 | [99] |
| NEAT1 | Resistance | 5-FU | miR-150-5p/CPSF4 | SW480, HCT116 | [101] |
| H19 | Resistance | 5-FU | miR-194-5p/SIRT1 | HCT8, HCT116 | [100] |
| H19 | Resistance | MTX | Wnt/β-catenin | HT29 | [118] |
| LINC00957 | Resistance | 5-FU | NA | HCT8, LOVO | [102] |
| PVT1 | Resistance | 5-FU | NA | HCT8, HCT116 | [103] |
| PVT-1 | Resistance | Cisplatin | NA | RKO, LOVO | [108] |
| SnaR | Resistance | 5-FU | NA | SNU-C4, SNU-C, 5HCT116 | [104] |
| GIHCG | Resistance | 5-FU | NA | LOVO, SW480 | [106] |
| GIHCG | Resistance | Oxaliplatin | NA | LOVO, SW480 | [106] |
| LINC00261 | Sensitivity | Cisplatin | Wnt/β-catenin | SW480 | [107] |
| SNHG14 | Resistance | Cisplatin | miR-186/ATG14 | SW620, SW480 | [109] |
| CACS15 | Resistance | Oxaliplatin | miR-145/ABCC1 | HT29, HCT116 | [110] |
| KCNQ1OT1 | Resistance | Oxaliplatin | miR-34a/Atg4B | HCT116, SW480 | [111] |
| KCNQ1OT1 | Resistance | MTX | miR760/PPP1R1B | HT29, Caco2 | [116] |
| MEG3 | Resistance | Oxaliplatin | miR-141/PDCD4 | HT29, HCT116 | [112] |
| CRNDE | Resistance | Oxaliplatin | miR-136/E2F1 | SW480, HCT116 | [113] |
| LINC00525 | Resistance | Oxaliplatin | miR-507/ELK3 | HCT116, HT29 | [114] |
| MALAT1 | Resistance | Oxaliplatin | EZH2 | HT29, SW480, SW620 | [70] |
| MALAT1 | Resistance | Radiotherapy | miR-101-3p | CCL244, LOVO | [124] |
| CCAL | Resistance | Oxaliplatin | Wnt/β-catenin | SW480, HCT116 | [115] |
| LUCAT1 | Resistance | Oxaliplatin | RPL40/MDM2/p53 | SW480,RKO, HCT116 | [105] |
| LUCAT1 | Resistance | 5-FU | RPL40/MDM2/p53 | SW480,RKO, HCT116 | [105] |
| BANCR | Resistance | ADR | miR-203/CSE1L | HCT116, LOVO | [119] |
| XIST | Resistance | DOX | miR-124/SGK1 | HCT116, LOVO | [120] |
| MIR100HG | Resistance | Cetuximab | Wnt/β-catenin | CRC cells | [121] |
| CRART16 | Resistance | Cetuximab | miR-371a-5p/ERBB3/MAPK | Caco-2 | [122] |
| OIP5-AS1 | Sensitivity | Radiotherapy | miR-369-3p/ DYRK1A | LOVO, SW480 | [123] |
| p21 | Sensitivity | Radiotherapy | Wnt/β-catenin | SW1116, SW620, LS174T, HT29, LOVO | [125] |
2.5.1. LncRNAs and resistance to 5-FU
Some lncRNAs modulate resistance to 5-FU by interacting with miRNAs; and the list of these lncRNA include HOTAIR[93], UCA1[94], SNHG6[95], TUG1[96], HOTAIRM1[97], HAND2-AS1[98], LINC00152[99], H19 [100]and NEAT1[101]. For instance, HOTAIR contributes to 5-FU resistance through suppression of miR-218 and activation of NF-κB signaling in CRC[93]. On the other hand, UCA1 enhances cell proliferation and 5-FU resistance in CRC by inhibiting miR-204–5p[94], while SNHG6 acts through ULK1-induced autophagy by sponging miR-26a-5p[95]. In addition, the suppression of LINC00957 can reverse 5-FU resistance in CRC by down-regulation of P-gP[102]. Other reports have indicated that inhibition of PVT-1 can reverse 5-FU resistance through MDR related proteins, including MRP1, P-gp, mTOR and Bcl-2[103]. The lncRNAs snaR[104], LUCAT1[105] and GIHCG[106] also have impact on 5-FU resistance in CRC. For instance, knockdown of snaR decreases cell death following 5-FU treatment, which indicates that snaR loss decreases in-vitro sensitivity to this chemotherapeutic drug[104]. Similarly, GIHCG has been confirmed to facilitate cell survival following 5-FU treatment[106], while LUCAT1 induces cell cycle arrest and resistance to 5-FU by binding UBA52 and activating the RPL40-MDM2-p53 pathway in CRC cells[105].
2.5.2. LncRNAs and resistance to platinum drugs
Like a double-edged sword, lncRNAs can not only increase sensitivity to platinum drugs, but also induce resistance to these compounds in CRC cells. As for cisplatin, LINC00261 sensitizes HCT116, HCT8, HT29, SW480 human colon cancer cells to cisplatin therapy via β-catenin pathway[107]. Another report showed that PVT-1 inhibits cisplatin sensitivity in HT29, SW480, HCT116, RKO, LOVO cells[108]. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of SW620, SW480 cells by regulating miR-186/ATG14 axis[109]. With regards to treatment with oxaliplatin, a number of lncRNAs promote resistance to this drug by sponging specific miRNAs, including CACS15[110], KCNQ1OT1[111], MEG3[112], CRNDE[113] and LINC00525[114]. Additionally, MALAT1 can promote oxaliplatin-based chemotherapeutic resistance in CRC patients[70]. Likewise, CCAL promotes oxaliplatin resistance in CRC cells by increasing β-catenin expression levels[115], while LUCAT1 can influence sensitivity to this drug treatment in CRC via p53 signaling pathway[105]. Finally, lncRNA GIHCG has been reported to induce chemoresistance to oxaliplatin and indicates poor prognosis in CRC[106].
2.5.3. LncRNAs and resistance to other chemotherapeutic and targeted drugs
In addition to specific lncRNAs that have been reported to associate with 5-FU and platinum-based drugs, others have been related to various other chemotherapies and targeted drugs. In this regard, lncRNA KCNQ1OT1[116] and TUG1[117] have been reported to mediate methotrexate resistance in CRC through the sponging of miR-760 and miR-186. In another report, H19 was shown to mediate methotrexate resistance in CRC through Wnt/β-catenin pathway[118]. LncRNA BANCR enhances adriamycin resistance in CRC by acting as a ceRNA[119], while suppression of XIST can inhibit doxorubicin resistance by upregulation of miR-124 and downregulation of SGK1[120]. Other lncRNAs such as MIR100HG and CRART16 have been reported to mediate cetuximab resistance in CRC by interacting with specific miRNAs[121][122]. In this context, MIR100HG-derived miR-100 and miR-125 have been reported to modulate cetuximab resistance via Wnt/β-catenin signaling[121], while CRART16 overexpression contributes towards cetuximab resistance through miR-371a-5p/ERBB3/MAPK axis[122].
2.5.4. LncRNAs and resistance to radiotherapy
Radiotherapy is a localized treatment for CRC, which is primarily used in combination with chemotherapy. LncRNAs are important determinants for resistance to radiotherapy. For example, lncRNA OIP5-AS1 regulates radiotherapeutic resistance by targeting DYRK1A through miR-369–3p in CRCcells[123]. Similarly, MALAT1 modulates radioresistance to CRC via miR-101–3p sponging[124]. Other reports have shown that lncRNA-p21 enhances the sensitivity of radiotherapy in CRC by targeting the Wnt/β-catenin signaling pathway[125], while downregulation of UCA1 and HOTAIR enhances the radiosensitivity in this malignancy[126][127].
3. LncRNA-associated novel oncogenic mechanisms
3.1. LncRNAs and cell autophagy
Autophagy is a common biological process during environmental changes such as starvation where cells degrade macromolecules to provide energy for fundamental biological processes[128]. The formation of the autophagosome comprises of three main steps: initiation, nucleation, and elongation. The molecular mechanisms underlying each step consist of several conserved autophagy-related genes (ATGs). Previous studies have suggested that autophagy plays a dual role in oncogenesis, including regulation of oncogenes and tumor suppressor gene expression and participating in response to chemoradiotherapy[129]. The expression of lncRNAs can regulate autophagy in CRC and play a vital role in tumor proliferation, EMT and chemoradiotherapy. The lncRNA SLCO4A1-AS1[130], HAGLROS[131], UCA1[132] and MALAT1[133] promote CRC cell proliferation by enhancing autophagy via miRNA related axis. In addition, lncRNA CPS1-IT1 suppresses EMT and metastasis in CRC by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α[134]. Likewise, GAS5 promotes autophagy by targeting miR-222–3p through the GAS5/PTEN-signaling pathway in CRC[135]. Other reports have shown that H19 confers resistance to 5-FU in CRC by promoting SIRT1-mediated autophagy[100]. In addition, NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity by targeting miR-34a[136], while SNHG14 stimulates cellular autophagy to facilitate cisplatin resistance in CRC by interacting with miR-186/ATG14 axis[109].
3.2. LncRNAs and cancer stem cells
The cancer stem cells (CSCs) are defined as a small subpopulation of cancer cells with embryonic stem cell (ESC) characteristics. CSCs have been confirmed to initiate tumor growth, slow-cycling cellular turnover, as well as the resistance to therapy[137]. In CRC, CSCs can be modulated by several lncRNAs through different pathways. For instance, suppression of HOTAIR inhibits the malignant characteristics of CD133(+) CSCs in CRC[138]. The lncRNAs BCAR4[139], cCSC1[140], LINC00525[114], ROR[141], LINC00657[142] and MALAT1[143] modulate CSC phenotype of CRC cells by targeting miRNAs related signaling. Similarly, GAS5[144] and GATA6[145] are critical for maintaining stemness, as evidenced by high expression of GATA6 in the cancer stem cells in CRC, leading to recruitment of the NURF complex onto the Ehf promoter and mediate its transcription, promote tumor initiation and progression[145]. Additionally, lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity in colorectal neoplasia[146].
