Abstract
A modified version of the Bacteroides fragilis transposon Tn4400, designated Tn4400′, enabling rapid isolation and analysis of B. fragilis mutants has been constructed. To identify potential virulence factors, Tn4400′-generated mutants were screened by a new method; this resulted in the isolation of 21 mutant strains with impaired growth characteristics on tissue culture monolayers but normal growth in rich medium anaerobically.
Tn4400′ mutagenesis.
Bacteroides fragilis is the most frequently isolated anaerobic species from human intraperitoneal and intra-abdominal infections involving anaerobes (2, 3, 9). Compound transposon Tn4400 was isolated from B. fragilis plasmid pBFTM10 by Robillard et al. (7). A similar transposon, Tn4351, studied in the laboratories of Smith (11) and Salyers (5, 10), was used to generate B. fragilis and Bacteroides thetaiotaomicron mutants. Here we describe further modifications of Tn4400 which have increased its transposition frequency as well as facilitated the rapid isolation of B. fragilis chromosomal fragments abutting the inserted transposon.
The entire transposon Tn4400, including a clindamycin-resistant (Clnr) determinant active in B. fragilis, was cloned as a single BglII fragment from a pBFTM10-F′ lac fusion plasmid, pOX446R1 (7), into a plasmid based on pDG5, which contains the pBR322 replicon, the bla gene for ampicillin resistance (Ampr) in Escherichia coli, and the origin of transfer (oriT) from the IncP plasmid RK2 (4). The resulting plasmid, pNJR609, apparently underwent an oriT-mediated rearrangement upon prolonged storage and consequently lost the oriT region. This plasmid was named pNJR609Δ (Table 1).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli HB101 | F− Δ(mcrC-mrr) recA13 rpsL20 | Laboratory stock |
| B. fragilis TM4000 | Clinical isolate, Rifr, aerotolerant | 8 |
| B. fragilis Tn4400′ mutants | ||
| JD3a.2 | Arginine auxotroph | This study |
| JD3a.3 | Arginine auxotroph | This study |
| JD4.9 | Arginine auxotroph | This study |
| YT44.2.1 | Tryptophan auxotroph | This study |
| YT58.1.3 | MGD bafA mutant | This study |
| YT65.2.10 | MGD, less aerotolerant than TM4000 | This study |
| YT112.2.16 | MGD | This study |
| YT118.1.13 | MGD | This study |
| YT120.1.7 | MGD | This study |
| YT123.2.18 | MGD | This study |
| YT129.2.20 | MGD, less aerotolerant than TM4000 | This study |
| YT135.2.8 | MGD, less aerotolerant than TM4000 | 13 |
| Plasmids | ||
| pDG5 | Ampr, E. coli plasmid carrying oriT from RP4 | 4 |
| pFD544 | E. coli replicon, tetQ gene | 6 |
| pJST51 | pDG5 with endR1 gene from E. coli | 14 |
| pNJR609 | BglII fragment containing Tn4400 from pOX446R1 cloned into pDG5 | This study |
| pNJR609Δ | pNJR609 with a spontaneous deletion, Ampr Tetr in E. coli | This study |
| pOX446R1 | Derived from pOX446 by homologous recombination, contains a single copy of Tn4400 | 7 |
| pRG23 | SacI tetQ fragment from pFD544 cloned into pSP72 | This study |
| RK231 | RP4 derivative, Kanr Tetr Tra+ cloning vector, Ampr | 4 |
| pSP72 | Promega, Madison, Wis. | |
| pYT644A and -B | oriT of RP4 from pJST51 cloned into the PstI site of pNJR609Δ, Amps Tetr (pYT644A and -B differ in the orientation of oriT) | This study |
| pYT645A and -B | tetQ is cloned as a BglII-BamHI fragment into the unique BglII sites of pYT644A and pYT644B | This study |
| pYT646A and -B | 1.6-kb PstI fragment from pJST51 cloned into the PstI sites of pYT645A and pYT645B, restoring the bla gene; Ampr Tetr in E. coli | This study |
Rifr, rifampin resistant; Kanr, kanamycin resistant; Tra+, transfer proficient.
