Abstract
Aims
Previous studies investigated the associations between sleep traits and cardiac diseases, but the evidence for the causal inferences was unclear. This study aimed to explore the causal relationship between sleep and cardiac diseases by virtue of Mendelian randomization (MR).
Methods and results
Summary‐level data for exposure variables (sleep duration, chronotype, and insomnia) and outcome variables (ischaemic heart disease, atrial fibrillation, myocardial infarction, and heart failure) were derived from UK Biobank. Data from the FinnGen consortium was used as a robustness check. In MR analysis, the inverse variance weighted (IVW) method was applied to infer causality between exposure and outcome. MR‐Egger regression was used to identify pleiotropy, and MR‐PRESSO outlier test was used to remove the pleiotropy of the genetic instruments. Based on UK Biobank, MR analysis suggested that sleep duration was weakly associated with atrial fibrillation (OR = 0.9999, 95% CI: 0.9998–0.9999) and ischaemic heart disease (OR = 0.9997, 95% CI: 0.9995–0.9998). Insomnia was associated with ischaemic heart disease (OR = 1.0117, 95% CI: 1.0051–1.0183) and myocardial infarction (OR = 1.0049, 95% CI: 1.0019–1.0079). No associations were found between chronotype and cardiac diseases (P > 0.05). We did not find pleiotropy except for insomnia with ischaemic heart disease and myocardial infarction using MR‐Egger regression, and MR‐PRESSO analysis consistent with IVW. Finally, we obtained the same direction as with UK Biobank using the FinnGen data.
Conclusions
Sleep duration and insomnia might be the potential causal risk factors of cardiac diseases. As the OR was small, these associations are probably not clinically relevant. Further validation studies are needed.
Keywords: Sleep, Cardiac disease, Mendelian randomization, Causal relationship
Introduction
As a complex phenotype driven by genetic and lifestyle factors, sleep disturbance has led to many health problems. 1 Over the past decades, there has been a growing interest in exploring the extent of impaired sleep patterns on various adverse health outcomes. 2 Some observational studies reported U‐shaped associations between sleep duration and cardiovascular diseases, 3 , 4 and the harmful effects of insomnia on the heart have been confirmed. 5 In addition, there are some reports on the association between circadian rhythm disorders and disease, 6 but there is limited evidence on the role of chronotype on disease. 7
Pathogenic mechanisms linking sleep duration to adverse health outcomes might include reciprocal changes in circulating levels of leptin and hunger hormone. 8 , 9 Changes in hormone levels might affect appetite, promote obesity and increase the risk of cardiovascular disease. 10 In addition, increased cortisol secretion and altered growth hormone metabolism were associated with low‐level inflammation, which was also activated during short sleep periods and might have implications for cardiovascular diseases. 11 The duration of sleep was inextricably linked to the quality of sleep. Furthermore, poor sleep quality might disrupt circadian rhythms. 12 There was evidence that changes in the timing of circadian rhythms might contribute to the development of disease, particularly metabolic disease. 13 For example, independent of sleep disturbance, evening people had a high risk of obesity 14 and type 2 diabetes. 15 However, the evidence was limited for a causal relationship between chronotype and cardiac diseases. Apart from, the evidence of the association between insomnia and all cardiovascular diseases was found. 16 A possible explanation for the increased risk of cardiovascular disease in people with insomnia was that insomnia was a hyperarousal disorder that chronically activates the stress response, resulting in increased metabolic rate, increased heart rate and decreased heart rate variability. 16 Whereas, it is difficult to distinguish whether abnormal sleep patterns cause adverse health outcomes or risk factors like poor health result in both abnormal sleep patterns and adverse health outcomes.
