Abstract
Ethnopharmacological relevance
Traditional Chinese medicines (TCMs) have made great contributions to the prevention and treatment of human diseases in China, and especially in cases of COVID-19. However, due to quality problems, the lack of standards, and the diversity of dosage forms, adverse reactions to TCMs often occur. Moreover, the composition of TCMs makes them extremely challenging to extract and isolate, complicating studies of toxicity mechanisms.
Aim of the review: The aim of this paper is therefore to summarize the advanced applications of mass spectrometry imaging (MSI) technology in the quality control, safety evaluations, and determination of toxicity mechanisms of TCMs.
Materials and methods
Relevant studies from the literature have been collected from scientific databases, such as “PubMed”, “Scifinder”, “Elsevier”, “Google Scholar” using the keywords “MSI”, “traditional Chinese medicines”, “quality control”, “metabolomics”, and “mechanism”.
Results
MSI is a new analytical imaging technology that can detect and image the metabolic changes of multiple components of TCMs in plants and animals in a high throughput manner. Compared to other chemical analysis methods, such as liquid chromatography-mass spectrometry (LC-MS), this method does not require the complex extraction and separation of TCMs, and is fast, has high sensitivity, is label-free, and can be performed in high-throughput. Combined with chemometrics methods, MSI can be quickly and easily used for quality screening of TCMs. In addition, this technology can be used to further focus on potential biomarkers and explore the therapeutic/toxic mechanisms of TCMs.
Conclusions
As a new type of analysis method, MSI has unique advantages to metabolic analysis, quality control, and mechanisms of action explorations of TCMs, and contributes to the establishment of quality standards to explore the safety and toxicology of TCMs.
Keywords: MSI, TCMs, Quality control, Spatial metabolomics, Pharmacology and toxicology
Graphical abstract
Abbreviations:
- TCMs
traditional Chinese medicines
- MSI
mass spectrometry imaging
- IHC
immunohistochemistry
- LC-MS
liquid chromatography-mass spectrometry
- NMR
nuclear magnetic resonance
- m/z
mass to charge ratio
- MALDI
matrix-assisted laser desorption/ionization
- DESI-MSI
desorption electrospray ionization-mass spectrometry imaging
- SIMS
secondary ion mass spectrometry
- LA-ICP-MSI
laser ablation-inductively coupled plasma-mass spectrometry imaging
- CMC
carboxymethyl cellulose
- GC
gas chromatography
- LC
liquid chromatography
- TLC
thin-layer chromatography
- CE
capillary electrophoresis
- FT-IR
fourier-transform infrared
- PTFE
polytetrafluoroethylene
- SA
sinapic acid
- CHCA
α-cyano-4-hydroxycinnamicacid
- 2-MBT
2-mercaptobenzothiazole
- DHB
2,5-dihydroxybenzoicacid
- DHAP
2,5-dihydroxyacetopheno
- 9-AA
9-aminoacridine
- DAN
1,5-diaminonaphthalene
- MCA
3,4-dimethoxycinnamic acid
- PLL
poly-L-lysine
- MCAEF
matrix coating assisted by an electric field
- AP-SMALDI
atmospheric pressure-scanning microprobe matrix-assisted laser desorption/ionization
- PALDI-MS
plasma assisted laser desorption ionization mass spectrometry
- GALDI
colloidal graphite-assisted laser desorption/ionization
- TIAs
terpenoid indole alkaloids
- ICs
idioblast cells
- LCs
laticifer cells
- Q-makers
quality markers
- PCA
principal component analysis
- OPLS-DA
orthogonal partial least squares-discriminant analysis
- LDA
linear discriminate analysis
- LLS
local least square
- HELP
heuristic evolving latent projections
- OPA
orthogonal projection analysis
- LLF
ligustri lucidi fructus
- QWBA
quantitative whole body autoradiography
- GD
graphite dot
- PNGL
notoginseng leaf triterpenes
- NG-R1
notoginsenoside R1
- MCAO/R
middle cerebral artery occlusion/reperfusion
- ATP
adenosine triphosphate
1. Introduction
Traditional Chinese medicines (TCMs) have been used in the clinic for thousands of years and have shown good therapeutic effects. Due to the complexity of components and the characteristics of multi-target actions, TCMs can be used for broad opportunities, but face severe challenges. Given their various types, qualities, and efficacies, the key to the modernization of TCMs is to study their material bases, discover their therapeutic or toxic components, control their qualities, and clarify their targets and mechanisms of action.
A variety of analytical methods have been used for the identification and mechanistic evaluation of individual TCM components, and can be mainly divided into two categories: chromatographic methods (including gas chromatography (GC) and hyphenated techniques (Zhang et al., 2013), liquid chromatography (LC) and hyphenated techniques (Wang et al., 2021a), thin-layer chromatography (TLC)(Chen et al., 2021), capillary electrophoresis (CE)) and spectroscopic methods (fourier-transform infrared (FT-IR)(Mukrimin et al., 2019), near-infrared spectroscopy (NIR)(Li et al., 2017) and nuclear magnetic resonance (NMR)(Zhao et al., 2020)). GC-MS and LC-MS in chromatographic methods are two very popular chromatographic detection methods with high resolutions and sensitivities. The GC is appropriate for the determination of volatile components and LC is suitable for the identification of liquid ingredients in TCMs. However, the premise of these two methods requires complex pre-processing of samples, which will not only destroy information on the distribution of compounds in tissues, but may cause the loss of substances in low abundance (Prideaux and Stoeckli, 2012). FT-IR and NIR are spectroscopic methods that are non-invasive, rapid, and require simple sample preparations. However, their accuracies are lower than that of GC-MS and LC-MS. NMR has high accuracy and stability, but its sensitivity is poor, which renders it incapable of analyzing a large number of low abundance metabolites (Jiang et al., 2010). As an emerging analytical method, mass spectrometry imaging (MSI) overcomes the above technical defects. Without requiring complicated sample pre-processing steps, MSI can detect known or unknown compounds in high-throughput, while achieving high sensitivities and resolutions. In addition, this technology can convert a large volume of mass spectral data into images, retaining in situ information to show the distribution of drugs and small molecule metabolites (Nilsson et al., 2012; Nimesh et al., 2013; Prideaux and Stoeckli, 2012).
In recent years, spatially resolved metabolomics derived from MSI technology has been widely used in quality control and mechanistic studies of TCMs, and was first proposed by Sumner's research group at the Joint Annual Meeting of the American-Fern-Society in 2007 (Watson et al., 2007). Compared to traditional MS methods (LC-MS/GC-MS), MSI can retain the in situ spatial information of metabolites. The former “Spatially” of spatially resolved metabolomics can be used to accurately identify and locate the differential distribution of various metabolites in Chinese herbal medicines in tissues and cells, and perform rapid quality screening of drugs. The latter “metabolomics” can be used for in-depth metabolic analyses of target micro-regions to identify the types and contents of metabolites and discover potential efficacy or toxicity biomarkers of various components of TCMs. Such studies lay the foundation for understanding the possible medicinal and toxic mechanisms of TCMs (Bjarnholt et al., 2014; Ganesh et al., 2021).
This article reviews the principles and characteristics of MSI technology, as well as its application to the identification, distribution, quality control, the discovery of efficacy/toxicity biomarkers, and possible mechanisms of action of TCM components. This review aims to promote the application of MSI technology in Chinese herbal medicine and provide new directions for the discovery of drugs and the establishment of quality control standards for TCMs.
