Abstract
As a discipline, structural biology has been transformed by the three-dimensional electron microscopy (3DEM) “Resolution Revolution” made possible by convergence of robust cryo-preservation of vitrified biological materials, sample handling systems, and measurement stages operating a liquid nitrogen temperature, improvements in electron optics that preserve phase information at the atomic level, direct electron detectors (DEDs), high-speed computing with graphics processing units, and rapid advances in data acquisition and processing software. 3DEM structure information (atomic coordinates and related metadata) are archived in the open-access Protein Data Bank (PDB), which currently holds more than 11,000 3DEM structures of proteins and nucleic acids, and their complexes with one another and small-molecule ligands (~ 6% of the archive). Underlying experimental data (3DEM density maps and related metadata) are stored in the Electron Microscopy Data Bank (EMDB), which currently holds more than 21,000 3DEM density maps. After describing the history of the PDB and the Worldwide Protein Data Bank (wwPDB) partnership, which jointly manages both the PDB and EMDB archives, this review examines the origins of the resolution revolution and analyzes its impact on structural biology viewed through the lens of PDB holdings. Six areas of focus exemplifying the impact of 3DEM across the biosciences are discussed in detail (icosahedral viruses, ribosomes, integral membrane proteins, SARS-CoV-2 spike proteins, cryogenic electron tomography, and integrative structure determination combining 3DEM with complementary biophysical measurement techniques), followed by a review of 3DEM structure validation by the wwPDB that underscores the importance of community engagement.
Keywords: Electron microscopy, Protein Data Bank, PDB, PDB-Dev, Electron Microscopy Data Bank, EMDB, Electron microscopy data resource, Resolution Revolution, Cryo-electron microscopy, Cryo-electron tomography, Sub-tomogram averaging, Electron crystallography, Micro-electron diffraction, Icosahedral viruses, Ribosomes, Integral membrane proteins, SARS-CoV-2 spike proteins, Integrative or hybrid methods, Structure validation, Q-score
Introduction
The Protein Data Bank (PDB) was the first open-access digital data resource in biology (Berman 2008). Now in its 51st year of continuous operations, it was established in 1971 with just seven protein structures (Protein Data Bank 1971; Burley et al. 2022c). As of mid-2022, the PDB archive housed > 190,000 3D structures of proteins and nucleic acids (DNA and RNA) and their complexes with one another and with small-molecule ligands (e.g., enzyme co-factors, drugs). Throughout its history, the PDB has been regarded as a pioneer in the open-access data movement. Nearly 60,000 structural biologists working on every permanently inhabited continent have generously deposited 3D structure information (atomic coordinates, experimental data, and related metadata) to the archive over more than 50 years. Currently supported experimental methods include macromolecular crystallography (MX), nuclear magnetic resonance spectroscopy (NMR), 3D electron microscopy (3DEM), Electron Crystallography (EC), and micro-electron diffraction (micro-ED). Today, millions of PDB data consumers worldwide working in fundamental biology, biomedicine, bioengineering, biotechnology, and energy sciences enjoy no-cost access to 3D biostructure information with no limitations on data usage.
Since 2003, the PDB archive has been jointly managed by the Worldwide Protein Data Bank (wwPDB, wwpdb.org) partnership (Berman et al. 2003; wwPDB consortium 2019). wwPDB Full Members include the US-funded Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB, RCSB.org, (Berman et al. 2000; Burley et al. 2021; Burley et al. 2022c; Burley et al. 2022b)); the Protein Data Bank in Europe (PDBe, PDBe.org, (Armstrong et al. 2020)); Protein Data Bank Japan (PDBj, PDBj.org, (Bekker et al. 2022)); the Electron Microscopy Data Bank (EMDB, emdb-empiar.org, (Tagari et al. 2002; Lawson et al. 2016)); and the Biological Magnetic Resonance Bank (BMRB, bmrb.io, (Ulrich et al. 2008)). Protein Data Bank China (PDBc) was recently admitted to the wwPDB as an Associate Member. wwPDB partners are committed to the FAIR (Findability, Accessibility, Interoperability, and Reusability) (Wilkinson et al. 2016) and FACT (Fairness, Accuracy, Confidentiality, and Transparency) (van der Aalst et al. 2017) Principles emblematic of responsible data stewardship in the modern era.
The RCSB PDB is headquartered at Rutgers, The State University of New Jersey with additional performance sites at the University of California San Diego and the University of California San Francisco. Within the wwPDB, RCSB PDB serves as the designated PDB Archive Keeper, responsible for archiving ~ 100 TB of digital information and a physical archive that includes correspondence and other artifacts accumulated since the early 1970s. Based on a conservative estimate of US$100,000 for the replacement cost of an individual PDB structure, replacement of the entire archive would cost about US$20 billion (Sullivan et al. 2017).
This review article, published in a special issue of Biophysical Reviews honoring Professor Haruki Nakamura on the occasion of his 70th birthday, is focused on 3DEM structures archived within the PDB and the impact of the 3DEM “Resolution Revolution” (Kuhlbrandt 2014; Herzik 2020) on basic and applied research across fundamental biology, biomedicine, energy sciences, and bioengineering and biotechnology. Nakamura served as founding Director of Protein Data Bank Japan (PDBj) from 2000 to 2017. In 2003, he was one of the co-founders of the wwPDB partnership (Berman et al. 2012; Berman et al. 2003). Under Nakamura’s leadership, PDBj assumed PDB data-in responsibility for all structure depositions coming from Asia and the Middle East. PDBj data-out activities play a unique role within the wwPDB partnership, delivering information from the PDB archive on its PDBj.org website in English, Japanese, Korean, and Mandarin (both Traditional and Modern) (Kinjo et al. 2018, 2017, 2012, 2010).
Growth, resolution, and composition/complexity of 3DEM structures in PDB
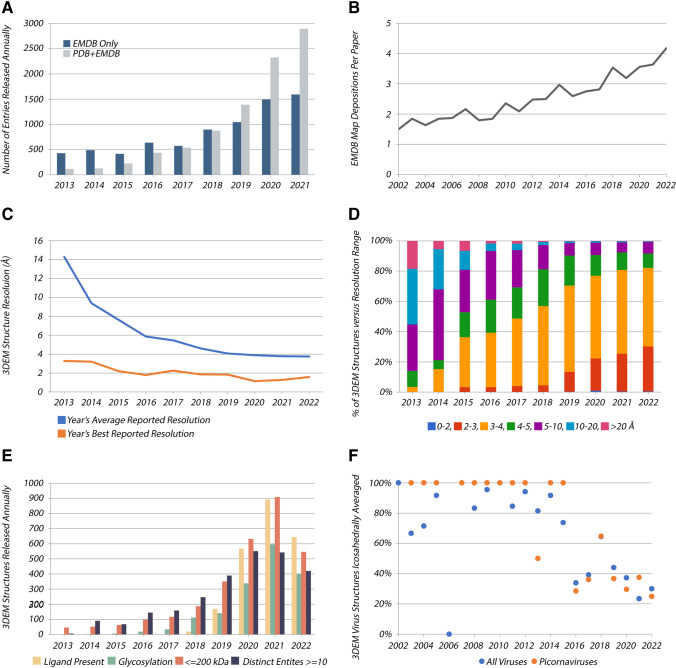
Transformation of cryogenic electron microscopy (cryo-EM) into a mainstream structural biology technique is evidenced by (i) dramatic growth in the number of 3DEM structures over the past decade (Fig. 1A), and (ii) award of the 2017 Nobel Prize in Chemistry to Jacques Dubochet, Joachim Frank, and Richard Henderson. Deposition of atomic coordinates to the PDB and experimental density maps to EMDB is mandatory for publication of newly determined 3DEM structures in all major scientific journals. Prior to 2013, the field was limited to a small number of expert laboratories. Structure determinations were hampered by methodological bottlenecks, and only 385 3DEM structures were deposited to the PDB in this era. This paucity of structures stands in stark contrast to the present. As of mid-2022, PDB archive holdings included a total of 11,309 3DEM structures determined using single-particle cryo-EM methods, cryogenic electron tomography (cryo-ET), EC, and microED.
Fig. 1.
Selected annual metrics for 3DEM structures in PDB and density maps in EMDB. A 3DEM structures (PDB and EMDB) and density maps (EMDB only) versus time. B Average number of 3DEM density map depositions reported per primary publication versus time. C Average reported resolution (blue) and best reported resolution (orange) for 3DEM structures versus time. D Percentage of 3DEM structures versus resolution range versus time. E PDB 3DEM structures wherein ligands are present; glycosylation is evident; size of the sample macromolecule or macromolecular complex is 200,000 Da; and the number of distinct molecular entities comprising the sample is 10. F Percentage of 3DEM virus structure depositions to PDB relying on icosahedral averaging versus time
Single-particle Cryo-EM has become one of the most sought-after experimental methods in structural biology thanks to a host of technical advances, including modern electron microscopes (Weis and Hagen 2020), DEDs (Campbell et al. 2012), software for image acquisition (Weis and Hagen 2020; Cheng et al. 2018; Mastronarde 2018), and maturation of subclassification (Scheres 2016) and other in silico “purification” methods powered by graphics processing unit (GPU) computing. These developments have revolutionized how cell and molecular biologists are working to understand important biochemical processes. Not surprisingly, the average number of 3DEM density maps reported in a single published paper has increased over time (Fig. 1B). Twenty years ago, publications describing 3DEM structural studies encompassed an average of 1.5 3DEM density maps deposited into EMDB. Today, that metric has increased to an average of > 4 deposited 3DEM density maps per primary publication. 3D characterization of the flexible pantograph-like motion of transcribing RNA polymerase coupled to the translating ribosome involved deposition of 24 3DEM density maps to EMDB and deposition to the PDB of the same number of structures (i.e., atomic coordinates) (Wang et al. 2020).
Coinciding with exponential growth in the number of 3DEM structures in PDB, the average resolution of 3DEM structures archived in the PDB has improved dramatically. Since 2013, the average resolution of 3DEM structures in PDB has steadily improved from being slightly worse than 14 Å to better than 4 Å (Fig. 1C). Moreover, during this same time period, 43 PDB structures with resolution better than 2 Å have been deposited to PDB. As the resolution revolution continues, macromolecular crystallography (MX) software developers are increasingly crossing the “methodology barrier” to improve high-resolution 3DEM density maps even further (e.g., (Terwilliger et al. 2020)). As of mid-2022, the PDB archive housed > 7600 3DEM structures determined at near-atomic resolution (2–4 Å), with most recent depositions falling within the 3–4 Å resolution range (Fig. 1D). At this resolution, individual β strands and bulky amino acid residue sidechains are well resolved, both of which are essential for building atomic coordinate structure models into 3DEM density maps. Reaching 2–3 Å resolution is becoming somewhat routine for 3DEM structures deposited to PDB. The level of detail present in accompanying 3DEM density maps often permits accurate definition of the atomic coordinates without prior structural knowledge of individual macromolecular constituents from previously determined experimental structures or computed structure models (i.e., either from PDB or from AlphaFold 2 (Jumper et al. 2021), RoseTTAFold (Baek et al. 2021)).
