Abstract
Background
Patients with acute myocardial infarction (MI) are at high risk of upcoming events, in particular heart failure (HF), but reliable stratification methods are lacking. Our goal was to evaluate the potential role of circulating miRNAs as prognostic biomarkers in patients presenting with MI.
Methods and results
We conducted a prospective study among 311 consecutive patients hospitalized with MI (65% ST‐segment elevation MI & median age of 55 years) with long‐term follow‐up. An initial screening was conducted to select candidate miRNAs, with subsequent study of 14 candidate miRNAs. The primary outcome was the composite of hospital admission for HF or cardiovascular death. During a mean follow‐up of 2.1 years miR‐21‐5p, miR‐23a‐3p, miR27b‐3p, miR‐122‐5p, miR210‐3p, and miR‐221‐3p reliably predicted the primary outcome. Multivariate Cox regression analyses highlighted that miR‐210‐3p [hazard ratio (HR) 2.65 per 1 SD increase, P < 0.001], miR‐23a‐3p (HR 2.11 per 1 SD increase, P < 0.001), and miR‐221‐3p (HR 2.03 per 1 SD increase, P < 0.001) were able to accurately predict the primary outcome, as well as cardiovascular death, HF hospitalizations, and long‐term New York Heart Association (NYHA) functional class. These three miRNAs clearly improved the performance of multivariate clinical models: ΔC‐statistic = 0.10 [95% confidence interval (CI), 0.03–0.17], continuous net reclassification index = 34.8% (95%CI, 5.8–57.4%), and integrated discrimination improvement (P < 0.001).
Conclusions
This is the largest study evaluating the prognostic value of circulating miRNAs for HF‐related events among patients with MI. We show that several miRNAs predict HF hospitalizations, cardiovascular mortality, and poor long‐term NYHA status and improve current risk prediction methods.
Keywords: MicroRNA, Myocardial infarction, Heart failure, Biomarker, Prognosis
Introduction
Cardiovascular diseases remain as the first cause of death worldwide. Short‐term mortality of myocardial infarction (MI) has improved over the last decades, shifting the focus towards the long‐term consequences of these events. Heart failure (HF) is the most frequent complication of MI and a crucial prognostic factor. The chronic incidence of HF after MI stands above 30%. 1 The importance of HF lies in the frequent need of hospitalizations due to decompensations, the overwhelming costs for healthcare systems, its great impact on quality of life, and high mortality rate. 2
There are not reliable methods to assess the risk of chronic HF after MI, despite of its relevance. Clinical risk scores (i.e. GRACE and TIMI) were designed to evaluate short‐term mortality risk at MI presentation but remain untested for chronic HF‐related events. Neither cardiac imaging techniques nor biochemical markers (i.e. troponins and natriuretic peptides) allow the early identification of a long‐term maladaptive myocardial response. Consequently, novel strategies are being pursued to improve risk stratification of patients and recognize those who might benefit from closer follow‐up and early HF treatment.
MicroRNAs (miRNAs) are small (20–25 nucleotides), non‐coding endogenous regulatory RNA molecules that post‐transcriptionally regulate gene expression. miRNAs play critical roles in heart development, function, and response to injury, such as ischemia/reperfusion damage. 3 There is increasing evidence suggesting that miRNAs may have a key role in the pathogenesis of HF through regulation of genes involved in adverse ventricular remodelling. 4 While their effect is mainly intracellular, they can be detected in cell‐free body fluids such as serum, conceiving them as suitable biomarkers for clinical practice.
In this prospective study, we aimed to identify and validate a panel of serum miRNAs differentially expressed in acute MI, testing their ability to stratify the risk of long‐term HF hospitalizations and cardiovascular mortality.
Methods
Study design
This is a prospective study with enrolment of consecutive patients with type 1 MI admitted at the Cardiac ICU of a large tertiary hospital. The main inclusion criterion was type 1 MI, presenting as ST‐segment elevation MI or non‐ST‐elevation MI. No explicit exclusion criteria were defined to avoid selection bias. A comprehensive study of clinical and imaging data, invasive angiography, and blood testing for established and novel biomarkers was performed. 5 Blood samples for determination of serum miRNAs were extracted within the first 24 h of hospital admission and stored at the local Biobank. All participants provided written informed consent.
A control group was also established, to serve as additional reference for normal miRNA expression levels. Samples from 30 healthy individuals adjusted for age and sex with MI patients were obtained from the Biobank blood donor. Study protocols were approved by the Local Ethics Committee (references 175/13 and 061/16) and complied with the Declaration of Helsinki.
Outcomes
The primary outcome was defined as the composite of hospital admission for HF or cardiovascular mortality. Several secondary outcomes were established: (i) cardiovascular mortality; (ii) hospital admission for HF, including visits to the Emergency Department for HF with administration of intravenous diuretics; and (iii) long‐term New York Heart Association (NYHA) functional class. All outcomes were reviewed by two cardiologists blinded to miRNAs results. Follow‐up was performed on site every 6 months. Covariate definitions were standardized for analysis.
MicroRNAs extraction from serum and retrotranscription reaction
Prior to RNA isolation, a synthetic RNA (Spike‐in) was added to serum samples and served as a technical control of extraction homogeneity by further Spike‐in amplification. Isolation of total RNA enriched in miRNAs was performed using the mini RNAeasy kit (Qiagen). As a control of complementary DNA synthesis efficiency, an external RNA (cel‐miR‐39) was added and further amplified. For complementary DNA synthesis, Universal RT miRNA PCR System (Qiagen) was used.
MicroRNA array profiling
In order to identify serum miRNAs differentially expressed in patients with an acute MI, a screening assay for 752 miRNAs using quantitative reverse transcription PCR arrays was performed in 6 patients with MI and 10 healthy controls adjusted for age, sex, and smoking habit (Supporting Information, Table S1 ). The miRCURY LNA miRNA miRNome PCR Panels I + II (YAHS‐312 YG‐8, Qiagen) were used. Array data were normalized using the average of the expression of all miRNAs exhibiting CT equal to or less than 34. Data were analysed using GenEx v.6 software.
The following step, in order to achieve a panel of miRNAs representative of the underlying biological processes, was to perform a further depuration based on two criteria: (i) statistical and (ii) functional: miRNAs with biological significance relevant for MI were selected based on bioinformatic analysis (refer to Supporting Information, Methods section and Figure S 1 ). Data are available in GEO repository: accession number GSE168856 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168856).
Validation of selected microRNAs by individual quantitative reverse transcription PCR
Complementary DNA was diluted 1/11 with nuclease‐free sterile water, and 4 μL of dilution were used as template for PCR. PCR detection was conducted using SYBR Green and specific LNA probes for each selected miRNA (Qiagen). All reactions were performed in triplicate using a Light Cycler 480 instrument (Roche), and C t values were computed using a second derivative method (Light Cycler 480 Software 1.5, Roche). miRNA expression values are presented as ΔC T , calculated as follows: ΔC T = miRNA C t − housekeeping C t . Normfinder and Bestkeeper software were used to determine the most stable housekeeping miRNAs. Finally, we found that miR‐103 expression showed the most stable values in our population and was selected as housekeeping for data normalization. Refer to Supporting Information, Methods section for more detailed information.