2. MSI: insights into the principles, indicators, and experimental processing
As a new type of molecular imaging technology, MSI performs mass spectrometry analysis and image visualization with high sensitivity, wide coverage, and strong identification ability. A variety of ions on the surface of tissue samples can be ionized point-by-point according to the spatial and multi-dimensional data of the mass to charge ratio (m/z), intensity, and position of ionized molecules obtained by mass spectrometry. Such data can be reconstructed and visualized using software (such as MassImager (He et al., 2018)) with the MSI functions of qualitative, quantitative, and positioning (Qin et al., 2018; Römpp and Spengler, 2013; Takahashi et al., 2015). Compared to LC-MS and immunohistochemistry (IHC), MSI can perform high-throughput detection of substances (endogenous and exogenous metabolites) in tissue sections, without requiring special labeling or complex pre-treatment, which can not only identify and analyze substances but also reveal their spatial distributions and relative contents in tissues (Schwamborn and Caprioli, 2010).
MSI was originally developed based on matrix-assisted laser desorption/ionization (MALDI). Therefore, MALDI-MSI is the most widely used mass spectrometry imaging method (Caprioli et al., 1997). In addition, related technologies include desorption electrospray ionization-mass spectrometry imaging (DESI-MSI), secondary ion mass spectrometry (SIMS) imaging, and laser ablation-inductively coupled plasma-mass spectrometry imaging (LA-ICP-MSI), etc.(de Souza et al., 2020; Oppenheimer and Drexler, 2011; Parrot et al., 2018). These technologies are mainly classified according to their ionization mode: SIMS imaging uses a primary ion beam to bombard the surface of the sample, and then introduces secondary ions sputtered from the surface into the mass spectrometer for mass separation and determination (Yoon and Lee, 2018). MALDI-MSI mainly makes use of a matrix to absorb the laser energy and then transfers energy to sample molecules for ionization (Knochenmuss, 2006). DESI-MSI uses atomized charged droplets to hit the surface of the sample. After being hit by high-speed droplets, the sample is sputtered and then subjected to the mass spectrometer (Ifa et al., 2007; Takáts et al., 2004). The ion source type, spatial resolution, sample preparation requirements, and other information of these three mass spectrometry imaging technologies are summarized in Table 1 .
Table 1.
Comparison of the three most commonly used MSI techniques.
| Ionization type | Ionization source | Environment | Resolution | Characteristic | Ref. |
|---|---|---|---|---|---|
| MALDI | IR/UV | High vacuum/low vacuum | IR:150 μm UV: 10–250 μm |
Need matrix, wide detection range | Heyman and Dubery (2016) |
| SIMS | Primary ion beam | High vacuum | 50 nm–5 μm | High resolution, high vacuum, easy to produce fragments of ions | Behrens et al. (2012) |
| DESI | Charged corpuscle | Atmospheric pressure | 100–200 μm | No matrix, atmospheric pressure | Ifa et al. (2007); Takáts et al. (2004); Wiseman et al. (2006) |
2.1. Critical indicators
Speed, spatial resolution, and sensitivity are critical indicators of MSI(Vestal et al., 2020). Speed is the main factor affecting the experimental time, and the scanning rate mainly depends on the influence of the laser frequency, mobile platform speed, and signal acquisition. The increase in scanning speed leads to a decrease in ionized ions (Tillner et al., 2017). In this case, high sensitivity is key to ensuring the imaging results of low abundance ions. Spatial resolution and sensitivity are negatively correlated and an improvement in sensitivity will inevitably lead to a decrease in the mass resolution (Vestal et al., 2020). Sensitivity is also closely related to ionization efficiency, ion transport efficiency, and ion detection (Merdas et al., 2021), while the mass resolution is mainly dependent on the specific desorption/ionization method used (Handberg et al., 2015; Römpp and Spengler, 2013) (Table 1). Therefore, MSI is a systematic project, in which the limit value of indicators should be selected according to the experiment.
2.2. Experimental process
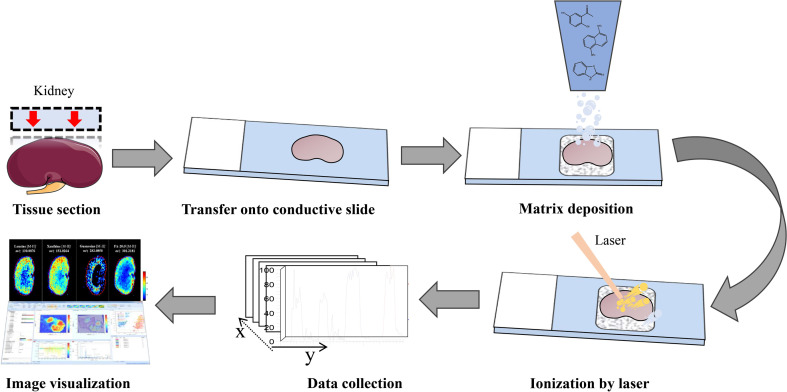
We will use the most widely used technology, MALDI-MSI, as an example to describe the specific experimental process. First, the appropriate sample preparation method is selected according to the nature of the animal/plant tissue sample; a suitable matrix is selected for spraying based on the type and nature of the test object; a laser beam is used to desorb and ionize each sampling point. Subsequently, the analyte ions are separated and detected by the mass spectrometer to obtain the mass spectra associated with the sample space position. Finally, the MSI map is obtained by matching and reorganizing all of the mass spectral data with their corresponding two-dimensional spatial positions using software (Fig. 1 ) (Dong et al., 2016; Grassl et al., 2011; Sturtevant et al., 2016). The following is an additional introduction to the key experimental steps to enhance the readers’ understanding.
Fig. 1.
The Experimental process of MALDI-MSI (kidney).
2.2.1. Sample preparation
Sample processing is the most critical step in MSI and the material basis for obtaining experimental results. The pretreatment method varies according to the type and location of the sample. For plant samples, a section of the roots, stems, and fruits is generally sliced using a cryostat microtome. Generally, such samples must also be embedded with gelatin (Beck and Stengel, 2016; Gemperline et al., 2014), 2% carboxymethyl cellulose (CMC) (Enomoto, 2020; Li et al., 2020b), or ice (Gorzolka et al., 2014), and frozen in liquid nitrogen prior to slicing into frozen sections (5–20 μm) at −20 °C. However, for plant stem slices with higher water contents or a young and small surface area, the sample is easily deformed or migration of the material occurs due to the blowing of spray gas. Thus, imprinting can be used for sample pretreatment in such situations. This technology utilizes external pressure to transfer a thin layer of plant tissue cell contents in situ to an adsorbent TLC plate (Liao et al., 2019) or the polytetrafluoroethylene (PTFE) membrane (Thunig et al., 2011) for imaging. For the petals and leaves, the surface must be kept as flat as possible, which can be directly blown or imprinted for indirect imaging.
2.2.2. Matrix selection
In MALDI-MS analysis, the image quality depends in large part on the establishment and optimization of the matrix system, and thus, the choice and spray type for the matrix is very important. Commonly used matrices include sinapic acid (SA)(Chaurand et al., 2008), α-cyano-4-hydroxycinnamicacid (CHCA)(Grassl et al., 2011; Lemaire et al., 2006), 2-mercaptobenzothiazole (2-MBT)(Astigarraga et al., 2008), 2,5-dihydroxybenzoicacid (DHB)(Li et al., 2016b), 2,5-dihydroxyacetopheno (DHAP)(Jovanović and Peter-Katalinić, 2016), 9-aminoacridine (9-AA)(Morikawa-Ichinose et al., 2019), and 1,5-diaminonaphthalene (DAN)(Korte and Lee, 2014). Among them, SA and DHAP are suitable for the detection of high molecular weight biomolecules (proteins, oligosaccharides, etc.), CHCA and 2-MBT are fit for the detection of medium molecular weight analytes (peptides, lipids), and DHB, DAN, and 9-AA are preferred for the detection of low molecular weight molecules (fatty acids, amino acids, nucleotides, etc.). In addition, some novel matrices such as quercetin (Wang et al., 2014), N-phenyl-2-naphthylamine (Liu, H. et al., 2018), graphene oxide (Wang et al., 2017), 3,4-dimethoxycinnamic acid (DMCA)(He, H. et al., 2019) and poly-L-lysine (PLL)(He, Y. et al., 2019) have been successfully used for MALDI-MSI. After selecting the suitable matrix according to the sample type, it is necessary to evenly cover the matrix solution on the surface of the tissue section to form good co-crystallization with the tissue surface molecules. There are three main methods of matrix covering, including manual spraying, automatic spraying, and vacuum sublimation (Bjarnholt et al., 2014). Furthermore, matrix coating assisted by an electric field (MCAEF) has also been proven to enhance tissue imaging (Wang et al., 2015).