Traditional observational studies are prone to be biased, and no experimental studies explain causal associations between sleep traits and cardiac diseases, especially in healthy participants. Mendelian randomization (MR) analysis is used to explore causality between exposure and outcome, which can empower causal explanation by using genetic instruments (IVs) associated with exposure, such as sleep duration. 17 , 18 MR reduces the effect of both reverse causality and bias as the IVs are assumed to be randomly allocated at the meiosis and unrelated to confounder. 19 Previous studies have analysed the association of sleep duration and cardiovascular diseases, including atrial fibrillation (AF), ischaemic heart disease (IHD) and myocardial infarction (MI), 20 but research on the associations of insomnia and chronnotype with cardiac diseases are lacking. Recently, larger genome wide association studies (GWAS) for insomnia, 21 chronotype, 22 and cardiac diseases from UK biobank and FinnGen consortium have become available. Therefore, this study aimed to investigate the causal associations of sleep traits (sleep duration, short sleep, long sleep, chronotype, and insomnia) with cardiac diseases [IHD, heart failure (HF), MI, and AF] by MR analysis.
Material and methods
Genetic instruments for sleep traits
Single nucleotide polymorphisms (SNPs) were used as genetic instrumental variable (IVs) in MR analysis. SNPs associated with five sleep traits were used in our study, including sleep duration, short sleep, long sleep, chronotype, and insomnia. The GWAS data of self‐reported habitual sleep traits were derived from the UK Biobank and performed by Dashti et al., 23 involving in sleep duration (n = 446 118), short sleep (n = 106 192 cases with <7 h of sleep relative to 305 742 controls with 7–8 h of sleep), and long sleep (n = 34 184 cases with ≥9 h of sleep to 305 742 controls with 7–8 h of sleep). Chronotype often referred to as circadian preference, is a physical and behavioural manifestation of the coupling between internal circadian cycles and the need for sleep, driven by sleep homoeostasis. 22 Genetic variants associated with chronotype (n = 252 287 cases with morning chronotype and n = 150 908 controls with evening chronotype) were obtained from Jones SE's study. 22 The GWAS data of frequent insomnia symptoms (n = 129 270 cases and n = 108 357 controls) were based on the UK biobank. 21 Study subjects reported whether they had symptoms of insomnia (‘never/rarely’ vs. ‘usually’ insomnia symptoms). The detailed information of GWAS data were listed in Table 1 .
Table 1.
Summary of genome‐wide association studies (GWAS) datasets for sleep‐related phenotypes
| Phenotype | GWAS data source | PMID | Sample size | Study | Race | No. of SNPs |
|---|---|---|---|---|---|---|
| Sleep duration | Dashti HS et al. 2019 | 30 846 698 | 446 118 people | UK Biobank study | European | 78 |
| Short sleep | Dashti HS et al. 2019 | 30 846 698 | 106 192 cases and 305 742 controls | UK Biobank study | European | 27 |
| Long sleep | Dashti HS et al. 2019 | 30 846 698 | 34 184 cases and 305 742 controls | UK Biobank study | European | 8 |
| Chronotype | Lane JM et al. 2019 | 30 696 823 | 252 287 cases and 150 908 controls | UK Biobank study | European | 153 |
| Insomnia | Jones SE et al. 2019 | 30 804 566 | 129 270 cases and 108 357 controls | UK Biobank study | European | 48 |
No. of SNPs, the number of single nucleotide polymorphisms.
Genome wide association studies data sources for cardiac diseases
The GWAS data for cardiac diseases were released by the UK Biobank. 24 UK Biobank enrolled around 500 000 adults, aged from 37 to 73 years, from 22 assessment centres across the United Kingdom during 2006 and 2010. For this study, cardiac diseases were categorized into IHD (20 857 cases and 340 337 controls), HF (360 106 cases and 361 194 controls), MI (7018 cases and 354 176 controls) and AF (5669 cases and 457 341 controls). Cardiac diseases were defined according to ICD‐10 (international classification of diseases, 10th revisions, respectively) codes. Apart from, FinnGen consortium (https://www.finngen.fi/fi) was used as a robustness check, including IHD (11 139 cases and 85 360 controls), HF (8016 cases and 75 137 controls), MI (4065 cases and 85 760 controls), and AF (7 244 cases and 56 378 controls).