Bioinformatics studies
miRGate database was used for obtaining the predicted target genes of those miRNAs associated with the study outcomes. Target genes were selected based on the criteria that they had at least two positive computational predictions using different algorithms, or biological evidence had been reported. Functional enrichment analysis was performed using the clusterProfiler package for R, with a P value threshold of 0.001 for filtering. 6 Bipartite networks were built using the selected miRNAs as source and putative regulated genes as targets. Networks were represented using the ForceAtlas2 layout algorithm implemented in Gephi™. 7
Statistical analysis
Clinical characteristics of study population are described according to the occurrence of the main composite outcome. Partial Spearman rank correlation coefficients were calculated to describe the relationship among miRNAs. Survival analysis was performed as follows: the relation of miRNA concentrations with the primary endpoint was assessed by Cox proportional hazards analyses adjusted for age and gender. In additional models, the age‐adjustment and sex‐adjustment was extended on conventional risk factors: hypertension, smoking status, diabetes, dyslipidaemia, and history of MI (Model 2); cardiac injury with troponin I levels (Model 3); natriuretic peptides with NT‐proBNP (Model 4); the validated GRACE risk score (Model 5); and the combination of left ventricular ejection fraction and Killip class (Model 6). No violation was found for using Cox regression. The hazard ratios P values were corrected for multiple testing (number of miRNAs) using the Benjamini and Hochberg method. For all regression models, the C‐index was computed at the mean follow‐up time (2 years). Confidence intervals for the C‐index and their increase with the addition of each miRNA were computed using a perturbation‐resampling method using 1000 iterations.
The multivariate clinical model with the highest C‐index was compared with vs. without miRNAs using the integrated discrimination improvement (IDI) index method, which compares the average difference in correctly predicting the risk for patients who have the primary outcome event with those who do not, and the continuous net reclassification improvement (cNRI), detailed in methods of the supporting information. 8 The area under the receiver‐operating curve was computed to display effect strength. Relation between miRNAs and NYHA class at the last follow‐up was evaluated using logistic regression. The statistical analyses were performed using SPSS software v.22.0 (SPSS Inc, Chicago, Illinois, USA) and R v.4.0.0.
Results
Patient profile
Baseline characteristics of the 311 patients enrolled are detailed in Table 1 . Median age was 55 years, 81% were male, and ST‐segment elevation MI was the predominant clinical presentation in 65% of cases. Successful revascularization of the culprit lesion was achieved in 94% of cases, and 1 year follow‐up was achieved in 90% of patients.
Table 1.
Clinical characteristics according to the occurrence of primary outcome
| Total (n = 311) | No event (n = 268) | Event (n = 43) | P value | |
|---|---|---|---|---|
| Age (years) a | 55 (48, 71) | 54 (47, 68) | 77 (55, 85) | <0.001 |
| Male sex (%) | 81 | 82 | 79 | 0.382 |
| Cardiovascular risk factors | ||||
| BMI, (kg/m2) a | 27.3 (25, 30) | 27.5 (25, 30) | 26.7 (24, 29) | 0.540 |
| Hypertension (%) | 57 | 55 | 72 | 0.018 |
| Diabetes (%) | 26 | 23 | 42 | 0.005 |
| Dyslipidaemia (%) | 51 | 50 | 58 | 0.458 |
| Active smoker (%) | 40 | 43 | 26 | 0.390 |
| MI index event | ||||
| ST‐elevation MI (%) | 65 | 66 | 61 | 0.908 |
| Successful revascularization (TIMI 3) (%) | 94 | 95 | 84 | 0.002 |
| Killip class ≥ II (%) | 23 | 15 | 67 | <0.001 |
| GRACE risk score a | 133 (114, 171) | 129 (111, 159) | 196 (144, 238) | <0.001 |
| Charlson Comorbidity Index a | 4 (2, 5) | 3 (2, 5) | 6 (5, 7) | <0.001 |
| Laboratory parameters | ||||
| NT‐proBNP (pg/mL) a | 903 (363, 2415) | 617 (215, 1541) | 5701 (2115, 14 714) | <0.001 |
| Troponin‐I (at 24 h) a | 8.8 (1, 38) | 6.9 (1, 32) | 29.1 (6, 88) | <0.001 |
| GFR (mL/min/1.73 m2) a | 91 (73, 103) | 93 (78, 104) | 52 (33, 78) | <0.001 |
| Total cholesterol (mg/dL) a | 177 (147, 206) | 179 (148, 209) | 158 (137, 192) | 0.028 |
| Triglycerides (mg/dL) a | 122 (88, 169) | 122 (88, 169) | 123 (89, 169) | 0.999 |
| Echocardiographic findings | ||||
| LVEF at discharge (%) a | 57 (46, 65) | 58 (48, 65) | 43 (36, 58) | <0.001 |
| LVEF < 50% (%) | 33 | 27 | 67 | <0.001 |
| Medications at discharge | ||||
| β‐Blocker (%) | 83 | 83 | 78 | 0.556 |
| ACE inhibitor or ARB (%) | 82 | 82 | 83 | 0.967 |
| Mineralocorticoid antagonist (%) | 37 | 10 | 33 | <0.001 |
| High intensity statin (%) | 97 | 97 | 92 | 0.025 |
ACE, angiotensin‐converting enzyme; ARB, angiotensin‐II receptor blocker; BMI, body mass index; GFR, glomerular filtration rate; LVEF, left ventricular ejection fraction; MI, myocardial infarction.
Median (25th and 75th quartile cut‐point).
During a mean follow‐up of 2.1 years, 43 patients (13.8%) presented the primary outcome, including 24 patients (7.7%) with hospitalizations for HF and 25 (8.0%) cardiovascular deaths. The mean time to the first primary outcome event was 6 months. At the last follow‐up, 25% of patients had a NYHA functional class II or higher.
Serum microRNAs differentially expressed in myocardial infarction
The profiling study identified a subset of miRNAs that are differentially expressed in MI. Following a sequential approach, based on their statistical significance and biological function assessed with bioinformatic target prediction tools, 14 miRNAs were selected for further validation in the 311 MI patients and 30 healthy controls adjusted for age and sex. During the global validation process, 11 out of the 14 candidate miRNAs statistically confirmed their difference between MI patients and healthy controls (Table S 2 ).
The expression levels of the majority of studied miRNAs were associated with a more severe clinical presentation of the index event. Briefly, the baseline expression levels of miR‐20a‐5p, miR‐21‐5p, miR‐23a‐3p, miR‐27b‐3p, miR‐30b‐5p, miR‐106b‐5p, miR‐107, miR‐148b‐3p, miR‐210‐3p, and miR‐221‐3p were significantly increased in subjects with higher Killip class, GRACE risk score, troponin I, NT‐proBNP, and poorer left ventricular ejection fraction. Distribution details are provided in Figures S 2 and S 3 .
In order to exclude redundancy among the selected miRNAs, partial Spearman rank correlation coefficients (R) were calculated for the 14 miRNAs after normalization (Table 2 ). Analyses yielded a weak correlation of most pairs of miRNAs, reflecting low collinearity among the selected miRNAs.
Table 2.