2.2.3. Data processing
MSI will obtain large volumes of mass spectral data during high-throughput detection, which can be reconstructed and visualized into image information using imaging software (such as MassImager (He et al., 2018), R Packages (Ràfols et al., 2020), MSiReader (Desbenoit et al., 2018), etc.). Imaging software can image the ions individually or simultaneously to show the distribution of the target molecule in the sectioned tissue. The identification of target molecules can be based on the accurate mass value in commonly used mass spectrometry databases such as METLIN (http://metlin.scripps.edu/), HMDB (http://hmdb.ca/), MassBank (https://massbank.eu/MassBank/), and Lipid Maps (http://www.lipidmaps.org/.) for preliminary search matching. Then the verification of the compound is performed according to the specific fragment ions of the compound in the MS/MS experiment and other experimental support materials (such as nuclear magnetic or ultraviolet spectroscopy). In addition, the mass spectral data can be screened according to the experimental design and compared with KEGG (https://www.kegg.jp/) and other databases to explore the drug mechanisms of action.
MSI has a wide detection range from exogenous drugs to endogenous metabolites (lipids, peptides, etc.) and metals (Aichler and Walch, 2015). Sample preparation, parameter settings, data processing, and other MSI operations are detailed in the literature (Gessel et al., 2014; Kaletaş et al., 2009; Schulz et al., 2019). To date, MSI has been widely used in the fields of medicine (Schulz et al., 2019; Végvári, 2015), environment (Böhme et al., 2015), food (Morisasa et al., 2019), and plant biology (Kaspar et al., 2011; Korte et al., 2015; Qin et al., 2018). The MSI methods, research drugs, tissue types, and imaged molecules involved in this article are summarized in their order of appearance in Table 2 .
Table 2.
Published literature showing the application of MSI for the composition, quality control, and mechanisms of action of TCMs and natural products.
| Drug | Tissue type | Technical method | Imaged molecules | Ref. | |
|---|---|---|---|---|---|
| Salvia miltiorrhiza | Whole plant | MALDI-MSI | Functional metabolites | Sun et al. (2020) | |
| Salvia miltiorrhiza | Roots, stems and leaves | MALDI-MSI | Phenolic acids and tanshinones | Li et al. (2020b) | |
| Tripterygium | Roots | MALDI-MSI | Triterpenoids and sesquiterpene alkaloids | Lange et al. (2017) | |
| Paeonia lactiflora | Roots | AP-SMALDI MSI | Gallotannins and monoterpene glucosides | Li et al. (2016a) | |
| Maple | Xylem | TOF-SIMS imaging | Syringyl and guaiacyl lignin | Saito et al. (2012) | |
| Putterlickia pyracantha | Stems and roots | MALDI-MSI | Maytansinoids | Eckelmann et al. (2016) | |
| Scutellaria baicalensis | Roots | PALDI-based MSI | Baicalein and wogonin | Feng et al. (2014) | |
| Asclepias curassavica | Injury site | 3D-surface MALDI MSI | Plant defensive cardiac glycosides | Dreisbach et al. (2021) | |
| Glycyrrhiza glabra | Rhizome | AP-MALDI-MSI(Koestler et al., 2008) | Flavonoids, flavonoid glycosides and saponins | Li et al. (2014) | |
| Ginkgo biloba L. | Leaves | AP-MALDI-MSI | Flavonoid glycosides and biflavonoids | Beck and Stengel (2016) | |
| Catharanthus roseus | Stem tissue | MALDI-MSI | TIAs | Yamamoto et al. (2016) | |
| Catharanthus roseus | Leaves | MALDI-MSI | TIAs and precursors | Yamamoto et al. (2019) | |
| Panax ginseng | Roots | MALDI-MSI | Ginsenosides | Bai et al. (2016); Lee et al. (2017); Taira et al. (2010) | |
| Ginseng | Roots | DESI-MSI | Ginsenosides | Yang et al. (2021) | |
| Panax ginseng, Panax quinquefolius, and Panax notoginseng | Roots | MALDI-MSI | Saponins | Wang et al. (2016) | |
| Aconitum carmichaeli Debx | Roots | MALDI-MSI | Aconitum alkaloids | Wang et al. (2009) | |
| Paeonia suffruticosa and Paeonia lactiflora | Roots | MALDI-MSI | Monoterpene and paeonol glycosides, tannins, flavonoids, saccharides and lipids | Li et al. (2021) | |
| Ligustri Lucidi Fructus (LLF) | LLF fruits | MALDI-MSI | Q-markers | Li et al. (2020a) | |
| Vinblastine | The whole body of rats | MALDI-IMS-MSI | Sinblastine and metabolites | Trim et al. (2008) | |
| Salidroside | Multiple organs | MALDI-MSI | Salidroside | Meng et al. (2020) | |
| Puerarin | Mice kidney tissue | GD-4-assisted MSI | Puerarin and its two metabolites (daidzein and dihydrodaidzein) |
Shi et al. (2017) | |
| Scutellarin | Mice kidney tissue | MALDI-MSI | Scutellarin and scutellarein | Wang et al. (2021c) | |
| Notoginseng leaf triterpenes (PNGL) | Rat brain | MALDI-MSI | Endogenous metabolites | Wang et al. (2021b) | |
| Notoginsenoside R1 | Rat brain | MALDI-MSI | Endogenous metabolites | Zhu et al. (2020) | |
| Thymoquinone | Rat brain | MALDI-MSI | Endogenous metabolites | Tian et al. (2020) | |
| Radix Aconiti Lateralis Preparata extracts | Rat heart | MALDI-MSI | Endogenous metabolites | Wu et al. (2019) | |
3. MSI: A camera for showing the distribution of multiple components in a plant
Investigations of the basal metabolism of TCMs are the premise for identifying new drug candidates, increasing the clinical range of drugs, and improving quality control. Secondary metabolites (such as flavonoids, mushrooms, alkaloids, etc.) are the main components of TCMs that can prevent or cure diseases. The types, contents, and relative proportions of secondary metabolites are key to determining the effectiveness and quality of TCMs (Zhang et al., 2018) and MSI is suitable for detecting the content and distribution of primary/secondary metabolites in various plant structures (petals, roots, stems, leaves, seeds, seedlings)(Enomoto, 2020; Enomoto and Nirasawa, 2020; Qin et al., 2018; Sagara et al., 2019).
The conventional mass spectrometry method used to study the multiple components of TCMs is LC-MS. Complex pretreatment is generally required for LC-MS, including solvent extraction and chromatographic column separation before structural characterization. Such work not only requires substantial investigator energy and wastes a considerable amount of chemical reagents for sample preparation, but may also cause the loss of analytes or damage to the active ingredient (Wu et al., 2007). Furthermore, LC-MS fails to provide location information for the analyte in the tissue. Conversely, MSI can directly analyze the solid sections of plant tissues, without labeling and pre-processing and many studies have confirmed the advantages of direct analysis of plant tissues (Talaty et al., 2005; Wu et al., 2007). MSI can detect and identify the metabolic distribution of various components of TCMs while retaining in situ information, which is especially suitable for showing the material differences among different tissue parts of TCMs and the distribution characteristics of multiple components in the tissue (Hemalatha and Pradeep, 2013).