Instrumental variable selection
Ideal IVs were selected based on three important characteristics. Firstly, the IVs should be closely related to exposure. Secondly, the IVs needed to be independent of possible confounding factors. Thirdly, the IVs did not affect the outcome directly, only possibly indirectly via the exposure. 25 We used four steps to select ideal IVs. 26 First, only SNPs achieved genome‐wide significant (P < 5 × 10−8) were included. We identified 78 SNPs for sleep duration, 27 SNPs for short sleep, 8 SNPs for long sleep, 153 SNPs for sleep chronotype, and 48 SNPs or frequency insomnia. Second, we clumped SNPs for linkage disequilibrium (LD) 27 (we only kept the SNPs with the smaller association P‐value for all pairs of SNPs violating the assumption with r 2 > 0.001) based on European ancestry reference data from the 1000 Genomes Project. Third, if the interest exposure SNPs were not found in the cardiac diseases outcome, we used proxy SNPs that were in high LD with the interest exposure SNPs. 28 Summary information of instruments achieved genome‐wide significant for sleep traits was presented in Supporting Information, Data S1 . Finally, in order to perform MR analysis, the effect of a SNP on an outcome and exposure must be harmonized to be relative to the same allele.
Statistical analysis
We tested the causality of different sleep traits with IHD, HF, MI, and AF using inverse variance weighted (IVW) method, 29 which assumes that all SNPs are valid instruments or are invalid in such a way that the overall bias is zero. We assume that there are J genetic variants that are valid instrumental variables (IVs), the effect of the exposure on the outcome is linear with no effect modification, and the associations of the genetic variants with the exposure and with the outcome are linear without effect modification. The model relating the variables is
where X is the exposure, G1 are the genetic variants, Y is the outcome, U is an unmeasured confounder, do (X = x) is Pearl's do‐operator meaning that the value of the exposure is set to x by intervention, and the causal effect parameter for all j = 1, …, J. This means that the ratio estimates for each genetic variant are consistent estimates of the same causal parameter. The IVW estimate can be expressed as
Then, MR‐Egger regression analysis was used to examine whether there was violation of the main MR assumptions due to directional pleiotropy. 30 In addition, we performed MR pleiotropy residual sum and outlier (MR‐PRESSO) analysis as comparable causal estimators after adjustment for pleiotropy. 31 Last, leave‐one‐out sensitivity analysis was performed to confirm that the causal relationship was not driven by a single IV. 32
The statistical analyses were conducted using the Two SampleMR and MR‐PRESSO packages in the R Version 4.0.5 software platform. Results were reported as odds ratio (OR) with corresponding 95% confidence intervals (CI) and P‐value.
Results
Mendelian randomization analyses
The potential causal associations between sleep traits and cardiac diseases were tested using MR analysis. Table 2 and Figure 1 showed MR analyses results for sleep duration, short sleep, long sleep, chronotype, and insomnia with IHD, HF, MI, and AF using IVW methods. Positive results of MR analyses detailed information could be found in Supporting Information, Data S2 .
Table 2.
Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in UK Biobank
| Exposure | Method | Outcome | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IHD | HF | MI | AF | ||||||
| SNPs | Odds ratio (95% CI) | SNPs | Odds ratio (95% CI) | SNPs | Odds ratio (95% CI) | SNPs | Odds ratio (95% CI) | ||
| Sleep duration | IVW | 59 | 0.9997 (0.9995–0.9998) | 59 | 0.9999 (0.9999–1.0000) | 59 | 0.9999 (0.9998–1.0000) | 58 | 0.9999 (0.9998–0.9999) |
| MR‐PRESSO | 0.9997 (0.9995–0.9998) | 0.9999 (0.9999–1.0000) | 0.9999 (0.9998–1.0000) | 0.9999 (0.9998–0.9999) | |||||
| Short sleep | IVW | 23 | 1.0094 (0.9983–1.0206) | 23 | 1.0012 (0.9988–1.0036) | 23 | 1.004 (0.9989–1.0091) | 22 | 1.0006 (0.997–1.0043) |
| MR‐PRESSO | 1.0099 (0.9995–1.0204) | 1.001 (0.9986–1.0034) | 1.004 (0.9993–1.0088) | 1.0011 (0.9976–1.0046) | |||||
| Long sleep | IVW | 7 | 0.9944 (0.9866–1.0023) | 7 | 0.9994 (0.9971–1.0018) | 7 | 0.9971 (0.991–1.0031) | 6 | 0.9955 (0.9919–0.9992) |
| MR‐PRESSO | 0.9944 (0.9872–1.0017) | 0.9994 (0.9971–1.0018) | 0.9971 (0.991–1.0031) | 0.9955 (0.9929–0.9982) | |||||
| Chronotype | IVW | 111 | 1.0005 (0.9934–1.0077) | 111 | 0.9993 (0.9978–1.0008) | 111 | 0.9992 (0.9957–1.0026) | 101 | 0.9984 (0.9957–1.0012) |
| MR‐PRESSO | 1.0007 (0.9937–1.0077) | 0.9992 (0.9977–1.0006) | 0.9993 (0.9959–1.0027) | 0.9987 (0.996–1.0014) | |||||
| Insomnia | IVW | 35 | 1.0117 (1.0051–1.0183) | 35 | 1.0005 (0.999–1.0021) | 35 | 1.0049 (1.0019–1.0079) | 33 | 1.0007 (0.9986–1.0029) |
| MR‐PRESSO | 1.0129 (1.0064–1.0195) | 1.0008 (0.9993–1.0024) | 1.0053 (1.0024–1.0081) | 1.0007 (0.9987–1.0028) | |||||
IVW, inverse variance weighted; MR‐PRESSO, MR pleiotropy residual sum and outlier; IHD, ischaemic heart disease; HF, heart failure; MI, myocardial infarction; AF, atrial fibrillation; CI, confidence intervals; SNPs, the number of single nucleotide polymorphisms.
Figure 1.
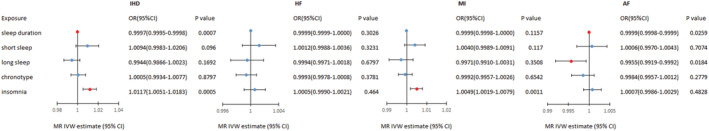
Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in UK Biobank. IVW, inverse variance weighted; MR‐PRESSO, IHD, ischaemic heart disease; HF, heart failure; MI, myocardial infarction; AF, atrial fibrillation; CI, confidence intervals.
For the causality between sleep duration and cardiac diseases, MR analyses discovered that genetically determined sleep duration was weakly associated with IHD (OR = 0.9997, 95% CI: 0.9995–0.9998, P < 0.001) and AF (OR = 0.9999, 95% CI: 0.9998–0.9999, P = 0.026), while no associations were observed for sleep duration with HF (OR = 0.9999, 95% CI: 0.9999–1.0000, P = 0.303) and MI (OR = 0.9999, 95% CI: 0.9999–1.0000, P = 0.116). For the causality between short sleep and cardiac diseases, MR analyses did not show evidence for associations with IHD (OR = 1.0094, 95% CI: 0.9983–1.0206, P = 0.096), HF (OR = 1.0012, 95% CI: 0.9988–1.0036, P = 0.323), MI (OR = 1.004, 95% CI: 0.9989–1.0091, P = 0.117), and AF (OR = 1.0006, 95% CI: 0.997–1.0043, P = 0.707). For the causality between long sleep and cardiac diseases, we observed an weak causal association of long sleep with AF (OR = 0.9955, 95% CI: 0.9919–0.9992, P = 0.018) using IVW method, but there were no associations with IHD (OR = 0.9944, 95% CI: 0.9866–1.0023, P = 0.169), HF (OR = 0.9994, 95% CI: 0.9971–1.0018, P = 0.680) and MI (OR = 0.9971, 95% CI: 0.991–1.0031, P = 0.351). For the causality between chronotype and cardiac diseases, MR analyses of the chronotype instrument did not show evidence for associations with IHD (OR = 1.0005, 95% CI: 0.9934–1.0077, P = 0.880), HF (OR = 0.9993, 95% CI: 0.9978–1.0008, P = 0.378), MI (OR = 0.9992, 95% CI: 0.9957–1.0026, P = 0.654), and AF (OR = 0.9984, 95% CI: 0.9957–1.0012, P = 0.278) using IVW method. For the causality between insomnia and cardiac diseases, the IVs used for insomnia indicated that insomnia increased the risk of IHD (OR = 1.0117, 95% CI: 1.0051–1.0183, P < 0.001) and MI (OR = 1.0049, 95% CI: 1.0019–1.0079, P = 0.001). No association was observed for insomnia with HF (OR = 1.0005, 95% CI: 0.9990–1.0021, P = 0.464) and AF (OR = 1.0007, 95% CI: 0.9986–1.0029, P = 0.483). MR‐Egger regression was used to identify pleiotropy, and the results were found in Supporting Information, Data S3 . It showed there was potential pleiotropy for insomnia with IHD and MI. However, the MR‐PRESSO showed the same results as IVW analyses and did not identified SNPs as outlier. Apart from, using MR‐PRESSO we obtained consistent results with IVW methods on other causal association analyses.
Sensitivity analysis
In the MR leave‐one‐out analysis using UK Biobank, we found that the risk estimates of genetically predicted sleep duration and AF slightly changed after excluding single SNP of rs13088093. Excluding SNP of rs17688916, we found the causal association between long sleep and AF disappeared. However, the causal associations of sleep duration with IHD, insomnia with IHD and insomnia with MI were shown to be robust. Specific information could be found in Supporting Information, Data S4 .
Robustness check
In the validation analysis using the FinnGen data, genetically determined sleep duration was weakly associated with four cardiac diseases, including IHD (OR = 0.9915, 95% CI: 0.9852–0.9978, P = 0.009), AF (OR = 0.9906, 95% CI: 0.9825–0.9989, P = 0.027), MI (OR = 0.9895, 95% CI: 0.9809–0.9982, P = 0.019), and HF (OR = 0.9915, 95% CI: 0.9853–0.9977, P = 0.008). Short sleep was associated with IHD (OR = 1.3453, 95% CI: 1.0156–1.7821, P = 0.039) and MI (OR = 1.7133, 95% CI: 1.1292–2.5995, P = 0.011). Chronotype was associated with IHD (OR = 1.2821, 95% CI: 1.0583–1.5531, P = 0.011), and no association was found between long sleep and insomnia with four cardiac diseases. Specific information could be found in Supporting Information, Data S5 .
Discussion
In this study, causal associations of sleep duration, long sleep, short sleep, insomnia and chronotype with IHD, HF, MI, and AF were investigated using MR analyses. Genetically predicted sleep duration was weakly associated with AF and IHD, and the results were confirmed in the FinnGen consortium. Otherwise, the other results in UK Biobank and FinnGen consortium were consistent in direction.
Cappuccinos et al. 33 found that both short and long duration of sleep were predictors for cardiovascular outcomes, and indicated that people with short sleep duration were at higher risk for coronary cardiac disease than those who sleep 7–8 h per day (relative risk = 1.48, 95% CI: 1.22–1.80, P < 0.001). Wang et al. 34 found that the risk of coronary heart disease increased by 11% for each hour of sleep reduction and by 7% for each hour of sleep increase, compared with 7 h of sleep per day. In contrast, Hoevenaar‐Blom et al. 35 suggested no association between long sleep duration and cardiovascular disease, and the results were debated. Epidemiological studies demonstrated an association between sleep and cardiovascular disease 8 , 9 , 35 but were unable to prove whether there was a causal link between them.
Previous MR studies suggested that short sleep may be a risk factor for MI 36 and coronary artery disease. 20 Furthermore, recent MR studies obtained inconsistent results regarding the causal relationship between sleep duration and coronary heart disease. 37 , 38 Most of these studies focused on the effect of single factor such as sleep duration on cardiovascular diseases. However, the purpose of our study was to investigate the effects of different sleep traits, including sleep duration, chronotype, and insomnia, on IHD, HF, MI, and AF. Our study found sleep traits and cardiac diseases had weak associations, with OR of approximately 1, which was consistent with the study of Larsson SC. 39 Compared with Ai et al., 20 Our study found similar results that genetically predicted sleep duration was negatively associated with AF and IHD, and the IVs for sleep duration, short sleep and long sleep were consistent. We did not obtain non‐linear MR result because of lacking individual data. Although definitive conclusions are hard to be drawn, the possibility of false‐positive or reverse causation would be very low in our study because of the application of strict IV selection procedure and MR test.