Partial Spearman rank correlation coefficients of circulating microRNAs
| hsa‐miR‐20a‐5p ΔC t | hsa‐miR‐21‐5p ΔC t | hsa‐miR‐23a‐3p ΔC t | hsa‐miR‐27b‐3p ΔC t | hsa‐miR‐30b‐5p ΔC t | hsa‐miR‐93‐5p ΔC t | hsa‐miR‐106b‐5p ΔC t | hsa‐miR‐107 ΔC t | hsa‐miR‐122‐5p ΔC t | hsa‐miR‐144‐3p ΔC t | hsa‐miR‐148b‐3p ΔC t | hsa‐miR‐210‐3p ΔC t | hsa‐miR‐221‐3p ΔC t | hsa‐let‐7a‐5p ΔC t | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa‐miR‐20a‐5p ΔC t | — | 0.31** | −0.18** | 0.03** | 0.20** | 0.51** | 0.54** | −0.32** | −0.12 | 0.08** | −0.10** | −0.01** | 0.01** | −0.03** |
| hsa‐miR‐21‐5p ΔC t | — | 0.46** | −0.20** | −0.24** | 0.06** | −0.28** | 0.20** | 0.32** | 0.12** | 0.31** | 0.21** | 0.00** | 0.26** | |
| hsa‐miR‐23a‐3p ΔC t | — | 0.59** | 0.18** | 0.22** | 0.28** | −0.42** | −0.15 | −0.26** | −0.25** | −0.04** | 0.36** | −0.16** | ||
| hsa‐miR‐27b‐3p ΔC t | — | 0.10** | 0.11** | −0.23** | 0.43** | 0.06** | 0.30** | 0.23** | −0.20** | 0.14** | −0.01** | |||
| hsa‐miR‐30b‐5p ΔC t | — | −0.08** | −0.06** | 0.17** | 0.07 | −0.01** | 0.34** | −0.10** | −0.04** | 0.37** | ||||
| hsa‐miR‐93‐5p ΔC t | — | 0.01** | 0.15** | 0.10 | 0.17** | 0.23** | −0.12** | −0.23** | −0.22** | |||||
| hsa‐miR‐106b‐5p ΔC t | — | 0.41** | 0.15 | 0.10** | 0.15** | 0.27** | −0.09** | 0.14** | ||||||
| hsa‐miR‐107 ΔC t | — | −0.19** | 0.02** | −0.09** | 0.13** | 0.08** | −0.01 | |||||||
| hsa‐miR‐122‐5p ΔC t | — | −0.20** | −0.49** | 0.24** | 0.03** | 0.15 | ||||||||
| hsa‐miR‐144‐3p ΔC t | — | −0.11** | 0.18** | −0.08 | −0.01 | |||||||||
| hsa‐miR‐148b‐3p ΔC t | — | 0.19** | 0.07** | 0.11** | ||||||||||
| hsa‐miR‐210‐3p ΔC t | — | 0.37** | −0.20** | |||||||||||
| hsa‐miR‐221‐3p ΔC t | — | 0.38** | ||||||||||||
| hsa‐let‐7a‐5p ΔC t | — |
Correlation is weak for most pairs of miRNAs, reflecting low collinearity. The only pairs of correlations with R > 0.5 were between miR‐23 & miR‐27, miR‐20 & miR‐106, and miR‐20 & miR‐93.
P value < 0.001.
Prognostic value of microRNAs
miR‐21‐5p, miR‐23a‐3p, miR‐27b‐3p, miR‐122‐5p, miR‐210‐3p, and miR‐221‐3p reliably predicted the occurrence of the primary endpoint (Figure 1 A, Table 3 , & Figure S 4 ), even in the advanced adjustment models. Cox regression analyses adjusted for age and gender highlighted in particular miR‐210‐3p (HR 2.65 per 1 SD increase, P < 0.001), miR‐23a‐3p (HR 2.11 per 1 SD increase, P < 0.001), miR‐221‐3p (HR 2.03 per 1 SD increase, P < 0.001), and miR‐21‐5p (HR 2.0 per 1 SD increase, P < 0.001) for precise prediction of the primary endpoint.
Figure 1.
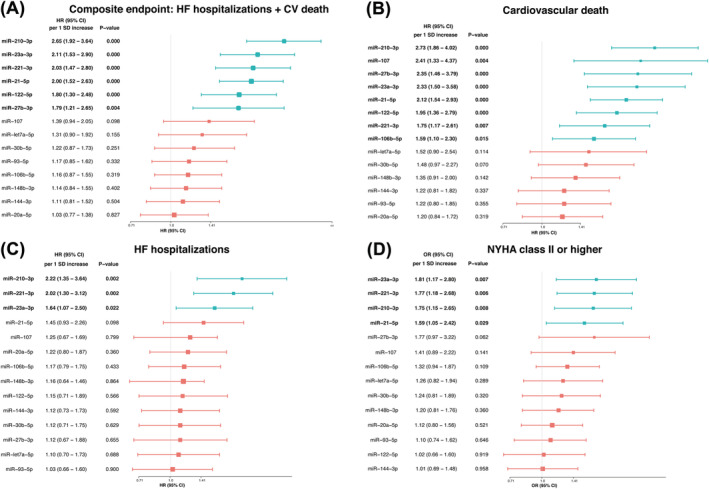
Summary of risk of the primary and secondary endpoints in the multivariate Cox survival model adjusted for age and sex. The boxes indicate the point estimates and the horizontal lines are 95% CIs. (A) Composite endpoint: HF hospitalizations + CV death, (B) cardiovascular death, (C) HF hospitalizations, and (D) NYHA class II or higher. CI, confidence interval; CV, cardiovascular; HF, heart failure; HR, hazard ratio; NYHA, New York Heart Association.
Table 3.
Association of serum miRNAs per 1 SD increase with the composite of heart failure hospitalizations or cardiovascular death during follow‐up among patients with myocardial infarction
| HR per 1 SD increase (95% CI) | P value | C‐statistic (95% CI) | Increase in C‐statistic (95% CI) | |
|---|---|---|---|---|
| miR‐20a‐5p a | 1.03 (0.77–1.38) | 0.827 | n/a | n/a |
| Model 2 | 1.07 (0.80–1.43) | 0.656 | n/a | n/a |
| Model 3 | 1.04 (0.77–1.40) | 0.815 | n/a | n/a |
| Model 4 | 1.26 (0.94–1.70) | 0.117 | n/a | n/a |
| Model 5 | 1.01 (0.78–1.32) | 0.920 | n/a | n/a |
| Model 6 | 1.01 (0.77–1.33) | 0.933 | n/a | n/a |
| miR‐21‐5p a | 2.00 (1.52–2.63) | 0.000 | 0.7828 (0.7141–0.8517) | 0.0517 (0.0011–0.1024) |
| Model 2 | 2.07 (1.56–2.74) | 0.000 | 0.7928 (0.7147–0.8709) | 0.0565 (0.0022–0.1109) |
| Model 3 | 1.87 (1.41–2.48) | 0.000 | 0.7962 (0.7258–0.8666) | 0.0376 (0–0.0807) |
| Model 4 | 1.82 (1.36–2.46) | 0.000 | 0.8403 (0.7618–0.9188) | 0.0143 (0–0.0393) |
| Model 5 | 1.52 (1.13–2.04) | 0.005 | 0.8110 (0.7389–0.8830) | 0.0181 (0–0.0458) |
| Model 6 | 1.42 (1.05–1.93) | 0.022 | 0.8316 (0.7677–0.8954) | 0.0104 (0–0.0296) |
| miR‐23a‐3p a | 2.11 (1.53–2.90) | 0.000 | 0.7909 (0.7246–0.8572) | 0.0598 (0.0135–0.1061) |
| Model 2 | 2.17 (1.56–3.01) | 0.000 | 0.7991 (0.7233–0.8749) | 0.0628 (0.0111–0.1145) |
| Model 3 | 2.00 (1.43–2.79) | 0.000 | 0.7969 (0.7323–0.8616) | 0.0383 (0–0.0827) |
| Model 4 | 1.89 (1.33–2.70) | 0.000 | 0.8529 (0.7934–0.9124) | 0.0270 (0–0.0584) |
| Model 5 | 1.59 (1.16–2.18) | 0.004 | 0.8233 (0.7619–0.8848) | 0.0305 (0–0.0653) |