To restore the ability of the plasmid to be mobilized by RK2, the oriT of RP4 was isolated as a blunt-ended 760-bp HaeII fragment from pJST51 (14) and cloned into the HincII site within the bla gene of pNJR609Δ. The resulting plasmids, pYT644A and pYT644B, were able to generate Clnr colonies by transposition upon transfer into B. fragilis. A tetQ gene cassette, which confers tetracycline resistance in B. fragilis but not E. coli (6), was cloned as a SacI fragment into a pSP72 vector plasmid, generating pRG23. A BglII-BamHI fragment from pRG23 containing the tetQ gene was cloned into the BglII sites of pYT644A and pYT644B. The resulting plasmids, pYT645A and pYT645B (Fig. 1A), were able to generate both Clnr and tetracycline-resistant (Tetr) B. fragilis strains. For conjugal transfer of the pYT644 or pYT645 plasmid from E. coli to B. fragilis, mid-log-phase broth cultures of an E. coli donor strain, a second E. coli strain containing the mobilizer RK231, and a B. fragilis recipient were mixed (2 ml-2 ml-5 ml) and concentrated by centrifugation, and the mixture was placed on a sterile filter (type HAWP 047; Millipore Corp., Bedford, Mass.) on the surface of a plate containing solidified brain heart infusion broth supplemented with 0.5% yeast extract and 5 μg of hemin per ml (BHIS). After overnight, aerobic incubation at 37°C, the bacteria were plated on BHIS plates containing gentamicin (50 μg/ml), rifampin (50 μg/ml), and tetracycline (2 μg/ml) or clindamycin (6 μg/ml).
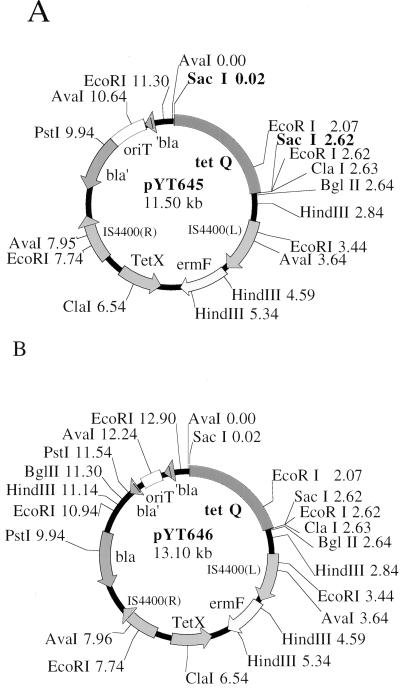
FIG. 1.
Schematics of pYT645 (A) and pYT646 (B), the Tn4400′ transposon delivery plasmids for B. fragilis. The tetX gene confers resistance to tetracycline only in aerobically grown E. coli (12). Parts of these plasmids have not yet been sequenced, and some of the endonuclease recognition sites may deviate slightly from the precise positions shown here. (L), left; (R), right.
To expedite isolation of DNA flanking the inserted transposon, a 1.6-kb PstI fragment from pJST51, which contains the 5′ half of the bla gene, was cloned into the PstI sites of pYT645A and pYT645B. The resulting Ampr plasmids, pYT646A and pYT646B (Fig. 1B), were used for large-scale mutagenesis of TM4000, originally a clinical isolate and the standard strain used in our pathogenesis studies. The two versions of pYT646 differ only in the orientation of RK2 oriT. The inverse transposon on pYT646 is named Tn4400′. A B. fragilis chromosome mutagenized by an inverse transposition event is depicted in Fig. 2A.
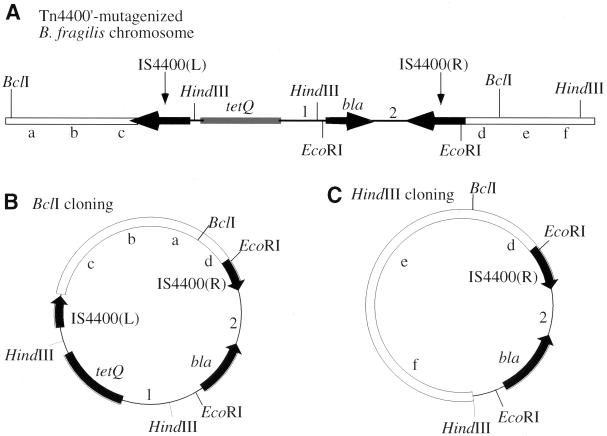
FIG. 2.