We observed slight discrepancy for the associations between sleep traits and cardiac diseases in FinnGen consortium and UK Biobank, which might be caused by higher prevalence of cardiac diseases in UK Biobank compared with Finn Gen consortium. In leave‐one‐out MR analysis, we found rs13088093 may dominate the estimate of the causal effect between sleep duration and AF, and rs17688916 may dominate the causal effect between long sleep and AF. Theoretically, if causality is proved by only one variant, then the validity of the reasoning depends only on that variant. Therefore, the results of sleep duration and long sleep with AF may be unstable.
The main strength of our study lied in the MR analysis, using multiple SNPs as IVs for sleep traits, which minimized confounding and reverse causal relationship. Although the results were not identical using UK Biobank and FinnGen consortium, they all suggested the effect of sleep traits on cardiac diseases. Furthermore, after excluding the effect of pleiotropy using MR‐PRESSO, sensitivity analyses showed the results were robust. Our study also had several limitations. First, sleep duration, short sleep, long sleep, chronotype, and insomnia were based on self‐reported answers, inevitably there were subjective bias. Secondly, only European participants were included in our study to control the influence of the genetic variation, so the results were less likely to be generalized to other population. Lastly, we were not able to assess the potential nonlinear associations of sleep traits with cardiac diseases due to lack of individual data.
This MR study indicated that genetically determined sleep traits were associated with modest risks of most cardiac diseases. As the effect size was small, these associations may not be clinically relevant. However, because couples of epidemiological studies reported quality of sleep was associated with incident of cardiovascular diseases, health benefits of improving the quality of sleep to reduce the risk of cardiac diseases still remained attractive.
Conflict of interest
The author reports no conflicts of interest in this work.
Funding
This research was supported by the National Natural Science Foundation of China (No. 82073670).
Supporting information
Data S1 Summary information of instruments identified for sleep traits.
Data S2 Specific information of Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in UK Biobank.
Data S3 Specific information of MR Egger results.
Data S4 Specific information of leave‐one‐out analysis results in UK Biobank.
Data S5 Specific information of Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in FinnGen.
Acknowledgements
This work was made possible by the generous sharing of GWAS summary statistics. The authors thank the UK Biobank study and the FinnGen consortium. The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Yang, Y. , Fan, J. , Shi, X. , Wang, Y. , Yang, C. , Lian, J. , Wang, N. , Zhao, C. , Zhao, Y. , and Jia, X. (2022) Causal associations between sleep traits and four cardiac diseases: a Mendelian randomization study. ESC Heart Failure, 9: 3160–3166. 10.1002/ehf2.14016.
References
- 1. Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, Emsley R, Gill S, Little MA, Luik AI, Loudon A, Scheer FAJL, Purcell SM, Kyle SD, Lawlor DA, Zhu X, Redline S, Ray DW, Rutter MK, Saxena R. Genome‐wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017; 49: 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. St‐Onge MP, Grandner MA, Brown D, Conroy MB, Jean‐Louis G, Coons M, Bhatt DL, American Heart Association Obesity, Behavior Change, Diabetes, and Nutrition Committees of the Council on Lifestyle and Cardiometabolic Health; Council on Cardiovascular Disease in the Young; Council on Clinical Cardiology; and Stroke Council . Sleep duration and quality: Impact on lifestyle behaviors and Cardiometabolic health: A scientific statement from the American Heart Association. Circulation. 