| Model 6 | 1.50 (1.09–2.06) | 0.013 | 0.8410 (0.7842–0.8978) | 0.0198 (0–0.0501) |
| miR‐27b‐3p a | 1.79 (1.21–2.65) | 0.004 | 0.7562 (0.6612–0.8512) | 0.0251 (0–0.0663) |
| Model 2 | 1.75 (1.18–2.59) | 0.005 | 0.7618 (0.6618–0.8618) | 0.0255 (0–0.0637) |
| Model 3 | 1.52 (1.01–2.28) | 0.043 | 0.7648 (0.6781–0.8514) | 0.0061 (0–0.0358) |
| Model 4 | 2.20 (1.39–3.48) | 0.001 | 0.8300 (0.7555–0.9044) | 0.0040 (0–0.0437) |
| Model 5 | 1.37 (0.97–1.95) | 0.075 | 0.8024 (0.7192–0.8857) | 0.0096 (0–0.0355) |
| Model 6 | 1.40 (0.99–2.00) | 0.059 | 0.8252 (0.7482–0.9021) | 0.0040 (0–0.0274) |
| miR‐30b‐5p a | 1.22 (0.87–1.73) | 0.251 | n/a | n/a |
| Model 2 | 1.25 (0.87–1.77) | 0.226 | n/a | n/a |
| Model 3 | 1.18 (0.84–1.67) | 0.342 | n/a | n/a |
| Model 4 | 1.46 (1.00–2.15) | 0.051 | n/a | n/a |
| Model 5 | 1.16 (0.85–1.58) | 0.351 | n/a | n/a |
| Model 6 | 1.13 (0.81–1.57) | 0.471 | n/a | n/a |
| miR‐106b‐5p a | 1.16 (0.87–1.55) | 0.319 | n/a | n/a |
| Model 2 | 1.19 (0.88–1.61) | 0.271 | n/a | n/a |
| Model 3 | 1.17 (0.87–1.56) | 0.294 | n/a | n/a |
| Model 4 | 1.39 (1.01–1.91) | 0.043 | n/a | n/a |
| Model 5 | 1.09 (0.85–1.41) | 0.491 | n/a | n/a |
| Model 6 | 1.06 (0.80–1.40) | 0.694 | n/a | n/a |
| miR‐107 a | 1.39 (0.94–2.05) | 0.098 | n/a | n/a |
| Model 2 | 1.46 (0.97–2.18) | 0.067 | n/a | n/a |
| Model 3 | 1.45 (0.98–2.15) | 0.062 | n/a | n/a |
| Model 4 | 1.48 (0.93–2.37) | 0.097 | n/a | n/a |
| Model 5 | 1.42 (0.96–2.10) | 0.081 | n/a | n/a |
| Model 6 | 1.28 (0.87–1.90) | 0.213 | n/a | n/a |
| miR‐122‐5p a | 1.80 (1.30–2.48) | 0.000 | 0.7427 (0.6528–0.8325) | 0.0115 (0–0.0579) |
| Model 2 | 1.81 (1.30–2.51) | 0.000 | 0.7589 (0.6655–0.8523) | 0.0226 (0–0.0722) |
| Model 3 | 1.70 (1.26–2.30) | 0.001 | 0.7747 (0.6888–0.8606) | 0.0161 (0–0.0544) |
| Model 4 | 1.64 (1.21–2.22) | 0.001 | 0.8236 (0.7474–0.8998) | 0.0024 (0–0.0186) |
| Model 5 | 1.31 (0.96–1.78) | 0.087 | 0.7880 (0.7096–0.8663) | 0.0049 (0–0.0105) |
| Model 6 | 1.30 (0.94–1.79) | 0.116 | 0.8181 (0.7437–0.8925) | 0.0031 (0–0.0107) |
| miR‐148b‐3p a | 1.14 (0.84–1.55) | 0.402 | n/a | n/a |
| Model 2 | 1.15 (0.85–1.56) | 0.363 | n/a | n/a |
| Model 3 | 1.16 (0.85–1.57) | 0.345 | n/a | n/a |
| Model 4 | 1.42 (0.98–2.06) | 0.650 | n/a | n/a |
| Model 5 | 1.03 (0.77–1.38) | 0.845 | n/a | n/a |
| Model 6 | 1.15 (0.83–1.60) | 0.409 | n/a | n/a |
| miR‐210‐3p a | 2.65 (1.92–3.64) | 0.000 | 0.8026 (0.7319–0.8732) | 0.0714 (0.0068–0.1360) |
| Model 2 | 2.70 (1.95–3.73) | 0.000 | 0.8050 (0.7194–0.8906) | 0.0687 (0–0.1420) |
| Model 3 | 2.42 (1.75–3.34) | 0.000 | 0.8079 (0.7322–0.8835) | 0.0492 (0–0.1074) |
| Model 4 | 2.21 (1.57–3.12) | 0.000 | 0.8443 (0.7742–0.9143) | 0.0183 (0–0.0511) |
| Model 5 | 1.88 (1.32–2.67) | 0.000 | 0.8257 (0.7579–0.8935) | 0.0329 (0–0.0793) |
| Model 6 | 1.84 (1.30–2.59) | 0.001 | 0.8432 (0.7743–0.9121) | 0.0220 (0–0.0562) |
| miR‐221‐3p a | 2.03 (1.47–2.80) | 0.000 | 0.7809 (0.7091–0.8526) | 0.0497 (0–0.1039) |
| Model 2 | 2.04 (1.45–2.86) | 0.000 | 0.7864 (0.7046–0.8682) | 0.0501 (0–0.1115) |
| Model 3 | 1.92 (1.40–2.64) | 0.000 | 0.7991 (0.7227–0.8756) | 0.0405 (0–0.0879) |
| Model 4 | 2.13 (1.45–3.13) | 0.000 | 0.8663 (0.8107–0.9219) | 0.0403 (0–0.0852) |
| Model 5 | 1.70 (1.23–2.37) | 0.001 | 0.8231 (0.7598–0.8864) | 0.0303 (0–0.0683) |
| Model 6 | 1.58 (1.14–2.19) | 0.006 | 0.8498 (0.7896–0.9101) | 0.0286 (0–0.0632) |
CI, confidence interval; HR, hazard ratio; n/a, not applicable; SD, standard deviation.
Model 2: adjusted for age, sex, hypertension, smoking status, diabetes, dyslipidaemia, and history of MI. Model 3: adjusted for age, sex, and log (troponin I). Model 4: adjusted for age, sex, and log (NT‐proBNP). Model 5: adjusted for age, sex, and GRACE risk score. Model 6: adjusted for age, sex, left ventricular ejection fraction (LVEF), and Killip class. P values presented are corrected for multiple comparisons (number of miRNAs) using the Benjamini and Hochberg correction. HRs and 95% CIs are presented without correction.
Age and sex adjusted.
When considered individually, each one of these miRNAs achieved high C‐statistic values (Figure S 5 ), while the combination of the three most prominent (i.e. miR‐210‐3p, miR‐23a‐3p, and miR‐221‐3p) to the baseline multivariate Cox model showed a significant improvement in the area under the curve as compared with the baseline multivariate Cox model: 0.86 vs. 0.77, P < 0.001 (Figure 2 ). The cNRI index was 34.8% (95% CI, 5.8–57.4%, P = 0.015) while the IDI showed a substantial incremental predictive ability (P < 0.001) when miRNAs were added to the clinical multivariate model (Figure S 6 ).
Figure 2.