3.3. LncRNAs and epigenetic mechanisms
Epigenetic alterations include reversible events within the human genome, and the primary processes include DNA methylation, histone modifications and chromatin remodeling[147]. In CRC, several epigenetic changes can be regulated by lncRNAs. The lncRNA HOTAIR contributes to histone H3 lysine 27 methylation and lysine 4 demethylation by interacting with both LSD1/CoREST/REST complex and PRC2[148]. This lncRNA is also reported to reprogram chromatin organization in CRC[149]. It has been reported that NEAT1[150] and MALAT1[151] contribute to chromatin remodeling by regulating chromatin modifiers. DACOR1 interacts with both chromatin and targets DNMT1, a DNA methyltransferase, to influence DNA methylation levels in colon cancer cells[152]. Furthermore, GAS5 inhibits progression of CRC by interacting with and triggering YAP phosphorylation and degradation, and is negatively regulated by the m6A reader YTHDF3[153]. Similarly, m6A-induced lncRNA RP11 triggers the dissemination of CRC cells via upregulation of ZEB1[154]. CCAT2 overexpression has been found to promote malignant transformation in colon cells through inducing chromosomal instability via BOP1/AURKB cascade[155].
3.4. LncRNAs and tumor microenvironment
Recent studies have suggested that lncRNAs derived from carcinoma-associated fibroblasts (CAF) promote CRC progression and therapeutic resistance. H19 enriched in CAFs activates the β-catenin pathway by serving as a competing endogenous RNA sponge for miR-141; thus enhancing the stemness and chemoresistance of CRC cells[156]. Likewise, CAF-derived CCAL interacts with human antigen R to upregulate β-catenin expression and thereby induce the resistance to oxaliplatin in CRC cells[115]. LncRNAs can also affect the cancer-related immune cells in tumor microenvironment. NKILA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death[157]. On the same line, lncRNA GM16343 overexpression in CD8 + T cells promotes the secretion of interferon γ and therefore enhances the antitumor immune response[158], while SATB2-AS1functions as a tumor suppressor by regulating TH1-type chemokines expression and immune cell density[159].
4. Potential of LncRNAs in clinical applications
In addition to their biological role in mediating cancer cell growth in various human malignancies, lncRNAs are also emerging as important substrates for the development of cancer biomarkers for the early detection, prognosis prediction, and predicting therapy response to various chemotherapies and developingtreatments (Figure 2). Subsequent sections will describe the current state of knowledge with regards to lncRNAs and their clinical significance in CRC.
Figure 2:
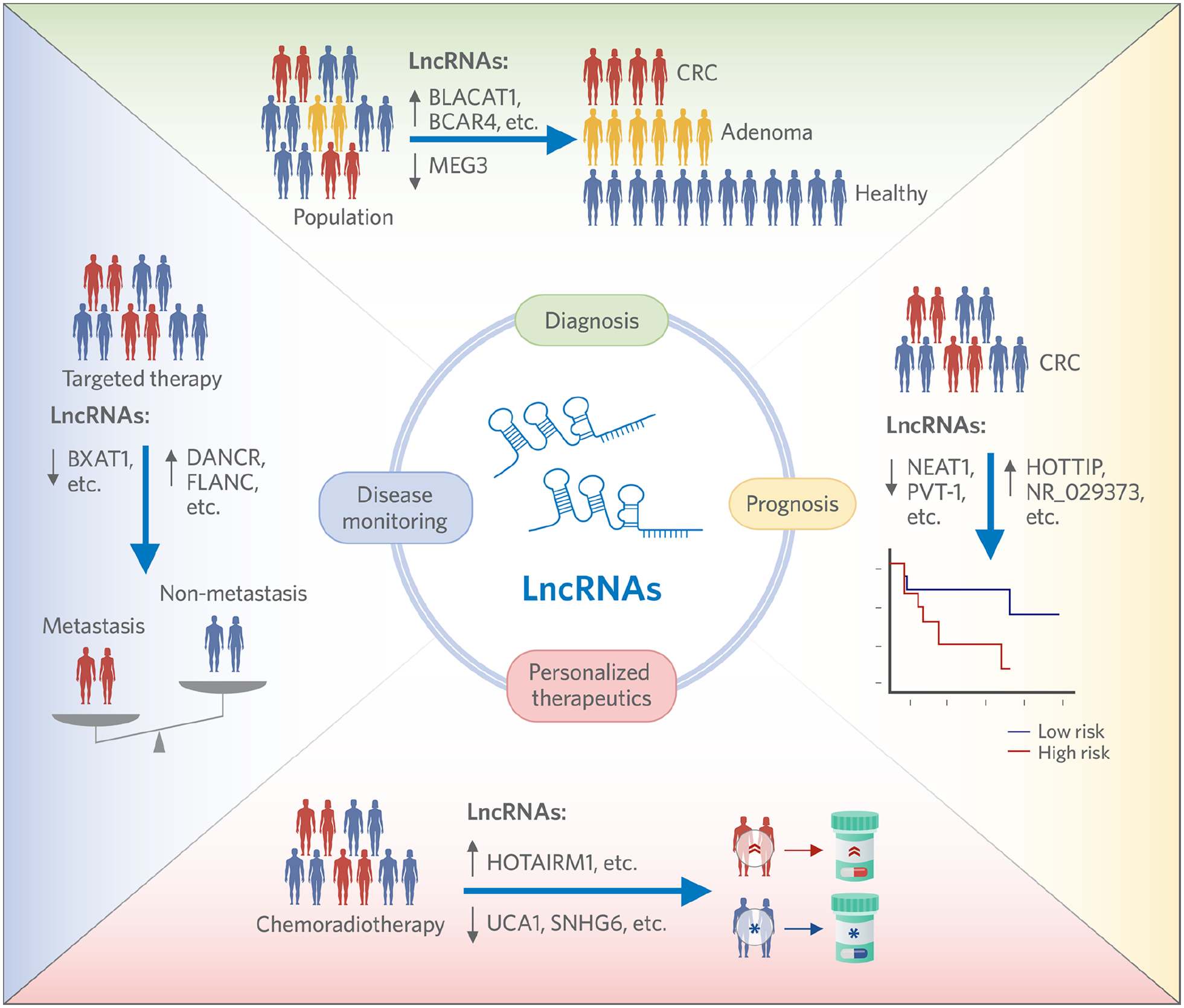
Schematic summary for the potential of lncRNA to serve as diagnostic, prognostic, personalized therapeutic and disease monitoring biomarkers in cancer patients.
4.1. LncRNAs as noninvasive biomarkers for early cancer diagnosis
Though invasive colonoscopy has significantly contributed to the early diagnosis of CRC, non-invasive biomarkers are imperative for improving the diagnostic accuracy for this disease, reducing the medical expense and increasing participation of average-risk population in cancer screening efforts. In this regard, accumulating evidence suggests that lncRNAs could act as novel noninvasive diagnostic biomarkers for CRC patients (Table 3). It has been reported that CCAT1 is overexpressed during the multistep normal-adenoma-carcinoma sequence, and can serve as a potential early diagnostic biomarker[160][161]. Similarly other lncRNAs, such as BLACAT1[162], BCAR4[163] and CRNDE[164] are frequently overexpressed in CRC with corresponding AUC values of 0.85, 0.936 and 0.921 respectively. Additionally, the combination of two mRNAs, KRTAP5–4 and MAGEA3, and lncRNA BCAR4, can discriminate CRC patients from healthy controls with an AUC value of 0.857[163].
Table 3.
Summary of the diagnostic performances of long non-coding RNAs in colorectal cancer.
| LncRNAs | Expression | CRC cases vs. controls | AUC | Sensitivity | Specificity | Sample | Reference |
|---|---|---|---|---|---|---|---|
| BLACAT1 | Up | 30 vs.30 | 0.858 | 83.3% | 76.7% | Serum | [162] |
| BCAR4 | Up | 76 vs.76 | 0.936 | NA | NA | Serum | [163] |
| KRTAP5-4, MAGEA3 and BCAR4 | - | 76 vs.76 | 0.857 | NA | NA | Serum | [163] |
| CRNDE-b | Up | 161 vs. 222 | 0.921 | 85% | 96% | Plasma | [164] |
| CRNDE-g | Up | 161 vs. 222 | 0.892 | 80% | 96% | Plasma | [164] |
| CRNDE-h | Up | 161 vs. 222 | 0.888 | 80% | 96% | Plasma | [164] |
| NEAT1 | Up | 100 vs. 100 | 0.787/0.871 | 69%/70%; | 79%/96% | Blood | [165] |
| 91H | Up | 76 vs.76 | 0.870 | 84.48% | 80.36% | Plasma | [166] |
| PVT-1 | Up | 76 vs.76 | 0.786 | 68.97% | 82.14% | Plasma | [166] |
| MEG3 | Down | 76 vs.76 | 0.819 | 75.86% | 82.14% | Plasma | [166] |
| GAS5 | Up | 76 vs.76 | 0.642 | 63.79% | 62.5% | Plasma | [166] |
| CCAT1-L | Up | 76 vs.76 | 0.748 | 75.86% | 73.21% | Plasma | [166] |
| 91H, PVT-1 and MEG3 | - | 76 vs.76 | 0.877 | 82.76% | 78.57% | Plasma | [166] |
| LINC00654 | Up | 114 vs.58 | 0.734 | NA | NA | Plasma | [167] |
| LINC00909 | Up | 114 vs.58 | 0.886 | NA | NA | Plasma | [167] |
| SNGH11 | Up | 114 vs.58 | 0.868 | NA | NA | Plasma | [167] |
| ZFAS1 | Up | 114 vs.58 | 0.850 | NA | NA | Plasma | [167] |
| ZFAS1, SNHG11, LINC00909 and | - | 0.937 | NA | NA | Plasma | [167] | |
| LINC00654 | |||||||
| LINC02418 | Up | 125 vs. 125 | 0.8978 | 95.2% | 66.4% | Serum | [169] |
| RP11-296E3.2 | Up | 60 CRC | 0.663 | 69.0% | 62.1% | Plasma | [171] |
| RP11-296E3.2 and CEA | - | 60 CRC | 0.722 | 72.4% | 71.4% | Plasma | [171] |
| BANCR, NR_026817, NR_029373, and NR_034119 | - | 120 vs. 120 | 0.881 | 89.17% | 75.83% | Serum and tissue | [168] |
Detecting NEAT1 expression in the whole blood sample has a diagnostic AUC of 0.787 (NEAT1_v1) and 0.871 (NEAT1_v2), with a corresponding sensitivity of 69% (NEAT1_v1) and 70% (NEAT1_v2) and a specificity of 79% (NEAT1_v1) and 96% (NEAT1_v2), respectively[165]. On similar lines, lncRNAs such as 91H, PVT-1 and MEG3 have been shown to be expressed at significantly higher levels in plasma from CRC patients and can be regarded as promising diagnostic biomarkers for early-stage CRC; especially considering that they have superior diagnostic accuracy compared to CEA and CA19–9[166]. Similarly, SNGH11, ZFAS1, LINC00909 and LINC00654 exhibit high diagnostic performance for CRC, especially in the early-stage disease[167]. Additionally, four lncRNAs (BANCR, NR_026817, NR_029373, and NR_034119) are significantly dysregulated in both tissue and serum samples of CRC patients and this distinctive 4-lncRNA panel has a better diagnostic performance than CEA[168]. Finally, LINC02418 is highly expressed in the serum of CRC patients and is considered as a promising, novel biomarker that can be used for the clinical diagnosis of CRC[169].