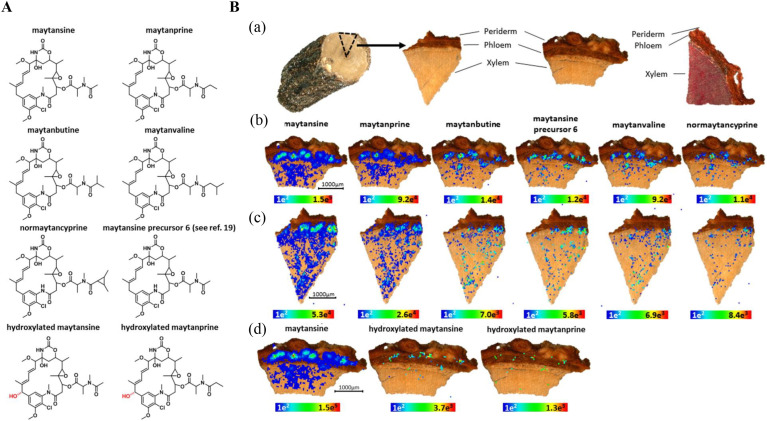
In studies of Salvia miltiorrhiza, MALDI-MSI was used to visualize the spatial dynamics of functional metabolites (such as amino acids, phenolic acids, fatty acids, oligosaccharides, cholines, etc.) (Sun et al., 2020) and MALDI-MSI was used to determine the distribution of metabolites in the tissue structures of roots, stems, and leaves. In this study, the characteristic constituents of the medicinal plant Salvia miltiorrhiza were identified as phenolic acids and tanshinones, which was consistent with the LC-MS data (Li et al., 2020b). MALDI-MSI was also used to identify and show the location of specific metabolites in Tripterygium roots (Lange et al., 2017). In a study of Paeonia lactiflora, atmospheric pressure-scanning microprobe matrix-assisted laser desorption/ionization mass spectrometry imaging (AP-SMALDI MSI, 10 μm/30 μm resolution) was used to detail the specific distribution of the major secondary metabolites, gallotannins and monoterpene glucosides, in root samples (Li et al., 2016a). SIMS imaging was used to characterize the morphological distribution of syringyl and guaiacyl lignin in the xylem of maple samples, which revealed a clear difference in the annual distribution of lignins between the fiber and vessel (Saito et al., 2012). Take Putterlickia Pyracantha as an example to illustrate in detail, the maytansinoids of Putterlickia pyracantha were visualized by AP-SMALDI MSI in the rhizome and were highly distributed in the vascular cambium region and the phloem. Such compounds were also widely distributed in the xylem and extremely low in the outer bark (periderm) of the stem. In addition, maytansine and maytanprine were also mainly detected in the central cylinder of the root (Fig. 2 )(Eckelmann et al., 2016).
Fig. 2.
MSI results from Putterlickia pyracantha stems (Eckelmann et al., 2016). A. Chemical structures of maytansinoids occurring in Putterlickia pyracantha. B. (a) Anatomical imaging of the cross section of Putterlickia pyracantha stems stained with phloroglucinol/HCL. (b–d) MALDI-imaging-HRMS of different Putterlickia pyracantha stem cuttings (spatial resolution: 40 μm; scan area: b: 3720 × 2600 μm; c: 3520 × 4120 μm; d: 3720 × 2600 μm). Localization of maytansine ([M+K]+; m/z 730.2503), maytanprine ([M+K]+; m/z 744.2659), maytanbutine ([M+K] +; m/z 758.2816), maytansine precursor 6 ([M+K] +; m/z 716.2347), maytanvaline ([M+K]+; m/z 772.2973), normaytancyprine ([M+K]+; m/z 770.2816), maytansine ([M+K]+; m/z 730.2503), hydroxylated maytansine ([M+K]+; m/z 746.2452), and hydroxylated maytanprine ([M+K]+; m/z 760.2609).
Due to high background noise in the low mass (<500 Da) region and the spatial inhomogeneity of matrix crystals formed on plant tissues, the application of MSI to the analysis of small molecule metabolites in plant tissues is more challenging than that in animal tissues. To improve the spatial resolution of MSI, some new ion sources were constructed for plant tissue imaging. Plasma assisted laser desorption ionization mass spectrometry (PALDI-MS) combines multiwavelength laser desorption and heated metastable plasma ionization of analytes, and does not require solvents to decrease ion suppression, reduce the pH effect, or simplify complicated spectra caused by adducts to a high spatial resolution of 60 μm × 60 μm. PALDI-based MSI for tissue section imaging of Scutellaria baicalensis showed that the two active components, baicalein, and wogonin, were mainly distributed in the epidermis of the root (Feng et al., 2014). To solve the problem of uneven matrix distribution in MDLDI-MSI, colloidal graphite was introduced as an alternative matrix that can be evenly distributed on the sample surface. Colloidal graphite-assisted laser desorption/ionization (GALDI) MS imaging was developed to analyze the metabolites of Arabidopsis, showing the specific distribution of flavonoids in Arabidopsis in the whole flower and a single petal (Cha et al., 2008). In addition, 3D-MSI has been developed as cutting-edge technology for plant imaging. The 3D-surface MALDI-MSI is the most recent instrumental approach in AP-SMALDI MSI and was developed to characterize the specific distribution of plant defensive cardiac glycosides at injury sites in Asclepias curassavica (Dreisbach et al., 2021).
Most of the MALDI imaging experiments performed on plant tissues have a spatial resolution of 50–200 μm. With high resolutions in mass and space, this technology has been applied to cell-level imaging in plants. The AP-MALDI-MSI approach (Koestler et al., 2008) that was independently developed by Li's laboratory achieves 10 μm resolution in cell level imaging in plants, thus, showing the distribution of the main natural products (flavonoids, flavonoid glycosides, and saponins) of Glycyrrhiza glabra (licorice)(Li et al., 2014). The technology was also used to detect and identify the distribution of flavonoid glycosides and biflavonoids in Ginkgo biloba L (Beck and Stengel, 2016). A study used MALDI-MSI based on the FT-ICR-MS detector (with a spatial resolution of 20 μm) to show that most of the terpenoid indole alkaloids (TIAs) in the stem tissue of Catharanthus roseus were accumulated in idioblast cells (ICs) and laticifer cells (LCs) (Yamamoto et al., 2016). Another study also used the FT-ICR-MS detector to image the leaves of Catharanthus roseus at a resolution of 10 μm, and was combined with single cell MS analysis to detail the biosynthesis of TIAs and determine the cell-specific localization of TIAs in leaf tissue (Yamamoto et al., 2019).
MSI technology can achieve high resolution cell and tissue imaging, showing the specific distribution of the functional metabolites of TCMs and laying a foundation for subsequent mechanistic exploration.
4. MSI: A simple and quick way to discover the quality markers of TCMs
Due to the polymorphism of medicinal plants, the quality control of drugs is a complicated process and includes a detailed characterization of the appearance, active ingredients, and physical and chemical properties of TCMs, as well as the quantification (absolute dry weight, yield, etc.), manufacturing (temperature, solvent, extraction and drying time), impurity testing, and chemical content determinations of the final active pharmaceutical ingredients (Liu, C. et al., 2018). In recent years, to improve the consistency and quality control of TCMs, quality markers (Q-makers) have been introduced ; (Guo, 2017; Liu et al., 2016). Q-markers of TCMs refer to substances that can be characterized qualitatively and quantitatively and are closely related to the function of the TCM in raw materials or during the processing and preparation of TCMs. The image of Q-makers plays an important role in the authenticity identification and quality assessment of TCMs (including raw materials, extracts, products, and compound preparations)(Yang et al., 2017). There are a variety of methods and strategies used for the discovery of Q-markers, including genomics, metabolomics, system pharmacology, pharmacokinetic analyses, and spider-web mode (Ren et al., 2020).