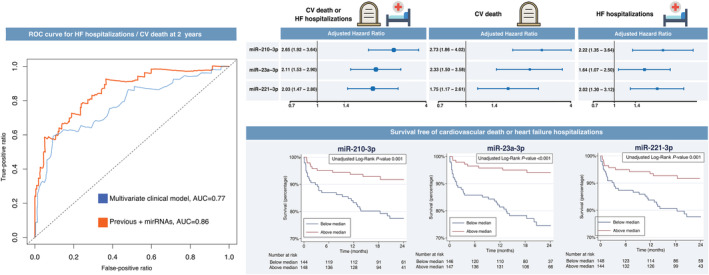
Prognostic reclassification with the addition of miR‐210‐3p, miR‐23a‐3p, and miR‐221‐3p. (Left) Receiver‐operating characteristic curves for the baseline clinical multivariate Cox model (adjusted for age and sex) in blue and with the addition of miR‐210‐3p, miR‐23a‐3p, and miR‐221‐3p, in red. (Top right) Forest plots for the study outcomes adjusted for age and sex. (Bottom right) Incidence of the composite of heart failure hospitalizations or cardiovascular death with the Kaplan–Meier method. AUC, area under the curve; CV, cardiovascular; HF, heart failure; ROC, receiver‐operating characteristic.
In addition, we evaluated the association of circulating miRNAs with secondary endpoints. Eight out of 14 miRNAs (miR‐21‐5p, miR‐23a‐3p, miR‐27b‐3p miR‐106b‐5p, miR‐107, miR‐122‐5p, miR‐210‐3p, and miR‐221‐3p) independently predicted cardiovascular death (Figure 1 B, Table S 3 , & Figure S 7 ). The predictive value of the miRNAs remained mainly unchanged, irrespective of adjustment for variables in the different models presented. Regarding HF hospitalizations, three miRNAs (i.e. miR‐23a‐3p, miR‐210‐3p, and miR‐221‐3p) were able to identify patients at higher risk (Figure 1 C & Table S 4 ). Finally, miR‐21‐5p, miR‐23a‐3p, miR‐210‐3p, and miR‐221‐3p were able to identify patients with poor functional class (NYHA II or higher) at the end of follow‐up (Figure 1 D and Table S , 5 ).
MicroRNAs functional significance
The biological significance of the eight circulating miRNAs with prognostic capability was explored by means of bioinformatic studies. Prediction of their target genes using appropriated databases and further clustering of the miRNAs based on common targets is shown in Figures 3 and S 8 . As it can be observed, all these miRNAs share common targets. Particularly, miR‐21‐5p, miR‐122‐5p, miR‐106b‐5p, and miR‐221‐3p exhibited stronger relations based on higher number of common targets, including PKD2, RhoA, HMGB3, TSC1, FNBP1, and PKM. The relationship exhibited among miRNAs suggests that they coordinately mediate through their targets the cardiac recovery after injury.
Figure 3.
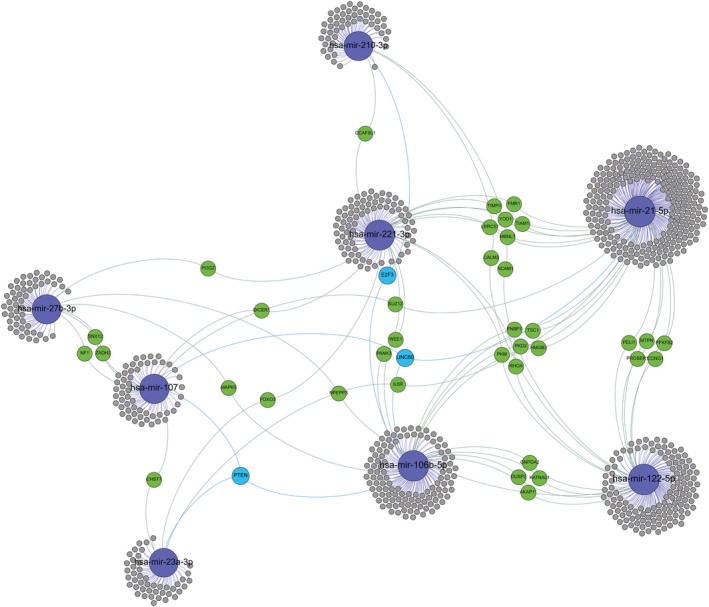
Target gene networks of microRNAs that exhibited prognostic capabilities. Only microRNAs that displayed independent association with outcome events are shown, highlighted in blue. Genes with at least two potential interactions are labelled. Genes with just one connection are unlabelled. Colour nodes of regulated genes are proportional to node degree (number of connections).
MicroRNAs as useful tools for prognostic reclassification
As circulating miRNAs accurately predicted cardiovascular death and HF hospitalizations in our study, we explored their capacity to restratify patient risk. A hierarchical cluster was built with the six miRNAs associated with the primary outcome (Figure 4 ). Patients that presented the primary outcome exhibited different expression profile of miRNAs, particularly miR‐21‐5p and miR23a‐3p. This different miRNAs' expression is independent from other clinical characteristics, unveiling new molecular information that might be clinically useful and relevant.
Figure 4.
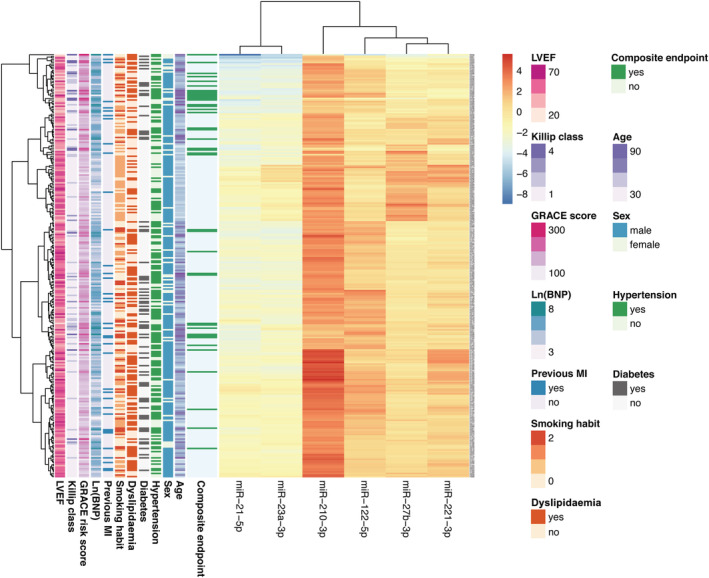
Heat map and hierarchical cluster of the validated microRNAs reclassifying MI patients according to the risk of the composite of heart failure hospitalization/cardiovascular death. The six validated microRNAs analysed by quantitative reverse transcription PCR in 311 patients with myocardial infarction reclassify patients according to their risk, independently of clinical features and based on miRNA pattern expression. MI, myocardial infarction; LVEF, left ventricular ejection fraction.
Circulating miRNAs also exhibited different grades of relation among them: miR‐21‐5p and miR23a‐3p were more related, exhibiting similar tendency in all patients attending to endpoints but lower relation with the rest of miRNAs that clustered in the other group, being miR‐27b‐3p and miR‐221‐3p tightly related. This hierarchical cluster indicates that circulating miRNAs reclassify patients independently of baseline clinical data, but with closer association with outcomes, thus enhancing the appeal of these molecular markers.
Discussion
In this study, we have demonstrated, for the first time, the prognostic value of serum miRNAs to identify patients at higher risk of HF‐related events after an acute MI. This prospective study with patients hospitalized for MI identified several miRNAs that were independently associated with HF hospitalizations, cardiovascular death, and poorer functional status. The novel information provided by miRNAs was independent but complementary to other clinical and biochemical parameters, and significantly improved conventional clinical risk prediction models when used as single variables or in combination.
Currently, robust evidence regarding the prognostic role of miRNAs in MI patients is lacking. Based on a rigorous screening selection among 752 miRNAs, we found independent association between six miRNAs and the primary composite endpoint of HF requiring hospital admission or cardiovascular mortality. Hence, the event rates doubled per 1 SD increase of miR‐21‐5p, miR‐23a‐3p, miR‐210‐3p, and miR‐221‐3p and increased by more than 50% for miR‐27b‐3p and miR‐122‐5p. Furthermore, the combination of several miRNAs consistently improved all the different multivariate models that were tested, certifying their independent prognostic ability regardless of adjustment for clinical variables, as assessed by marked improvements in IDI and cNRI. This was likely achieved due to the low correlation among the selected miRNAs (reflected by a low Spearman's rho), derived from the selection criteria applied and the different functional pathways that each one of miRNAs is involved.
Considering the secondary endpoints, we also found a significant association between eight miRNAs and cardiovascular mortality rates, with more than two‐fold increases per 1 SD for miR‐21‐5p, miR‐23a‐3p, miR‐27b‐3p, miR‐107, and miR‐210‐3p in all multivariate‐adjusted models. Additionally, levels of three miRNAs (miR‐23a‐3p, miR‐210‐3p, and miR‐221‐3p) were independently associated with hospitalizations for HF and predicted a poorer functional status at the end of follow‐up.
It is noteworthy to highlight that the prognostic value offered by these miRNAs was additive and independent of other parameters. Current clinical risk scores in MI are of limited value for the prediction of long‐term complications related to HF. Therefore, the possibility to improve recognition of individuals with an acute MI that are at a higher risk of HF‐related events represents an actionable goal to tailor more adequate therapy.
MicroRNAs in large‐scale studies
Ours is the first prospective study with circulating miRNAs in MI that evaluates long‐term HF‐related complications (including cardiovascular mortality). To date, only one other large‐scale study has been performed in secondary prevention, where miR‐197 and miR‐223 predicted cardiovascular death in a population that included stable angina 9 ; a substudy of the same cohort that used a literature‐based selection of candidate miRNAs subsequently identified another eight miRNAs, including miR‐210, that reached statistical significance for the prediction of cardiovascular mortality, but with high collinearity. 10 Smaller studies such as Maciejak et al. evaluated several miRNAs after MI using a profiling‐validation approach, but the small sample size and the use of left ventricular ejection fraction as surrogate of HF limit the value of their observations. 11 Other studies using a case–control approach detected that several miRNAs identify patients at higher risk of recurrent events after MI and improve GRACE and clinical risk prediction models. 12 , 13 Because there is not a generally accepted risk stratification tool for HF events after MI, our data emphasized the potential role of the determination of miRNAs circulating levels, particularly the analysis of miR‐210‐3p, miR‐221‐3p, and miR23a‐3p, in the prognosis stratification and the assessment of the risk of developing HF after an acute coronary event.
Pathophysiology
Hypoxic conditions stimulate the miRNome, and a growing number of miRNAs have been included in the group of oxygen‐responsive miRNAs, as miRNAs are common targets of transcription factors. 14 However, long‐term pathophysiological consequences of MI are a multifaceted process due to the complex molecular mechanisms involved in adverse ventricular remodelling. The focus of our study was to identify, among all miRNAs differentially expressed in MI, those with prognostic value.
In our study, the most relevant and influential was miR‐210‐3p, regulated by hypoxia through the HIF‐1 pathway, which was independently associated with HF, cardiovascular mortality and functional class during follow‐up, suggesting a pivotal role in cardiac regeneration. miR‐210‐3p expression increases shortly (<3 h) after MI, 15 and it has been found to inhibit apoptosis of cardiomyocytes. In our study, patients with higher expression levels of miR‐210‐3p presented more frequently HF decompensations and adverse ventricular remodelling, in line with results from experimental models. 16
Similarly, we found that miR‐221‐3p is a marker of more severe index event and worse long‐term outcomes. miR‐221 is linked with endothelial dysfunction through regulation of endothelial nitric oxide synthase, and it has been successfully evaluated as part of a miRNA prosurvival cocktail to improve cell engraftment of transplanted cardiac progenitor cells in mice with MI. 17 Interestingly, miR‐23, a member of the miR‐23/27/24 cluster, has been proposed as protective regulator after MI both in vitro and in vivo experimental models, linked with ventricular remodelling and HF 18 ; miR‐27b has a pro‐angiogenic effect and its targeted overexpression induced recruitment of bone marrow derived cells to the neovasculature, decreased fibrosis, and increased ejection fraction in experimental models. 19 miR‐21‐5p is a well‐known miRNA induced by hypoxia that modulates cardiomyocyte survival, and it has been associated in animal studies with adverse ventricular remodelling and HF after MI, 18 potentially reversible by the use of an antimiR‐21. 20 Likewise, a crucial cardiac role has also been implicated for miR‐122 by modulating cardiomyocyte cell death in vitro 21 ; elevated levels of miR‐122 have been found in patients with acute HF. 22 To our knowledge, none of the abovementioned miRNAs have been previously identified as predictors of survival free of events after an MI.
Functional analyses of the eight miRNAs reliably associated to adverse events, indicate that at least four of them, that is, miR‐21‐5p, miR‐122‐5p, miR‐106b‐5p, and miR‐221‐3p share common targets, including PKD2, RhoA, HMGB3, TSC1, FNBP1, and PKM. These genes have been proved relevant for heart function and remodelling after injury, such as PKD2, an intracellular calcium channel expressed in cardiomyocytes that has been associated with dilated cardiomyopathy. 23 RhoA, a key gene for cytoskeleton dynamics, and the RhoK/RhoA pathway have been found to play crucial roles in response to ischemia, leading to their current evaluation as therapeutic target in cardiovascular medicine. 24 HMGB3, belonging to the HMGB transcription factors family, is involved in skeletal muscle regeneration and elicits both harmful and beneficial responses during and after cardiac injury 25 ; TSC1 is a specific gene of myocytes connected with arterial injury and the development of HF and ventricular hypertrophy 26 ; FNBP1, involved in degradation through lysosomal pathway and PKM2, a key enzyme for cell metabolism involved in Warburg effect, that it is involved in embryonic cardiac development, has been recently associated to HF through cell metabolism impairment. 27 Based on these relevant common targets, it is conceivable that circulating miRNAs detected in our population could be part of the molecular and cellular pathways cooperatively acting in the cardiac injury and repair process.
Conversely, several miRNAs linked with cardiovascular events did not show prognostic capability in our population. Thus, miR‐106b‐5p and miR‐144‐3p did not provide prognostic information despite they have been associated with cardiovascular mortality in healthy population. 28 As for miR‐30b‐5p and miR‐let‐7a‐5p, none of them showed differential expression among patients with MI during the validation, nor did miR‐20a‐5p, miR‐93‐5p, and miR‐148b‐3p provide any prognostic information.
Limitations
This study has some limitations. First, the selection of candidate miRNAs was independent and based on our own initial observations; while this approach was chosen to avoid repetitive testing of same miRNAs previously described in the literature, this might have led to exclusion of potentially relevant miRNAs. Second, several Cox regression analyses are presented in order to evaluate the behaviour of miRNAs in different settings, although caution should be paid for interpretation due to the possibility of over‐adjustment.
Conclusions
We have demonstrated, for the first time, the capability of several miRNAs to independently predict long‐term hospitalizations for HF and cardiovascular mortality in patients after an acute MI. Based on an independent profiling‐validation process, our study identifies a set of circulating miRNAs, in particular miR‐210‐3p, miR‐221‐3p, and miR‐23a‐3p, with a strong and independent prognostic value.
Translational perspective
Patients who suffer an MI are at high risk of developing long‐term complications, but mechanisms leading to poor recovery are largely unknown. miRNAs specifically mediate pathophysiological processes and can be easily detectable as circulating biomarkers. This work positively evaluates the prognostic value of circulating miRNAs to identify patients at higher risk of future hospitalizations of HF, cardiovascular death and poor functional status. The novel information provided by miRNAs was independent to other parameters and improved conventional risk prediction models. Future studies will determine whether miRNAs might constitute as new therapeutic targets or be useful to guide secondary therapy.
Competency in patient care and procedural skills
Long‐term complications including HF and cardiovascular death are common among patients with MI, and circulating miRNAs have the capability to identify those at higher risk of upcoming events.
Translational outlook
Additional studies are required in order to elucidate whether circulating miRNAs might be useful to guide secondary therapy, based on the stratification of prognostic risk.
Conflict of interest
Authors report no conflicts of interest.
Funding
This study was supported by the Instituto de Salud Carlos III (PI15/00667), the CIBERCV CB16/11/00250), and the Spanish Society of Cardiology (2015/CC).
Supporting information
Table S1. Main clinical features of patients used for screening analysis. Age, group, clinical presentation and smoking habit are shown. MI, myocardial infarction; STEMI: ST‐elevation myocardial infarction; NSTEMI: non‐ST‐elevation myocardial infarction; NA: not applicable.
Table S2. Panel with selected candidate miRNAs and their expression levels among patients with myocardial infarction and healthy matched‐controls.
Table S3. Association of serum miRNAs per 1 standard deviation increase with cardiovascular mortality during follow‐up among the 311 patients with myocardial infarction.
Table S4. Association of serum miRNAs per 1 standard deviation increase with hospitalization for heart failure during follow‐up among the 311 patients with myocardial infarction.
Table S5. Association of serum miRNAs per 1 standard deviation increase with NYHA functional class II or higher at the end of follow‐up among the 311 patients with myocardial infarction*.
Table S6. Main clinical characteristics of the “healthy” population (without CV disease) adjusted for age and sex established as control group during the validation process (n = 30).
Table S7. Collection, processing and storage processes and conditions in the samples from cohort of patients with myocardial infarction and controls.
Figure S1. Heat map and hierarchical clustering of miRNAs in venous samples from 6 myocardial infarction and 10 healthy controls, performed as part of the miRNA profiling process. 752 miRNAs were analysed, and the top 42 miRNAs with highest mean differentiation between groups are shown. Normalized ΔCT values were used for the analysis.
Figure S2. Statistical association of some of the selected miRNAs with severity of index event: a) Killip‐Kimbal class; b) GRACE risk score at admission and c) troponin I levels at 24 h. miRNAs levels are expressed as delta crossing thresholds (DCT) and only those exhibiting a statistically significant association are shown.
Figure S3. Association of miRNAs with parameters indicative of acute heart failure/congestion: a) BNP levels (logarithmic), b) left ventricular ejection fraction at discharge.
Figure S4. Incidence of the composite of heart failure hospitalizations or cardiovascular mortality with the Kaplan–Meier method of miRs‐21, −23, −27, −122, −210 and −221 for values above and below the median.
Figure S5. Receiver‐operating characteristic (ROC) curves for prediction of the primary endpoint (heart failure hospitalization or cardiovascular death) at 2 years.
Figure S6. Assessing of IDI and cNRI with the addition of miR‐210‐3p, miR‐23a‐3p and miR‐221‐
Figure S7. Incidence of cardiovascular mortality over time with the Kaplan–Meier method of miRs‐21, −23, −27, −107, −210 and −221 for values above and below the median.
Figure S8. Functional enrichment analysis of miRNAs associated with cardiovascular death or heart failure hospitalizations. Barplot of statistically significant GO terms. GO categories are divided: BP (Biological process) and MF (Molecular function).
Figure S9. Boxplots showing the non‐normalized threshold cycle (Ct) values for the 14 miRNAs evaluated in the study.
Figure S10. Boxplots showing the non‐normalized threshold cycle (Ct) values for the 14 miRNAs by the occurrence of the primary (A) and secondary (B and C) endpoints.
Rincón, L. M. , Rodríguez‐Serrano, M. , Conde, E. , Lanza, V. F. , Sanmartín, M. , González‐Portilla, P. , Paz‐García, M. , Del Rey, J. M. , Menacho, M. , García Bermejo, M.‐L. , and Zamorano, J. L. (2022) Serum microRNAs are key predictors of long‐term heart failure and cardiovascular death after myocardial infarction. ESC Heart Failure, 9: 3367–3379. 10.1002/ehf2.13919.
Tweet: Circulating #miRNAs are strong predictors of #heartfailure, cardiovascular death and NYHA functional status in patients with #myocardialinfarction.
Luis M. Rincón and María‐Laura García Bermejo equally contributed as corresponding authors.
Contributor Information
Luis M. Rincón, Email: lmrincon@secardiologia.es.
María‐Laura García Bermejo, Email: garciabermejo@gmail.com.
References
- 1. Velagaleti RS, Pencina MJ, Murabito JM, Wang TJ, Parikh NI, D'Agostino RB, Levy D, Kannel WB, Vasan RS. Long‐term trends in the incidence of heart failure after myocardial infarction. Circulation. 2008; 118: 2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli‐Ducci C, Bueno HH, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, Hindricks G, Kastrati A, Lenzen MJ, Prescott E, Roffi M, Valgimigli M, Varenhorst C, Vranckx P, Widimský P, ESC Scientific Document Group , Widimsky P. 2017 ESC guidelines for the management of acute myocardial infarction in patients presenting with ST‐segment elevation: the task force for the management of acute myocardial infarction in patients presenting with ST‐segment elevation of the European Society. Eur Heart J England. 2018; 39: 119–177. [DOI] [PubMed] [Google Scholar]
- 3. Maegdefessel L. The emerging role of microRNAs in cardiovascular disease. J Intern Med. 2014; 276: 633–644. [DOI] [PubMed] [Google Scholar]
- 4. Liu X, Meng H, Jiang C, Yang S, Cui F, Yang P. Differential microRNA expression and regulation in the rat model of post‐infarction heart failure. PLoS ONE. 2016; 11: e0160920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rincón LM, Sanmartín M, Alonso GL, Rodríguez JA, Muriel A, Casas E, Navarro M, Carbonell A, Lázaro C, Fernández S, González P, Rodríguez M, Jiménez‐Mena M, Fernández‐Golfín C, Esteban A, García‐Bermejo ML, Zamorano JL. A genetic risk score predicts recurrent events after myocardial infarction in young adults. Rev Española Cardiol (English Ed.). 2020; 73: 623–631. [DOI] [PubMed] [Google Scholar]
- 6. Yu G, Wang L‐G, Han Y, He Q‐Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Proc Third Int AAAI Conf Weblogs Soc Media, San Jose, CA, USA. 2009; 17–20: 361–362. [Google Scholar]
- 8. Uno H, Tian L, Cai T, Kohane IS, Wei LJ. A unified inference procedure for a class of measures to assess improvement in risk prediction systems with survival data. Stat Med. 2013; 32: 2430–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, Hartmann T, Lackner KJ, Westermann D, Schnabel RB, Blankenberg S, Zeller T. MiRNA‐197 and miRNA‐223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS ONE. 2015; 10: e0145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karakas M, Schulte C, Appelbaum S, Ojeda F, Lackner KJ, Münzel T, Schnabel RB, Blankenberg S, Zeller T. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease‐results from the large AtheroGene study. Eur Heart J. 2017; 38: 516–523. [DOI] [PubMed] [Google Scholar]