The B. fragilis chromosome after Tn4400′ insertion and the clone-out strategy. (A) After Tn4400′ insertion, the B. fragilis cell becomes Tetr and Clns. The letters a through f designate arbitrary markers in the B. fragilis chromosome. The number 1 indicates oriT from RP4; the number 2 indicates rep from pBR322. (B) Schematic of the cloning of B. fragilis chromosomal fragments adjacent to Tn4400′. The Tn4400′-containing B. fragilis chromosome is digested with BclI. Self-ligation of the Tn4400′-containing BclI fragment generates an Ampr E. coli plasmid. (C) Schematic of the clone-out technique. Tn4400′-containing B. fragilis chromosomal DNA is cut with HindIII and religated to transform E. coli to Ampr. Plasmids purified from these Ampr colonies contain the right-hand half of Tn4400′ plus a fragment of B. fragilis chromosomal DNA. When a high concentration of chromosomal fragments is used in a clone-out ligation, the resulting plasmids may contain more than one HindIII fragment. This causes little difficulty because the additional HindIII sites will appear in the sequencing results. The multiple HindIII fragments contained in the same clone-out plasmid usually come from different loci on the B. fragilis chromosome (data not shown). (R), right; (L), left.
In B. fragilis the inverse transposition events occurred more frequently than direct Tn4400 transposition or cointegrate formation events (Table 2). (Curiously, pYT645A and pYT645B, which differ only in the orientation of RK2 oriT, gave different ratios of Tetr to Clnr colonies.) A typical conjugation experiment with HB101/pYT646B as the donor gave rise to Tetr and clindamycin-sensitive (Clns) mutants at a frequency between 10−7 and 10−6 per input donor cell. Since the conjugation experiments were done aerobically and B. fragilis does not replicate under these conditions, many transposon mutants could be isolated from one single mating. Among 50 Tn4400′-generated mutants analyzed in detail, only three (5%) were siblings.
TABLE 2.
Enumeration of the transposition events after transfer of pYT645 into TM4000a
| Mutagen | Mating | Tetr coloniesb
|
Clnr coloniesc
|
|||
|---|---|---|---|---|---|---|
| Total no. of colonies tested | Clns | Clnr | Tets | Tetr | ||
| pYT645A | 1 | 24 | 19 | 5 | 3 | 0 |
| 2 | 20 | 2 | 0 | |||
| 3 | 24 | 2 | 0 | |||
| 4 | 30 | 28 | 2 | 2 | 0 | |
| 5 | 36 | 4 | 0 | |||
| pYT645B | 6 | 103 | 88 | 15 | 31 | 4 |
| 7 | 19 | 3 | 0 | |||
| 8 | 50 | 9 | 0 | |||
| 9 | 45 | 41 | 4 | 9 | 0 | |
| 10 | 23 | 4 | 3 | |||
Each mating filter was resuspended in 3 ml of MPBS buffer. Aliquots of 0.2 ml were added to BHIS-gentamicin-rifampin-tetracycline or BHIS-gentamicin-rifampin-clindamycin plates.
Colonies on one tetracycline plate from each mating were counted, and the numbers are shown. Four plates, from matings 1, 4, 6, and 9, were replica plated onto BHIS-clindamycin plates to count Clns and Clnr colonies among the Tetr colonies.
The number of colonies on one clindamycin plate from each mating is shown. Clnr colonies were transferred to tetracycline plates individually with sterile toothpicks. There is a discrepancy between the numbers of Tetr Clnr colonies obtained in this way and the numbers obtained after an initial tetracycline selection. This may suggest that the structures of the cointegrates allow the loss of the tetQ gene by homologous recombination.
It is relatively simple to “clone out” the chromosomal fragment adjacent to the inverse transposon, as shown in Fig. 2. The mutant chromosome can be cut with HindIII, and the fragments religated to transform E. coli, selecting for Ampr colonies. Alternatively, when a HindIII site occurs in the B. fragilis chromosome very close to the right end of Tn4400′, the mutant chromosome can be cut with PstI and the resulting fragments can be ligated with the 1.6-kb PstI band from pJST51 before being transformed into E. coli with selection for Ampr colonies. The ligation regenerates a complete bla gene. Chromosomal sequences abutting the left end of Tn4400′ are more difficult to obtain. The mutant chromosomal DNA can be cut with BclI, which does not cut within the entire Tn4400′ transposon, and then religated and used to transform E. coli to Ampr. The resulting plasmid recovered from E. coli will contain chromosomal DNA flanking both transposon ends (Fig. 2B). The clone-out technique is unsuitable if the mutant is a Tetr Clnr cointegrate. However, very few (4%) of the Tetr colonies generated by pYT646 were cointegrates. When pYT645 was used as the mutagen, the frequencies of cointegrate formation were slightly higher (Table 2).