As mentioned, MSI can detect the content and distribution of multiple components of TCMs in a high throughput manner. As a new analytical method, this technique has been used to discover the quality markers of TCMs. In this application, massive volumes of mass spectral data are generated and subsequently analyzed and processed by chemometric methods. Such methods mainly include principal component analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA), linear discriminate analysis (LDA), local least square (LLS), heuristic evolving latent projections (HELP), and orthogonal projection analysis (OPA)(Bansal et al., 2014). Compared to other chemical analyses such as LC-MS and UV, MSI does not require complicated sample extraction and separation steps, and does not lose low-abundance components. Thus, MSI quickly distinguishes the active ingredients and metabolic characteristics of different drugs, as well as readily identifies Q-markers. All such capabilities are suitable to rapidly and semi-quantitatively perform quality screening of TCMs (Huang et al., 2016).
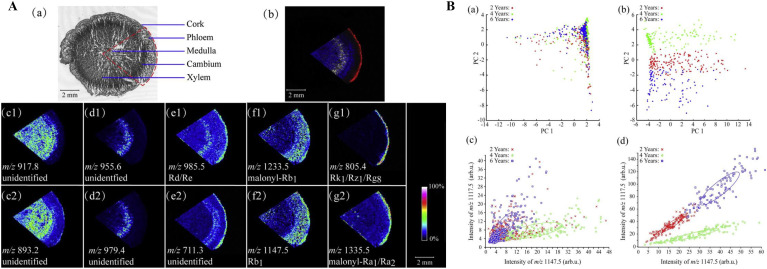
Panax ginseng is a type of precious Chinese medicine, known as the king of medicines. However, as there are multiple species of Panax ginseng, the origin, age, efficacy, and nutritional value of ginseng medicines are also different, and counterfeit or substandard products often exist in the market. Ginsenoside is the main active component in Panax ginseng, and the content of ginsenoside increases with plant age. Many studies have used MSI to reveal that ginsenosides are mainly distributed in the sebaceous layer and part of the cortex of Panax ginseng tissue located in the center of the root. Dozens of ginsenoside analytes have been identified by MS/MS as specific markers for quickly distinguishing different varieties, ages, and organs of Panax ginseng based on their specific distributions in tissues (Fig. 3 )(Bai et al., 2016; Lee et al., 2017; Taira et al., 2010; Wang et al., 2016; Yang et al., 2021). In one study, UPLC-QTOF MS and DESI-MSI were simultaneously used to detect and characterize the age and parts of ginseng to identify the common biomarkers across different age groups using the OPLS-DA method. The results showed that compared to UPLC-QTOF MS, DESI-MSI was a novel and stable method for the rapid evaluation of ginseng root slices (Yang et al., 2021). In addition, LC-MS and MALDI-MSI were also used to analyze Aconitum alkaloids in the Chinese herbal medicine, Aconitum carmichaeli Debx. The results between the two analytical methods were consistent and revealed significant differences in the contents of alkaloids between different samples. The comparative study using two analytical methods showed that MALDI-MSI was a more rapid and robust analytical method than LC-MS for semi-quantitative analyses of high concentration alkaloids (Wang et al., 2009). In addition, spatial metabolomics based on MALDI-MSI was also used to comprehensively and accurately detect the differential distribution of metabolites in Paeonia suffruticosa and Paeonia lactiflora (both belonging to genus Paeonia), including monoterpenes and paeonol glycosides, tannins, flavonoids, carbohydrates, and lipids, and it was also used to further visualize the gallotannins biosynthesis pathway in the roots of Paeonia suffruticosa and Paeonia lactiflora (Li et al., 2021). Most TCMs are crude drugs and the majority of which must be processed to reduce their toxicity in clinical medications. A strategy integrating multi-component characterization, non-target metabolomics, and MSI was proposed for quality control during processing. MSI was used to visualize the spatial distribution of four main biomarkers in the Ligustri Lucidi Fructus (LLF) based on steaming time (Li et al., 2020a).
Fig. 3.
MALDI-MSI distinguishing ginseng of different ages based on the localization of ginsenosides (Bai et al., 2016) A. (a) Optical scan image of ginseng. (b) Overlay of ion images: red, m/z 805.5 (Rg8/Rk/Rz1); yellow, m/z 955.6 (unidentified); blue, m/z 917.8 (unidentified). (c–g) Five localization modes of signals: xylem-medulla type (c1 and c2); xylem-only type (d1 and d2); cork-xylem type (e1 and e2); cork-phloem-cambium-medulla type (f1 and f2); and cork-only type (g1 and g2). B. PCA score plot and 2D peak distribution plot of m/z 1117.5 and m/z 1147.5: a and c, whole tissue; b and d, cork. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
MSI technique was used to discover Q-makers, providing a new direction and insights for the quality control of TCMs. As a rapid evaluation method, MSI has a broad applicability for the quality control of TCMs.
5. MSI: A tool for studying the metabolic distribution and therapeutic/toxic mechanisms of TCMs
MSI can be applied to entire animal bodies or multiple tissue sections to observe the distribution of metabolites of active components in each organ, and to determine the target organ and toxicity.
A study on the anticancer drug, vinblastine, performed MALDI-IMS-MS whole body imaging. The results showed that most of the product ions of vinblastine were highly distributed in the liver, renal cortex, and surrounding the gastric intestinal tract. The accuracy of the MSI results was verified by quantitative whole body autoradiography (QWBA)(Trim et al., 2008). By collecting multiple organ samples from mice at various time points after intravenous administration of salidroside, MALDI-MSI visualized the temporal and spatial distribution of salidroside showing that salidroside was heterogeneously distributed throughout the kidney and heart, and could be quickly eliminated with 5 min (Meng et al., 2020).
The distribution of drugs in tumor tissues or organs is heterogeneous. The possible metabolic pathways of TCMs can also be predicted by MSI to analyze the distribution of active components and their metabolites in microregions of tissues or organs. It has been found that hydroxyl-group-dominated graphite dots (GD) are an ideal matrix with extremely low background noise and ultra-high sensitivity. GD-4-assisted MSI has been used to show the distribution characteristics of puerarin and its metabolites in renal microregions showing that puerarin was primarily distributed in the renal pelvis and major calyx. However, its metabolites (daidzein and dihydrodaidzein) were also detected in the renal pelvis, major calyx, and partly in the minor calyx, but were nearly absent in the medulla (Shi et al., 2017). In another study, MALDI-MSI was also used to identify the in situ localization of scutellarin (traditional Chinese botanic drug of Erigeron breviscapus extract) and its metabolites to show metabolic differences in the kidney (Wang et al., 2021d). Imaging the distribution characteristics after drug administration facilitates an understanding of the biological activity and metabolism of drugs in various animal organs.
In recent years, various cutting-edge omics technologies (genomics, transcriptomics, proteomics, metabolomics, lipidomics) have been applied to diverse fields of TCM research, including screening, quality control, research and development, mechanistic research, and clinical verification. Taking metabolomics as an example, metabolism reflects the changes of small molecule metabolites in the body. Metabolomics with high-throughput monitoring can identify the metabolic network of molecules following drug administration, which has become a powerful tool effectively breaking through the application bottleneck of the study of the multi-component mechanisms of TCMs. The discovery of metabolic markers provides a foundation for the early identification of toxicity, quality control, and clinical utility of TCMs (Han et al., 2020; Shi et al., 2016; Sun et al., 2012; Wang et al., 2021a). As MSI is a high-throughput and label-free technology, it can obtain drug metabolism distribution information and also endogenous small molecule metabolism information (that is, metabonomics data) from the same animal tissue. Compared to traditional metabolomics methods, spatial high resolution metabolomics studies based on MSI can preserve tissue integrity and visualize the distribution of metabolites. Researchers can also superimpose MS images with optical/HE scanning images and focus on the tissue microregions or lesions of interest to accurately extract mass spectral data for the target area for metabolic research; thus, avoiding the challenges associate with difficult separations of research specimens.