2016; 134: e367–e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C, Bangdiwala SI, Rangarajan S, Lear SA, AlHabib KF, Mohan V, Teo K, Poirier P, Tse LA, Liu Z, Rosengren A, Kumar R, Lopez‐Jaramillo P, Yusoff K, Monsef N, Krishnapillai V, Ismail N, Seron P, Dans AL, Kruger L, Yeates K, Leach L, Yusuf R, Orlandini A, Wolyniec M, Bahonar A, Mohan I, Khatib R, Temizhan A, Li W, Yusuf S. Association of estimated sleep duration and naps with mortality and cardiovascular events: A study of 116 632 people from 21 countries. Eur Heart J. 2019; 40: 1620–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim Y, Wilkens LR, Schembre SM, Henderson BE, Kolonel LN, Goodman MT. Insufficient and excessive amounts of sleep increase the risk of premature death from cardiovascular and other diseases: The multiethnic cohort study. Prev Med. 2013; 57: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng B, Yu C, Lv J, Guo Y, Bian Z, Zhou M, Yang L, Chen Y, Li X, Zou J, Ning F, Chen J, Chen Z, Li L, China Kadoorie Biobank Collaborative Group . Insomnia symptoms and risk of cardiovascular diseases among 0.5 million adults: A 10‐year cohort. Neurology. 2019; 93: e2110–e2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wright KP Jr, Drake AL, Frey DJ, Fleshner M, Desouza CA, Gronfier C, Czeisler CA. Influence of sleep deprivation and circadian misalignment on cortisol, inflammatory markers, and cytokine balance. Brain Behav Immun. 2015; 47: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skarke C, Lahens NF, Rhoades SD, Campbell A, Bittinger K, Bailey A, Hoffmann C, Olson RS, Chen L, Yang G, Price TS, Moore JH, Bushman FD, Greene CS, Grant GR, Weljie AM, FitzGerald GA. A pilot characterization of the human Chronobiome. Sci Rep. 2017; 7: 17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004; 141: 846–850. [DOI] [PubMed] [Google Scholar]
- 9. Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004; 1: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009; 5: 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller MA, Cappuccio FP. Inflammation, sleep, obesity and cardiovascular disease. Curr Vasc Pharmacol. 2007; 5: 93–102. [DOI] [PubMed] [Google Scholar]
- 12. Bin YS. Is sleep quality more important than sleep duration for public health? Sleep. 2016; 39: 1629–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmid SM, Hallschmid M, Schultes B. The metabolic burden of sleep loss. Lancet Diabetes Endocrinol. 2015; 3: 52–62. [DOI] [PubMed] [Google Scholar]
- 14. Patterson F, Malone SK, Grandner MA, Lozano A, Perkett M, Hanlon A. Interactive effects of sleep duration and morning/evening preference on cardiovascular risk factors. Eur J Public Health. 2018; 28: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merikanto I, Lahti T, Puolijoki H, Vanhala M, Peltonen M, Laatikainen T, Vartiainen E, Salomaa V, Kronholm E, Partonen T. Associations of chronotype and sleep with cardiovascular diseases and type 2 diabetes. Chronobiol Int. 2013; 30: 470–477. [DOI] [PubMed] [Google Scholar]
- 16. Roth T. Insomnia: Definition, prevalence, etiology, and consequences. J Clin Sleep Med. 2007; 3: S7–S10. [PMC free article] [PubMed] [Google Scholar]
- 17. Smith GD, Ebrahim S. Mendelian randomization: Prospects, potentials, and limitations. Int J Epidemiol. 2004; 33: 30–42. [DOI] [PubMed] [Google Scholar]
- 18. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008; 27: 1133–1163. [DOI] [PubMed] [Google Scholar]
- 19. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013; 37: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ai S, Zhang J, Zhao G, Wang N, Li G, So HC, Liu Y, Chau SWH, Chen J, Tan X, Jia F, Tang X, Shi J, Lu L, Wing YK. Causal associations of short and long sleep durations with 12 cardiovascular diseases: Linear and nonlinear Mendelian randomization analyses in UK biobank. Eur Heart J. 2021; 42: 3349–3357. [DOI] [PubMed] [Google Scholar]