- 11. Maciejak A, Kostarska‐Srokosz E, Gierlak W, Dluzniewski M, Kuch M, Marchel M, Opolski G, Kiliszek M, Matlak K, Dobrzycki S, Lukasik A, Segiet A, Sygitowicz G, Sitkiewicz D, Gora M, Burzynska B. Circulating miR‐30a‐5p as a prognostic biomarker of left ventricular dysfunction after acute myocardial infarction. Sci Rep. 2018; 8: 9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jakob P, Kacprowski T, Briand‐Schumacher S, Heg D, Klingenberg R, Stähli BE, Jaguszewski M, Rodondi N, Nanchen D, Räber L, Vogt P, Mach F, Windecker S, Völker U, Matter CM, Lüscher TF, Landmesser U. Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST‐segment elevation myocardial infarction. Eur Heart J. 2017; 38: 511–515. [DOI] [PubMed] [Google Scholar]
- 13. Hromádka M, Černá V, Pešta M, Kučerová A, Jarkovský J, Rajdl D, Rokyta R, Moťovská Z. Prognostic value of microRNAs in patients after myocardial infarction: a substudy of PRAGUE‐18. Dis Markers. 2019; 2019: 2925019–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2015; 12: 135–142. [DOI] [PubMed] [Google Scholar]
- 15. Shalaby SM, El‐Shal AS, Shoukry A, Khedr MH, Abdelraheim N. Serum miRNA‐499 and miRNA‐210: a potential role in early diagnosis of acute coronary syndrome. IUBMB Life. 2016; 68: 673–682. [DOI] [PubMed] [Google Scholar]
- 16. Zhu J, Lu K, Zhang N, Zhao Y, Ma Q, Shen J, Lin Y, Xiang P, Tang Y, Hu X, Chen J, Zhu W, Webster KA, Wang J, Yu H. Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA‐210 in an nSMase2‐dependent way. Artif Cells, Nanomed Biotechnol. 2018; 46: 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu S, Huang M, Nguyen PK, Gong Y, Li Z, Jia F, Lan F, Liu J, Nag D, Robbins RC, Wu JC. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation. 2011; 124: S27–S34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charrier H, Cuvelliez M, Dubois‐Deruy E, Mulder P, Richard V, Bauters C, Pinet F. Integrative system biology analyses identify seven microRNAs to predict heart failure. Non‐coding RNA. 2019; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veliceasa D, Biyashev D, Qin G, Misener S, Mackie AR, Kishore R, Volpert OV. Therapeutic manipulation of angiogenesis with miR‐27b. Vasc Cell. 2015; 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hinkel R, Ramanujam D, Kaczmarek V, Howe A, Klett K, Beck C, Dueck A, Thum T, Laugwitz K‐L, Maegdefessel L, Weber C, Kupatt C, Engelhardt S. AntimiR‐21 prevents myocardial dysfunction in a pig model of ischemia/reperfusion injury. J Am Coll Cardiol. 2020; 75: 1788–1800. [DOI] [PubMed] [Google Scholar]
- 21. Huang X, Huang F, Yang D, Dong F, Shi X, Wang H, Zhou X, Wang S, Dai S. Expression of microRNA‐122 contributes to apoptosis in H9C2 myocytes. J Cell Mol Med. 2012; 16: 2637–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating microRNA‐208b and microRNA‐499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010; 3: 499–506. [DOI] [PubMed] [Google Scholar]
- 23. Paavola J, Schliffke S, Rossetti S, Kuo IY‐T, Yuan S, Sun Z, Harris PC, Torres VE, Ehrlich BE. Polycystin‐2 mutations lead to impaired calcium cycling in the heart and predispose to dilated cardiomyopathy. J Mol Cell Cardiol. 2013; 58: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimokawa H, Sunamura S, Satoh K. RhoA/rho‐kinase in the cardiovascular system. Circ Res. 2016; 118: 352–366. [DOI] [PubMed] [Google Scholar]
- 25. Raucci A, Di MS, Scavello F, D'Ambrosio A, Bianchi ME, Capogrossi MC. The Janus face of HMGB1 in heart disease: a necessary update. Cell Mol Life Sci. 2019; 76: 211–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tham YK, Bernardo BC, Ooi JYY, Weeks KL, McMullen JR. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol. 2015; 89: 1401–1438. [DOI] [PubMed] [Google Scholar]
- 27. Rees ML, Subramaniam J, Li Y, Hamilton DJ, Frazier OH, Taegtmeyer H. A PKM2 signature in the failing heart. Biochem Biophys Res Commun. 2015; 459: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bye A, Røsjø H, Nauman J, Silva GJJ, Follestad T, Omland T, Wisløff U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals – The HUNT study. J Mol Cell Cardiol. 2016; 97: 162–168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Main clinical features of patients used for screening analysis. Age, group, clinical presentation and smoking habit are shown. MI, myocardial infarction; STEMI: ST‐elevation myocardial infarction; NSTEMI: non‐ST‐elevation myocardial infarction; NA: not applicable.
Table S2. Panel with selected candidate miRNAs and their expression levels among patients with myocardial infarction and healthy matched‐controls.
Table S3. Association of serum miRNAs per 1 standard deviation increase with cardiovascular mortality during follow‐up among the 311 patients with myocardial infarction.
Table S4. Association of serum miRNAs per 1 standard deviation increase with hospitalization for heart failure during follow‐up among the 311 patients with myocardial infarction.
Table S5. Association of serum miRNAs per 1 standard deviation increase with NYHA functional class II or higher at the end of follow‐up among the 311 patients with myocardial infarction*.
Table S6. Main clinical characteristics of the “healthy” population (without CV disease) adjusted for age and sex established as control group during the validation process (n = 30).
Table S7. Collection, processing and storage processes and conditions in the samples from cohort of patients with myocardial infarction and controls.
Figure S1. Heat map and hierarchical clustering of miRNAs in venous samples from 6 myocardial infarction and 10 healthy controls, performed as part of the miRNA profiling process. 752 miRNAs were analysed, and the top 42 miRNAs with highest mean differentiation between groups are shown. Normalized ΔCT values were used for the analysis.
Figure S2. Statistical association of some of the selected miRNAs with severity of index event: a) Killip‐Kimbal class; b) GRACE risk score at admission and c) troponin I levels at 24 h. miRNAs levels are expressed as delta crossing thresholds (DCT) and only those exhibiting a statistically significant association are shown.
Figure S3. Association of miRNAs with parameters indicative of acute heart failure/congestion: a) BNP levels (logarithmic), b) left ventricular ejection fraction at discharge.
Figure S4. Incidence of the composite of heart failure hospitalizations or cardiovascular mortality with the Kaplan–Meier method of miRs‐21, −23, −27, −122, −210 and −221 for values above and below the median.
Figure S5. Receiver‐operating characteristic (ROC) curves for prediction of the primary endpoint (heart failure hospitalization or cardiovascular death) at 2 years.
Figure S6. Assessing of IDI and cNRI with the addition of miR‐210‐3p, miR‐23a‐3p and miR‐221‐
Figure S7. Incidence of cardiovascular mortality over time with the Kaplan–Meier method of miRs‐21, −23, −27, −107, −210 and −221 for values above and below the median.
Figure S8. Functional enrichment analysis of miRNAs associated with cardiovascular death or heart failure hospitalizations. Barplot of statistically significant GO terms. GO categories are divided: BP (Biological process) and MF (Molecular function).
Figure S9. Boxplots showing the non‐normalized threshold cycle (Ct) values for the 14 miRNAs evaluated in the study.
Figure S10. Boxplots showing the non‐normalized threshold cycle (Ct) values for the 14 miRNAs by the occurrence of the primary (A) and secondary (B and C) endpoints.