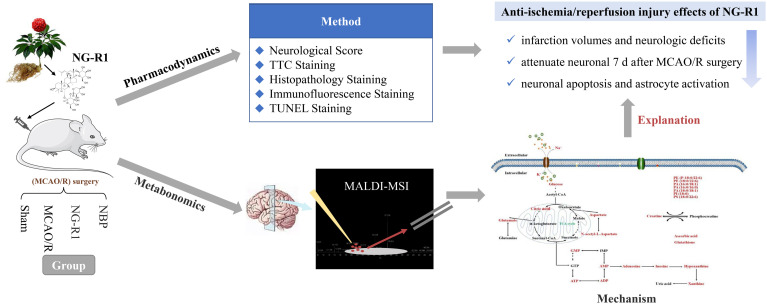
Panax notoginseng is a traditional Chinese medicine and is widely used for the treatment and prevention of ischemic cerebrovascular diseases (Yan et al., 2018). Notoginseng leaf triterpenes (PNGL) and notoginsenoside R1 (NG-R1, Fig. 4 ) extracted from Panax notoginseng were visualized by MALDI-MSI to study the effect on small molecule metabolism after perfusion injury. According to the results, the two drugs had a callback effect on the tricarboxylic acid (TCA) cycle and adenosine triphosphate (ATP) metabolism pathway, and also played a role in improving the malate-aspartate shuttle; thus, improving the antioxidant capacity and maintaining the homeostasis of Na+ and K+ (Wang et al., 2021b; Zhu et al., 2020). In the similar disease model, Fang et al. also used MALDI-MSI to explore the role of Thymoquinone, the main active ingredient in Nigella sativa, in regulating abnormal metabolism in injured brain areas by promoting the aerobic oxidation of glucose, regulating intracellular energy metabolism, improving the phospholipid molecular level, increasing the content of small antioxidant molecules, and balancing sodium homeostasis (Tian et al., 2020). According to the results of metabolomics studies in the same model, it is known that the mechanisms of related drugs for the treatment of stroke and other central nervous system diseases begin with mitochondrial oxidative damage, energy metabolism, lipid metabolism disorders, and Na+ homeostasis. In another study, MALDI-MSI was used to study anti-myocardial infarction effects of Radix Aconiti Lateralis Preparata extracts. Pharmacodynamics results showed that Radix Aconiti Lateralis Preparata extracts can improve the hemodynamic status and organ weight index and inhibit myocardial injury of rats with myocardial infarction. The corresponding MALDI-MSI results elucidated the possible mechanism of action by presenting Radix Aconiti Lateralis Preparata extracts to reverse metabolic changes of related small molecules (energy metabolism-related molecules, phospholipids, potassium ions, and glutamine in the heart) to produce anti-myocardial infarction effects (Wu et al., 2019). The identification of potential biomarkers of TCMs based on changes in the metabolic networks of small molecules in vivo, thus, lays a foundation for further exploration of the mechanisms of action.
Fig. 4.
Spatially resolved metabolomics based on MSI to elucidate the pharmacodynamic mechanisms of NG-R1 (Zhu et al., 2020). The rats in this study were divided into four groups: Sham, MCAO/R, NG-R1 (20 mg/kg, 7 days), and NBP (20 mg/kg, 7 days). Pharmacodynamic studies (included neurological score, TTC staining, histopathology staining, immunofluorescence staining, and TUNEL staining) conducted 7 days after ischemic-reperfusion showed that NG-R1 can reduce infarction volumes and neurologic deficits in MCAO/R rats and attenuate neuronal loss 7 d after MCAO/R surgery, while also inhibiting neuronal apoptosis and astrocyte activation. To clarify the mechanisms by which those events occur, the study further used spatially resolved metabolomics based on MALDI-MSI and found that NG-R1 can regulate the abnormal accumulation of glucose and citric acid, increase the content of glutamate and malate-aspartic acid shuttle components, increase antioxidant content, increase ATP metabolism, and maintain the homeostasis of Na+ and K+ to achieve anti-ischemia/reperfusion injury effects.
Spatial metabolomics based on MSI can detail the interactions between metabolites, and further screen and identify biomarkers with significant changes by comparing the correlation between metabolomics spectra and histopathological/biochemical indicators. Finally, the analysis of related metabolic pathways can reveal the possible effects or toxic mechanisms of TCMs. The above studies illustrate that spatial metabolomics analyses based on MSI methods are powerful in exploring the therapeutic effects of TCMs and provide insights into the potential mechanisms of action of TCMs.
6. Summary and conclusion
In recent years, MSI has attracted the attention of many researchers and was rapidly developed. Currently, the quality control of most TCMs is limited to the identification and analysis following extraction and separation, and the process is cumbersome and time-consuming. The ingredients with lower concentrations are often overlooked and are not the focus of studies. MSI provides a new method for the rapid screening and control of the quality of TCMs. The understanding of modern medicine in TCMs has developed from macroscopic to microscopic considerations. In particular, the discovery and identification of active components of TCMs in the body is a key research topic. MSI technology has become a powerful tool for the analysis of metabolites in animal/plant tissues, as well as single cells, providing a means to study transport pathways, metabolic pathways, and the accumulation of exogenous drugs in animal tissues and endogenous metabolites in plant tissues. The multi-component and multi-target synergistic characteristics of TCMs have been advantageous for the treatment of certain chronic diseases. Extracting active ingredients from TCMs and isolating monomers is a key approach to the identification of new drugs. MSI also offers a new visual perspective and provides multi-dimensional information for metabolomics analysis. However, MSI technology has faced many challenges, such as its limited spatial resolution and insufficient sensitivity. By improving sample preparation methods, matrix replacement, algorithm optimization, and instrument improvements (Abdelmoula et al., 2018; Alexandrov et al., 2011; He et al., 2015; Morikawa-Ichinose et al., 2019; Song et al., 2017), MSI technology has achieved substantial breakthroughs in its sensitivity, resolution and sample suitability. With the integration of MSI with other technologies (Porta Siegel et al., 2018), such as LC-MS(Desbenoit et al., 2013), microscopic imaging (Tian et al., 2019; Van de Plas et al., 2015), Raman spectroscopy (Bocklitz et al., 2015), and magnetic resonance imaging (Verbeeck et al., 2017), the application of MSI technology to TCMs research will also become broader.
Declaration of competing interests
The authors report no conflicts of interest.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81773996, 81773678, 81973476, and 82074104]; the National Major Scientific and Technological Special Project for “Major New Drugs Development” [grant numbers 2018ZX09735006, 2018ZX09711001-002-001]; and the Research Project of Clinical Toxicology Transformation from the Chinese Society of Toxicology [grant number CST2019CT105].
Declaration of interest
The authors declare no conflict of interests.
CRediT authorship contribution statement
Haiyan Jiang: conceived the idea of the topic scope, wrote the manuscript, performed the literature search, and analyzed the data. Yaxin Zhang: wrote the manuscript, performed the literature search, and analyzed the data. Zhigang Liu: performed experiments and data collection. Xiangyi Wang: performed experiments and data collection. Jiuming He: conceived the idea of the topic scope, performed data analysis and critically revised the manuscript. Hongtao Jin: conceived the idea of the topic scope, performed data analysis and critically revised the manuscript.