4.2. LncRNAs as prognostic and predictive biomarkers for clinical outcomes in cancer
LncRNAs can be considered as prognostic biomarkers of colorectal cancer for predicting patient survival and therapeutic response, as depicted in Table 4. Retrospective clinical studies have revealed that various lncRNAs including LUADT1[170], LEF1-AS1[171], H19, MIR31HG, HOTAIR, WT1-AS, LINC00488[172], XIST[173], CCAT1,2[174], DANCR[175], PVT-1[176], SBDSP1[177], BANCR[178], UCA1[179] and CRNDE[180] are frequently over-expressed in CRC, and their expression significantly correlates with shorter survival time in these patients. Additionally, high expression LUADT1[170] and XIST[173] correlates with tumor size, metastasis, and TNM staging in CRC. Similarly, the expression of CRNDE associates with high tumor stage and lymph node metastasis[180]. Other reports have shown that SBDSP1 expression is significantly associated with tumor differentiation, depth of invasion and TNM stage. Patients with higher SBDSP1 expression have significantly shorter disease-free survival and overall survival, compared to those with lower expression of this lncRNA[177]. Upregulation of BANCR may be associated with the lymph node metastasis and poor survival in CRC patients[178]. Similarly, the overexpression of UCA1 is associated with poor prognosis and more advanced clinicopathological features, including tumor differentiation, lymph node metastasis, distant metastasis, tumor node metastasis, stage, tumor invasion depth and tumor size[179]. In contrast, low expression HOTTIP is an independent prognostic marker for overall survival in CRC patients[181]. Dysregulated expression of AFAP1-AS1 has been confirmed to associate with oncogenesis and tumor progression, and can serve as a novel potential molecular biomarker in tumor prognosis[182][183]. Low expression of NR_029373 and NR_034119 have both been identified as independent unfavorable prognostic factors affecting the disease-specific survival rate of CRC patients[168]. Finally, the expression of NEAT1 has been associated with tumor recurrence and unfavorable prognosis in CRC [184].
Table 4.
Summary of the prognostic significance of long non-coding RNAs in colorectal cancer.
| LncRNAs | Expression | Patient No. | Univariate analysis HR (95%CI) | Multivariate analysis HR (95%CI) | Reference | ||
|---|---|---|---|---|---|---|---|
| OS | PFS/DFS/RFS | OS | PFS/DFS/RFS | ||||
| NEAT1_v1 | Up | 191 | 0.415(0.227–0.758) | NA | NA | NA | [165] |
| NEAT1_v2 | Up | 191 | 0.426(0.231–0.784) | NA | 0.519 (0.280–0.965) | NA | [165] |
| NEAT1 | Up | 239 | 1.88(1.32–2.69) | 1.93(1.38–2.71) | 1.70(1.18–2.45) | 1.80(1.27–2.55) | [184] |
| PVT-1 | Up | 210 | 2.07(1.42–3.06) | 2.01(1.26–2.64) | 1.76(1.16–2.37) | 2.15(1.39–2.76) | [176] |
| HOTTIP | Down | 52 | NA | NA | 4.949(1.685–11.984) | NA | [181] |
| XIST | Up | 196 | 1.284(1.143–1.442) | 1.242(1.115–1.384) | 1.197(1.064–1.346) | 1.165(1.044–1.300) | [173] |
| CCAT1 | Up | 135 | 4.06(1.47–16.8) | 3.88(1.67–8.39) | 5.90(2.09–24.7) | 2.52(1.07–5.56) | [174] |
| CCAT2 | Up | 135 | 2.04(1.05–3.84) | 2.55(1.19–5.31) | 2.40(1.22–4.59) | 2.39(1.10–5.08) | [174] |
| CCAT1+CCAT2 | Up | 135 | NA | NA | 8.38(2.68–37.0) | 2.60(1.04–6.06) | [174] |
| DANCR | Up | 104 | 2.491(1.335–7.264) | 2.614(1.572–7.715) | 2.131(1.157–7.058) | 2.397(1.385–7.279) | [175] |
| SBDSP1 | Up | 171 | 2.673(1.562–5.632) | 2.523(1.429–5.013) | 2.137(1.238–4.672) | 2.132(1.123–4.239) | [177] |
| BANCR | Up | 106 | 2.74(1.52–4.96) | NA | 2.24(1.22–4.16) | NA | [178] |
| UCA1 | Up | 715 | 2.65(1.93–3.63) | NA | 2.39(1.80–3.16) | NA | [179] |
| AFAP1-AS1 | Up | 52 | 2.780(1.308–5.909) | 2.389(1.162–4.913) | 2.358(1.110–5.008) | 2.120(1.032–4.353) | [182][183] |
| NR_029373 | Down | 94 | NA | 0.477(0.261–0.870) | NA | 0.457(0.246–0.848) | [168] |
| NR_034119 | Down | 94 | NA | 0.561(0.316–0.996) | NA | 0.517(0.277–0.964) | [168] |
4.3. LncRNAs as primary targets for new anti-cancer drug development
Considering the crucial role of lncRNAs in cancer biology, it is reasonable to identify appropriate lncRNAs as primary targets for developing novel anti-cancer drugs. Several experimental attempts have been made in other cancers based on advancing biomaterial techniques. Nanoliposomes delivering Let-7b into esophageal cancer stem-like cells significantly inhibited the Wnt signaling-mediated self-renewal and enhanced cell sensitivity to 5-fluorouracil or docetaxel[185]. An engineered nanoparticle platform that delivered lncAFAP1-AS1 siRNA effectively reversed radioresistance of triple-negative breast cancer (TNBC)[186]. Similarly, nanoparticles carrying RNA interference of DANCR dramatically inhibited the growth of TNBC xenografts without inducing obvious toxic side-effects[187]. In CRC, 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine nanoparticles carrying lncFLANC siRNA restrained the metastasis of CRC cells in vivo without inducing evident toxic or inflammatory responses[188]. In addition to nanoparticle based drug delivery systems, recent studies have proposed the therapeutic potential of the CRISPR/Cas9 system to edit the expression of LncRNAs in CRC patients and related explorations are starting[189].Despite of these effects, we noted that rare clinical trials are currently available for lncRNA-based targeted drugs, which may be attributed to inherent factors such as lack of well-validated targets and uncertain safety of novel drugs.
5. Challenges and perspectives
In this review, we focused on the role of lncRNAs as hallmarks of CRC including their ability to impact proliferative signaling, EMT-mediated invasion and migration, glycolysis metabolism, tumor angiogenesis and resistance to chemoradiotherapy. The mechanistic investigations have revealed that lncRNAs exert their oncogenic role either through regulation of canonical signaling pathways (such as Wnt/β-catenin and PI3K/Akt) or by serving as ceRNAs for miRNAs. These novel studies have closely linked lncRNAs with cell autophagy, cancer stem phenotype, epigenetic regulation and tumormicroenvironment, where further explorations are still needed. For instance, the regulatory function of lncRNAs in m6A modifications during CRC progression remains poorly studied. Since immune checkpoint inhibitors (ICI) have held the promise for late-stage CRC, the potential correlation of lncRNAs with anti-cancer efficacy of ICIs may be worth studying. Furthermore, accumulating evidence has pointed to the crucial oncogenic role of gut microbiota in CRC and whether the expression of oncogenic lncRNAs is regulated by specific bacteria, which remains unknown. On the other hand, in addition to their oncogenic role, the biological functions of lncRNAs in normal intestinal epithelial development, barrier function protection and homeostasis maintenance should be clarified, which will provide pivotal information for the clinical translation of lncRNAs into CRC diagnosis and treatment.
In this review, we also discussed the clinical potential of lncRNAs as diagnostic or prognostic biomarkers. Encouragingly, a number of circulating lncRNAs have shown superior performance in discriminating CRC patients from healthy controls, implying their attractive potential to serve as noninvasive diagnostic biomarkers. However, several challenges remain to date that must be overcome prior to their clinical translation. First, rare diagnostic lncRNA signatures are currently constructed, which may help improve the performance of a single lncRNA detection. Second, the levels of circulating lncRNAs in patients with stage I-II CRC, advanced adenoma and inflammatory bowel disease should be clarified, which will provide better clues for their potential as early diagnosis and risk population screening. Finally, whether dynamic detection of postoperative circulating lncRNAs for their ability to diagnose CRC recurrence and progression should also be studied. On the other hand, mounting evidence points to their predictive role as tissue-derived lncRNAs in patient survival and disease recurrence, suggesting detection of these lncRNAs may help identify high risk populations from patients within the same TNM stage. However, the prognosis of CRC is affected by various clinical factors and it will be promising to integrate lncRNAs with traditional clinical and pathological indicators to establish and validate multifactorial risk-stratification models. Finally, considering the crucial role of lncRNAs in CRC progression and therapeutic response, it is possible to develop them into therapeutic targets, where selection of most appropriate candidate lncRNAs and developing matched drug delivery systems would be of tremendous clinical significance.
6. Conclusions
In summary, emerging and accumulating evidence has demonstrated that lncRNAs play a crucial role in hallmarks of CRC through various molecular mechanisms and therefore have potential to be utilized for targeted drug development. However, identification and validation of reliable targets, and developing stable and safe drug delivery systems remain to be challenging. In addition, although clinical evidence suggests that lncRNAs can serve as promising diagnostic and prognostic biomarkers in CRC patients, although multicenter validations based on sufficient samples are still necessary. Meanwhile, quality control of samples and standardized detecting techniques of LncRNAs should be emphasized in the following translational work. With our increasing knowledge of lncRNAs in CRC, it is hopeful that some of these may benefit the biomarker-directed precision medicine approaches in improving survival outcomes in CRC patients.
Highlights.
LncRNAs extensively participate in various hallmarks of CRC.
LncRNAs affect the efficacy of chemotherapy and radiotherapy in CRC.
Detecting circulating LncRNAs benefits the early diagnosis of CRC.
Tumor-derived LncRNAs act as a reliable prognostic biomarker for CRC.
LncRNAs can be utilized as primary targets for anti-CRC drug development.
Funding:
The present work was supported by the grants CA72851, CA181572, CA184792, CA187956 and CA202797 from the National Institute of Health (NIH) to AG. This work was also supported by grants from the National Natural Science Foundation of China (Nos. 81920108026, 81871964), the National Ten Thousand Plan Young Top Talents (for Dr. Yanlei Ma), the Shanghai Young Top Talents (for Dr. Yanlei Ma. No. QNBJ1701), the Shanghai Science and Technology Development Fund (No.19410713300, No. 20XD1421200), the CSCO-Roche Tumor Research Fund (No. Y-2019Roche-079), Fudan University Excellence 2025 Talent Cultivation Plan (for Dr. Yanlei Ma), the National Natural Science Foundation of China (for Xuebing Yan, No.81902422), Program of Jiangsu Commission of Health (for Xuebing Yan, No.M2020024); Social Development Program of Yangzhou Science and Technology Bureau (for Xuebing Yan, No.YZ2020079)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
None of the authors have any conflicts of interest to disclose.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019., CA. Cancer J. Clin 69 (2019) 7–34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- [2].Brenner H, Kloor M, Pox CP, Colorectal cancer., Lancet (London, England) 383 (2014) 1490–1502. 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- [3].Dy GK, Hobday TJ, Nelson G, Windschitl HE, O’Connell MJ, Alberts SR, Goldberg RM, Nikcevich DA, Sargent DJ, Long-term survivors of metastatic colorectal cancer treated with systemic chemotherapy alone: a north central cancer treatment group review of 3811 patients, n0144., Clin. Colorectal Cancer 8 (2009) 88–93. 10.3816/CCC.2009.n.014. [DOI] [PubMed] [Google Scholar]
- [4].Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X, Evaluation of Serum CEA, CA19–9, CA72–4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer., Sci. Rep 8 (2018) 2732. 10.1038/s41598-018-21048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M, The emerging role of noncoding RNAs in colorectal cancer chemoresistance., Cell. Oncol. (Dordr) 42 (2019) 757–768. 10.1007/s13402-019-00466-8. [DOI] [PubMed] [Google Scholar]
- [6].Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B, Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer., Int. J. Mol. Sci 20 (2019). 10.3390/ijms20225758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sun W, Ren S, Li R, Zhang Q, Song H, LncRNA, a novel target biomolecule, is involved in the progression of colorectal cancer., Am. J. Cancer Res 9 (2019) 2515–2530. [PMC free article] [PubMed] [Google Scholar]
- [8].Hu W, Liu C, Bi Z-Y, Zhou Q, Zhang H, Li L-L, Zhang J, Zhu W, Song Y-Y-Y, Zhang F, Yang H-M, Bi Y-Y, He Q-Q, Tan G-J, Sun C-C, Li D-J, Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology., Mol. Cancer 19 (2020) 102. 10.1186/s12943-020-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bermúdez M, Aguilar-Medina M, Lizárraga-Verdugo E, Avendaño-Félix M, Silva-Benítez E, López-Camarillo C, Ramos-Payán R, LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer., Front. Oncol 9 (2019) 1008. 10.3389/fonc.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pandya G, Kirtonia A, Sethi G, Pandey AK, Garg M, The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential., Biochim. Biophys. Acta. Rev. Cancer 1874 (2020) 188423. 10.1016/j.bbcan.2020.188423. [DOI] [PubMed] [Google Scholar]
- [11].Yousefi H, Maheronnaghsh M, Molaei F, Mashouri L, Reza Aref A, Momeny M, Alahari SK, Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance., Oncogene 39 (2020) 953–974. 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- [12].Farooqi AA, de la Roche M, Djamgoz MBA, Siddik ZH, Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights., Semin. Cancer Biol 58 (2019) 65–79. 10.1016/j.semcancer.2019.01.001. [DOI] [PubMed] [Google Scholar]
- [13].Yuan S, Tao F, Zhang X, Zhang Y, Sun X, Wu D, Role of Wnt/β-Catenin Signaling in the Chemoresistance Modulation of Colorectal Cancer., Biomed Res. Int 2020 (2020) 9390878. 10.1155/2020/9390878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P, Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta., Curr. Biol 8 (1998) 573–581. 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- [15].MacDonald BT, Tamai K, He X, Wnt/beta-catenin signaling: components, mechanisms, and diseases., Dev. Cell 17 (2009) 9–26. 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Samuels Y, Velculescu VE, Oncogenic mutations of PIK3CA in human cancers., Cell Cycle 3 (2004) 1221–1224. 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- [17].Kim SH, Park KH, Shin SJ, Lee KY, Il Kim T, Kim NK, Rha SY, Ahn JB, CpG Island Methylator Phenotype and Methylation of Wnt Pathway Genes Together Predict Survival in Patients with Colorectal Cancer., Yonsei Med. J 59 (2018) 588–594. 10.3349/ymj.2018.59.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ma Y, Yang Y, Wang F, Moyer M-P, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H, Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α., Gut 65 (2016) 1494–1504. 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- [19].Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song M-A, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, V Davuluri R, Mimori K, Mori M, Sieuwerts AM, Martens JWM, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA, CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer., Genome Res 23 (2013) 1446–1461. 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y, Zhang F, Lu Y, Zheng L, Zhang W, Li X, Li X, Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer., Cancer Lett 376 (2016) 62–73. 10.1016/j.canlet.2016.03.022. [DOI] [PubMed] [Google Scholar]
- [21].Han P, Li J-W, Zhang B-M, Lv J-C, Li Y-M, Gu X-Y, Yu Z-W, Jia Y-H, Bai X-F, Li L, Liu Y-L, Cui B-B, The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling., Mol. Cancer 16 (2017) 9. 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lv S-Y, Shan T-D, Pan X-T, Tian Z-B, Liu X-S, Liu F-G, Sun X-G, Xue H-G, Li X-H, Han Y, Sun L-J, Chen L, Zhang L-Y, The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/β-catenin signaling., Cell Cycle 17 (2018) 1245–1254. 10.1080/15384101.2018.1471317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang W, Ning N, Jin X, The lncRNA H19 Promotes Cell Proliferation by Competitively Binding to miR-200a and Derepressing β-Catenin Expression in Colorectal Cancer., Biomed Res. Int 2017 (2017) 2767484. 10.1155/2017/2767484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li S, Wu T, Zhang D, Sun X, Zhang X, The long non-coding RNA HCG18 promotes the growth and invasion of colorectal cancer cells through sponging miR-1271 and upregulating MTDH/Wnt/β-catenin., Clin. Exp. Pharmacol. Physiol 47 (2020) 703–712. 10.1111/1440-1681.13230. [DOI] [PubMed] [Google Scholar]
- [25].Jia G-Q, Zhang M-M, Wang K, Zhao G-P, Pang M-H, Chen Z-Y, Long non-coding RNA PlncRNA-1 promotes cell proliferation and hepatic metastasis in colorectal cancer., J. Cell. Biochem 119 (2018) 7091–7104. 10.1002/jcb.27031. [DOI] [PubMed] [Google Scholar]
- [26].Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y, LncRNA HOTAIR is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ß-Catenin Signaling Pathway., Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 46 (2018) 1275–1285. 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- [27].Sun N, Zhang G, Liu Y, Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway., Gene 665 (2018) 141–148. 10.1016/j.gene.2018.04.014. [DOI] [PubMed] [Google Scholar]
- [28].Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, Li D, Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/β-catenin signaling pathway., Oncotarget 8 (2017) 111715–111727. 10.18632/oncotarget.22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F, Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/β-catenin signaling pathway., Biomed. Pharmacother 96 (2017) 877–883. 10.1016/j.biopha.2017.10.033. [DOI] [PubMed] [Google Scholar]
- [30].Zhu Y, Bian Y, Zhang Q, Hu J, Li L, Yang M, Qian H, Yu L, Liu B, Qian X, LINC00365 promotes colorectal cancer cell progression through the Wnt/β-catenin signaling pathway., J. Cell. Biochem 121 (2020) 1260–1272. 10.1002/jcb.29359. [DOI] [PubMed] [Google Scholar]