3DEM structure determination beyond ~ 2 Å resolution appears unlikely to become routine. Bona fide atomic resolution (better than ~ 1.5 Å), wherein the positions of individual non-hydrogen atoms are discernable as isolated peaks in 3DEM density maps, has only been achieved to date for one biological specimen of exceptional stability and 3D structural homogeneity (human apoferritin), and the best-to-date being a ~ 1.15 Å resolution structure (PDB ID 7a6a (Yip et al. 2020)). Most samples of biological macromolecules embedded in vitreous ice may not have extremely well-ordered structures. It is remarkable that the average resolution of PDB MX structures plateaued at ~ 2.0 Å in ~ 1990 (S.K. Burley et al. 2022a), which probably reflects inherent both limitations in our ability to prepare well-ordered crystals of biological material and structural heterogeneity of many proteins found in nature.
As both quantity and quality of 3DEM structures in the PDB archive increase, it is gratifying to see that the boundaries of the method continue to be pushed (Fig. 1E). Since 2013, there has been a steady growth in 3DEM PDB structures wherein ligands have been modeled, which is a promising trend for the biopharmaceutical industry. In many cases, 3DEM can resolve structures of challenging/impossible-to-crystallize, high-value targets (e.g., integral membrane proteins (Robertson et al. 2022), see Area of Focus No. 3) for use in structure-guided drug discovery. Another interesting development of late is that 3DEM methods can now resolve sugars covalently bound to extracellular proteins. For example, PDB archive holdings of 3DEM structures of the highly glycosylated SARS-CoV-2 spike protein exceed ~ 1000 (see Area of Focus No. 4). Some of these data and earlier PDB structures of SARS-CoV spike proteins have informed design of vaccines (Goodsell and Burley 2022) and development of monoclonal antibodies to combat the COVID-19 pandemic (Gilliland et al. 2012; Chiu and Gilliland 2016). Equally impressive is the fact that previously encountered technical limitations of cryo-EM methods with respect to size (e.g., smaller < 200 kDa macromolecules) and complexity (e.g., distinct molecular entities 10) have been largely overcome. Finally, the future of cryo-ET combined with sub-tomogram averaging is looking very bright (see Area of Focus No. 5). As of mid-2022, the highest resolution cryo-ET structure in the PDB archive was PDB ID 7bzt (3.3 Å RuBisCO visualized within native Halothiobacillus neapolitanus carboxysomes (Cui et al. 2020)).
“Resolution Revolution” areas of focus
Notwithstanding the milestones described above and the impressive metrics illustrated in Fig. 1, the impact of the resolution revolution can only be fully appreciated by delving into the structural biology literature. Six representative areas of focus are presented below, beginning with icosahedral viruses that served as “pioneer” samples in development of 3DEM methods and culminating with integrative or hybrid methods (I/HM) structural studies of complex macromolecular machines. These structures are so large and/or complicated that their determination necessitated combining 3DEM with complementary biophysical measurements. In many cases, I/HM structure determination was buttressed by structural information archived in the PDB and computed structure models coming from comparative protein structure modeling or artificial intelligence/machine learning methods.
Area of Focus No. 1: 60-fold symmetric icosahedral viruses
The largest class of biological assemblies with regular non-crystallographic symmetry in the PDB archive is icosahedral viruses (as of mid-2022, count > 1100 structures). Throughout the 1980s and early-to-mid 1990s, such structures were determined exclusively using MX. In the late 1990s, however, the PDB began to receive depositions of atomic coordinates for icosahedral viruses obtained by fitting previously determined structures (already archived in the PDB) into low-resolution (20–30 Å) 3DEM density maps. Subsequently, vitrified icosahedral virus particles proved to be ideal specimens for 3DEM reconstruction software development, because the presence of 60 identical asymmetric units facilitated rapid and accurate orientation determination (Kaelber et al. 2017). The first 3DEM icosahedral virus structure made publicly available in the archive PDB was that of Spiroplasma virus in 1999 (PDB ID 1kvp (Chipman et al. 1998)). It was quickly followed by several 3DEM structures of viruses bound to cellular receptors or antibody fragments (e.g., rhinovirus: PDB ID 1d3i (Kolatkar et al. 1999) and1d3e (Kolatkar et al. 1999); poliovirus: PDB ID 1dgi (He et al. 2000); Foot and Mouth Disease virus: PDB ID 1qgc (Hewat et al. 1997)). Other early exemplars provided first demonstrations that high symmetry combined with 3DEM reconstruction could yield higher resolution structures, including Semliki Forest virus at 9 Å resolution (PDB ID 1dyl (Mancini et al. 2000)), and bacteriophages HK97 and PRD1, both determined at 12 Å resolution (PDB IDs 1if0 (Conway et al. 2001) and 1hb5 (Martin et al. 2001)).
All of the early 3DEM structures deposited to PDB were based on density maps reconstructed from single-particle images laboriously recorded and manually processed using photographic film. In the mid-2000s, charge-coupled device (CCD) detectors enabled automated 3DEM data collection (e.g., (Potter et al. 1999; Zhang et al. 2009; Mastronarde 2005)). Many virus-focused laboratories did not, however, embrace this new technology, primarily because CCD detectors were limited in terms of size and electron-detection sensitivity. Direct electron detectors were the key technology development that enabled rapid growth in numbers and increased resolution for icosahedral virus structures. Imaging virus particles in “movie mode” allows individual frames to be aligned, substantially reducing blurring from particle motion on the cryogenic sample stage that occurs during electron beam exposure (Campbell et al. 2012).
During the mid-2000’s, the wwPDB undertook the task of remediating virus structures with high non-crystallographic symmetry (Lawson et al. 2008). Three major issues were addressed, including (i) missing or erroneous sets of transformation operations, (ii) inconsistency in coordinate-frame representations, and (iii) overly complex instructions for building the virus assembly. A simplified uniform notation was adopted, yielding completely machine-readable instructions for building full virus assemblies in all icosahedral virus structures going forward.
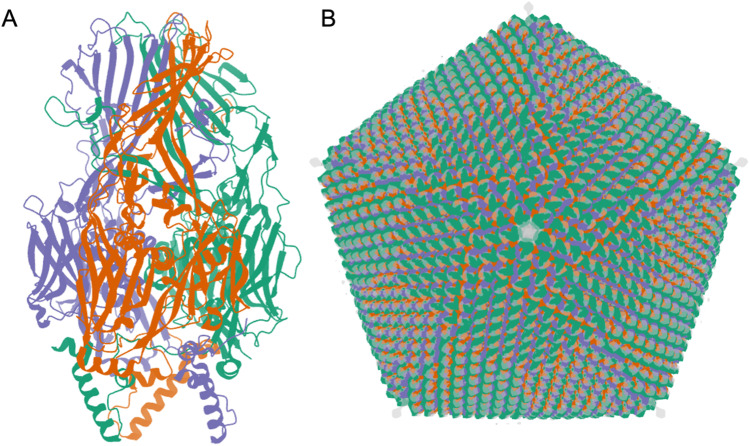
As of mid-2022, 3DEM methods have been used to determine ~ 70% (~ 760) of all icosahedral virus structures archived in the PDB. Over the past 3 years, more than 100 new structures of icosahedral viruses have been publicly released by the PDB annually, with an average resolution of ~ 3.5 Å. The best resolved structure in this class is that of an adeno-associated virus, a gene therapy viral vector candidate, determined at 1.56 Å resolution (PDB ID 7kfr (Xie et al. 2020)). The “heaviest” icosahedral virus structure available in the PDB is that of Faustovirus (PDB ID 5j7v (Klose et al. 2016), Fig. 2). The Faustovirus is a 240-nm-diameter double-stranded DNA virus that infects amoebae. It consists of 5,340,600 amino acid residues weighing in at ~ 594,000 KDa. On the public health front, 3DEM structures of icosahedral viruses are informing our understanding of important viral pathogens. For example, atomic coordinates of Dengue, West Nile, and Zika flaviviruses have enabled mapping sites of glycosylation, amino-acid sequence variation, and neutralizing antibody binding and vaccine design (Goodsell and Burley 2022; Hasan et al. 2018).
Fig. 2.
Faustovirus (Klose et al. 2016). A. 3DEM atomic coordinates in the PDB (entry PDB ID 5j7v) consist of the capsid protein trimer illustrated in ribbon representation. B Atomic coordinates for the full icosahedral capsid are generated by applying 2760 transformation matrices to the trimer atomic coordinates. The superimposed 15 Å resolution 3DEM density map (entry EMD-8144, in semi-transparent grey) reveals additional spike-like features for which atomic coordinates are not available extending from each fivefold vertex. Images generated using Mol* (Sehnal et al. 2021)
While icosahedral viruses are of substantial biological and biomedical importance in their own right, they continue to serve as real-world samples with which to improve 3DEM structure determination methodologies. These viruses are, as a rule, not perfectly icosahedral, although deviations from 60-fold symmetry are typically not observed until relatively large datasets and sophisticated approaches to image processing are employed (Kaelber et al. 2017). For example, newer methods have allowed visualization of the entire Levivirus genome asymmetrically packaged inside a capsid shell, which itself has icosahedral symmetry (Gorzelnik et al. 2016; Koning et al. 2016). Figure 1F documents that within the structural virology community, reliance on icosahedral averaging during structure determination declined markedly in 2016. This shift cannot be explained solely by increased emphasis on less symmetric viruses; it is even seen for members of the family Picornaviridae—a prototypically icosahedral virus. Moreover, intrinsic and induced asymmetries in viral capsids may become apparent when symmetry-breaking or symmetry-free reconstructions are used during structure determination. In one dramatic example, the structure of a Coxsackie virus capsid bound to nanodisc-embedded receptor exhibited receptor-induced asymmetry due to deformation of the ostensibly icosahedral capsid (PDB ID 3jd7 (Lee et al. 2016)). Looking to the future of structural virology, we can expect to learn a great deal more about dynamic, transient, and local conformational heterogeneity and flexibility that would otherwise remain obscured if we were limited to using MX with non-crystallographic symmetry restraints or 3DEM with icosahedral averaging for virus structure determination.
Area of Focus No. 2: asymmetric ribosomal subunits and ribosomes
Ribosomes are responsible for messenger RNA template-directed protein synthesis in all living cells. They are also the targets of numerous antibiotics (Wilson 2014; Lin et al. 2018). The ribosome is a very large (2.5 MDa or more), highly intricate molecular machine comprising 40 or more distinct polypeptide chains apportioned between large and small subunits, plus 3 to 4 ribosomal RNA chains (totaling 1000s of RNA nucleotides). This picture is further complicated by the fact that ribosomes also form transient complexes with messenger RNAs (mRNAs), transfer RNAs (tRNAs), and numerous additional components responsible for regulation of translation throughout the processes of initiation, elongation, and termination. As structural studies of translation have evolved, they have been extended beyond the ribosome alone to capture mechanistic views of the ribonucleoprotein machine as the central player in many of the discrete biochemical steps occurring during protein synthesis, including translocation along mRNA, binding of tRNAs to their A, P, and E binding sites, and interactions with regulatory proteins, such as elongation factor-G or EF-G (Carbone et al. 2021; Noller et al. 2017).