Acknowledgments
The authors are grateful to International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
References
- Abdelmoula W.M., Pezzotti N., Hölt T., Dijkstra J., Vilanova A., McDonnell L.A., Lelieveldt B.P.F. Interactive visual exploration of 3D mass spectrometry imaging data using hierarchical stochastic neighbor embedding reveals spatiomolecular structures at full data resolution. J. Proteome Res. 2018;17(3):1054–1064. doi: 10.1021/acs.jproteome.7b00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichler M., Walch A. MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab. Invest. 2015;95(4):422–431. doi: 10.1038/labinvest.2014.156. [DOI] [PubMed] [Google Scholar]
- Alexandrov T., Meding S., Trede D., Kobarg J., Balluff B., Walch A., Thiele H., Maass P. Super-resolution segmentation of imaging mass spectrometry data: solving the issue of low lateral resolution. J Proteomics. 2011;75(1):237–245. doi: 10.1016/j.jprot.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Astigarraga E., Barreda-Gómez G., Lombardero L., Fresnedo O., Castaño F., Giralt M.T., Ochoa B., Rodríguez-Puertas R., Fernández J.A. Profiling and imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal. Chem. 2008;80(23):9105–9114. doi: 10.1021/ac801662n. [DOI] [PubMed] [Google Scholar]
- Bai H., Wang S., Liu J., Gao D., Jiang Y., Liu H., Cai Z. Localization of ginsenosides in Panax ginseng with different age by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry imaging. J. Chromatogr. B. 2016;1026:263–271. doi: 10.1016/j.jchromb.2015.09.024. [DOI] [PubMed] [Google Scholar]
- Bansal A., Chhabra V., Rawal R.K., Sharma S. Chemometrics: a new scenario in herbal drug standardization. J Pharm Anal. 2014;4(4):223–233. doi: 10.1016/j.jpha.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S., Stengel J. Mass spectrometric imaging of flavonoid glycosides and biflavonoids in Ginkgo biloba L. Phytochemistry. 2016;130:201–206. doi: 10.1016/j.phytochem.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Behrens S., Kappler A., Obst M. Linking environmental processes to the in situ functioning of microorganisms by high-resolution secondary ion mass spectrometry (NanoSIMS) and scanning transmission X-ray microscopy (STXM) Environ. Microbiol. 2012;14(11):2851–2869. doi: 10.1111/j.1462-2920.2012.02724.x. [DOI] [PubMed] [Google Scholar]
- Bjarnholt N., Li B., D'Alvise J., Janfelt C. Mass spectrometry imaging of plant metabolites – principles and possibilities. Nat. Prod. Rep. 2014;31(6):818–837. doi: 10.1039/C3NP70100J. [DOI] [PubMed] [Google Scholar]
- Bocklitz T., Bräutigam K., Urbanek A., Hoffmann F., von Eggeling F., Ernst G., Schmitt M., Schubert U., Guntinas-Lichius O., Popp J. Novel workflow for combining Raman spectroscopy and MALDI-MSI for tissue based studies. Anal. Bioanal. Chem. 2015;407(26):7865–7873. doi: 10.1007/s00216-015-8987-5. [DOI] [PubMed] [Google Scholar]
- Böhme S., Stärk H.J., Kühnel D., Reemtsma T. Exploring LA-ICP-MS as a quantitative imaging technique to study nanoparticle uptake in Daphnia magna and zebrafish (Danio rerio) embryos. Anal. Bioanal. Chem. 2015;407(18):5477–5485. doi: 10.1007/s00216-015-8720-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli R.M., Farmer T.B., Gile J. Molecular imaging of biological Samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Cha S., Zhang H., Ilarslan H.I., Wurtele E.S., Brachova L., Nikolau B.J., Yeung E.S. Direct profiling and imaging of plant metabolites in intact tissues by using colloidal graphite-assisted laser desorption ionization mass spectrometry. Plant J. 2008;55(2):348–360. doi: 10.1111/j.1365-313X.2008.03507.x. [DOI] [PubMed] [Google Scholar]
- Chaurand P., Latham J.C., Lane K.B., Mobley J.A., Polosukhin V.V., Wirth P.S., Nanney L.B., Caprioli R.M. Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: bypassing formalin fixation. J. Proteome Res. 2008;7(8):3543–3555. doi: 10.1021/pr800286z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li L., Xu R., Li F., Gu L., Liu H., Wang Z., Yang L. Characterization of natural herbal medicines by thin-layer chromatography combined with laser ablation-assisted direct analysis in real-time mass spectrometry. J. Chromatogr. A. 2021;1654:462461. doi: 10.1016/j.chroma.2021.462461. [DOI] [PubMed] [Google Scholar]
- de Souza L.P., Borghi M., Fernie A. Plant single-cell metabolomics-challenges and perspectives. Int. J. Mol. Sci. 2020;21(23):8987. doi: 10.3390/ijms21238987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbenoit N., Schmitz-Afonso I., Baudouin C., Laprévote O., Touboul D., Brignole-Baudouin F., Brunelle A. Localisation and quantification of benzalkonium chloride in eye tissue by TOF-SIMS imaging and liquid chromatography mass spectrometry. Anal. Bioanal. Chem. 2013;405(12):4039–4049. doi: 10.1007/s00216-013-6811-7. [DOI] [PubMed] [Google Scholar]
- Desbenoit N., Walch A., Spengler B., Brunelle A., Römpp A. Correlative mass spectrometry imaging, applying time-of-flight secondary ion mass spectrometry and atmospheric pressure matrix-assisted laser desorption/ionization to a single tissue section. Rapid Commun. Mass Spectrom. : RCM (Rapid Commun. Mass Spectrom.) 2018;32(2):159–166. doi: 10.1002/rcm.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Li B., Malitsky S., Rogachev I., Aharoni A., Kaftan F., Svatoš A., Franceschi P. Sample preparation for mass spectrometry imaging of plant tissues: a review. Front. Plant Sci. 2016;7(60) doi: 10.3389/fpls.2016.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisbach D., Petschenka G., Spengler B., Bhandari D.R. 3D-surface MALDI mass spectrometry imaging for visualising plant defensive cardiac glycosides in Asclepias curassavica. Anal. Bioanal. Chem. 2021;413(8):2125–2134. doi: 10.1007/s00216-021-03177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelmann D., Kusari S., Spiteller M. Occurrence and spatial distribution of maytansinoids in Putterlickia pyracantha, an unexplored resource of anticancer compounds. Fitoterapia. 2016;113:175–181. doi: 10.1016/j.fitote.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Enomoto H. Mass spectrometry imaging of flavonols and ellagic acid glycosides in ripe strawberry fruit. Molecules. 2020;25(20) doi: 10.3390/molecules25204600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H., Nirasawa T. Localization of flavan-3-ol species in peanut testa by mass spectrometry imaging. Molecules. 2020;25(10):2373. doi: 10.3390/molecules25102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Zhang J., Chang C., Li L., Li M., Xiong X., Guo C., Tang F., Bai Y., Liu H. Ambient mass spectrometry imaging: plasma assisted laser desorption ionization mass spectrometry imaging and its applications. Anal. Chem. 2014;86(9):4164–4169. doi: 10.1021/ac403310k. [DOI] [PubMed] [Google Scholar]
- Ganesh S., Hu T., Woods E., Allam M., Cai S., Henderson W., Coskun A.F. Spatially resolved 3D metabolomic profiling in tissues. Sci Adv. 2021;7(5) doi: 10.1126/sciadv.abd0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperline E., Jayaraman D., Maeda J., Ané J.-M., Li L. Multifaceted investigation of metabolites during nitrogen fixation in Medicago via high resolution MALDI-MS imaging and ESI-MS. J. Am. Soc. Mass Spectrom. 2014;26(1):149–158. doi: 10.1007/s13361-014-1010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessel M.M., Norris J.L., Caprioli R.M. MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery. J Proteomics. 2014;107:71–82. doi: 10.1016/j.jprot.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzolka K., Bednarz H., Niehaus K. Detection and localization of novel hordatine-like compounds and glycosylated derivates of hordatines by imaging mass spectrometry of barley seeds. Planta. 2014;239(6):1321–1335. doi: 10.1007/s00425-014-2061-y. [DOI] [PubMed] [Google Scholar]
- Grassl J., Taylor N.L., Millar A. Matrix-assisted laser desorption/ionisation mass spectrometry imaging and its development for plant protein imaging. Plant Methods. 2011;7(1):21. doi: 10.1186/1746-4811-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.A. Quality marker concept inspires the quality research of traditional Chinese medicines. Chin. Herbal Med. 2017;9(1):1–2. doi: 10.1016/S1674-6384(17)60069-8. [DOI] [Google Scholar]
- Han Y., Sun H., Zhang A., Yan G., Wang X.J. Chinmedomics, a new strategy for evaluating the therapeutic efficacy of herbal medicines. Pharmacol. Ther. 2020;216:107680. doi: 10.1016/j.pharmthera.2020.107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg E., Chingin K., Wang N., Dai X., Chen H. Mass spectrometry imaging for visualizing organic analytes in food. Mass Spectrom. Rev. 2015;34(6):641–658. doi: 10.1002/mas.21424. [DOI] [PubMed] [Google Scholar]
- He H., Qin L., Zhang Y., Han M., Li J., Liu Y., Qiu K., Dai X., Li Y., Zeng M., Guo H., Zhou Y., Wang X. 3,4-Dimethoxycinnamic acid as a novel matrix for enhanced in situ detection and imaging of low-molecular-weight compounds in biological tissues by MALDI-MSI. Anal. Chem. 2019;91(4):2634–2643. doi: 10.1021/acs.analchem.8b03522. [DOI] [PubMed] [Google Scholar]
- He J., Huang L., Tian R., Li T., Sun C., Song X., Lv Y., Luo Z., Li X., Abliz Z. MassImager: a software for interactive and in-depth analysis of mass spectrometry imaging data. Anal. Chim. Acta. 2018;1015:50–57. doi: 10.1016/j.aca.2018.02.030. [DOI] [PubMed] [Google Scholar]
- He J., Luo Z., Huang L., He J., Chen Y., Rong X., Jia S., Tang F., Wang X., Zhang R., Zhang J., Shi J., Abliz Z. Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Anal. Chem. 2015;87(10):5372–5379. doi: 10.1021/acs.analchem.5b00680. [DOI] [PubMed] [Google Scholar]
- He Y., Guo W., Luo K., Sun Q., Lin Z., Cai Z. Poly-l-lysine-based tissue embedding compatible with matrix-assisted laser desorption ionization-mass spectrometry imaging analysis of dry and fragile aristolochia plants. J. Chromatogr. A. 2019;1608:460389. doi: 10.1016/j.chroma.2019.460389. [DOI] [PubMed] [Google Scholar]
- Hemalatha R.G., Pradeep T. Understanding the molecular signatures in leaves and flowers by desorption electrospray ionization mass spectrometry (DESI MS) imaging. J. Agric. Food Chem. 2013;61(31):7477–7487. doi: 10.1021/jf4011998. [DOI] [PubMed] [Google Scholar]
- Heyman H.M., Dubery I.A. The potential of mass spectrometry imaging in plant metabolomics: a review. Phytochemistry Rev. 2016;15(2):297–316. doi: 10.1007/s11101-015-9416-2. [DOI] [Google Scholar]
- Huang Y., Wu Z., Su R., Ruan G., Du F., Li G. Current application of chemometrics in traditional Chinese herbal medicine research. J. Chromatogr. B. 2016;1026:27–35. doi: 10.1016/j.jchromb.2015.12.050. [DOI] [PubMed] [Google Scholar]
- Ifa D.R., Wiseman J.M., Song Q., Cooks R.G. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI) Int. J. Mass Spectrom. 2007;259(1):8–15. doi: 10.1016/j.ijms.2006.08.003. [DOI] [Google Scholar]
- Jiang Y., David B., Tu P., Barbin Y. Recent analytical approaches in quality control of traditional Chinese medicines—a review. Anal. Chim. Acta. 2010;657(1):9–18. doi: 10.1016/j.aca.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Jovanović M., Peter-Katalinić J. Negative ion MALDI-TOF MS, ISD and PSD of neutral underivatized oligosaccharides without anionic dopant strategies, using 2,5-DHAP as a matrix. J. Mass Spectrom. 2016;51(2):111–122. doi: 10.1002/jms.3727. [DOI] [PubMed] [Google Scholar]
- Kaletaş B.K., van der Wiel I.M., Stauber J., Dekker L.J., Güzel C., Kros J.M., Luider T.M., Heeren R.M.A. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- Kaspar S., Peukert M., Svatos A., Matros A., Mock H.P. MALDI-imaging mass spectrometry - an emerging technique in plant biology. Proteomics. 2011;11(9):1840–1850. doi: 10.1002/pmic.201000756. [DOI] [PubMed] [Google Scholar]
- Knochenmuss R. Ion formation mechanisms in UV-MALDI. Analyst. 2006;131(9):966–986. doi: 10.1039/b605646f. [DOI] [PubMed] [Google Scholar]
- Koestler M., Kirsch D., Hester A., Leisner A., Guenther S., Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun. Mass Spectrom. : RCM (Rapid Commun. Mass Spectrom.) 2008;22(20):3275–3285. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- Korte A.R., Lee Y.J. MALDI-MS analysis and imaging of small molecule metabolites with 1,5-diaminonaphthalene (DAN) J. Mass Spectrom. 2014;49(8):737–741. doi: 10.1002/jms.3400. [DOI] [PubMed] [Google Scholar]
- Korte A.R., Yagnik G.B., Feenstra A.D., Lee Y.J. Multiplex MALDI-MS imaging of plant metabolites using a hybrid MS system. Methods Mol. Biol. 2015;1203:49–62. doi: 10.1007/978-1-4939-1357-2_6. [DOI] [PubMed] [Google Scholar]
- Lange B.M., Fischedick J.T., Lange M.F., Srividya N., Šamec D., Poirier B.C. Integrative approaches for the identification and localization of specialized metabolites in Tripterygium roots. Plant Physiol. 2017;173(1):456–469. doi: 10.1104/pp.15.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Ji S.-H., Lee Y.-S., Choi D.J., Choi B.-R., Kim G.-S., Baek N.-I., Lee D.Y. Mass spectrometry based profiling and imaging of various ginsenosides from Panax ginseng roots at different ages. Int. J. Mol. Sci. 2017;18(6):1114. doi: 10.3390/ijms18061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire R., Tabet J.C., Ducoroy P., Hendra J.B., Salzet M., Fournier I. Solid ionic matrixes for direct tissue analysis and MALDI imaging. Anal. Chem. 2006;78(3):809–819. doi: 10.1021/ac0514669. [DOI] [PubMed] [Google Scholar]
- Li B., Bhandari D.R., Janfelt C., Römpp A., Spengler B. Natural products in Glycyrrhiza glabra (licorice) rhizome imaged at the cellular level by atmospheric pressure matrix-assisted laser desorption/ionization tandem mass spectrometry imaging. Plant J. 2014;80(1):161–171. doi: 10.1111/tpj.12608. [DOI] [PubMed] [Google Scholar]
- Li B., