Dear Editor,
N6-methyladenosine (m6A) is the most abundant and prevalent internal modification in mRNA.1 In mammals, m6A exerts pivotal roles in posttranscriptional regulation and its dysregulation is implicated in various diseases including cancer.2 m6A is installed by a multicomponent methyltransferase complex (MTC, also known as the m6A writer complex).3,4 The mammalian MTC is composed of the core m6A methyltransferase METTL3–METTL14 complex (MTC core) and several regulatory proteins including WTAP, the adaptor responsible for METTL3–METTL14 localization and proper substrate recruitment,5 and VIRMA (KIAA1429), the specificity mediator that mediates preferential m6A modification at the 3′ untranslated regions (UTRs, Fig. 1a).6 Dysregulation of MTC components results in the disruptions of m6A.2 Compared to the other currently identified regulators (HAKAI, ZC3H13 and RBM15), WTAP and VIRMA are reported to have greater impacts on total mRNA m6A levels upon knockdown.5,6 Despite the advances in understanding the roles of individual MTC components and the structural determination of MTC core,7–9 the overall molecular architecture of the m6A writer holocomplex is missing. Here, we report the cryogenic electron microscopy (cryo-EM) structure of human WTAP–VIRMA (3.1 Å) in the METTL3–METTL14–WTAP–VIRMA (M–M–W–V) complex and modeled a structure of the quaternary M–M–W–V complex based on AlphaFold2 predictions and structural restraints from intermolecular chemical crosslinking mass spectrometry (CXMS).
Fig. 1. Structure of the human m6A writer complex.
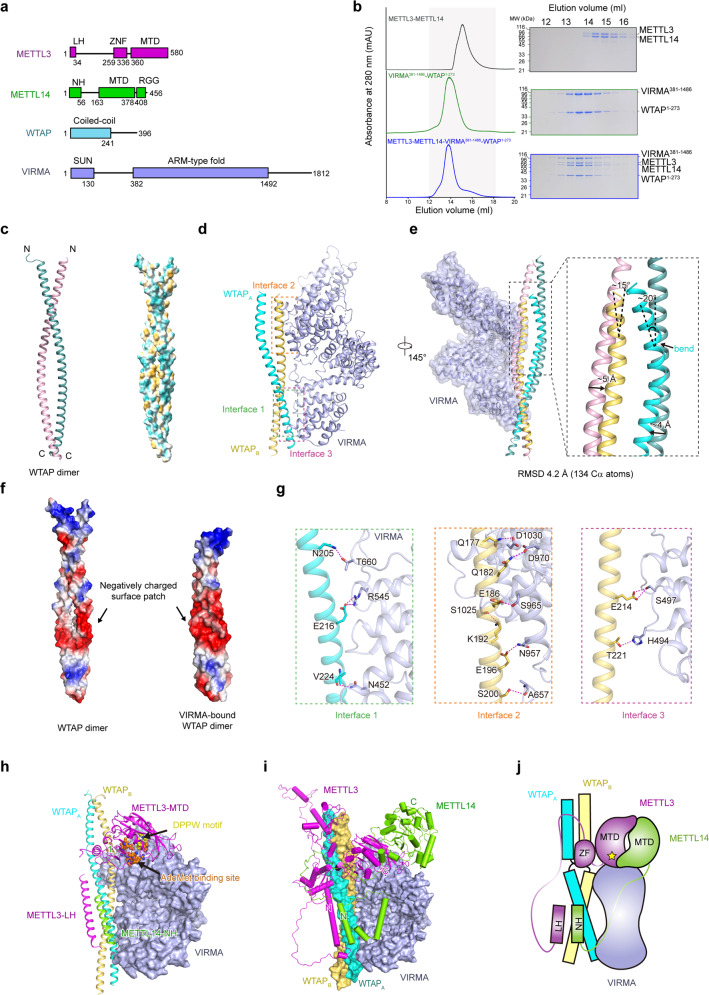
a Schematic diagram of the domain information of METTL3, METTL14, WTAP and VIRMA. LH leader helix, ZFD zinc finger domain, MTD MTase domain, NH N-terminus helix. b Gel filtration analysis of the human METTL3–METTL14–WTAP1–273–VIRMA381–1486 complex. c The crystal structure of WTAP alone. Two molecules of WTAP form a symmetric parallel α-helical coiled-coil, with the two chains colored light pink and light teal, respectively. d The cryo-EM structure of WTAP–VIRMA complex in the METTL3–METTL14–WTAP1–273–VIRMA381–1486 quaternary complex. Three interaction interfaces can be observed between WTAPs and VIRMA. WTAPA, cyan; WTAPB, orange. e Comparison of the crystal structure of WTAP alone and the cryo-EM structure of VIRMA-bound WTAPs. VIRMA-bound WTAPs bend around the observed N-termini. f Electrostatic surfaces of WTAP alone (PDB: 7YFJ) and VIRMA-bound WTAPs (PDB: 7YG4). g Details of the observed interaction interfaces between WTAP and VIRMA. h, i Structural model of the METTL3–METTL141–399–WTAP148–237–VIRMA342–1292 complex based on CXMS measurement and AI prediction. METTL3 and METTL14 are shown in green and magenta, respectively. METTL3 LH is docked close to WTAP, and the MTC core catalytic center is docked close to VIRMA, with the DPPW motif and AdoMet binding site positioned towards a cleft between WTAP and VIRMA. j Schematic diagram of the modeled METTL3–METTL141–399–WTAP148–237–VIRMA342–1292 complex.
Utilizing a baculovirus-insect cell expression system, we obtained the METTL3–METTL14 complex (Fig. 1b) and investigated the interaction between METTL3–METTL14, WTAP, and VIRMA through co-expression and purification. WTAP was observed to form a complex with the MTC core, while VIRMA can only form a stable complex with the MTC core in the presence of WTAP (Supplementary information, Fig. S1). The methyl transfer activity of the quaternary M–M–W–V complex was significantly higher than that of the MTC core, yet a quaternary M–M–W–V complex containing a truncated VRIMA381–1486 with N- and C-terminal disordered regions removed showed comparable activity to the MTC core (Supplementary information, Fig. S2). In parallel, we obtained the WTAP protein alone (residues 130–241, WTAP130–241) using the Escherichia coli expression system and determined the crystal structure of the WTAP (2.40 Å, Supplementary information, Table S1). Two molecules of WTAP (with the assigned density of residues 150–241, WTAP150–241) formed a symmetric parallel α-helical coiled-coil homodimer through hydrophobic interactions between two identical surface patches. Specifically, the side chains of residues M158, I165, L183, L204, I208, L211, V215, M218, I222, L225, L229 and L236 from the two WTAPs formed a hydrophobic interaction network (Fig. 1c; Supplementary information, Fig. S3a).
We further purified the METTL3–METTL14–WTAP1–273–VIRMA381–1486 complex for gradient fixation and cryo-EM analysis (Fig. 1b). Unexpectedly, in the cryo-EM structure, only WTAP171–237 and VIRMA381–1292 (with the stoichiometric ratio 2:1) and an unassigned density close to WTAP were observed (Supplementary information, Figs. S4, S5). The density for the rest of the quaternary complex was missing, suggesting high flexibility (Supplementary information, Table S2). The observed WTAP171–237–VIRMA381–1292 in the quaternary complex adopted a flag-and-pole-like architecture with dimensions of ~116 Å × 103 Å × 80 Å (Fig. 1d). Two WTAP171–237 molecules (designated WTAPA and WTAPB) formed an asymmetric homodimer (the “flag pole”) while VIRMA381–1292 that contains 17 armadillo-like (ARML) repeats (designated ARML 1–17) formed a twisted α-solenoid-like superhelix (the “flag”). Of the ARML repeats, each of ARML 2–4, 6, 7, 9, 12, 15, and 17 consists of three helices while the rest are two-helix units. Two long helices in between of ARML 4–5 and of ARML 7–8 mediate the turning of the VIRMA α-solenoid, possibly accounting for the formation of the twisted architecture (Supplementary information, Figs. S6, S7).