Understanding mRNA translation at the atomic level became a reality in 2000 with determination of ribosomal subunit structures using MX: PDB IDs 1ffk (Ban et al. 2000), 1fka (Schluenzen et al. 2000), and 1fjg (Carter et al. 2000). Leaders of the research groups responsible for these landmark structures (Ada Yonath, Venki Ramakrishnan, and Thomas E. Steitz) shared the 2009 Nobel Prize in Chemistry. As of mid-2022, the PDB archive housed approximately 1600 structures identified as ribosomes. Of these, nearly 1000 were determined using 3DEM, versus ~ 600 determined using MX. 3DEM has become the method of choice for studying ribosome structure and function. Benefitting from technical advances described above, 3DEM structures of ribosomes are being determined at better than 2.0 Å resolution (e.g., PDB ID 7k00 (Watson et al. 2020)), and the average resolution of 3DEM ribosome structures archived in the PDB from the beginning of 2018 onwards is ~ 3.6 Å (versus an average of ~ 3.1 Å for PDB MX structures of ribosome from the same period). 3DEM ribosome structures in PDB come from all of the kingdoms of life: ~ 31% eukaryotic, ~ 63% eubacterial, and ~ 6% archaebacterial.
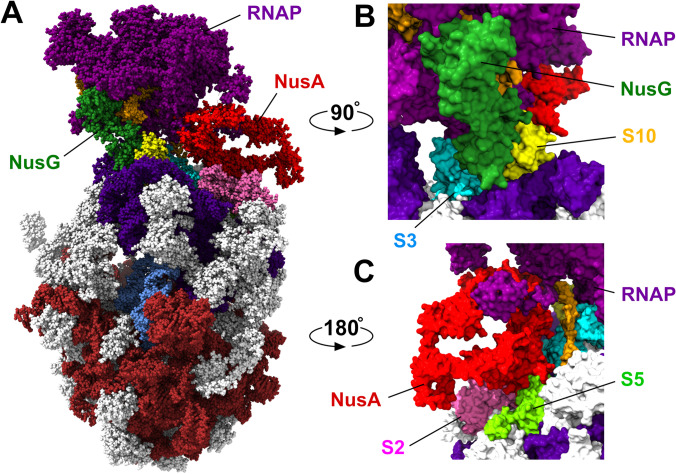
Illustrative of the advantages offered by 3DEM for functional studies is the recent work of Wang et al. (2020) studying transcription-translation coupling in bacteria. In prokaryotes, a transcription-translation complex (TTC) performs transcription of mRNA from DNA by RNA polymerase (RNAP) whilst that same mRNA transcript is being translated a ribosome yielding the polypeptide chain encoded by the gene. Formation of a TTC involves physical contact between RNAP and the ribosome. Previously published work demonstrated that coupling can be mediated by transcription elongation factors, such as NusG (Burmann et al. 2010) and NusA (Strauss et al. 2016). Wang et al. assembled E. coli TTCs comprising the DNA, RNAP, mRNA, ribosome, and tRNAs, both in the presence and absence of NusG and NusA. Their 3DEM structural studies of 24 distinct DNA, protein, RNA complexes detected four distinct TTC classes. One such class, TTC-B, observed in the presence of mild detergent with 8, 9, or 10 mRNA codons separating the RNAP and ribosome active sites, was identified as the functional complex (Fig. 3). Taking advantage of the fact that single-particle cryo-EM methods are not unduly compromised by sample heterogeneity or the presence of multiple conformational and/or configurational states present in electron images obtained from a single sample (Frank 2017), Wang et al. were able to subdivide TTC-B into subclasses (TTC-B1, TTC-B2, and TTC-B3) in which RNAP is differentially rotated relative to NusA and the ribosome. In all three TTC-B subclasses, NusG acts as a molecular bridge, binding to RNAP and bringing both itself and RNAP into contact with ribosomal proteins S10 and S3, respectively, in the 30S subunit. NusA in turn acts as a bridge, binding to RNAP and ribosomal proteins S2 and S5, while making no contacts with NusG. Uncovering of the structural bases of transcription-translation coupling by three research groups working independently highlights the convergence of single-particle cryo-EM of biochemically defined molecular species (Webster et al. 2020; Wang et al. 2020) with cryo-ET for in situ analyses (O’Reilly et al. 2020).
Fig. 3.
A E. coli transcription-translation complex (Wang et al. 2020) TTC-B2 (PDB ID 6x 7f), color coding: RNAP-purple, DNA-orange; ribosomal RNAs: large subunit-brown, small subunit-indigo; ribosomal proteins-grey (also see below); tRNA-blue; and transcription elongation factors NusG-dark green and NusA-red. The mRNA transcript is not visible in this representation. B Interaction of NusG (dark green) with RNAP (purple) and ribosomal proteins S3 (cyan) and S10 (yellow). C Interaction of NusA (red) with ribosomal proteins S2 (pink) and S5 (light green). Images generated using ChimeraX (Pettersen et al. 2021)
Area of Focus No. 3: 3DEM studies of integral membrane proteins using single-particle methods
Membrane proteins represent nearly 60% of currently validated drug targets. Historically, determination of membrane protein structures posed significant technical challenges related to the complex lipid environment surrounding and stabilizing the protein. Bottlenecks in sample preparation for high-resolution structure determination by MX or 3DEM included sample extraction from biological membranes and solubilization and transfer into lipid vesicles in a manner that preserved both structure and function. Resulting samples were often aggregated and/or highly heterogeneous, rendering them unsuitable for crystallization or cryo-EM single-particle imaging. Recent breakthroughs in sample preparation largely overcame such challenges, making near atomic resolution structure determination of integral membrane proteins appear somewhat routine.
The first integral membrane protein structure deposited to the PDB was that of bacteriorhodopsin, determined using EC at 3.5 Å resolution by Henderson and coworkers (PDB ID 1brd (Henderson et al. 1990)). Subsequent discovery of detergents with low critical micellar concentration permitted extraction and transfer of individual membrane proteins from native membranes into lipid vesicles. This early approach was fraught with difficulties owing to variation in lipid vesicle size, complicating specimen vitrification, particle picking, classification, and alignment, and ultimately 3DEM structure determination. The first successful application of single-particle cryo-EM methods to studying the structure of an integral membrane protein yielded 3DEM density maps of the BK potassium channel at 17–20 Å resolution (EMD-5114 and EMD-5121 (Wang and Sigworth 2009)). Introduction of protein-lipid nanodiscs has enabled custom design of homogenous populations of lipid vesicle carriers for integral membrane proteins. Membrane scaffold protein (or MSP) nanodiscs (cube-biotech.com) are among the most popular at present. They consist of two copies of the amphipathic membrane scaffold protein composed of repeated -helix-forming segments. Two MSPs together form adjacent “belts” surrounding the aliphatic tails on the periphery of the disc-like lipid bilayer, thereby stabilizing the high-density lipoprotein-like assembly. Variations in the number of -helical repeats allow for control of nanodisc diameter, governing the size of membrane proteins that it can receive. The thickness of the lipid bilayer can also be controlled using homogeneous preparations of lipids with acyl chains of differing lengths. The composition of the nanodisc can be further optimized by including signaling phosphoinositides or cholesterol to create lipid raft-like environments.
Direct electron detectors (DEDs) were first used for structural studies of integral membrane proteins in 2013, yielding a 3.4 Å resolution structure of a mammalian TRP channel, TRPV1 (PDB ID 3j5q (Cao et al. 2013)). As of mid-2022, PDB holdings included 10,001 membrane protein structures (6591 determined by MX, 3370 by 3DEM, and 40 by EC). Approximately 99% of the 3DEM structures now archived in PDB relied on the use of DEDs. At the time of writing, the highest resolution 3DEM PDB structure of a membrane protein is that of a human gamma-aminobutyric acid receptor, the GABA(A)R- 3 homopentamer bound to histamine and megabody Mb25 (Nakane et al. 2020) determined at an overall resolution of 1.73 Å (as judged by the Fourier Shell Coefficient or FSC = 0.143 sigma criterion, see below). Locally, the resolution of the 3DEM density map ranges between ~ 1.6 Å and ~ 2.3 Å. As of mid-2022, the largest membrane protein-containing structure in the PDB was that of the 7546 kDa mitochondrial ATP synthase hexamer from Toxoplasma gondii (PDB ID 6tml (Muhleip et al. 2021)), which was determined at 4.8 Å resolution. With improving sample preparation methods, improved instrumentation, and advances in structure determination software many more exciting new 3DEM structures of integral membrane proteins will be deposited into the PDB in the coming years.
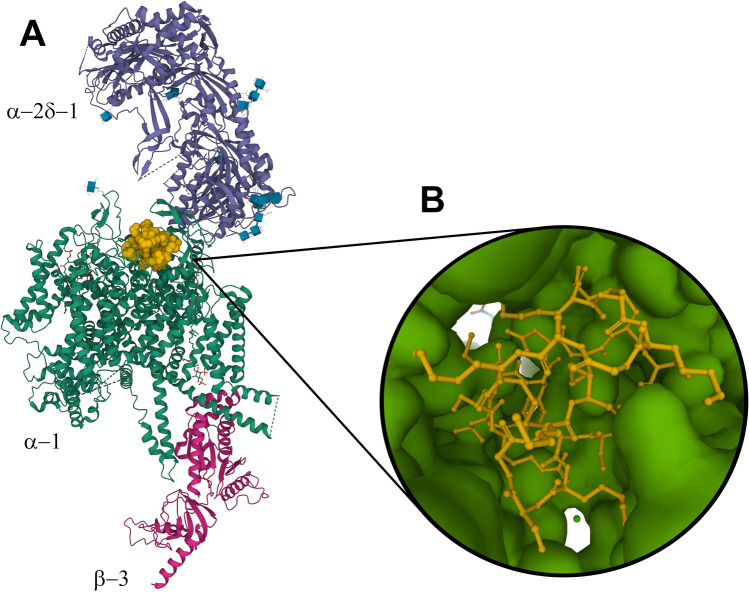
Of particular importance in structure-based drug discovery is the advent of 3DEM studies of small-molecules bound to membrane proteins that represent potential drug discovery targets. Figure 4, for example, illustrates the structure of human Cav2.2, a neuronal-type voltage-gated calcium channel bound to ziconotide, a United States (US) Food and Drug Administration (FDA)-approved drug prescribed for treating intractable pain. Yan and co-workers used a single-particle cryo-EM to reveal how the drug blocks the ion-conducting pore of Cav2.2 (Gao et al. 2021). Ziconotide is a biologic, a neurotoxic peptide derived from the cone snail Conus magus, comprising 25 amino acids with three disulfide bridges. It is administered via intrathecal injection into the spinal canal. Understanding the mode of action of this biologic agent at the atomic level could enable structure-guided discovery of orally bioavailable small-molecule organic compounds for effective management of chronic pain without exposing patients to the risk of opiate addiction.
Fig. 4.
A Mol* ribbon representation of the 3DEM structure of the human Cav2.2 bound to ziconotide (PDB ID 7mix (Gao et al. 2021). Color coding: ziconotide-orange (space-filling representation); -2 -1 subunit-purple; -1 subunit-green; -3 subunit-red. Glycosyl groups covalently bound to the -2 -1 and -1 subunits are displayed as blue cubes with atomic stick figures using the GlycanBuilder representation described in (Shao et al. 2021). B Rotated Mol* closeup representation of the interaction of ziconotide (ball-and-stick) with -1 (surface)
Area of Focus No. 4: SARS-CoV-2 spike proteins
Coronaviruses were named for their appearance in negative stain electron micrographs showing a “corona,” which we now know to be due to the presence of spike proteins arrayed on the surfaces of individual virions. These spikes are transmembrane glycoproteins responsible for mediating viral entry into host cells and inducing neutralizing immune responses. They are also the antigens presented to the immune system by the two mRNA vaccines now in common use to combat the COVID-19 pandemic (Goodsell and Burley 2022).
