Abstract
Porphyromonas gingivalis is a gram-negative, black-pigmented, oral anaerobe strongly associated with adult periodontitis. Previous transposon mutagenesis studies with this organism were based on the Bacteroides transposon Tn4351. Characterization of Tn4351-disrupted genes by cloning has not been an efficient way to analyze large numbers of mutants and is further complicated by the high rate of cointegration of the suicide delivery vector containing Tn4351. In this study, we mutagenized P. gingivalis with a modified version of the Bacteroides fragilis transposon Tn4400. Plasmid pYT646B carrying the transposon was mobilized from Escherichia coli to P. gingivalis ATCC 33277 by conjugation. Both normal and inverse transposition frequencies were similar (3 × 10−8). However, the inverse transposon (Tn4400′) contains a pBR322 replicon and a β-lactamase gene; thus, the cloning of disrupted genomic DNAs from inverse transposition mutants was easily accomplished after ligation of genomic fragments and transformation into E. coli. Thousands of transconjugants could be obtained in a single mating experiment, and inverse transposition was random as demonstrated by Southern hybridization. By this procedure the disrupted genes from P. gingivalis pleiotropic mutants were quickly cloned, sequenced, and identified.
The ability to isolate mutations in bacterial virulence genes is a powerful technique for defining their role in pathogenesis. Transposon mutagenesis is the method of choice because mutations are ideally random and mutants can be readily selected and identified. Plasmid vectors and transposons developed for colonic Bacteroides spp. have proved invaluable for genetic studies with Porphyromonas gingivalis, a gram-negative, black-pigmented, oral anaerobe. Tn4351, the first transposon developed for mutagenesis of P. gingivalis, has been used to generate mutants of several strains (2, 12), but its use has limitations. For example, in a separate study with strain ATCC 33277 (1), only 20% of mutations were simple transposon insertions, the rest being cointegrates containing the delivery vector and the transposon. While mutant phenotypes appeared stable, the cointegration of 53 kb of extra plasmid DNA complicated the molecular characterization of mutant genes.
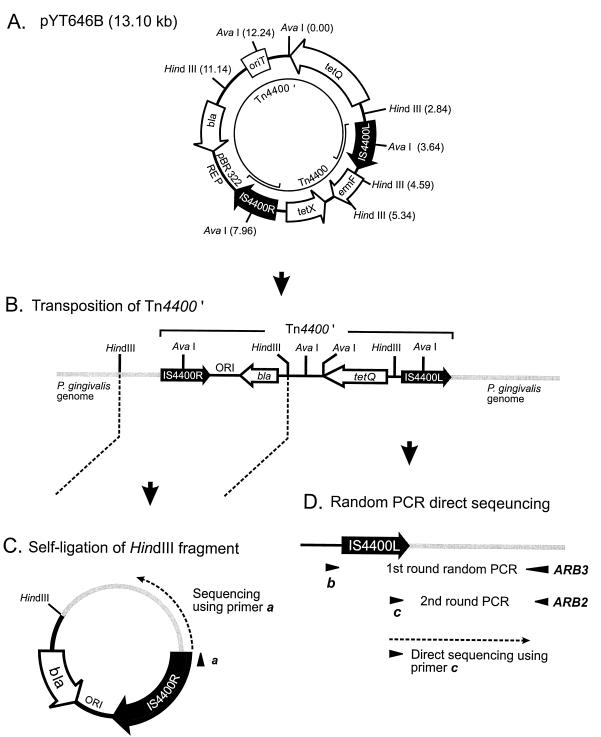
In this report we describe a new mutagenesis system for P. gingivalis based on Tn4400′, a modified version of the Bacteroides transposon Tn4400 (10). The transposon delivery vector, pYT646B (11), shown in Fig. 1A, carries on its Escherichia coli vector backbone a conjugational transfer origin, oriT, from the broad-host-range IncP plasmid RK2 (3) and the following tetracycline resistance genes: tetX, which is expressed in aerobically grown E. coli, and tetQ, which is expressed in P. gingivalis (6). The vector pYT646B does not contain a P. gingivalis replicon; therefore, upon conjugal transfer, tetracycline-resistant (Tcr) transconjugants can be obtained only from either inverse transposition of Tn4400 (Fig. 1B) or cointegration of pYT646B (see below). Because pYT646B contains the pBR322 replicon and a β-lactamase gene, mutations