The structure of the asymmetric WTAP171–237 dimer in the quaternary complex is similar to the crystal structure of the symmetric WTAP150–241 dimer with a root-mean-square deviation (RMSD) of 4.2 Å over 134 Cα atoms (Fig. 1e). However, compared to the WTAPs alone, VIRMA-bound WTAPs bent around the observed N-termini (around WTAPA residue 188 and WTAPB residue 185) and displayed a more compact and dense conformation with distinct intramolecular interactions (Fig. 1e). Hydrophobic interactions were found between WTAPA and WTAPB residues 180–236 through the side-chain interactions of WTAPA–WTAPB residue pairs L183A–I180B (L183 from WTAPA and I180 from WTAPB), L187A–L183B, I208A–L204B, L211A–I208B, V215A–L211B, M218A–V215B, I222A–M218B, L225A–I222B, L229A–L225B, and L236A–L229B. WTAP K191A and Q201A side chains formed hydrogen bonds with Q190B and Q201B side chains, respectively. Hydrogen bond interactions were also observed between WTAP Q201A side chain and K198B main chain and between N205A main chain and Q201B side chain. These together contributed to the formation of the compact VIRMA-bound WTAP dimer that harbors a contiguous negatively charged surface patch involved in the interaction with VIRMA (Fig. 1e, f). Some of these residues involved in WTAPA–WTAPB interactions (around residues 180–236) are also conserved in Danio rerio and Drosophila melanogaster WTAPs (Supplementary information, Fig. S3c).
Three intermolecular interaction interfaces, designated Interfaces 1, 2, and 3, could be observed between the WTAP171–237 dimer and VIRMA381–1292 with buried areas of ~773.9 Å2, ~1155.6 Å2, and ~101 Å2, respectively (Fig. 1d). Interface 1 is located in between WTAPA (residues 205–224) and VIRMA (residues 452–660). Specifically, the side chains of WTAP N205A and E216A and VIRMA T660 and R545 formed hydrogen bonds, and E216A and VIRMA R545 formed a salt bridge. V244A main chain and VIRMA N452 side chain formed a hydrogen bond. Interfaces 2 and 3 are in between of WTAPB (residues 177–200, 214–221) and VIRMA (residues 657–1030, 494–497). The side chains of WTAP Q177B, Q182B, E186B, K192B, E196B, E214B, and T221B formed hydrogen bonds with the side chains of VIRMA D1030, D970, S965, S1025, N957, S497, and H494, respectively. WTAP S200B and E203B side chains formed hydrogen bonds with VIRMA A657 and M656 main chains, respectively (Fig. 1g; Supplementary information, Fig. S8a, b).
We further investigated the importance of the residues involved in the formation of Interfaces 1–3 via alanine substitutions and subsequent co-expression and purification assays (Supplementary information, Fig. S8). Specifically, Flag-tagged WTAP1–273 and His-tagged VIRMA381–1486 were co-expressed in the human embryonic kidney (HEK) 293F cells. The protein mixtures from the whole-cell extracts were subjected to western blot analysis against the tagged proteins and purification via anti-Flag beads. The single WTAP mutations (Q177A, Q182A, Q192A, E196A, E203A, N205A, or E216A) had little influence on WTAP–VIRMA381–1486 interaction, whereas the multi-alanine substitution mutant WTAPQ177A/Q182A/K192A/E196A/E203A lost VIRMA381–1486-binding capacity (Supplementary information, Fig. S8c). The VIRMA381–1486 mutants containing individual N452A, T660A, N957A, or S1025A mutations retained binding to WTAP1–273, while the single alanine substitution of VIRMA381–1292 residue R545 or D970 abolished its interaction with WTAP1–273 (Supplementary information, Fig. S8d).
Despite that gel filtration analysis clearly indicates the presence of METTL3 and METTL14 in the METTL3–METTL14–WTAP1–273–VIRMA381–1486 quaternary complex, the cryo-EM density for the MTC core is largely missing in the cryo-EM structure, indicating that the MTC core is highly dynamic. This is also supported by previous reports that both METTL3 and METTL14 contain highly flexible intrinsically disordered regions (IDRs).10 Thus, we further conducted in vitro CXMS and AI-based prediction to model the structure of the quaternary M–M–W–V complex based on the solved structures. In total, we identified 227 (61 unique inter-subunit and 166 intra-subunit) bis(sulfosuccinimidyl) suberate (BS3)-crosslinked residue pairs with the false discovery rate (FDR) < 5% (Supplementary information, Fig. S9a). Notably, crosslinked pairs between WTAP coiled-coil and METTL3 N-terminal region (METTL3 residues 1–251, including METTL3–WTAPA crosslinked pairs K13–K192 K132–K192, K241–K155, and K241–K160), VIRMA and the METTL14 N-terminal helix (NH, including METTL14–VIRMA crosslinked pair K38–K399), and VIRMA and MTC core catalytic center (including METTL3–VIRMA crosslinked pairs K576–K899, K576–K978, and K578–K899; METTL14–VIRMA crosslinked pair K148–K887) were identified (Supplementary information, Fig. S9b). No crosslinked pairs were identified between the METTL3 ZFD and METTL14 RGG domains with WTAP or VIRMA. The final structural model of the quaternary complex can account for all intermolecular crosslinks between METTL3/METTL14 core domains and the two regulatory subunits, except for two crosslinks involving flexible loop residues. In the highest-ranking model, the METTL3 N-terminal leader helix (LH) region (residues 1–34) was docked in close proximity to WTAP, accounting for the unassigned cryo-EM density (Fig. 1h, i; Supplementary information, Fig. S5c). This is in line with a previous report that the METTL3 LH is crucial for WTAP–METTL3 interaction.11 The rest of the METTL3 N-terminal region was also predicted to be close to WTAP. The N-terminus of METTL14 was docked close to VIRMA and potentially interacts with WTAP. Although the MTC core is likely dynamically positioned, its catalytic center was docked next to VIRMA with the catalytic center facing VIRMA ARML 9–10 (Fig. 1h, i). This model thus represents a possible resting state of the quaternary complex.
WTAP and VIRMA are important subunits of the m6A writer complex.1,3 We determined the structure of WTAP–VIRMA in the M–M–W–V complex at atomic resolution and modeled the quaternary complex structure using an AI-empowered integrative approach (Fig. 1j). Consistent with the previously reported adaptor role of WTAP,3 the observed WTAP dimer likely serves as a linker connecting VIRMA (and/or other MTC regulatory subunits) with the MTC core. VIRMA mediates preferential m6A mRNA methylation in 3′ UTR, but the exact mechanism is unknown.6 Like other ARML-containing proteins,12 the observed VIRMA superhelix contains extensive solvent-accessible surfaces that can accommodate RNA substrate6 and other regulator proteins.6,13 Moreover, in the model the VIRMA ARML 9–10 region (containing several positively charged residues) is close to the catalytic center of METTL3, suggesting a potential VIRMA–RNA interaction. During our manuscript revision, Su and colleagues also reported the structure of the regulatory subunit m6A-METTL-associated complex (MACOM) containing WTAP–VIRMA–ZC3H13–HAKAI.14 Our WTAP–VIRMA structure in the M–M–W–V complex is almost identical to the counterpart in the MACOM. Moreover, our AI-empowered integrative structural model indicates the interaction between the N-terminal helix of METTL3 and WTAP coiled-coil, and proximity between the MTC catalytic core and a VIRMA positively charged surface. As such, VIRMA N- and C-terminal disordered regions likely interact with the MTC core or substrate RNA, giving rise to the higher methyltransferase activity for the full-length VIRMA-containing quaternary complex. The two structural models provide a framework for future study on the molecular architecture and catalytic process of the m6A writer holocomplex, and offer new insights into potential therapeutic manipulation of m6A modification through the design of small molecules or peptides targeting WTAP–VIRMA interaction and modulating m6A catalytic specificity/activity.15 We anticipate that future structural and biochemical characterization of the complete m6A writer complex and its substrate-bound form will unveil the specificity and regulatory details underlying mRNA m6A methylation.