- [31].Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, Xu M, Sun H, Liu C, Wei P, Du X, The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5., J. Hematol. Oncol 11 (2018) 113. 10.1186/s13045-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xie F, Xiang X, Huang Q, Ran P, Yuan Y, Li Q, Qi G, Guo X, Xiao C, Zheng S, Reciprocal control of lncRNA-BCAT1 and β-catenin pathway reveals lncRNA-BCAT1 long non-coding RNA acts as a tumor suppressor in colorectal cancer., Oncotarget 8 (2017) 23628–23637. 10.18632/oncotarget.15466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA, Portrait of the PI3K/AKT pathway in colorectal cancer., Biochim. Biophys. Acta 1855 (2015) 104–121. 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- [34].Wang Y, Kuang H, Xue J, Liao L, Yin F, Zhou X, LncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway., Biomed. Pharmacother 93 (2017) 1230–1237. 10.1016/j.biopha.2017.07.024. [DOI] [PubMed] [Google Scholar]
- [35].Shao Q, Xu J, Deng R, Wei W, Zhou B, Yue C, Zhu M, Huang X, Zhu H, Long non-coding RNA-422 acts as a tumor suppressor in colorectal cancer., Biochem. Biophys. Res. Commun 495 (2018) 539–545. 10.1016/j.bbrc.2017.10.076. [DOI] [PubMed] [Google Scholar]
- [36].Song W, Mei J-Z, Zhang M, Long Noncoding RNA PlncRNA-1 Promotes Colorectal Cancer Cell Progression by Regulating the PI3K/Akt Signaling Pathway., Oncol. Res 26 (2018) 261–268. 10.3727/096504017X15031557924132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cui Z, Han B, Wang X, Li Z, Wang J, Lv Y, Long Non-Coding RNA TTN-AS1 Promotes the Proliferation and Invasion of Colorectal Cancer Cells by Activating miR-497-Mediated PI3K/Akt/mTOR Signaling., Onco. Targets. Ther 12 (2019) 11531–11539. 10.2147/OTT.S229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Du W, Hong J, Wang Y-C, Zhang Y-J, Wang P, Su W-Y, Lin Y-W, Lu R, Zou W-P, Xiong H, Fang J-Y, Inhibition of JAK2/STAT3 signalling induces colorectal cancer cell apoptosis via mitochondrial pathway., J. Cell. Mol. Med 16 (2012) 1878–1888. 10.1111/j.1582-4934.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Costa-Pereira AP, Bonito NA, Seckl MJ, Dysregulation of janus kinases and signal transducers and activators of transcription in cancer., Am. J. Cancer Res 1 (2011) 806–816. [PMC free article] [PubMed] [Google Scholar]
- [40].Park S-Y, Lee C-J, Choi J-H, Kim J-H, Kim J-W, Kim J-Y, Nam J-S, The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance., J. Exp. Clin. Cancer Res 38 (2019) 399. 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu B, Liu Q, Pan S, Huang Y, Qi Y, Li S, Xiao Y, Jia L, The HOTAIR/miR-214/ST6GAL1 crosstalk modulates colorectal cancer procession through mediating sialylated c-Met via JAK2/STAT3 cascade., J. Exp. Clin. Cancer Res 38 (2019) 455. 10.1186/s13046-019-1468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang G, Wu X, Li S, Xu X, Zhu H, Chen X, The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer., Sci. Rep 6 (2016) 26524. 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Levine AJ, p53, the cellular gatekeeper for growth and division., Cell 88 (1997) 323–331. 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- [44].Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, Wen X, Parvathaneni S, Kazandjian D, Jenkins LM, Tang W, Elloumi F, Martindale JL, Huarte M, Zhu Y, Robles AI, Frier SM, Rigo F, Cam M, Ambs S, Sharma S, Harris CC, Dasso M, V Prasanth K, Lal A, Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer., Cell Rep 20 (2017) 2408–2423. 10.1016/j.celrep.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li H, Jiang X, Niu X, Long Non-Coding RNA Reprogramming (ROR) Promotes Cell Proliferation in Colorectal Cancer via Affecting P53., Med. Sci. Monit. Int. Med. J. Exp. Clin. Res 23 (2017) 919–928. 10.12659/msm.903462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L, He B, Pan Y, Sun H, Wang S, The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154–5p., Mol. Cancer 17 (2018) 141. 10.1186/s12943-018-0894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fu Y, Yin Y, Peng S, Yang G, Yu Y, Guo C, Qin Y, Zhang X, Xu W, Qin Y, Small nucleolar RNA host gene 1 promotes development and progression of colorectal cancer through negative regulation of miR-137., Mol. Carcinog 58 (2019) 2104–2117. 10.1002/mc.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yang W, Redpath RE, Zhang C, Ning N, Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway., Oncol. Lett 16 (2018) 3365–3372. 10.3892/ol.2018.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bin J, Nie S, Tang Z, Kang A, Fu Z, Hu Y, Liao Q, Xiong W, Zhou Y, Tang Y, Jiang J, Long noncoding RNA EPB41L4A-AS1 functions as an oncogene by regulating the Rho/ROCK pathway in colorectal cancer., J. Cell. Physiol (2020). 10.1002/jcp.29880. [DOI] [PubMed] [Google Scholar]
- [50].Ye C, Shen Z, Wang B, Li Y, Li T, Yang Y, Jiang K, Ye Y, Wang S, A novel long non-coding RNA lnc-GNAT1–1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit., J. Exp. Clin. Cancer Res 35 (2016) 187. 10.1186/s13046-016-0467-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Greenburg G, Hay ED, Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells., J. Cell Biol 95 (1982) 333–339. 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Thiery JP, Acloque H, Huang RYJ, Nieto MA, Epithelial-mesenchymal transitions in development and disease., Cell 139 (2009) 871–890. 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- [53].O’Brien SJ, V Carter J, Burton JF, Oxford BG, Schmidt MN, Hallion JC, Galandiuk S, The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review., Int. J. Cancer 142 (2018) 2501–2511. 10.1002/ijc.31282. [DOI] [PubMed] [Google Scholar]
- [54].Shibue T, Weinberg RA, EMT, CSCs, and drug resistance: the mechanistic link and clinical implications., Nat. Rev. Clin. Oncol 14 (2017) 611–629. 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen D-L, Chen L-Z, Lu Y-X, Zhang D-S, Zeng Z-L, Pan Z-Z, Huang P, Wang F-H, Li Y-H, Ju H-Q, Xu R-H, Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer., Cell Death Dis 8 (2017) e3011. 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Liang W-C, Fu W-M, Wong C-W, Wang Y, Wang W-M, Hu G-X, Zhang L, Xiao L-J, Wan DC-C, Zhang J-F, Waye MM-Y, The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer., Oncotarget 6 (2015) 22513–22525. 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen D-L, Lu Y-X, Zhang J-X, Wei X-L, Wang F, Zeng Z-L, Pan Z-Z, Yuan Y-F, Wang F-H, Pelicano H, Chiao PJ, Huang P, Xie D, Li Y-H, Ju H-Q, Xu R-H, Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression., Theranostics 7 (2017) 4836–4849. 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, Gu C, Yan Q, Fang Y, Zhao X, Liu S, LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1., Int. J. Med. Sci 16 (2019) 51–59. 10.7150/ijms.27359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang L, Wei Z, Wu K, Dai W, Zhang C, Peng J, He Y, Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203., Aging (Albany. NY) 10 (2018) 3662–3682. 10.18632/aging.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ji L, Li X, Zhou Z, Zheng Z, Jin L, Jiang F, LINC01413/hnRNP-K/ZEB1 Axis Accelerates Cell Proliferation and EMT in Colorectal Cancer via Inducing YAP1/TAZ1 Translocation., Mol. Ther. Nucleic Acids 19 (2020) 546–561. 10.1016/j.omtn.2019.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tao Y, Han T, Zhang T, Ma C, Sun C, LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway., Oncotarget 8 (2017) 36410–36422. 10.18632/oncotarget.16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jiang H, Li T, Qu Y, Wang X, Li B, Song J, Sun X, Tang Y, Wan J, Yu Y, Zhan J, Zhang H, Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression., Cancer Lett 425 (2018) 78–87. 10.1016/j.canlet.2018.03.038. [DOI] [PubMed] [Google Scholar]
- [63].Sun J, Hu J, Wang G, Yang Z, Zhao C, Zhang X, Wang J, LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer., J. Exp. Clin. Cancer Res 37 (2018) 106. 10.1186/s13046-018-0771-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [64].Ding D, Li C, Zhao T, Li D, Yang L, Zhang B, LncRNA H19/miR-29b-3p/PGRN Axis Promoted Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Acting on Wnt Signaling., Mol. Cells 41 (2018) 423–435. 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C, LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway., J. Exp. Clin. Cancer Res 37 (2018) 222. 10.1186/s13046-018-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M, A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer., Mol. Ther 26 (2018) 1287–1298. 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yuan Z, Yu X, Ni B, Chen D, Yang Z, Huang J, Wang J, Chen D, Wang L, Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicts favorable prognosis., Int. J. Oncol 48 (2016) 2675–2685. 10.3892/ijo.2016.3447. [DOI] [PubMed] [Google Scholar]
- [68].Li T, Zhu J, Wang X, Chen G, Sun L, Zuo S, Zhang J, Chen S, Ma J, Yao Z, Zheng Y, Chen Z, Liu Y, Wang P, Long non-coding RNA lncTCF7 activates the Wnt/β-catenin pathway to promote metastasis and invasion in colorectal cancer., Oncol. Lett 14 (2017) 7384–7390. 10.3892/ol.2017.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M, Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis., Oncogene 31 (2012) 2062–2074. 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C, MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2., Mol. Cancer Ther 16 (2017) 739–751. 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- [71].Ren W, Gao L, Song J, Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation., Genes (Basel) 9 (2018). 10.3390/genes9120620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu X, Cui L, Hua D, Long Noncoding RNA XIST Regulates miR-137-EZH2 Axis to Promote Tumor Metastasis in Colorectal Cancer., Oncol. Res 27 (2018) 99–106. 10.3727/096504018X15195193936573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [73].Li X, Zhao X, Yang B, Li Y, Liu T, Pang L, Fan Z, Ma W, Liu Z, Li Z, Long non-coding RNA HOXD-AS1 promotes tumor progression and predicts poor prognosis in colorectal cancer., Int. J. Oncol 53 (2018) 21–32. 10.3892/ijo.2018.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhi H, Lian J, LncRNA BDNF-AS suppresses colorectal cancer cell proliferation and migration by epigenetically repressing GSK-3β expression., Cell Biochem. Funct 37 (2019) 340–347. 10.1002/cbf.3403. [DOI] [PubMed] [Google Scholar]