Since Tn4400′ is a transposon designed for generalized mutagenesis, its target sequence specificity is of great importance. Tn4400′ apparently has some preference for AT-rich regions (data not shown). However, its insertion sites are numerous on the B. fragilis chromosome. The sequences of more than 50 separate mutants have been analyzed, and except for 3 which are likely to be siblings (since they were isolated from the same mating), no two mutants had Tn4400′ inserted in the same place. Mutants were also screened for the presence of auxotrophs. The percentages of auxotrophs among the Tn4400′ mutants obtained from each mating were consistently slightly above 1%, regardless of whether pYT646A or pYT646B was used for the mutagenesis. Overall, a total of more than 11,000 mutants were screened this way, and 1.2% of the mutants were found to be auxotrophs.
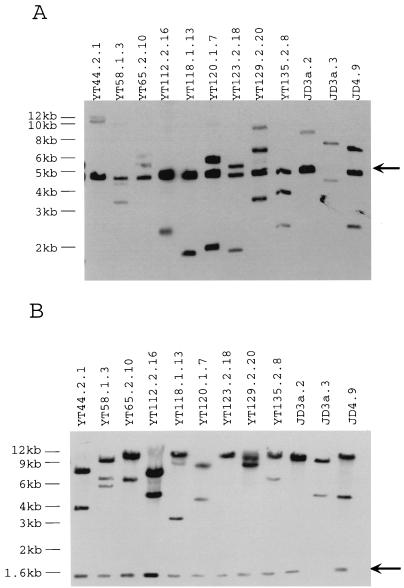
Under the experimental conditions just described, most B. fragilis mutants received only one copy of the inverse transposon. The results of Southern blot analysis of 12 Tetr Clns mutants are shown in Fig. 3. Southern hybridization was carried out with an ECL Direct kit (Amersham Life Science, Little Chalfont, Buckinghamshire, England). Plasmid pYT646B was used to make probes. Two mutants, YT58.1.3 and YT129.2.20, appeared to contain more than one copy of Tn4400′. In one of the mutants, JD3a.3, the Tn4400′ transposon appeared to have undergone rearrangement, with the loss of part of its sequence.
FIG. 3.
Southern blot analysis of the chromosomes of 12 Tetr Clns mutants. The entire pYT646B plasmid was used to generate hybridization probes. (A) Southern blot analysis after complete HindIII digestions of the mutant chromosomal DNAs. The arrow indicates the fragment internal to Tn4400′. Each copy of Tn4400′ inserted in a mutant chromosome will give rise to two junction fragments. Mutant YT129.2.20 appears to contain two copies of Tn4400′. The transposon in the chromosome of mutant JD3a.3 has undergone rearrangement. (B) Southern blot analysis after complete PstI digestions. The arrow indicates the PstI fragment internal to Tn4400′ (as defined by the structure of pYT646). The DNA from mutant YT58.1.3 clearly showed more than two junction fragments. YT58.1.3 may have suffered more than one transposition event.
In agreement with previous reports on the closely related Tn4351 (5, 11), Tn4400′ transposition resulted in a 3-bp duplication at the target site (data not shown). Tn4400′ insertion may also cause deletions at the target site. However, the transposon apparently did not cause mutations in unrelated sites in the bacterial chromosome at a high frequency. To date, eight transposon mutants with discernible phenotypes have been complemented with B. fragilis chromosomal DNA fragments. All of the complementing fragments overlap the regions of the chromosome where Tn4400′ insertions occurred in the corresponding mutants.
Once the inverse transposon is in the chromosome, it appears to be very stable: reversion events do not occur at any observable frequency. A number of auxotrophs were isolated after Tn4400′ mutagenesis, but none reverted to prototrophy at frequencies greater than 10−9. Occasionally, Tets derivatives of Tn4400′ mutants were isolated, but the mutant phenotypes were not reversed. Apparently, internal portions of the inverse transposon, including the tetQ gene, were lost.
Screen for monolayer growth-deficient (MGD) mutants.
The tissue culture monolayer system for B. fragilis pathogenecity (1a) was used to screen for mutants which grow poorly on animal cell monolayers incubated aerobically but which grew normally in an anaerobic chamber. These mutants may be defective in virulence factors such as aerotolerance, attachment to animal cells, and interaction between the bacteria and the animal cells.