- 21. Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, Strand LB, Winsvold BS, Wang H, Bowden J, Song Y, Patel K, Anderson SG, Beaumont RN, Bechtold DA, Cade BE, Haas M, Kathiresan S, Little MA, Luik AI, Loudon AS, Purcell S, Richmond RC, Scheer FAJL, Schormair B, Tyrrell J, Winkelman JW, Winkelmann J, HUNT All In Sleep , Hveem K, Zhao C, Nielsen JB, Willer CJ, Redline S, Spiegelhalder K, Kyle SD, Ray DW, Zwart JA, Brumpton B, Frayling TM, Lawlor DA, Rutter MK, Weedon MN, Saxena R. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019; 51: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS, Tuke MA, Yaghootkar H, Sharp SA, Jie Y, Thompson WD, Harrison JW, Dawes A, Byrne EM, Tiemeier H, Allebrandt KV, Bowden J, Ray DW, Freathy RM, Murray A, Mazzotti DR, Gehrman PR, Lawlor DA, Frayling TM, Rutter MK, Hinds DA, Saxena R, Weedon MN. Genome‐wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019; 10: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dashti HS, Jones SE, Wood AR, Lane JM, van Hees VT, Wang H, Rhodes JA, Song Y, Patel K, Anderson SG, Beaumont RN, Bechtold DA, Bowden J, Cade BE, Garaulet M, Kyle SD, Little MA, Loudon AS, Luik AI, Scheer FAJL, Spiegelhalder K, Tyrrell J, Gottlieb DJ, Tiemeier H, Ray DW, Purcell SM, Frayling TM, Redline S, Lawlor DA, Rutter MK, Weedon MN, Saxena R. Genome‐wide association study identifies genetic loci for self‐reported habitual sleep duration supported by accelerometer‐derived estimates. Nat Commun. 2019; 10: 1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y, Briesacher BA. Use of instrumental variable in prescription drug research with observational data: A systematic review. J Clin Epidemiol. 2011; 64: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan J, Shi X, Jia X, Wang Y, Zhao Y, Bao J, Zhang H, Yang Y. Birth weight, childhood obesity and risk of hypertension: A Mendelian randomization study. J Hypertens. 2021; 39: 1876–1883. [DOI] [PubMed] [Google Scholar]
- 27. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR‐base platform supports systematic causal inference across the human phenome. Elife. 2018; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Machiela MJ, Chanock SJ. LDlink: A web‐based application for exploring population‐specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015; 31: 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, EPIC‐ InterAct Consortium . Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015; 30: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through egger regression. Int J Epidemiol. 2015; 44: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018; 50: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017; 28: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cappuccio FP, Cooper D, D'Elia L, Strazzullo P, Miller MA. Sleep duration predicts cardiovascular outcomes: A systematic review and meta‐analysis of prospective studies. Eur Heart J. 2011; 32: 1484–1492. [DOI] [PubMed] [Google Scholar]
- 34. Wang D, Li W, Cui X, Meng Y, Zhou M, Xiao L, Ma J, Yi G, Chen W. Sleep duration and risk of coronary heart disease: A systematic review and meta‐analysis of prospective cohort studies. Int J Cardiol. 2016; 219: 231–239. [DOI] [PubMed] [Google Scholar]
- 35. Hoevenaar‐Blom MP, Spijkerman AM, Kromhout D, van den Berg JF, Verschuren WM. Sleep duration and sleep quality in relation to 12‐year cardiovascular disease incidence: The MORGEN study. Sleep. 2011; 34: 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daghlas I, Dashti HS, Lane J, Aragam KG, Rutter MK, Saxena R, Vetter C. Sleep duration and myocardial infarction. J Am Coll Cardiol. 2019; 74: 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao LZ, Li WD, Liu Y, Li JP, Zhuang XD, Liao XX. Causal assessment of sleep on coronary heart disease. Sleep Med. 2020; 67: 232–236. [DOI] [PubMed] [Google Scholar]
- 38. Zhuang Z, Gao M, Yang R, Li N, Liu Z, Cao W, Huang T. Association of physical activity, sedentary behaviours and sleep duration with cardiovascular diseases and lipid profiles: A Mendelian randomization analysis. Lipids Health Dis. 2020; 19: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsson SC, Markus HS. Genetic liability to insomnia and cardiovascular disease risk. Circulation. 2019; 140: 796–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Summary information of instruments identified for sleep traits.
Data S2 Specific information of Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in UK Biobank.
Data S3 Specific information of MR Egger results.
Data S4 Specific information of leave‐one‐out analysis results in UK Biobank.
Data S5 Specific information of Mendelian randomization estimates for the associations of genetically predicted sleep traits with cardiac diseases in FinnGen.