- [75].Lin J, Shi Z, Yu Z, He Z, LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129–5p and DNMT3A., Biomed. Pharmacother 98 (2018) 433–439. 10.1016/j.biopha.2017.12.058. [DOI] [PubMed] [Google Scholar]
- [76].Xue J, Liao L, Yin F, Kuang H, Zhou X, Wang Y, LncRNA AB073614 induces epithelial- mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway., Cancer Biomark 21 (2018) 849–858. 10.3233/CBM-170780. [DOI] [PubMed] [Google Scholar]
- [77].Wu K, Xu K, Liu K, Huang J, Chen J, Zhang J, Zhang N, Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer., Int. J. Biochem. Cell Biol 99 (2018) 219–225. 10.1016/j.biocel.2018.04.001. [DOI] [PubMed] [Google Scholar]
- [78].Jahangiri B, Khalaj-Kondori M, Asadollahi E, Sadeghizadeh M, Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1., J. Cell Commun. Signal 13 (2019) 53–64. 10.1007/s12079-018-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen X, Zeng K, Xu M, Hu X, Liu X, Xu T, He B, Pan Y, Sun H, Wang S, SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150–5p/VEGFA axis., Cell Death Dis 9 (2018) 982. 10.1038/s41419-018-0962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation., Science 324 (2009) 1029–1033. 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sfakianaki M, Papadaki C, Tzardi M, Trypaki M, Manolakou S, Messaritakis I, Saridaki Z, Athanasakis E, Mavroudis D, Tsiaoussis J, Gouvas N, Souglakos J, PKM2 Expression as Biomarker for Resistance to Oxaliplatin-Based Chemotherapy in Colorectal Cancer., Cancers (Basel) 12 (2020). 10.3390/cancers12082058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, Yao S, Jin G, Du J, Han W, Yin Y, Huang S, Fei B, Zou J, Huang Z, LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling., Clin. Cancer Res. an Off. J. Am. Assoc. Cancer Res 24 (2018) 4808–4819. 10.1158/1078-0432.CCR-17-2967. [DOI] [PubMed] [Google Scholar]
- [83].Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X, Tian X, Guo F, Liang Q, Liu Q, Zhong M, Chen J, Ge Z, Li X, Chen X, Cui Y, Chen Y, Zou W, Chen H, Hong J, Fang J-Y, LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc., Nat. Commun 10 (2019) 3499. 10.1038/s41467-019-11447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang Y, Lu J-H, Wu Q-N, Jin Y, Wang D-S, Chen Y-X, Liu J, Luo X-J, Meng Q, Pu H-Y, Wang Y-N, Hu P-S, Liu Z-X, Zeng Z-L, Zhao Q, Deng R, Zhu X-F, Ju H-Q, Xu R-H, LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer., Mol. Cancer 18 (2019) 174. 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen C, Wei M, Wang C, Sun D, Liu P, Zhong X, Yu W, Long noncoding RNA KCNQ1OT1 promotes colorectal carcinogenesis by enhancing aerobic glycolysis via hexokinase-2., Aging (Albany. NY) 12 (2020) 11685–11697. 10.18632/aging.103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Guo X, Zhang Y, Liu L, Yang W, Zhang Q, HNF1A-AS1 Regulates Cell Migration, Invasion and Glycolysis via Modulating miR-124/MYO6 in Colorectal Cancer Cells., Onco. Targets. Ther 13 (2020) 1507–1518. 10.2147/OTT.S231249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Goradel NH, Asghari MH, Moloudizargari M, Negahdari B, Haghi-Aminjan H, Abdollahi M, Melatonin as an angiogenesis inhibitor to combat cancer: Mechanistic evidence., Toxicol. Appl. Pharmacol 335 (2017) 56–63. 10.1016/j.taap.2017.09.022. [DOI] [PubMed] [Google Scholar]
- [88].Zhang Y, Sun J, Qi Y, Wang Y, Ding Y, Wang K, Zhou Q, Wang J, Ma F, Zhang J, Guo B, Long non-coding RNA TPT1-AS1 promotes angiogenesis and metastasis of colorectal cancer through TPT1-AS1/NF90/VEGFA signaling pathway., Aging (Albany. NY) 12 (2020) 6191–6205. 10.18632/aging.103016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, Li G, Wang G, Song J, Li Z, Zhang Z, Yuan W, Li X, YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126–5p in colorectal cancer., Oncogene 38 (2019) 2627–2644. 10.1038/s41388-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [90].Zheng S, Lin F, Zhang M, Mu N, Ge X, Fu J, Long non-coding RNA AK001058 regulates tumor growth and angiogenesis in colorectal cancer via methylation of ADAMTS12., Am. J. Transl. Res 11 (2019) 6117–6123. [PMC free article] [PubMed] [Google Scholar]
- [91].Song J, Shu H, Zhang L, Xiong J, Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/β-catenin signaling pathway., J. Cell. Biochem (2019). 10.1002/jcb.27743. [DOI] [PubMed] [Google Scholar]
- [92].Roselló S, Papaccio F, Roda D, Tarazona N, Cervantes A, The role of chemotherapy in localized and locally advanced rectal cancer: A systematic revision., Cancer Treat. Rev 63 (2018) 156–171. 10.1016/j.ctrv.2018.01.001. [DOI] [PubMed] [Google Scholar]
- [93].Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C, lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer., Mol. Ther. Nucleic Acids 8 (2017) 356–369. 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z, LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204–5p., Sci. Rep 6 (2016) 23892. 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wang X, Lan Z, He J, Lai Q, Yao X, Li Q, Liu Y, Lai H, Gu C, Yan Q, Fang Y, Zhang Y, Li A, Liu S, LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells., Cancer Cell Int 19 (2019) 234. 10.1186/s12935-019-0951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang M, Hu H, Wang Y, Huang Q, Huang R, Chen Y, Ma T, Qiao T, Zhang Q, Wu H, Chen Q, Han D, Wang G, Wang X, Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197–3p in colorectal cancer., J. Cancer 10 (2019) 4603–4613. 10.7150/jca.32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ren T, Hou J, Liu C, Shan F, Xiong X, Qin A, Chen J, Ren W, The long non-coding RNA HOTAIRM1 suppresses cell progression via sponging endogenous miR-17–5p/ B-cell translocation gene 3 (BTG3) axis in 5-fluorouracil resistant colorectal cancer cells., Biomed. Pharmacother 117 (2019) 109171. 10.1016/j.biopha.2019.109171. [DOI] [PubMed] [Google Scholar]
- [98].Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou T, Li Y, Chen S, LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer., Cell. Signal 66 (2020) 109483. 10.1016/j.cellsig.2019.109483. [DOI] [PubMed] [Google Scholar]
- [99].Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D, Huang Z, Correction: Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139–5p., Oncogenesis 7 (2018) 63. 10.1038/s41389-018-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, Gao X, Wang X, Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy., Cell Death Dis 9 (2018) 1149. 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang X, Jiang G, Ren W, Wang B, Yang C, Li M, LncRNA NEAT1 Regulates 5-Fu Sensitivity, Apoptosis and Invasion in Colorectal Cancer Through the MiR-150–5p/CPSF4 Axis., Onco. Targets. Ther 13 (2020) 6373–6383. 10.2147/OTT.S239432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhang LH, Li LH, Zhang PF, Cai YF, Hua D, LINC00957 Acted as Prognostic Marker Was Associated With Fluorouracil Resistance in Human Colorectal Cancer., Front. Oncol 9 (2019) 776. 10.3389/fonc.2019.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fan H, Zhu J-H, Yao X-Q, Knockdown of long non-coding RNA PVT1 reverses multidrug resistance in colorectal cancer cells., Mol. Med. Rep 17 (2018) 8309–8315. 10.3892/mmr.2018.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lee H, Kim C, Ku J-L, Kim W, Yoon SK, Kuh H-J, Lee J-H, Nam SW, Lee EK, A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells., Mol. Cells 37 (2014) 540–546. 10.14348/molcells.2014.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun Y, Yuan X, LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52., Cancer Sci 110 (2019) 1194–1207. 10.1111/cas.13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jiang X, Li Q, Zhang S, Song C, Zheng P, Long noncoding RNA GIHCG induces cancer progression and chemoresistance and indicates poor prognosis in colorectal cancer., Onco. Targets. Ther 12 (2019) 1059–1070. 10.2147/OTT.S192290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang ZK, Yang L, Wu LL, Mao H, Zhou YH, Zhang PF, Dai GH, Long non-coding RNA LINC00261 sensitizes human colon cancer cells to cisplatin therapy., Brazilian J. Med. Biol. Res. = Rev. Bras. Pesqui. Medicas e Biol 51 (2017) e6793. 10.1590/1414-431X20176793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ping G, Xiong W, Zhang L, Li Y, Zhang Y, Zhao Y, Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer., Am. J. Transl. Res 10 (2018) 138–149. [PMC free article] [PubMed] [Google Scholar]
- [109].Han Y, Zhou S, Wang X, Mao E, Huang L, SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis., Biomed. Pharmacother 121 (2020) 109580. 10.1016/j.biopha.2019.109580. [DOI] [PubMed] [Google Scholar]
- [110].Gao R, Fang C, Xu J, Tan H, Li P, Ma L, LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145., Arch. Biochem. Biophys 663 (2019) 183–191. 10.1016/j.abb.2019.01.005. [DOI] [PubMed] [Google Scholar]
- [111].Li Y, Li C, Li D, Yang L, Jin J, Zhang B, lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway., Onco. Targets. Ther 12 (2019) 2649–2660. 10.2147/OTT.S188054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wang H, Li H, Zhang L, Yang D, Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis., Biomed. Pharmacother 106 (2018) 1607–1615. 10.1016/j.biopha.2018.07.131. [DOI] [PubMed] [Google Scholar]
- [113].Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H, Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer., Onco. Targets. Ther 10 (2017) 205–216. 10.2147/OTT.S116178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang S, Li J, Yang X, Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis., Int. J. Stem Cells 12 (2019) 347–359. 10.15283/ijsc19041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Zhang X, Kong F, Guan M, Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells., Int. J. Cancer 146 (2020) 1700–1716. 10.1002/ijc.32608. [DOI] [PubMed] [Google Scholar]
- [116].Xian D, Zhao Y, LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway., J. Cell. Mol. Med 23 (2019) 3808–3823. 10.1111/jcmm.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Li C, Gao Y, Li Y, Ding D, TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis., Biochem. Biophys. Res. Commun 491 (2017) 552–557. 10.1016/j.bbrc.2017.03.042. [DOI] [PubMed] [Google Scholar]
- [118].Wu K-F, Liang W-C, Feng L, Pang J-X, Waye MM-Y, Zhang J-F, Fu W-M, H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway., Exp. Cell Res 350 (2017) 312–317. 10.1016/j.yexcr.2016.12.003. [DOI] [PubMed] [Google Scholar]
- [119].Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y, Yang S, Zhang W, Zhang R, LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer., Aging (Albany. NY) 10 (2018) 2062–2078. 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhu J, Zhang R, Yang D, Li J, Yan X, Jin K, Li W, Liu X, Zhao J, Shang W, Yu T, Knockdown of Long Non-Coding RNA XIST Inhibited Doxorubicin Resistance in Colorectal Cancer by Upregulation of miR-124 and Downregulation of SGK1., Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 51 (2018) 113–128. 10.1159/000495168. [DOI] [PubMed] [Google Scholar]
- [121].Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, Wei T, Yang M, Yeatman TJ, Lee E, Saito-Diaz K, Hinger S, Patton JG, Chung CH, Emmrich S, Klusmann J-H, Fan D, Coffey RJ, lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling., Nat. Med 23 (2017) 1331–1341. 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu J, Yue T, Wang J, Zhu J, Bu D, Wang X, The novel long noncoding RNA CRART16 confers cetuximab resistance in colorectal cancer cells by enhancing ERBB3 expression via miR-371a-5p., Cancer Cell Int 20 (2020) 68. 10.1186/s12935-020-1155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan X, Rao J, Xiong H, Yu S, Yuan X, Zhu F, Hu G, Wang Y, Xiong H, LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369–3p in colorectal cancer cells., Eur. J. Cell Biol 97 (2018) 369–378. 10.1016/j.ejcb.2018.04.005. [DOI] [PubMed] [Google Scholar]
- [124].Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X, Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101–3p sponging., Exp. Mol. Pathol 115 (2020) 104448. 10.1016/j.yexmp.2020.104448. [DOI] [PubMed] [Google Scholar]
- [125].Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X, Ma Z, Li X, Zhang Y, LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway., Oncol. Rep 31 (2014) 1839–1845. 10.3892/or.2014.3047. [DOI] [PubMed] [Google Scholar]
- [126].Yang X, Liu W, Xu X, Zhu J, Wu Y, Zhao K, He S, Li M, Wu Y, Zhang S, Cao J, Ye Z, Xing C, Downregulation of long non-coding RNA UCA1 enhances the radiosensitivity and inhibits migration via suppression of epithelial-mesenchymal transition in colorectal cancer cells., Oncol. Rep 40 (2018) 1554–1564. 10.3892/or.2018.6573. [DOI] [PubMed] [Google Scholar]
- [127].Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R, Ge H, Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer., Cell Death Dis 11 (2020) 175. 10.1038/s41419-020-2268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yin Z, Pascual C, Klionsky DJ, Autophagy: machinery and regulation., Microb. Cell (Graz, Austria) 3 (2016) 588–596. 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Yun CW, Lee SH, The Roles of Autophagy in Cancer., Int. J. Mol. Sci 19 (2018). 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wang Z, Jin J, LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508–3p/PARD3 axis., Aging (Albany. NY) 11 (2019) 4876–4889. 10.18632/aging.102081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zheng Y, Tan K, Huang H, Long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression., J. Cell. Biochem 120 (2019) 3922–3933. 10.1002/jcb.27676. [DOI] [PubMed] [Google Scholar]
- [132].Song F, Li L, Liang D, Zhuo Y, Wang X, Dai H, Knockdown of long noncoding RNA urothelial carcinoma associated 1 inhibits colorectal cancer cell proliferation and promotes apoptosis via modulating autophagy., J. Cell. Physiol 234 (2019) 7420–7434. 10.1002/jcp.27500. [DOI] [PubMed] [Google Scholar]
- [133].Si Y, Yang Z, Ge Q, Yu L, Yao M, Sun X, Ren Z, Ding C, Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer., Cell. Mol. Biol. Lett 24 (2019) 50. 10.1186/s11658-019-0175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zhang W, Yuan W, Song J, Wang S, Gu X, LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α., Biochimie 144 (2018) 21–27. 10.1016/j.biochi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- [135].Liu L, Wang H-J, Meng T, Lei C, Yang X-H, Wang Q-S, Jin B, Zhu J-F, lncRNA GAS5 Inhibits Cell Migration and Invasion and Promotes Autophagy by Targeting miR-222–3p via the GAS5/PTEN-Signaling Pathway in CRC., Mol. Ther. Nucleic Acids 17 (2019) 644–656. 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [136].Liu F, Ai F-Y, Zhang D-C, Tian L, Yang Z-Y, Liu S-J, LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a., Cancer Med 9 (2020) 1079–1091. 10.1002/cam4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Munro MJ, Wickremesekera SK, Peng L, Tan ST, Itinteang T, Cancer stem cells in colorectal cancer: a review., J. Clin. Pathol 71 (2018) 110–116. 10.1136/jclinpath-2017-204739. [DOI] [PubMed] [Google Scholar]
- [138].Dou J, Ni Y, He X, Wu D, Li M, Wu S, Zhang R, Guo M, Zhao F, Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells., Am. J. Transl. Res 8 (2016) 98–108. [PMC free article] [PubMed] [Google Scholar]
- [139].Ouyang S, Zhou X, Chen Z, Wang M, Zheng X, Xie M, LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer., Cancer Cell Int 19 (2019) 72. 10.1186/s12935-019-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z, LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway., J. Cell. Biochem 121 (2020) 2510–2524. 10.1002/jcb.29473. [DOI] [PubMed] [Google Scholar]
- [141].Yan ZY, Sun XC, [LincRNA-ROR functions as a ceRNA to regulate Oct4, Sox2, and Nanog expression by sponging miR-145 and its effect on biologic characteristics of colonic cancer stem cells]., Zhonghua bing li xue za zhi = Chinese J. Pathol 47 (2018) 284–290. 10.3760/cma.j.issn.0529-5807.2018.04.011. [DOI] [PubMed] [Google Scholar]
- [142].Zhao L, Liu C, Yan S, Hu G, Xiang K, Xiang H, Yu H, LINC00657 promotes colorectal cancer stem-like cell invasion by functioning as a miR-203a sponge., Biochem. Biophys. Res. Commun 529 (2020) 500–506. 10.1016/j.bbrc.2020.04.049. [DOI] [PubMed] [Google Scholar]
- [143].Tang D, Yang Z, Long F, Luo L, Yang B, Zhu R, Sang X, Cao G, Wang K, Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis., J. Cell. Physiol 234 (2019) 20816–20828. 10.1002/jcp.28687. [DOI] [PubMed] [Google Scholar]
- [144].Zhou X, Xiao D, Long non-coding RNA GAS5 is critical for maintaining stemness and induces chemoresistance in cancer stem-like cells derived from HCT116., Oncol. Lett 19 (2020) 3431–3438. 10.3892/ol.2020.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, He L, Ye B, Wang S, Meng S, Fan D, Wang J, Yang L, Qin X, Du Y, Li C, He L, Ren W, Wu X, Tian Y, Fan Z, LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis., Nat. Cell Biol 20 (2018) 1134–1144. 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
- [146].Zhang Y, Fang Z, Guo X, Dong H, Zhou K, Huang Z, Xiao Z, lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity., J. Cell. Physiol 234 (2019) 18524–18534. 10.1002/jcp.28489. [DOI] [PubMed] [Google Scholar]
- [147].El Bairi K, Tariq K, Himri I, Jaafari A, Smaili W, Kandhro AH, Gouri A, Ghazi B, Decoding colorectal cancer epigenomics., Cancer Genet 220 (2018) 49–76. 10.1016/j.cancergen.2017.11.001. [DOI] [PubMed] [Google Scholar]
- [148].Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY, Long noncoding RNA as modular scaffold of histone modification complexes., Science 329 (2010) 689–693. 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M, Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers., Cancer Res 71 (2011) 6320–6326. 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- [150].Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB, An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles., Mol. Cell 33 (2009) 717–726. 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Chujo T, Yamazaki T, Hirose T, Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies., Biochim. Biophys. Acta 1859 (2016) 139–146. 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [152].Merry CR, Forrest ME, Sabers JN, Beard L, Gao X-H, Hatzoglou M, Jackson MW, Wang Z, Markowitz SD, Khalil AM, DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer., Hum. Mol. Genet 24 (2015) 6240–6253. 10.1093/hmg/ddv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J, Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3., Mol. Cancer 18 (2019) 143. 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, Du J, Shan H, Liu H, Wang H, m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1., Mol. Cancer 18 (2019) 87. 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Chen B, Dragomir MP, Fabris L, Bayraktar R, Knutsen E, Liu X, Tang C, Li Y, Shimura T, Ivkovic TC, De Los Santos MC, Anfossi S, Shimizu M, Shah MY, Ling H, Shen P, Multani AS, Pardini B, Burks JK, Katayama H, Reineke LC, Huo L, Syed M, Song S, Ferracin M, Oki E, Fromm B, Ivan C, Bhuvaneshwar K, Gusev Y, Mimori K, Menter D, Sen S, Matsuyama T, Uetake H, Vasilescu C, Kopetz S, Parker-Thornburg J, Taguchi A, Hanash SM, Girnita L, Slaby O, Goel A, Varani G, Gagea M, Li C, Ajani JA, Calin GA, The Long Noncoding RNA CCAT2 induces chromosomal instability through BOP1 - AURKB signaling., Gastroenterology (2020). 10.1053/j.gastro.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y, Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19., Theranostics 8 (2018) 3932–3948. 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, Chen F, Su F, Yao H, Liu Q, Su S, Song E, NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death., Nat. Immunol 19 (2018) 1112–1125. 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- [158].Mao D, Hu C, Zhang J, Feng C, Zhang Z, Wang J, Man Z, Zhu Z, Wang Y, Zhao H, Zhu X, Ouyang J, Dong X, Zhao X, Long Noncoding RNA GM16343 Promotes IL-36β to Regulate Tumor Microenvironment by CD8(+)T cells., Technol. Cancer Res. Treat 18 (2019) 1533033819883633. 10.1177/1533033819883633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K, Liu X, Xu T, Sun L, Qin J, He B, Pan Y, Sun H, Wang S, LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2., Mol. Cancer 18 (2019) 135. 10.1186/s12943-019-1063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B, Gure AO, Nissan A, Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence., BMC Cancer 13 (2013) 196. 10.1186/1471-2407-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A, Dayanc BE, Ritter G, Gomceli I, Bostanci EB, Akoglu M, Chen Y-T, Old LJ, Gure AO, Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues., Int. J. Cancer 130 (2012) 1598–1606. 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- [162].Dai M, Chen X, Mo S, Li J, Huang Z, Huang S, Xu J, He B, Zou Y, Chen J, Dai S, Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation., Sci. Rep 7 (2017) 46572. 10.1038/srep46572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Dong L, Lin W, Qi P, Xu M-D, Wu X, Ni S, Huang D, Weng W-W, Tan C, Sheng W, Zhou X, Du X, Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer., Cancer Epidemiol. Biomarkers Prev. a Publ. Am. Assoc. Cancer Res. Cosponsored by Am. Soc. Prev. Oncol 25 (2016) 1158–1166. 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- [164].Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, Lapointe LC, Molloy PL, Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas., Genes Cancer 2 (2011) 829–840. 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z, Xu Y, Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer., Mol. Cancer 14 (2015) 191. 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Liu H, Ye D, Chen A, Tan D, Zhang W, Jiang W, Wang M, Zhang X, A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers., Clin. Chem. Lab. Med 57 (2019) 1073–1083. 10.1515/cclm-2019-0052. [DOI] [PubMed] [Google Scholar]
- [167].Xu W, Zhou G, Wang H, Liu Y, Chen B, Chen W, Lin C, Wu S, Gong A, Xu M, Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer., Int. J. Cancer 146 (2020) 2901–2912. 10.1002/ijc.32747. [DOI] [PubMed] [Google Scholar]
- [168].Wang R, Du L, Yang X, Jiang X, Duan W, Yan S, Xie Y, Zhu Y, Wang Q, Wang L, Yang Y, Wang C, Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer., J. Cancer Res. Clin. Oncol 142 (2016) 2291–2301. 10.1007/s00432-016-2238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Zhao Y, Du T, Du L, Li P, Li J, Duan W, Wang Y, Wang C, Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer., Cell Death Dis 10 (2019) 568. 10.1038/s41419-019-1804-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Zhang X-F, Zhang Y, Shen Z, Yang G-G, Wang H-D, Li L-F, Liu D-C, Qiu J-M, LncRNALUADT1 is overexpressed in colorectal cancer and its expression level is related to clinicopathology., Eur. Rev. Med. Pharmacol. Sci 22 (2018) 2282–2286. 10.26355/eurrev_201804_14816. [DOI] [PubMed] [Google Scholar]
- [171].Shi Q, He Y, Zhang X, Li J, Cui G, Zhang X, Wang X, Two Novel Long Noncoding RNAs - RP11–296E3.2 and LEF1-AS1can - Separately Serve as Diagnostic and Prognostic Bio-Markers of Metastasis in Colorectal Cancer., Med. Sci. Monit. Int. Med. J. Exp. Clin. Res 25 (2019) 7042–7051. 10.12659/MSM.916314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Zhang H, Wang Z, Wu J, Ma R, Feng J, Long noncoding RNAs predict the survival of patients with colorectal cancer as revealed by constructing an endogenous RNA network using bioinformation analysis., Cancer Med 8 (2019) 863–873. 10.1002/cam4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Zhang X-T, Pan S-X, Wang A-H, Kong Q-Y, Jiang K-T, Yu Z-B, Long Non-Coding RNA (lncRNA) X-Inactive Specific Transcript (XIST) Plays a Critical Role in Predicting Clinical Prognosis and Progression of Colorectal Cancer., Med. Sci. Monit. Int. Med. J. Exp. Clin. Res 25 (2019) 6429–6435. 10.12659/MSM.915329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A, CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 “gene desert”, serve as important prognostic biomarkers in colorectal cancer., Ann. Oncol. Off. J. Eur. Soc. Med. Oncol 28 (2017) 1882–1888. 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Liu Y, Zhang M, Liang L, Li J, Chen Y-X, Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer., Int. J. Clin. Exp. Pathol 8 (2015) 11480–11484. [PMC free article] [PubMed] [Google Scholar]
- [176].Fan H, Zhu J-H, Yao X-Q, Long non-coding RNA PVT1 as a novel potential biomarker for predicting the prognosis of colorectal cancer., Int. J. Biol. Markers 33 (2018) 415–422. 10.1177/1724600818777242. [DOI] [PubMed] [Google Scholar]
- [177].Zhou H-B, Li Q, Liu M, Cao Y-Q, Xu J-Y, Increased expression of long non-coding RNA SBDSP1 correlates with poor survival in colorectal cancer., Eur. Rev. Med. Pharmacol. Sci 21 (2017) 3837–3841. [PubMed] [Google Scholar]
- [178].Shen X, Bai Y, Luo B, Zhou X, Upregulation of lncRNA BANCR associated with the lymph node metastasis and poor prognosis in colorectal cancer., Biol. Res 50 (2017) 32. 10.1186/s40659-017-0136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Liu X, Liu X, Qiao T, Chen W, Prognostic and clinicopathological significance of long non-coding RNA UCA1 in colorectal cancer: Results from a meta-analysis., Medicine (Baltimore) 98 (2019) e18031. 10.1097/MD.0000000000018031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Xie H, Ma B, Gao Q, Zhan H, Liu Y, Chen Z, Ye S, Li J, Yao L, Huang W, Long non-coding RNA CRNDE in cancer prognosis: Review and meta-analysis., Clin. Chim. Acta 485 (2018) 262–271. 10.1016/j.cca.2018.07.003. [DOI] [PubMed] [Google Scholar]
- [181].Correction., RNA Biol 17 (2020) 309. 10.1080/15476286.2019.1675952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ji D, Zhong X, Jiang X, Leng K, Xu Y, Li Z, Huang L, Li J, Cui Y, The role of long non-coding RNA AFAP1-AS1 in human malignant tumors., Pathol. Res. Pract 214 (2018) 1524–1531. 10.1016/j.prp.2018.08.014. [DOI] [PubMed] [Google Scholar]
- [183].Wang F, Ni H, Sun F, Li M, Chen L, Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer., Biomed. Pharmacother 81 (2016) 152–159. 10.1016/j.biopha.2016.04.009. [DOI] [PubMed] [Google Scholar]
- [184].Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, Zhao Q, Li J, NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer., Oncotarget 6 (2015) 27641–27650. 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Pang Y, Liu J, Li X, Zhang Y, Zhang B, Zhang J, Du N, Xu C, Liang R, Ren H, Tang S-C, Sun X, Nano Let-7b sensitization of eliminating esophageal cancer stem-like cells is dependent on blockade of Wnt activation of symmetric division., Int. J. Oncol 51 (2017) 1077–1088. 10.3892/ijo.2017.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Bi Z, Li Q, Dinglin X, Xu Y, You K, Hong H, Hu Q, Zhang W, Li C, Tan Y, Xie N, Ren W, Li C, Liu Y, Hu H, Xu X, Yao H, Nanoparticles (NPs)-Meditated LncRNA AFAP1-AS1 Silencing to Block Wnt/β-Catenin Signaling Pathway for Synergistic Reversal of Radioresistance and Effective Cancer Radiotherapy., Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger 7 (2020) 2000915. 10.1002/advs.202000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, Sun D, Scheidt J, Qian V, He S, Gilmore H, Schiemann WP, Lu Z-R, Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy., Bioconjug. Chem 30 (2019) 907–919. 10.1021/acs.bioconjchem.9b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Pichler M, Rodriguez-Aguayo C, Nam SY, Dragomir MP, Bayraktar R, Anfossi S, Knutsen E, Ivan C, Fuentes-Mattei E, Lee SK, Ling H, Catela Ivkovic T, Huang G, Huang L, Okugawa Y, Katayama H, Taguchi A, Bayraktar E, Bhattacharya R, Amero P, He WR, Tran AM, Vychytilova-Faltejskova P, Klec C, Bonilla DL, Zhang X, Kapitanovic S, Loncar B, Gafà R, Wang Z, Cristini V, Hanash SM, Bar-Eli M, Lanza G, Slaby O, Goel A, Rigoutsos I, Lopez-Berestein G, Calin GA, Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer., Gut 69 (2020) 1818–1831. 10.1136/gutjnl-2019-318903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Ray SK, Mukherjee S, Genome editing with CRISPR-Cas9: A budding biological contrivance for colorectal carcinoma research and its perspective in molecular medicine., Curr. Mol. Med (2020). 10.2174/1566524020666201119143943. [DOI] [PubMed] [Google Scholar]