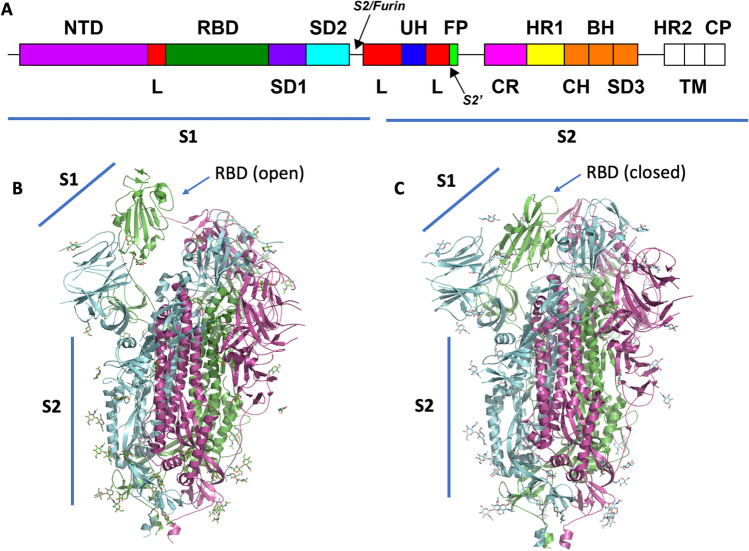
In SARS-CoV-2, spike proteins found on the surfaces of mature virions occur as clover-shaped homotrimers, with three receptor-binding S1 segments sitting atop a trimeric membrane-fusion S2 segment stalk (Wrapp et al. 2020; Walls et al. 2020). Each S1 segment contains a receptor-binding domain (RBD) and an N-terminal domain (NTD) (Fig. 5A). During viral entry, the RBD binds to one or more of its host receptors (e.g., angiotensin-converting enzyme 2, ACE2), mediating virion attachment (Shang et al. 2020). Short amino acid sequence motifs at the S1/S2 inter-segment boundary and/or S2′ site are cleaved specifically by host proteases (Xia et al. 2020). ACE2 binding and proteolytic cleavage together trigger S1 to dissociate (Benton et al. 2020) from the trimer. Then S2 undergoes a large structural change leading to fusion of the viral and host cell membranes, thereby allowing the genetic material of the virus to enter the host cell (Cai et al. 2020). 3DEM structures of SARS-CoV-2 spike protein hold the key to understanding the complex process of cell entry and evolution of immune evasion by variants of concern identified by the World Health Organization.
Fig. 5.
A Schematic view of SARS-CoV-2 spike protein sequence showing arrangement of polypeptide chain segments S1 and S2 and various domains. Proteolytic cleavage sites are indicated with arrows. B Mol* ribbon representation of the one-up-two-down RBD conformation of the spike protein (PDB ID 6vsb (Wrapp et al. 2020)). C Mol* ribbon representation of the all-down RBD conformation observed in PDB ID 6vxx (Walls et al. 2020). Individual trimers are color-coded magenta, green, and cyan, respectively. Covalently bound glycosyl groups are depicted as atomic stick figures
Technical advances described above enabled extremely rapid determination of spike protein structures within months of the onset of the COVID-19 pandemic and identification of the novel coronavirus SARS-CoV-2 as the causative infectious agent. The first PDB structure of the spike protein was determined at ~ 3.5 Å resolution via 3DEM by McLellan and colleagues (PDB ID 6vsb; Fig. 5B (Wrapp et al. 2020)) and made publicly available in late February 2020. This structure of the pre-fusion spike protein showed one RBD within the trimer adopted the “standing up” conformation ready to engage ACE2 on the cell surface, whereas the other two RBDs of the trimer adopted the “lying down” position, which sequesters the receptor-binding motif (one up-two down conformation). Within weeks, Zhuo and colleagues and Veesler and colleagues released the structures of ACE2-RBD complex (PDB ID 6m17; resolution 2.9 Å (Yan et al. 2020)) and the spike protein in the three-RBD-down state (PDB ID 6vxx; resolution 2.9 Å (Walls et al. 2020); Fig. 5C), respectively, both determined using single-particle cryo-EM. By July 2020, Chen and colleagues had determined the structure of the post-fusion conformation of the spike (PDB ID 6xra; resolution 3.0 Å (Cai et al. 2020)). Detailed comparison of pre- and post-fusion structures revealed conformational changes mediating fusion of viral and host cell membranes. The highest resolution structure of the prefusion spike as of mid-2022 was determined by Veesler and colleagues, who included NTD-binding antibodies to stabilize the structure of the trimer (PDB ID 7lxy; resolution 2.20 Å (McCallum et al. 2021)). Notably, Subramaniam and colleagues determined the structures of the Delta and Kappa variants of the spike protein at relatively high resolution (PDB IDs 7tey, 7tf3; resolution 2.25 Å (Saville et al. 2022)). Brunger and colleagues determined the structure of an intermediate conformation in the fusion process (PDB ID 7rzq, resolution 2.09 Å (Yang et al. 2022)). As of mid-2022, there were > 700 structures of the SARS-CoV-2 spike protein in the PDB. Many of these PDB IDs include bound monoclonal antibodies or other designed proteins, enabling mechanistic characterization of therapeutic agents and establishing opportunities discovery and development of new or modified biologics to combat emerging variants of concern (Hunt et al. 2022).
Area of Focus No. 5: cryo-electron tomography with sub-tomogram averaging
Over the past two decades, cryo-ET has emerged as an exciting new approach to structure determination of biological specimens in their native, hydrated states. Single-particle cryo-EM requires purification of biomolecules of interest, which may disrupt molecular interactions, modify environment-specific conformations, or even eliminate biologically relevant contextual information. Sample preparation for cryo-ET, in contrast, does not require isolating biological molecules from their native cellular or subcellular milieux. This in situ method provides unique opportunities for visualizing macromolecular machines exhibiting compositional and/or conformational heterogeneity, membrane-associated complexes, or high-order arrangements, all within complex native environments.
Once a 3D tomogram has been recorded by tilting the sample stage within the electron microscope and imaging projections at multiple angles, objects of interest in the electron micrographs can be extracted digitally and further processed by sub-tomogram averaging. To produce a high-resolution 3D cryo-ET density map, sub-tomograms of the same object are iteratively aligned and averaged to increase signal-to-noise ratio and address missing wedge artifacts intrinsic to cryo-ET due to mechanical tilt limitations of the sample stage. Because cryo-ET data collection is lower throughput and more computationally demanding than single-particle cryo-EM, sub-tomogram averaging is a less popular method of 3DEM structure determination. As of mid-2022, there were only 229 cryo-ET structures in the PDB (~ 2% of all 3DEM archival holdings).
The very first cryo-ET structures deposited into the PDB were those of rigor cross-bridges in insect flight muscle (PDB IDs 1m8q, 1mvw, 1o18, 1o19, 1o1a, 1o1b, 1o1c, 1o1d, 1o1e, 1o1f, 1o1g (Chen et al. 2002)) and cadherins visualized within desmosomes (PDB IDs 1q55, 1q5a, 1q5b, and 1q5c (He et al. 2003)). In these pioneering studies, tomograms of chemically preserved samples were used to generate density envelopes for positioning of known PDB structures. Despite being low resolution, both studies delineated domain organization and interactions between subunits. For the insect flight muscle case, atomic-level PDB structures of actin (PDB ID 1atn (Kabsch et al. 1990)) and myosin sub-fragment (PDB ID 2mys (Rayment et al. 1993)) positioned as rigid bodies by real-space refinement yielded new interaction information. These pioneering studies propelled cryo-ET towards the forefront of structural biology and laid the groundwork for integrative, multiscale structural biology combining data from complementary techniques to reveal more a complete “picture” of proteins visualized in their native environment.
Cryo-ET has come a long way since the early 2000s. Over the past two decades, technical advances in cryo-electron microscopy, DEDs, and state-of-the-art software for automated data acquisition and image processing (to name a few, M (Tegunov et al. 2021), emClarity (Himes and Zhang 2018), EMAN2 (Chen et al. 2019)) have led to significant improvements in 3D structure determination via sub-tomogram averaging. In 2020, structures of capsid domain (CA) in lentivirus equine infectious anemia virus (EIAV) immature Gag lattices assembled at two different pH conditions were resolved at sub-4 Å resolution via sub-tomogram averaging (PDB IDs 6t61, 6t64, and 6t63 (Dick et al. 2020)). In 2021, 3DEM structures of the 70S ribosome in Mycoplasma pneumoniae cells (PDB IDs 7ood and 7p6z (Xue et al. 2021)) were determined at 3.4 Å resolution. These PDB structures highlight the rapid growth and maturation of cryo-ET as a mainstream tool for structure determination at near-atomic resolutions and its potential for revealing complex structures involved in dynamic processes within organelles and cells.
Most early cryo-ET studies were focused on enriched organelles, membrane fractions, or small prokaryotic cells, because electron beams of transmission electron microscopes are unable to penetrate thicker eukaryotic cells. Sample milling using focused ion beams at cryogenic temperatures (cryo-FIB) was developed to prepare thinned samples without unduly damaging the specimen. This method can produce compression-free, electron-transparent lamellae, substantially extending the range of biological samples that can be investigated by cryo-ET. As of mid-2022, the highest resolution cryo-ET structure in the PDB employing cryo-FIB milling (3.3 Å) was that of RuBisCO visualized within native Halothiobacillus neapolitanus carboxysomes (PDB ID 7bzt (Cui et al. 2020)).
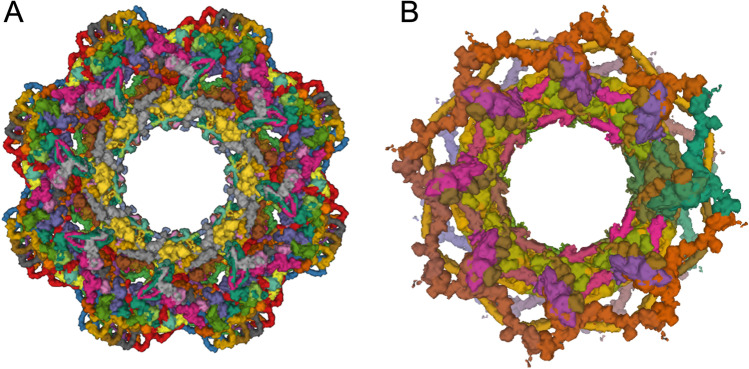
Immediate-term prospects for cryo-FIB milling followed by cryo-ET combined with sub-tomogram averaging brightened considerably with the advent of AlphaFold 2 (Jumper et al. 2021) and RoseTTAFold (Baek et al. 2021). For example, computed structure models of human nuclear pore complex (NPC) proteins from AlphaFold DB (Varadi et al. 2022) were combined with cellular cryo-ET and molecular dynamics simulations to generate composite 3DEM density maps of the human NPC in both dilated and constricted conformations (PDB IDs 7r5k, 7tbl, 7tbm, 7tbj, 7tbk, and 7tbi (Mosalaganti et al. 2022). Figure 6A illustrates a projection view of a nominal 12 Å resolution structure of the human NPC in its constricted state. The human NPC consists of ~ 1000 distinct polypeptide chains, nearly double that of the yeast NPC discussed in Area of Focus No. 6 and illustrated in Fig. 6B.
Fig. 6.
A Cryo-ET structure of the eightfold symmetric human NPC in its constricted state determined at 12 Å resolution (PDB ID 7r5k (Mosalaganti et al. 2022)). B Integrative structure of yeast NPC with eight spokes (PDBDEV_00000012) determined using the Integrative Modeling Platform (Kim et al. 2018). Images generated using Mol*
With advances in sample preparation, data processing, and protein structure prediction, cryo-ET has become a versatile tool for capturing the structural dynamics and spatial arrangements of macromolecular machines within cellular landscapes. Ongoing efforts in optimizing correlative light-electron microscopy will facilitate identification of macromolecular assemblies present in what are often crowded cellular environments. Additionally, use of artificial intelligence and machine learning-based algorithms to accurately annotate individual molecular assemblies within cells has already played a significant role in structure interpretation and bias-free, high-throughput sub-tomogram extraction for downstream sub-tomogram analysis (Chen et al. 2017; Che et al. 2018; Moebel et al. 2021). These exciting developments will open new opportunities for studying a broader range of biological systems in situ at unprecedented resolution.
Area of Focus No. 6: PDB-Dev integrative structures largely reliant on 3DEM
PDB-Dev (pdb-dev.wwpdb.org, (Vallat et al. 2018; Vallat et al. 2021; Burley et al. 2017)) is the wwPDB prototype repository for archiving integrative structures of large macromolecular assemblies. Integrative or hybrid methods structure determination involves measuring data with complementary experimental methods and combining this information with previously determined 3D structures or computed structure models of individual components to assemble a set of spatial restraints. “Integrative structures” are then determined through an iterative computational process known as satisfaction of spatial restraints (Rout and Sali 2019). The resulting integrative structures may include individual atomic coordinates and/or coarse-grained representations. Experimental tools commonly used in integrative structure determinations include cryo-EM and cryo-ET, small-angle scattering (SAS), chemical crosslinking mass spectrometry (CX-MS), hydrogen–deuterium exchange mass spectrometry, Forster resonance energy transfer, and electron paramagnetic resonance spectroscopy.
In the face of growing interest in integrative structural biology, the wwPDB established an integrative/hybrid methods task force to identify and recommend how best to address challenges involved in archiving, validating, visualizing, and disseminating these structures (Sali et al. 2015; Berman et al. 2019). The first order of business was creation of data representations for different kinds of experimental restraints. To support PDB-Dev, the PDBx/mmCIF data representation (Westbrook et al. 2022; Westbrook et al. 2005; Fitzgerald et al. 2005; John D. Westbrook and Fitzgerald 2009) that underpins the PDB archive was extended to represent integrative structures and associated experimental information. In addition, new software tools, a data harvesting system, and a website for data delivery were built (Vallat et al. 2021, 2019). At the time of writing, work is underway to develop methods for validating integrative structures based on the recommendations of the task force (Berman et al. 2019).
PDB-Dev was launched in 2016 with three integrative structures determined using the Integrative Modeling Platform (Russel et al. 2012) (Nup87 (PDBDEV_00000001 (Shi et al. 2014)); Exosome (PDBDEV_00000002 (Shi et al. 2015)); and Mediator (PDBDEV_00000003 (Robinson et al. 2015)). As of mid-2022, PDB-Dev holdings encompassed nearly 100 publicly released structures (or entries), plus some fully processed entries to be released on publication. The inaugural PDB-Dev integrative structure based largely on 3DEM data is that of the Mediator complex (PDBDEV_00000003 (Robinson et al. 2015)). Currently, there are 20 structures in PDB-Dev (including 19 released structures and 1 on hold pending publication) largely based on 3DEM. Ten of these 20 3DEM-based PDB-Dev structures also used distance restraints measured using CX-MS.
In 2018, an integrative structure of the yeast NPC consisting of 552 polypeptide chains was determined with the Integrative Modeling Platform (Fig. 6B) at sub-nanometer precision (Kim et al. 2018). The corresponding PDB-Dev submission consists of three related entries: PDBDEV_00000010 (single spoke), PDBDEV_00000011 (3 spokes), and PDBDEV_00000012 (8 spokes). Experimental restraints for the yeast NPC were obtained from single-particle cryo-EM, 2D EM class averages, SAS, and CX-MS. 3D structures of individual NPC components were obtained from PDB, PDB-Dev, or generated via comparative protein structure modeling. The integrative structure of the yeast NPC can be sub-divided into the membrane ring, inner and outer rings, a cytoplasmic export platform, the nuclear basket and the disordered FG repeats that fill the pore. The integrative structure of the yeast NPC provided insights into underlying architectural principles, mechanisms of transport across the nuclear membrane, functional regulation, and assembly/disassembly processes and broadened our understanding of the evolutionary origins of NPCs (Akey et al. 2022; Petrovic et al. 2022; Bley et al. 2022; Zimmerli et al. 2021; Allegretti et al. 2020; Mosalaganti et al. 2018).
3DEM structure validation by the wwPDB
As is the case for MX and NMR, validation standards for 3DEM structures archived in the PDB are being developed collaboratively by the wwPDB and community experts. The inaugural wwPDB 3DEM Validation Task Force (VTF) Workshop (Henderson et al. 2012) provided initial recommendations, including use of FSC for objective assessment of 3DEM density map resolution (Rosenthal and Henderson 2003). The wwPDB 3DEM VTF also recommended development of new criteria for evaluation of 3DEM density maps and emphasized the importance of statistically rigorous assessment of the fit of atomic coordinates to 3DEM density maps.
Based on the outcome of the 2011 Data Management Challenges in 3DEM Workshop (Patwardhan et al. 2012), new services for 3DEM data depositors were developed, including the EMPIAR archive (Iudin et al. 2016), standalone FSC and tilt-pair services (Patwardhan and Lawson 2016; Wasilewski and Rosenthal 2014), and Visual Analysis web pages (Abbott et al. 2018; Lagerstedt et al. 2013).
Following the onset of the 3DEM resolution revolution (Kuhlbrandt 2014), there was substantial growth in the number of moderate-to-high resolution 3DEM structures deposited to PDB, inducing community experts to update their recommendations for archiving and validating 3DEM structures and experimental 3DEM density maps and related metadata. The EMDataResource (EMDR) has organized highly influential community challenges to assess ongoing improvements in both structure determination software and 3DEM structure and density map validation (Lawson et al. 2020, 2016). In 2016, EMDR sponsored two separate challenges (Heymann et al. 2018). One evaluated density map generation while the other evaluated atomic coordinates-to-map fitting, concluding at the 2017 EMDR Joint Challenges Workshop (Lawson and Chiu 2018). At the Workshop, it became painfully apparent that different practitioners can arrive at very different 3DEM density map resolution estimates from exactly the same data. Community-wide adoption of a standardized method for estimating 3DEM density map resolution was identified as being critical for the well-being of the field. Thereafter, deposition of experimental half-maps became mandatory, enabling consistent, objective assessment of resolution using the FSC = 0.143 criterion by the wwPDB OneDep software system for every 3DEM structure deposited to PDB and every 3DEM density map deposited to EMDB.
At the time of writing, the FSC = 0.143 sigma criterion had been broadly adopted within the 3DEM community. Alternative cutoffs such as the FSC = 0.5 sigma criterion, and the half-bit criterion have been derived from first principles and/or empirically (van Heel and Schatz 2005). For example, FSC = 0.5 corresponds to the resolution at which the signal-to-noise ratio is unity (Baldwin and Lyumkis 2020). Although reducing resolution to a single, consistent number with FSC = 0.143 has clear practical value, no single number can fully encapsulate resolution when it comes to 3DEM density maps. Resolution may vary as a function of direction (Baldwin and Lyumkis 2020) or spatial location within the sample (Nakane et al. 2018), and frequency-dependent fall off in the signal-to-noise ratio can differ among density maps of similar nominal resolution. The most common reason for resolution to vary by direction is non-uniform sampling that occurs when the biological specimen assumes a preferred orientation on the planar EM grid, typically because of adherence to the air–water interface or substrate-water interface at the edge of the layer of cryopreserved liquid (Baldwin and Lyumkis 2020). Approaches to conveying more information concerning density map resolution include: depositing a full FSC curve for the entire density map, using a tool such as 3DFSC (Tan et al. 2017) to assess directional anisotropy, or using tools to assess local variations in resolution, such as ResMap (Kucukelbir et al. 2014) or MonoDir (Vilas et al. 2020).
Following the 2019 Cryo-EM model challenge, the 2019 Model Metrics Workshop was convened to assess findings and make recommendations (Lawson et al. 2021). Key workshop recommendations were as follows: (a) archives can independently estimate resolution by FSC from deposited unmasked, minimally filtered half-maps to eliminate differences observed in depositor-derived resolution estimates; (b) EMRinger (Barad et al. 2015) scores reflect map and model quality for certain amino acid residue sidechains; (c) Q-scores (Pintilie et al. 2020) reflect both local 3DEM density map quality and fit of atomic coordinates to these maps; (d) CaBLAM (Williams et al. 2018) and Molprobity (Chen et al. 2010) cis-peptide detection can be used to evaluate protein backbone conformation; (e) 3DEM map density-based cross-correlation scores (Farabella et al. 2015; Afonine et al. 2018; Joseph et al. 2017; Vasishtan and Topf 2011) and atom inclusion (Lagerstedt et al. 2013) can be used to evaluate atomic coordinate model-to-map fit; (f) Z-scores can be used instead of raw scores for several metrics evaluated; and (g) use of refined atomic displacement parameters (ADPs) or B-factors for 3DEM should be investigated.
At the time of writing, wwPDB validation reports for 3DEM structures include: (a) assessment of model geometry similar to that used for all MX and NMR structures (ClashScores, Ramachandran outliers, sidechain outliers, nucleic acid polymer backbone outliers); (b) orthogonal projections of map and map-model overlays; (c) half-map FSC plot based on mandatory half-maps collected at deposition; (d) voxel-value distribution and volume-estimation graph; (e) evaluation of map-model fit via atom-inclusion plot and residue inclusion analysis; and (f) finer evaluation of resolvability and atomic coordinate model-to-map fit, incorporating both overall and per residue Q-scores (Pintilie et al. 2020). EMDB also provides 3DEM density map and structure quality assessments on its website, including Q-scores (Z. Wang et al. 2022).
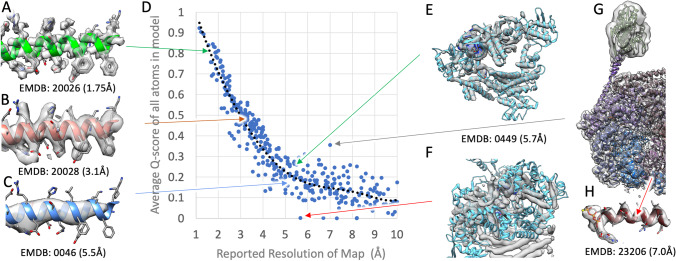
Now that Q-scores are being used more widely to assess 3DEM density maps archived in EMDB, experience has confirmed their utility as measures of both resolvability and atomic coordinate model-to-map fit. Figure 7 illustrates how resolvability of maps at different resolutions is reflected in overall Q-score values (Fig. 7A–C). Even at lower resolution, an atypical Q-score value near zero can signify an improper model-to-map fit, as shown by Fig. 7E (re-aligned corrected fit, Q-score ~ 0.27) and Fig. 7F (mis-aligned incorrect fit, Q-score ~ 0). Figure 7G and H illustrate a 3DEM structure and related experimental density map for which the reported resolution is ~ 7.0 Å, although overall resolvability is higher as indicated by a Q-score ~ 0.36, which is well above the expected Q-score at that resolution. Comparison of the 3DEM density map and the atomic model confirms that while some parts of the structure are not well resolved, much of the density map has resolvability approaching ~ 4 Å resolution (e.g., helix pitch is clearly visible in Fig. 7H). Overall Q-score values can, therefore, be used for independent assessment of depositor reported resolution (which may not reflect the value computed by the wwPDB using the FSC = 0.143 sigma criterion where 3DEM density half maps are available), and as an aid to identifying incorrectly fitted atomic coordinates.