It was empirically determined that, when a mixture of sodium dodecyl sulfate and p-nitrophenyl-conjugated substrate for N-acetyl-β-d-glucosaminidase (NAGase) was added, infected Monika cell (a murine fibroblast cell line) monolayers produced a much more intense yellow color than uninfected monolayers. Moreover, when monolayers were infected with TC2, a B. fragilis mutant that grows poorly on tissue culture monolayers (8), the yellow color was not as intense as when wild-type B. fragilis strains were used. These observations led to the development of the enzyme assay into a rapid screen for MGD B. fragilis mutants, although it is not known whether the NAGase activity detected on infected monolayers was a result of enzyme production by the bacteria or heightened enzyme production by the animal cells in response to the bacterial infection.
Monika cells were passaged as previously described (1), with the exception that 8% (vol/vol) fetal bovine serum (FBS; HyClone, Logan, Utah), instead of 10% FBS, was used in the medium. Confluent monolayers were diluted 1:5 and seeded in 24-well tissue culture plates (Corning Costar, Cambridge, Mass.) and incubated for 2 days before bacteria were introduced. Individual Tn4400′-containing, Tetr B. fragilis candidates were picked with sterile toothpicks and transferred onto a BHIS plate containing 2 μg of tetracycline per ml, and the same toothpicks were washed in wells of a 96-well plate, each containing 100 μl of MPBS buffer (0.01 M sodium phosphate buffer [pH 6.9], 0.85% NaCl, 0.1% gelatin). Fresh RPMI 1640 medium with 8% FBS was added to confluent Monika cell monolayers in 24-well tissue culture plates at 0.5 ml/well, along with 5 μl of the bacterial suspension. One well per plate was not inoculated with bacteria as a negative control. The infected monolayers were incubated at 37°C under a normal atmosphere containing 5% CO2 for 48 h. The supernatant in the wells was then aspirated, which left behind the animal cell monolayers and attached bacterial cells. A 0.5-ml aliquot of a substrate mixture containing 3 mM p-nitrophenyl N-acetyl-β-d-glucosaminide (N9376; Sigma, St. Louis, Mo.), 0.2 M sodium phosphate buffer (pH 6.0), and 0.5% sodium dodecyl sulfate was added to each well. After the plates were incubated at 37°C for 1 h, 1 ml of 1 M sodium carbonate was added to each well and the plates were inspected by eye.
One advantage of this screen lies in the fact that it was not necessary to keep the bacterial inocula constant to distinguish a growth-deficient mutant from a growth-competent mutant. For a growth-competent strain, initial inocula ranging from 103 to 107 viable cells per well would generate yellow colors of approximately the same intensity. Mutants with even a small defect in growth on tissue culture monolayers would generate a lighter color even when they were inoculated at 106 viable cells per well.
A total of 7,222 individual Tn4400′-generated mutants were screened; 44 strains persistently generated lighter colors in the screen. These candidates were tested for growth in rich medium in the anaerobic chamber, and about half of the strains were determined to be slow growers. The remaining mutant candidates were seeded onto Monika cell monolayers in 60-mm-diameter tissue culture dishes, and growth curves were determined. Twenty-one mutants showed defects when they grew on tissue culture monolayers but not in rich medium in the anaerobic chamber. These mutants were termed MGD mutants.
Four mutants were found to have normal growth kinetics when their growth on monolayers was measured directly. However, during the NAGase screen, the wells containing these four mutants had lighter color. These four mutants were termed the pale mutants. Wells containing two of the four pale mutants were slightly less yellow during the assay, whereas wells containing the remaining two were almost colorless.
The MGD mutants were subjected to the clone-out analysis. Potential open reading frames in the vicinity of the transposon insertions were analyzed by homology. Several of the disrupted open reading frames may potentially code for proteins with remarkable similarity to proteins of known functions from other organisms. For example, the disrupted gene in mutant YT22.1.22 codes for a protein with similarity to the LysA protein of Pseudomonas species. This mutant was confirmed to be a lysine auxotroph.
However, most of the disrupted potential open reading frames have no matches to proteins of known function in the databases. Three mutants, YT65.2.10, YT129.2.20, and YT135.2.8, when exposed to atmospheric oxygen were found to be less aerotolerant than TM4000 (data not shown). All three mutants have been complemented by a B. fragilis chromosomal fragment library. The complementing fragment for YT135.2.8 contains five open reading frames, which constitute the batI locus (13).