Supplementary information
Acknowledgements
We thank the Cryo-EM Facility of Hubei University for providing technical support during EM image acquisition and the Center for Protein Research and Public Laboratory of Electron Microscopy, Huazhong Agricultural University, for technical support, J. Cao for assistance during cryo-EM image acquisition. We thank National Center for Protein Sciences at Peking University in Beijing, China, for assistance with mass spectrometry experiments. C. Wang has improved this manuscript. This work was supported by funds from the National Key R&D Program of China (2018YFA0507700), the National Natural Science Foundation of China (31870753 and 91753132) and the Foundation of Hubei Hongshan Laboratory (2021hszd011).
Author contributions
P.Y. and C.T. conceived the project. X.Y. and P.Y. designed all the experiments. X.Y., F.L., K.P., J.Y., X.J., and M.H. performed the experiments. X.Y. and Q.W. collected the EM data. Z.G. determined the crystal and cryo-EM structures. K.P. and C.T. performed mass spectrometry analysis. All authors analyzed the data and contributed to manuscript preparation. X.Y., Q.W., C.T. and P.Y. wrote the manuscript.
Data availability
Atomic coordinates of the WTAP130–241 has been deposited in the Protein Data Bank (PDB) under accession number 7YFJ. The cryo-EM density map for the human METTL3–METTL14–WTAP1–273–VIRMA381–1486 complex has been deposited in EM Database under the accession code EMD-33807. The corresponding atomic coordinates have been deposited in the PDB under accession code 7YG4. The CXMS data have been deposited to the ProteomeXchange Consortium PRIDE: PXD036144.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xuhui Yan, Kai Pei, Zeyuan Guan.
Contributor Information
Chun Tang, Email: Tang_Chun@pku.edu.cn.
Ping Yin, Email: yinping@mail.hzau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00741-8.
References
- 1.Yang Y, Hsu PJ, Chen YS, Yang YG. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang H, Weng H, Chen J. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaccara S, Ries RJ, Jaffrey SR. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 4.Davalos V, Blanco S, Esteller M. Cell. 2018;174:498–498.e1. doi: 10.1016/j.cell.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 5.Ping XL, et al. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue Y, et al. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, et al. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Doxtader KA, Nam Y. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Śledź P, Jinek M. Elife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZH, et al. EMBO J. 2021;40:e106309. doi: 10.15252/embj.2020106309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schöller E, et al. RNA. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Z, et al. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bawankar P, et al. Nat. Commun. 2021;12:3778. doi: 10.1038/s41467-021-23892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su, S. et al. Cell Res. 10.1038/s41422-022-00725-8 (2022).
- 15.Yankova E, et al. Nature. 2021;593:597–601. doi: 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the WTAP130–241 has been deposited in the Protein Data Bank (PDB) under accession number 7YFJ. The cryo-EM density map for the human METTL3–METTL14–WTAP1–273–VIRMA381–1486 complex has been deposited in EM Database under the accession code EMD-33807. The corresponding atomic coordinates have been deposited in the PDB under accession code 7YG4. The CXMS data have been deposited to the ProteomeXchange Consortium PRIDE: PXD036144.