Fig. 7.
Extracted portions of 3DEM density maps and corresponding atomic models shown in Panels A–C, with arrows indicating their overall Q-score values in the plot of Q-score versus reported density map resolution plot (Panel D). The plot was based on 374 EMDB density maps released between 2018 and 2021, randomly chosen such that resolution is evenly distributed between ~ 1 and ~ 10 Å. Panels E and F show for PDB ID 6nme/EMD-0449 (H. Zhang et al. 2019) that even at a lower resolution (~ 5.5 Å), an atypical Q-score value near zero can indicate an improper global fit of the atomic coordinates to the 3DEM density map. Panels G and H show PDB ID 7l6n/EMD-23206 (Yin et al. 2021) for which the reported resolution is ~ 7.0 Å. Actual resolvability is higher as indicated by the Q-score of ~ 0.36 (versus the value of ~ 0.16 expected at the reported resolution)
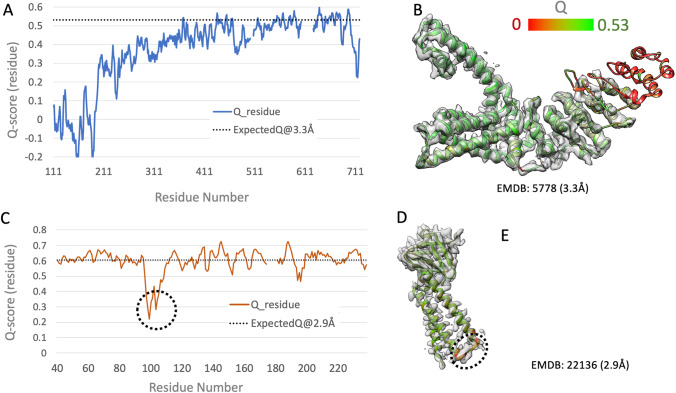
The relationship between Q-score and resolution depicted in Fig. 7D can be also used together with per-residue Q-score plots and 3DEM density map/atomic coordinate model visualization to identify regions of the structure wherein the density map is less well resolved than expected (Fig. 8A, B). Some samples studied using 3DEM are dynamic and conformationally heterogeneous under identical experimental conditions on the same EM grid. 3DEM density maps are typically less well resolved in such cases, particularly when averaging has been performed across large numbers of conformationally distinct particles. The FSC = 01.43 sigma criterion-based resolution of such 3DEM density maps may reflect the well-resolved portions of the sample. Fitted atomic coordinates can also be annotated with per-residue Q-scores to identify parts of structure that are less well resolved. Detailed comparison of 3DEM structures and related density maps, available respectively from PDB and EMDB, sometimes reveals that the atomic coordinates were not correctly fit to the experimental density map. Such cases typically exhibit lower-than-expected Q-scores, as shown in Fig. 8C–E for PDB ID 6xdc/EMD-22136. The plot of per residue Q-score versus residue number (Fig. 8C) reveals a deep minimum in the vicinity of residue number 100, wherein the computed Q-score is significantly lower than the value expected at 2.9 Å resolution (horizontal dotted line). Visual inspection of the 3DEM density map overlaid with the atomic model (Fig. 8D, E) reveals inconsistencies between the atomic coordinates and the experimental density map for the loop segment of the polypeptide chain connecting two long -helices. In contrast, the atomic coordinate model-to-map fit for the pair of -helices is entirely consistent (Fig. 7D). The two use cases presented in Fig. 8 exemplify the value of careful scrutiny of per-residue Q-scores and the fit of atomic coordinates to the density map prior to deposition of 3DEM structures to PDB and density maps to EMDB using the wwPDB OneDep software system. PDB depositors are strongly encouraged to use the wwPDB standalone validation system (https://validate.wwpdb.org) both during structure determination and before depositing any 3DEM structure to the PDB or 3DEM density maps to EMDB via OneDep.
Fig. 8.
Per-residue Q-scores can be used to assess resolvability (panels A and B) and identify opportunities to improve atomic coordinate model-to-map fit (panels C–E). Panels A and C show per-residue Q-scores versus residue number for PDB ID 3j5p/EMD-5778 (resolution 3.3 Å (Liao et al. 2013)) and PDB ID 6xdc/EMD-22136 (resolution 2.9 Å (Kern et al. 2021)), respectively. Average per-residue Q-score for reported resolution of the 3DEM density map resolution is shown as a horizontal dotted line, based on the dotted line fit to overall Q-scores versus reported density map resolution (Fig. 7D). Panels A and B illustrate how per-residue Q-scores falling below expected average values can be used to identify segments of the polypeptide chain that are not well resolved (given the reported density map resolution). Panels C–E illustrate how per-residue Q-scores falling below expected average values can be used to identify segments of the polypeptide chain wherein the atomic coordinate model-to-map fit is not consistent
Future perspectives
Looking ahead, the prospects of single-particle cryo-EM, cryo-ET, and microED as 3D biostructure detemination methods appear very bright. There is every reason to believe that the number of 3DEM structures and density maps deposited annually to the PDB and EMDB, respectively, will continue to increase year-on-year for some time. It also appears likely that the number of single-particle cryo-EM studies yielding only 3DEM density maps with no accompanying atomic-level PDB structure(s) will decline as instrumentation, methodology, and software continue to improve (and journal editors and referees “raise the bar” for publication). When, if ever, the number of annual PDB depositions of cryo-ET structures coming from sub-tomogram averaging will begin to rival the productivity of single-particle cryo-EM practitioners is not clear. But individual in situ structures could well have a much larger impact on our understanding of the inner workings of organelles and cells than many single-particle cryo-EM structures. Whatever the merits of cryo-ET versus single-particle cryo-EM structures, open access to computed structure models of essentially any protein can only serve to accelerate progress for both techniques with enormous potential benefits accruing to basic and applied research in fundamental biology, biomedicine, bioengineering, biotechnology, and energy sciences.
At the time of writing, price inflation had returned to many economies around the world, as “too many dollars chasing too few goods” made each dollar less valuable. Structural biology as a discipline and the “market for 3D structures of biological macromolecules,” in contrast, do not appear to obey the laws of macroeconomics. Between the beginning of 2013 and the end of 2021, the PDB more than doubled in terms of number of structures archived (growing from 86,184 to 185,472). In late 2022 or early 2023, the total number of structures stored in the PDB will almost certainly exceed 200,000. But structural biologists and the structures they deposit to the PDB are not perceived as declining in value. If anything, 3D biostructures being published in 2022 are viewed as more valuable, as are structural biologists (particularly those with major accomplishments in cryo-EM or cryo-ET).
3D structures of biological macromolecules are not, of course, the same as dollar bills. Relentless growth in the PDB has been accompanied by increased complexity of newly deposited 3D biostructures. Average PDB structure size (i.e., assessed by the number of amino acid and nucleotide residues comprising the sample) has increased (Burley et al. 2022a). The same is true for the average number of distinct polymer chains/PDB ID and the average number of small-molecule ligands/PDB ID (Burley et al. 2022a). Simply put, while the number of structures archived in the PDB has grown, newly deposited structures are becoming more “interesting” and, hence, more valuable to the research community. Some of the growth in structure size and structure complexity can be attributed to the success of the 3DEM “Resolution Revolution.” Structural biologists are no longer hostages to what can be crystallized or rendered adequately soluble in an NMR tube. They are putting ever larger, ever more complex macromolecular machines onto EM grids, and, in favorable cases, “seeing” at the atomic level how the individual protein and nucleic acid chains are arranged, and how individual macromolecules recognize one another and how they bind to small-molecule ligands, such as enzyme co-factors, substrates, inhibitors, investigational agents, and US FDA-approved drugs.
In due course, however, today’s electron microscopists are likely to find, as previous generations of structural biologists did with MX and NMR, that much of the “low-hanging fruit” has been picked. They will turn, as some have already done, to integrative or hybrid methods to tackle challenging systems wherein NMR, MX, or 3DEM are not by themselves sufficient for structure determination. Instead, these three mainstays of structural biology and the PDB will come to be viewed as important tools that must often be combined with other biophysical measurement techniques to determine 3D structures of larger and more complex experimental systems. This trend is already evidenced by the growing number of protein crystallographers reinventing themselves as electron microscopists, driven by the conviction that in biology “function follows form.” Beginning with the structures of the DNA duplex (Watson and Crick 1953) and sperm whale myoglobin (Kendrew et al. 1960), generations of structuralists have shown that it can pay handsomely to study systems in 3D at the atomic level in order to understand biological phenomena and it can also pay to be nimble.
Acknowledgements
The authors thank the tens of thousands of structural biologists who deposited structures to the PDB since 1971 and the many millions of researchers, educators, and students around the world who consume PDB data. We also gratefully acknowledge contributions to the success of the PDB archive made by past members of RCSB PDB and our Worldwide Protein Data Bank partners (PDBe, PDBj, EMDB, and BMRB).
Funding
RCSB PDB core operations are jointly funded by the National Science Foundation (NSF; DBI-1832184, PI: S.K. Burley), the US Department of Energy (DOE; DE-SC0019749, PI: S.K. Burley), and the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Institute of General Medical Sciences of the National Institutes of Health (NIH-NIGMS; R01GM133198, PI: S.K. Burley). Other funding awards to RCSB PDB by the NSF and to PDBe by the UK Biotechnology and Biological Research Council are jointly supporting development of a Next Generation PDB archive (DBI-2019297, PI: S.K. Burley; BB/V004247/1, PI: Sameer Velankar) and new Mol* features (DBI-2129634, PI: S.K. Burley; BB/W017970/1, PI: Sameer Velankar). EMDR was supported by NIH-NIGMS (R01GM079429, PI: W. Chiu). The Nucleic Acid Database is supported by NIH-NIGMS (R01 R01GM085328, PI: Craig Zirbel with subcontract to C.L. Lawson). PDB-Dev is supported by NSF (DBI-1756248 and DBI-2112966, PI:B. Vallat; DBI-1756250 and DBI-2112967, PI A. Sali). Sali received additional support from NIH-NIGMS (R01GM083960, PI:A. Sali; P41GM109824, PI: M.P. Rout). Dai received support from start-up funds provided by the Rutgers University Institute for Quantitative Biomedicine and NSF (MCB-2046180, PI:W. Dai). Kaelber received support from start-up funds provided by the Rutgers University Institute for Quantitative Biomedicine and NIH-NIGMS (R21GM140345, PI: J.T. Kaelber). Khare received support from NIH-NIGMS (R01GM132565, PI: S.D. Khare) and NSF (CBET-1929237, PI: S.D. Khare). Kulczyk a received support from start-up funds provided by the Rutgers University Institute for Quantitative Biomedicine, Busch Biomedical Grant (PI: A.W. Kulczyk), and NIH-NIAID (R01AI141635, PI: X-P. Li). Molecular graphics and analyses for Q-score were performed using UCSF Chimera v1.13 developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311, and MapQ Plugin v1.9.5 developed and maintained by G. Pintile (https://github.com/gregdp/mapq).
Declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
John D. Westbrook passed away during the preparation of this paper.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbott S, Iudin A, Korir PK, Somasundharam S, Patwardhan A. EMDB Web Resources. Curr Protoc Bioinformatics. 2018;61(1):5.10.1–5.10.12. doi: 10.1002/cpbi.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol. 2018;74(Pt 9):814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, et al. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell. 2022;185(2):361–378 e25. doi: 10.1016/j.cell.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, et al. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature. 2020;586(7831):796–800. doi: 10.1038/s41586-020-2670-5. [DOI] [PubMed] [Google Scholar]
- Armstrong DR, Berrisford JM, Conroy MJ, Gutmanas A, Anyango S, Choudhary P, et al. PDBe: improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020;48(D1):D335–D343. doi: 10.1093/nar/gkz990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin PR, Lyumkis D. Non-uniformity of projection distributions attenuates resolution in Cryo-EM. Prog Biophys Mol Biol. 2020;150:160–183. doi: 10.1016/j.pbiomolbio.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at a 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12(10):943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker GJ, Yokochi M, Suzuki H, Ikegawa Y, Iwata T, Kudou T, et al. Protein Data Bank Japan: Celebrating our 20th anniversary during a global pandemic as the Asian hub of three dimensional macromolecular structural data. Protein Sci. 2022;31(1):173–186. doi: 10.1002/pro.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588(7837):327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr A. 2008;64(1):88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The Protein Data Bank at 40: reflecting on the past to prepare for the future. Structure. 2012;20(3):391–396. doi: 10.1016/j.str.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Adams PD, Bonvin AA, Burley SK, Carragher B, Chiu W, et al. Federating structural models and data: outcomes from a workshop on archiving integrative structures. Structure. 2019;27(12):1745–1759. doi: 10.1016/j.str.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, et al. Architecture of the cytoplasmic face of the nuclear pore. Science. 2022;376(6598):eabm9129. doi: 10.1126/science.abm9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Kurisu G, Markley JL, Nakamura H, Velankar S, Berman HM, et al. PDB-Dev: a prototype system for depositing integrative/hybrid structural models. Structure. 2017;25(9):1317–1318. doi: 10.1016/j.str.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow G, et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acid Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Duarte JM, Feng Z, Flatt JW, Hudson BP, et al. Protein Data Bank: A comprehensive review of 3D Structure holdings and worldwide utilization by researchers, educators, and students. Biomolecules. 2022;12:1425. doi: 10.3390/biom12101425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, et al. RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Sci. 2022;31(1):187–208. doi: 10.1002/pro.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S. K., Bhikadiya, C., Bi, C., Bittrich, S., Chao, H., Chen, L et al (2022b) RCSB Protein Data Bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci e4482. 10.1002/pro.4482 [DOI] [PMC free article] [PubMed]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328(5977):501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Jr, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369(6511):1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504(7478):113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone CE, Loveland AB, Gamper HB, Jr, Hou YM, Demo G, Korostelev AA. Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat Commun. 2021;12(1):7236. doi: 10.1038/s41467-021-27415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Che C, Lin R, Zeng X, Elmaaroufi K, Galeotti J, Xu M. Improved deep learning-based macromolecules structure classification from electron cryo-tomograms. Mach vis Appl. 2018;29(8):1227–1236. doi: 10.1007/s00138-018-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Winkler H, Reedy MK, Reedy MC, Taylor KA. Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle. J Struct Biol. 2002;138(1–2):92–104. doi: 10.1016/S1047-8477(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Dai W, Sun SY, Jonasch D, He CY, Schmid MF, et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat Methods. 2017;14(10):983–985. doi: 10.1038/nmeth.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Bell JM, Shi X, Sun SY, Wang Z, Ludtke SJ. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat Methods. 2019;16(11):1161–1168. doi: 10.1038/s41592-019-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Eng ET, Alink L, Rice WJ, Jordan KD, Kim LY, et al. High resolution single particle cryo-electron microscopy using beam-image shift. J Struct Biol. 2018;204(2):270–275. doi: 10.1016/j.jsb.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman PR, Agbandje-McKenna M, Renaudin J, Baker TS, McKenna R. Structural analysis of the Spiroplasma virus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure. 1998;6(2):135–145. doi: 10.1016/s0969-2126(98)00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu ML, Gilliland GL. Engineering antibody therapeutics. Curr Opin Struct Biol. 2016;38:163–173. doi: 10.1016/j.sbi.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, et al. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292(5517):744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Cui Y, Peng R, Song H, Tong Z, Qu X, Liu S, et al. Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1. Proc Natl Acad Sci U S A. 2020;117(31):18711–18718. doi: 10.1073/pnas.2005341117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick RA, Xu C, Morado DR, Kravchuk V, Ricana CL, Lyddon TD, et al. Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly. PLoS Pathog. 2020;16(1):e1008277. doi: 10.1371/journal.ppat.1008277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabella I, Vasishtan D, Joseph AP, Pandurangan AP, Sahota H, Topf M. TEMPy: a Python library for assessment of three-dimensional electron microscopy density fits. J Appl Crystallogr. 2015;48(Pt 4):1314–1323. doi: 10.1107/S1600576715010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald PMD, Westbrook JD, Bourne PE, McMahon B, Watenpaugh KD, Berman HM (2005) 4.5 Macromolecular dictionary (mmCIF). In Hall SR, McMahon B (Eds.), International Tables for Crystallography G. Definition and exchange of crystallographic data (pp. 295–443). Dordrecht, The Netherlands: Springer
- Frank J (2017) The translation elongation cycle-capturing multiple states by cryo-electron microscopy. Philos Trans R Soc Lond B Biol Sci 372(1716). 10.1098/rstb.2016.0180 [DOI] [PMC free article] [PubMed]
- Gao S, Yao X, Yan N (2021) Structure of human Cav2.2 channel blocked by the painkiller ziconotide. Nature 596(7870):143–147. 10.1038/s41586-021-03699-6 [DOI] [PMC free article] [PubMed]
- Gilliland GL, Luo J, Vafa O, Almagro JC. Leveraging SBDD in protein therapeutic development: antibody engineering. Methods Mol Biol. 2012;841:321–349. doi: 10.1007/978-1-61779-520-6_14. [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Burley SK. RCSB Protein Data Bank resources for structure-facilitated design of mRNA Vaccines for existing and emerging viral pathogens. Structure. 2022;30:252–262.e4. doi: 10.1016/j.str.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzelnik KV, Cui Z, Reed CA, Jakana J, Young R, Zhang J. Asymmetric cryo-EM structure of the canonical allolevivirus Qbeta reveals a single maturation protein and the genomic ssRNA in situ. Proc Natl Acad Sci U S A. 2016;113(41):11519–11524. doi: 10.1073/pnas.1609482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Sevvana M, Kuhn RJ, Rossmann MG. Structural biology of Zika virus and other flaviviruses. Nat Struct Mol Biol. 2018;25(1):13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- He Y, Bowman VD, Mueller S, Bator CM, Bella J, Peng X, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci U S A. 2000;97(1):79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302(5642):109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20(2):205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzik MA., Jr Cryo-electron microscopy reaches atomic resolution. Nature. 2020;587(7832):39–40. doi: 10.1038/d41586-020-02924-y. [DOI] [PubMed] [Google Scholar]
- Hewat EA, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, et al. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 1997;16(7):1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Marabini R, Kazemi M, Sorzano COS, Holmdahl M, Mendez JH, et al. The first single particle analysis Map challenge: a summary of the assessments. J Struct Biol. 2018;204(2):291–300. doi: 10.1016/j.jsb.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BA, Zhang P. emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat Methods. 2018;15(11):955–961. doi: 10.1038/s41592-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AC, Case JB, Park YJ, Cao L, Wu K, Walls AC, et al. Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci Transl Med. 2022;14(646):eabn1252. doi: 10.1126/scitranslmed.abn1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nat Methods. 2016;13(5):387–388. doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
- Joseph AP, Lagerstedt I, Patwardhan A, Topf M, Winn M. Improved metrics for comparing structures of macromolecular assemblies determined by 3D electron-microscopy. J Struct Biol. 2017;199(1):12–26. doi: 10.1016/j.jsb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kaelber JT, Hryc CF, Chiu W. Electron cryomicroscopy of viruses at near-atomic resolutions. Annu Rev Virol. 2017;4(1):287–308. doi: 10.1146/annurev-virology-101416-041921. [DOI] [PubMed] [Google Scholar]
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, et al. Structure of myoglobin: a three-dimensional Fourier synthesis at 2 A. resolution. Nature. 1960;185(4711):422–7. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Kern DM, Sorum B, Mali SS, Hoel CM, Sridharan S, Remis JP, et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat Struct Mol Biol. 2021;28(7):573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555(7697):475–482. doi: 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo AR, Yamashita R, Nakamura H. PDBj Mine: design and implementation of relational database interface for Protein Data Bank Japan (Research Support, Non-U.S. Gov’t) Database (Oxford) 2010;2010:baq021. doi: 10.1093/database/baq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, et al. Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format (Research Support, Non-U.S. Gov’t) Nucleic Acids Res. 2012;40(Database issue):D453–60. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo AR, Bekker GJ, Suzuki H, Tsuchiya Y, Kawabata T, Ikegawa Y, et al. Protein Data Bank Japan (PDBj): updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res. 2017;45(D1):D282–D288. doi: 10.1093/nar/gkw962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo AR, Bekker GJ, Wako H, Endo S, Tsuchiya Y, Sato H, et al. New tools and functions in data-out activities at Protein Data Bank Japan (PDBj) Protein Sci. 2018;27(1):95–102. doi: 10.1002/pro.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T, Reteno DG, Benamar S, Hollerbach A, Colson P, La Scola B, et al. Structure of faustovirus, a large dsDNA virus. Proc Natl Acad Sci U S A. 2016;113(22):6206–6211. doi: 10.1073/pnas.1523999113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. Embo J. 1999;18(22):6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun. 2016;7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343(6178):1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Lagerstedt I, Moore WJ, Patwardhan A, Sanz-Garcia E, Best C, Swedlow JR, et al. Web-based visualisation and analysis of 3D electron-microscopy data from EMDB and PDB. J Struct Biol. 2013;184(2):173–181. doi: 10.1016/j.jsb.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Chiu W. Comparing cryo-EM structures. J Struct Biol. 2018;204(3):523–526. doi: 10.1016/j.jsb.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Dutta S, Westbrook JD, Henrick K, Berman HM. Representation of viruses in the remediated PDB archive. Acta Crystallogr D Biol Crystallogr. 2008;D64(Pt 8):874–882. doi: 10.1107/S0907444908017393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Patwardhan A, Baker ML, Hryc C, Garcia ES, Hudson BP, et al. EMDataBank unified data resource for 3DEM. Nucleic Acids Res. 2016;44(D1):D396–403. doi: 10.1093/nar/gkv1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Berman HM, Chiu W. Evolving data standards for cryo-EM structures. Struct Dyn. 2020;7(1):014701. doi: 10.1063/1.5138589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Kryshtafovych A, Adams PD, Afonine PV, Baker ML, Barad BA, et al. Cryo-EM model validation recommendations based on outcomes of the 2019 EMDataResource challenge. Nat Methods. 2021;18(2):156–164. doi: 10.1038/s41592-020-01051-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Shingler KL, Organtini LJ, Ashley RE, Makhov AM, Conway JF, et al. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv. 2016;2(8):e1501929. doi: 10.1126/sciadv.1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem. 2018;87:451–478. doi: 10.1146/annurev-biochem-062917-011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5(2):255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Martin CS, Burnett RM, de Haas F, Heinkel R, Rutten T, Fuller SD, et al. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9(10):917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. Advanced data acquisition from electron microscopes with SerialEM. Microsc Microanal. 2018;24(S1):864–865. doi: 10.1017/S1431927618004816. [DOI] [Google Scholar]
- McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–2347 e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebel E, Martinez-Sanchez A, Lamm L, Righetto RD, Wietrzynski W, Albert S, et al. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nat Methods. 2021;18(11):1386–1394. doi: 10.1038/s41592-021-01275-4. [DOI] [PubMed] [Google Scholar]
- Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salome PA, et al. In situ architecture of the algal nuclear pore complex. Nat Commun. 2018;9(1):2361. doi: 10.1038/s41467-018-04739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, et al. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science. 2022;376(6598):eabm9506. doi: 10.1126/science.abm9506. [DOI] [PubMed] [Google Scholar]
- Muhleip A, Kock Flygaard R, Ovciarikova J, Lacombe A, Fernandes P, Sheiner L, et al. ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria. Nat Commun. 2021;12(1):120. doi: 10.1038/s41467-020-20381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown P, et al. Single-particle cryo-EM at atomic resolution. Nature. 2020;587(7832):152–156. doi: 10.1038/s41586-020-2829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane, T., Kimanius, D., Lindahl, E., Scheres, S. H. (2018) Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife 7. 10.7554/eLife.36861 [DOI] [PMC free article] [PubMed]
- Noller HF, Lancaster L, Zhou J, Mohan S. The ribosome moves: RNA mechanics and translocation. Nat Struct Mol Biol. 2017;24(12):1021–1027. doi: 10.1038/nsmb.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly FJ, Xue L, Graziadei A, Sinn L, Lenz S, Tegunov D, et al. In-cell architecture of an actively transcribing-translating expressome. Science. 2020;369(6503):554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Lawson CL. Databases and archiving for CryoEM. Methods Enzymol. 2016;579:393–412. doi: 10.1016/bs.mie.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, et al. Data management challenges in three-dimensional EM. Nat Struct Mol Biol. 2012;19(12):1203–1207. doi: 10.1038/nsmb.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, et al. Architecture of the linker-scaffold in the nuclear pore. Science. 2022;376(6598):eabm9798. doi: 10.1126/science.abm9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie G, Zhang K, Su Z, Li S, Schmid MF, Chiu W. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat Methods. 2020;17(3):328–334. doi: 10.1038/s41592-020-0731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77(3–4):153–161. doi: 10.1016/S0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Protein Data Bank Crystallography: Protein Data Bank. Nature (London) New Biol. 1971;233(42):223–223. doi: 10.1038/newbio233223b0. [DOI] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Robertson MJ, Meyerowitz JG, Skiniotis G. Drug discovery in the era of cryo-electron microscopy. Trends Biochem Sci. 2022;47(2):124–135. doi: 10.1016/j.tibs.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, et al. Molecular architecture of the yeast Mediator complex. Elife. 2015;4:e08719. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rout MP, Sali A. Principles for integrative structural biology studies. Cell. 2019;177(6):1384–1403. doi: 10.1016/j.cell.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10(1):e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Berman HM, Schwede T, Trewhella J, Kleywegt G, Burley SK, et al. Outcome of the First wwPDB hybrid/integrative methods task force workshop. Structure. 2015;23(7):1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville JW, Mannar D, Zhu X, Srivastava SS, Berezuk AM, Demers JP, et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants. Nat Commun. 2022;13(1):742. doi: 10.1038/s41467-022-28324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. Processing of Structurally Heterogeneous Cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/S0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Sehnal D, Bittrich S, Deshpande M, Svobodova R, Berka K, Bazgier V, et al. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431–W437. doi: 10.1093/nar/gkab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Feng Z, Westbrook JD, Peisach E, Berrisford J, Ikegawa Y, et al. Modernized UNIFORM representation of carbohydrate molecules in the Protein Data Bank. Glycobiology. 2021;31:1204–1218. doi: 10.1093/glycob/cwab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, et al. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014;13(11):2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Pellarin R, Fridy PC, Fernandez-Martinez J, Thompson MK, Li Y, et al. A strategy for dissecting the architectures of native macromolecular assemblies. Nat Methods. 2015;12(12):1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M, Vitiello C, Schweimer K, Gottesman M, Rosch P, Knauer SH. Transcription is regulated by NusA:NusG interaction. Nucleic Acids Res. 2016;44(12):5971–5982. doi: 10.1093/nar/gkw423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K. P., Brennan-Tonetta, P., Marxen, LJ (2017) Economic Impacts of the research collaboratory for structural bioinformatics (RCSB) Protein Data Bank. 10.2210/rcsb_pdb/pdb-econ-imp-2017
- Tagari M, Newman R, Chagoyen M, Carazo JM, Henrick K. New electron microscopy database and deposition system. Trends Biochem Sci. 2002;27(11):589. doi: 10.1016/S0968-0004(02)02176-X. [DOI] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods. 2017;14(8):793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 A in cells. Nat Methods. 2021;18(2):186–193. doi: 10.1038/s41592-020-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, Afonine PV. Improvement of cryo-EM maps by density modification. Nat Methods. 2020;17(9):923–927. doi: 10.1038/s41592-020-0914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallat B, Webb B, Westbrook JD, Sali A, Berman HM. Development of a Prototype system for archiving integrative/hybrid structure models of biological macromolecules. Structure. 2018;26(6):894–904 e2. doi: 10.1016/j.str.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallat B, Webb B, Westbrook J, Sali A, Berman HM. Archiving and disseminating integrative structure models. J Biomol NMR. 2019;73:385–398. doi: 10.1007/s10858-019-00264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallat B, Webb B, Fayazi M, Voinea S, Tangmunarunkit H, Ganesan SJ, et al. New system for archiving integrative structures. Acta Crystallogr D Struct Biol. 2021;77(Pt 12):1486–1496. doi: 10.1107/S2059798321010871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aalst WMP, Bichler M, Heinzl A. Responsible Data Science (journal article) Bus Inf Syst Eng. 2017;59(5):311–313. doi: 10.1007/s12599-017-0487-z. [DOI] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151(3):250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasishtan D, Topf M. Scoring functions for cryoEM density fitting. J Struct Biol. 2011;174(2):333–343. doi: 10.1016/j.jsb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Vilas JL, Tagare HD, Vargas J, Carazo JM, Sorzano COS. Measuring local-directional resolution and local anisotropy in cryo-EM maps. Nat Commun. 2020;11(1):55. doi: 10.1038/s41467-019-13742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461(7261):292–295. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Molodtsov V, Firlar E, Kaelber JT, Blaha G, Su M, et al. Structural basis of transcription-translation coupling. Science. 2020;369(6509):1359–1365. doi: 10.1126/science.abb5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Patwardhan A, Kleywegt GJ. Validation analysis of EMDB entries. Acta Crystallogr D Struct Biol. 2022;78(Pt 5):542–552. doi: 10.1107/S205979832200328X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski S, Rosenthal PB. Web server for tilt-pair validation of single particle maps from electron cryomicroscopy. J Struct Biol. 2014;186(1):122–131. doi: 10.1016/j.jsb.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Watson, Z. L., Ward, F. R., Meheust, R., Ad, O., Schepartz, A., Banfield, J. F., et al (2020) Structure of the bacterial ribosome at 2 A resolution. Elife 9.10.7554/eLife.60482 [DOI] [PMC free article] [PubMed]
- Webster MW, Takacs M, Zhu C, Vidmar V, Eduljee A, Abdelkareem M, et al. Structural basis of transcription-translation coupling and collision in bacteria. Science. 2020;369(6509):1355–1359. doi: 10.1126/science.abb5036. [DOI] [PubMed] [Google Scholar]
- Weis F, Hagen WJH. Combining high throughput and high quality for cryo-electron microscopy data collection. Acta Crystallogr D Struct Biol. 2020;76(Pt 8):724–728. doi: 10.1107/S2059798320008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook JD, Fitzgerald PMD. Chapter 10 The PDB format, mmCIF formats, and other data formats. In: Bourne PE, Gu J, editors. Structural Bioinformatics. 2. Hoboken, NJ: John Wiley & Sons Inc; 2009. pp. 271–291. [Google Scholar]
- Westbrook JD, Young JY, Shao C, Feng Z, Guranovic V, Lawson C, et al. PDBx/mmCIF Ecosystem: Foundational semantic tools for structural biology. J Mol Biol. 2022;434:167599. doi: 10.1016/j.jmb.2022.167599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook, J. D., Yang, H., Feng, Z., Berman, H. M. (2005) 5.5 The use of mmCIF architecture for PDB data management. In S. R. Hall, & B. McMahon (Eds.), International Tables for Crystallography (pp. 539–543). Dordrecht, The Netherlands: Springer
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3(160018):1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27(1):293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wwPDB consortium Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47(D1):D520–D528. doi: 10.1093/nar/gky949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Yoshioka, C. K., Chapman, M. S. (2020) Adeno-Associated virus (AAV-DJ)-cryo-EM structure at 1.56 A resolution. Viruses 12(10). 10.3390/v12101194 [DOI] [PMC free article] [PubMed]
- Xue, L., Lenz, S., Zimmermann-Kogadeeva, M., Tegunov, D., Cramer, P., Bork, P., et al (2021) Visualizing translation dynamics at atomic detail inside a bacterial cell. bioRxiv 2021.12.18.473270. 10.1101/2021.12.18.473270
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang C, White KI, Pfuetzner RA, Esquivies L, Brunger AT. Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle. Proc Natl Acad Sci U S A. 2022;119(16):e2119467119. doi: 10.1073/pnas.2119467119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Feng X, Yu H, Fay A, Kovach A, Glickman MS, et al. Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system. Cell Rep. 2021;35(8):109166. doi: 10.1016/j.celrep.2021.109166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KM, Fischer N, Paknia E, Chari A, Stark H. Atomic-resolution protein structure determination by cryo-EM. Nature. 2020;587(7832):157–161. doi: 10.1038/s41586-020-2833-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, et al. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J Struct Biol. 2009;165(1):1–9. doi: 10.1016/j.jsb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Z, Daczkowski CM, Gabel C, Mesecar AD, Chang L. Structural basis for the inhibition of CRISPR-Cas12a by anti-CRISPR proteins. Cell Host Microbe. 2019;25(6):815–826 e4. doi: 10.1016/j.chom.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, et al. Nuclear pores dilate and constrict in cellulo. Science. 2021;374(6573):eabd9776. doi: 10.1126/science.abd9776. [DOI] [PubMed] [Google Scholar]