One mutant, YT58.1.3, showed a decreased level of attachment to glutaraldehyde-fixed Monika cell monolayers. A DNA fragment containing an open reading frame disrupted in this mutant, named the bafA gene, was amplified from the wild-type B. fragilis chromosome by PCR. This fragment, when introduced into YT58.1.3, enhanced its binding properties, as well as its growth on animal cell monolayers (our unpublished data).
Conclusion.
Transposons are widely used for general mutagenesis of bacteria. The improved transposon mutagen described above, Tn4400′, has several useful properties which make it an ideal reagent for mutagenesis of B. fragilis. Its insertion into the B. fragilis chromosome produced a large set of different mutants. By the aerobic mating protocol, most of the mutants were not siblings. Moreover, multiple insertions were not common. The insertion events were not readily reversible, which therefore obviated the need to constantly apply antibiotic pressure to retain the transposon. The ease of isolation of the DNA sequences flanking the insertions has made possible DNA sequence analysis of many MGD mutants. Furthermore, preliminary results showed that Tn4400′ insertion events can be detected in other Bacteroides strains (M. M. Dallas, M. F. Maiden, Y. P. Tang, M. J. Duncan, and M. H. Malamy, unpublished data) and in Porphrymonas gingivalis (1).
Acknowledgments
The research reported in this paper was supported by grant AI-19497 from the National Institutes of Health.
We are grateful to B. E. B. Claesson for providing the Monika cell line, A. A. Salyers for providing the plasmid containing tetQ, M. M. Dallas for performing the screen for auxotrophs in Tn4400′-mutagenized B. fragilis, R. A. Gallegos for providing plasmid pRG23, and J. DePonte III for isolating B. fragilis strains auxotrophic for arginine.
REFERENCES
- 1.Chen T, Dong H, Tang Y P, Dallas M M, Malamy M H, Duncan M J. Identification and cloning of genes from Porphyromonas gingivalis after mutagenesis with a modified Tn4400 transposon from Bacteroides fragilis. Infect Immun. 2000;68:421–424. doi: 10.1128/iai.68.1.420-423.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Claesson B E B, Gotthardsson I H. A tissue culture model for study of growth promotion and antimicrobial susceptibility in Bacteroides fragilis. J Antimicrob Chemother. 1988;21:17–26. doi: 10.1093/jac/21.1.17. [DOI] [PubMed] [Google Scholar]
- 2.Finegold S M. General aspects of anaerobic infection. In: Finegold S M, George W L, editors. Anaerobic infections in humans. New York, N.Y: Academic Press; 1989. pp. 137–153. [Google Scholar]
- 3.Gorbach S L, Bartlett J G. Anaerobic infections. N Engl J Med. 1974;290:1177–1184. doi: 10.1056/NEJM197405232902106. [DOI] [PubMed] [Google Scholar]
- 4.Guiney D G, Hasegawa P, Davis C E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci USA. 1984;81:7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwa V, Shoemaker N B, Salyers A A. Direct repeats flanking the Bacteroides transposon Tn4351 are insertion sequence elements. J Bacteriol. 1988;170:449–451. doi: 10.1128/jb.170.1.449-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikolich M P, Hong G, Shoemaker N B, Salyers A A. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl Environ Microbiol. 1994;60:3255–3260. doi: 10.1128/aem.60.9.3255-3260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robillard N J, Tally F P, Malamy M H. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J Bacteriol. 1985;164:1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo T A, Thompson J S, Godoy V G, Malamy M H. Cloning and expression of the Bacteroides fragilis TAL2480 neuraminidase gene, nanH, in Escherichia coli. J Bacteriol. 1990;172:2594–2600. doi: 10.1128/jb.172.5.2594-2600.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salyers A A. Bacteroides of the human lower intestinal tract. Annu Rev Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- 10.Shoemaker N B, Getty C, Gardner J F, Salyers A A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith C J. Nucleotide sequence analysis of Tn4551: use of ermFS operon fusions to detect promoter activity in Bacteroides fragilis. J Bacteriol. 1987;169:4589–4596. doi: 10.1128/jb.169.10.4589-4596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speer B S, Bedzyk L, Salyers A A. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol. 1991;173:176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y P, Dallas M M, Malamy M H. Characterization of the batI (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol Microbiol. 1999;32:139–149. doi: 10.1046/j.1365-2958.1999.01337.x. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J S, Malamy M H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase II. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]