generated by inverse transposition can be readily characterized after cloning and direct sequencing of the genomic DNAs adjacent to both sides of the inverse transposon Tn4400′ (Fig. 1). Cloning of genomic sequences flanking IS4400R in Tn4400′ is achieved by digestion of mutant genomic DNA with HindIII and self-ligation (Fig. 1C), followed by transformation of E. coli and selection of ampicillin-resistant clones. The interrupted gene can be identified by sequencing the cloned fragment with a primer derived from IS4400R. Genomic sequence adjacent to IS4400L is more difficult to clone due to the unavailability of restriction enzyme sites. For example, BclI does not cut Tn4400′ and only infrequently cuts within genomic DNA; thus, BclI-generated flanking fragments are often too large for efficient cloning and maintenance. Therefore, our alternative strategy was to amplify the DNA fragment adjacent to IS4400L by randomly primed PCR (1a) and then to directly sequence the PCR products (Fig. 1D).
FIG. 1.
Schematic structure of a Tn4400′-mutagenized P. gingivalis chromosome and strategies for identification of the transposition loci. A mutation generated by inverse transposition is shown, and relevant restriction sites are indicated. REP, replicon.
The transposon library was obtained with an E. coli-P. gingivalis conjugation system. E. coli HB101 carrying the broad-host-range mobilizing IncP plasmid RK231 (3) and pYT646B was used as the donor. Cells were grown overnight at 37°C on Luria-Bertani (LB) plates containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml) and then subcultured into LB without antibiotics and grown to an optical density at 600 nm of ca. 0.8 (ca. 108 CFU/ml). The recipient, P. gingivalis ATCC 33277, was grown anaerobically for 48 h on blood agar plates (1b) and suspended in brain heart infusion broth supplemented with 0.5% yeast extract and 5 μg of hemin per ml (BHIS) at a concentration of approximately 2.5 × 1010 CFU/ml (equivalent to 5 ml per plate cross-streaked with P. gingivalis). Donor and recipient were mixed in a 1:3 volume ratio, centrifuged, and resuspended in 0.5 ml of BHIS. The suspension was spread on a sterile HAWP 047 S0 membrane filter (Millipore Corporation, Bedford, Mass.) placed on the surface of a BHIS plate. After overnight aerobic incubation, bacteria were harvested in 10 ml of BHIS and centrifuged to reduce the volume to 2 ml. One-tenth-milliliter aliquots were spread on blood agar selection plates containing 50 μg of gentamicin per ml to counter-select E. coli and 1 μg of tetracycline per ml to select for transconjugants resulting from inverse transposition. After 7 days of anaerobic incubation, each plate contained more than 1,000 Tcr colonies and the transconjugation frequency was calculated as 3 × 10−8 per donor. Transconjugants were purified on blood plates containing gentamicin and tetracycline to confirm tetracycline resistance, and a subset were tested on blood plates containing 5 μg of erythromycin per ml to determine the frequency of cointegrate formation. Approximately 10% of the Tcr colonies were also erythromycin resistant (Emr), indicating the frequency of cointegration.
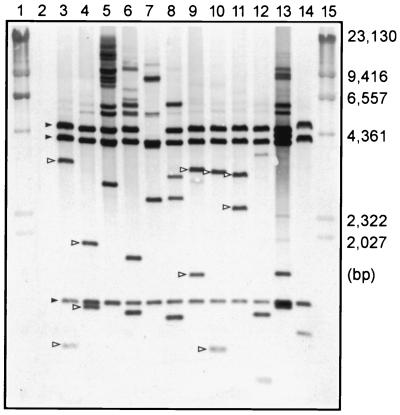
The randomness of transposition was examined by Southern hybridization. Genomic DNAs were isolated from 12 randomly picked Tcr transconjugants with a MasterPure DNA purification kit (Epicentre Technologies, Madison, Wis.). DNA (approximately 5 μg) was digested with AvaI, which cuts in the middle of the insertion elements flanking Tn4400′ (Fig. 1B). The probe was pGAT400ΔBglII, which contains pDG5 and Tn4400 (6, 13). Hybridization results are shown in Fig. 2. Three hybridizing fragments of 4.7, 3.6, and 1.1 kb were present in most of the mutants as predicted, since these three fragments were internal to Tn4400′. However, the 4.7-kb fragment was missing from one of the mutants (lane 7). Whether this mutant contains a truncated version of Tn4400′ remains to be determined. All strains contained at least two variably sized probe-reactive fragments, corresponding to the P. gingivalis genomic fragments on either side of Tn4400′ (Fig. 2). The results were indicative of inverse transposition of Tn4400. However, some strains had more than two variably sized hybridizing bands (Fig. 2, lanes 8 and 12); whether these contained multiple insertions of the transposon will be determined. The multiple cross-reacting fragments of high molecular weight (lanes 5, 6, 7, and 13) were apparently the result of incomplete digestion of genomic DNA. All 12 mutants were verified to be erythromycin-sensitive (Ems), ruling out the possibility of vector cointegration.
FIG. 2.
Southern hybridization of genomic DNA from Tcr transconjugants with a Tn4400 probe. DNA samples were digested with AvaI. Lanes 1 and 15, HindIII-digested bacteriophage λ marker; lane 2, parent strain ATCC 33277; lanes 3 to 14, 12 randomly picked transconjugants. Solid arrowheads indicate the three internal AvaI-hybridizing bands of Tn4400′; open arrowheads represent the AvaI bands containing the end of IS4400L or IS4400R and the adjacent genomic fragment.
As a test of the new mutagenesis system, approximately 4,500 transconjugants were screened for loss of pigmentation on blood agar plates containing tetracycline. Three white (nonpigmented) mutants were identified and purified. Mutant genomic DNA was digested with HindIII, fragments were self-ligated and transformed into E. coli DH5α competent cells, and ampicillin-resistant (Apr) transformants were selected on LB-ampicillin (100 μg/ml) plates. Plasmid DNA isolated from Apr colonies contained the DNA fragment adjacent to IS4400R and was used as the template for DNA sequencing with primer L78 (5′-CAATAATCGACCTCGTAAAAGACT-3′, derived from IS4400R and indicated as primer a in Fig. 1C). To confirm that the strains were true transpositional mutants of Tn4400′, the IS4400L flanking region was also sequenced by a randomly primed PCR method (Chen et al., submitted for publication; 9) shown in Fig. 1D. In brief, the first round of PCR used primer b, derived from a unique region of Tn4400′, and the semirandom primer ARB3. The latter consists of three segments: a 5′ arbitrary 10-bp sequence, a central 10-bp fully degenerate sequence, and a 5-bp 3′ arbitrary sequence. The second round of PCR used primer c, which is closer to the IS4400L genomic junction, and ARB2, which is derived from and primes within ARB3, to further amplify and truncate the IS4400L portion of the product. Both rounds of PCR were carried out in 50-μl reaction volumes containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 0.001% (wt/vol) gelatin, 250 μM deoxynucleoside triphosphates, 0.1 μM each primer, and 1.25 U of AmpliTaq Gold DNA polymerase. PCRs were performed in a Peltier thermal cycler (model PTC-200; MJ Research, Watertown, Mass.). Chromosomal DNAs were extracted from the mutants with a MasterPure DNA purification kit (Epicentre Technologies). Approximately 50 ng of chromosomal DNA was used as the template in the first-round PCR, and 1 μl of the first-round product was used directly as the template in the second. In the first round, primer UP58 (primer b in Fig. 1D) (5′-TTGAATCCCTTTTGTTT-3′) and a random primer, ARB3 (5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC-3′), were used and cycling conditions were 10 min at 95°C; 6 cycles of 30 s at 95°C, 30 s at 30°C, and 1.5 min at 72°C with 5-s increments per cycle; 30 cycles of 30 s at 95°C, 30 s at 45°C, and 2 min at 72°C with 5-s increments per cycle; and 5 min at 72°C. Primers R1030 (5′-TAGCAAACTTTATCCATTCAG-3′) (primer c in Fig. 1D) and ARB2 (5′-GGCCACGCGTCGACTAGTAC-3′) were used in the second round, and the cycling conditions were 10 min at 95°C; 35 cycles of 45 s at 95°C, 45 s at 55°C, and 1.5 min at 72°C with 5-s increments per cycle; and 10 min at 72°C. Second-round PCR products were purified with a Qiagen PCR purification kit and sequenced with primer R1030. DNA sequencing reactions were carried out with either dRhodamine or Big Dye Terminator cycle sequencing kits (Perkin-Elmer, Foster City, Calif.) with a PE 9700 thermocycler, and reactions were run on an ABI PRISM 377 DNA sequencer.
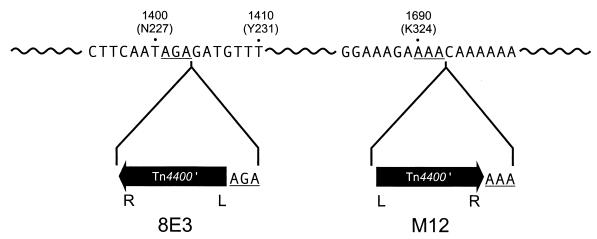
Both flanking regions from two white mutants of P. gingivalis were identified, and each one carried an insertion in the kgp gene encoding Lys-gingipain. Mutant 8E3 had an insertion at amino acid Asp229 and M12 at amino acid Thr325 (Fig. 3). In each mutant the target site of Tn4400′ was duplicated (Fig. 3), which is typical of transposon insertion. Thus, we confirmed that each mutation was generated by transposition of Tn4400′. The white colony phenotype of kgp mutants is in agreement with previous results which showed that 7 of 10 Tn4351-generated white mutants carried inserts in the kgp gene (1). Recently, it was demonstrated that HGP15, a cleavage product of the P. gingivalis-secreted activities Arg-gingipain A, Lys-gingipain, and HagA, could bind hemaglobin (5, 7, 8). The accumulation of heme from hemaglobin results in the black pigmentation of P. gingivalis colonies observed after growth on blood plates. Lys-gingipain activity was essential to process the HGP15 polypeptide from parent proteins, and kgp-negative mutants were white on blood plates. This study confirms these results, since two of the white mutants were defective in kgp. The DNA sequence flanking Tn4400′ in the third mutant mapped to an unidentified open reading frame which was found in the P. gingivalis W83 genome database.
FIG. 3.
Transposition loci of Tn4400′ in the kgp genes from two mutant strains. The orientations of Tn4400′ in two strains were as indicated. L denotes IS4400L, and R denotes IS4400R. Nucleotide numbering was based on the kgp gene from P. gingivalis 381 (NCBI accession no. D83258). Numbers in parentheses denote the corresponding amino acid positions. Underlined nucleotides are the duplication target sites resulting from the transposition event.
To determine transposon, and hence mutant, stability, four mutants were picked at random and grown anaerobically for 24 h (approximately three generations) in tryptic soy broth without antibiotics. After dilution, bacteria were plated on blood plates without tetracycline and grown anaerobically. From these plates, between 100 and 200 colonies were picked at random and streaked to blood plates with and without tetracycline. This procedure was repeated five times so that mutant stability over approximately 15 generations was determined. The numbers of colonies on plates with and without antibiotics were counted, and loss of Tcr was taken as an indication of the loss of Tn4400′. As shown in Table 1, three of the four mutants were extremely stable, with no loss after five subcultures and 15 generations without antibiotic; one mutant (15A5C) began to lose tetracycline resistance after four transfers. These results indicate that Tn4400′ mutants of P. gingivalis are stable, which makes them ideal candidates for use in animal models. The transposon was similarly stable in B. fragilis, since the frequency of reversion for auxotrophs was less than 10−9. Rare Tcs derivatives of B. fragilis Tn4400′ mutants retained their mutant phenotypes, suggesting that parts of the tetQ gene were deleted (11).
TABLE 1.
Stability of Tn4400′ mutants after growth without antibiotic
| Mutant strain | No.a of colonies which were Tcr after transfer:
|
||||
|---|---|---|---|---|---|
| 1b | 2 | 3 | 4 | 5 | |
| 15A1A | 101/101 | 190/190 | 177/177 | 50/50 | 100/100 |
| 15A3B | 122/122 | 152/152 | 134/134 | 54/54 | 101/101 |
| 15A5C | 152/152 | 133/133 | 158/158 | 39/39 | 118/131 |
| 15A7D | 167/167 | 183/183 | 108/108 | 41/41 | 131/131 |
The number of Tcr colonies/the total number streaked.
Each transfer represents three generations.
In summary, this is the first demonstration of transposon mutagenesis based on Tn4400′ in P. gingivalis. Large numbers of mutants were generated, and preliminary experiments showed that these mutants were stable for many generations in the absence of antibiotics. Mutants were the result of simple insertions, and inactivated loci were readily identified after simple cloning procedures and sequencing or direct random PCR sequencing as shown in this report. Currently, transposon libraries are being used to select and screen for mutations which alter P. gingivalis interactions with epithelial cells.
Acknowledgments
This work was supported by NIDCR grant R01 DE 10510 (M.J.D.) and NIAID grant R01AI19497 (M.H.M.).
REFERENCES
- 1.Chen, T., H. Dong, R. Yong, and M. J. Duncan. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microbial. Pathogen., in press. [DOI] [PubMed]
- 1a.Chen, T., R. Yong, H. Dong, and M. J. Duncan. 27 October 1999, posting date. A general method for direct sequencing of transposon mutants by randomly primed PCR, T01834. Elsevier Trends Journals Technical Tips Online. [Online.] http://tto.biomednet.com.
- 1b.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genco C A, Schifferle R E, Njoroge T, Forng R Y, Cutler C W. Resistance of a Tn4351-generated polysaccharide mutant of Porphyromonas gingivalis to polymorphonuclear leukocyte killing. Infect Immun. 1995;63:393–401. doi: 10.1128/iai.63.2.393-401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guiney D G, Hasegawa P, Davis C E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci USA. 1984;81:7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht D W, Malamy M H. Tn4399, a conjugal mobilizing transposon of Bacteroides fragilis. J Bacteriol. 1989;171:3603–3608. doi: 10.1128/jb.171.7.3603-3608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 6.Leng Z, Riley D E, Berger R E, Krieger J N, Roberts M C. Distribution and mobility of the tetracycline resistance determinant tetQ. J Antimicrob Chemother. 1997;40:551–559. doi: 10.1093/jac/40.4.551. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemaglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 9.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 10.Robillard N J, Tally F P, Malamy M H. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J Bacteriol. 1985;164:1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y P, Malamy M H. Isolation of Bacteroides fragilis mutants with in vivo growth defects by using Tn4400′, a modified Tn4400 transposition system, and a new screening method. Infect Immun. 2000;68:416–420. doi: 10.1128/iai.68.1.415-419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe-Kato T, Hayashi J I, Terazawa Y, Hoover C I, Nakayama K, Hibi E, Kawakami N, Ikeda T, Nakamura H, Noguchi T, Yoshimura F. Isolation and characterization of transposon-induced mutants of Porphyromonas gingivalis deficient in fimbriation. Microb Pathog. 1998;24:25–35. doi: 10.1006/mpat.1997.0170. [DOI] [PubMed] [Google Scholar]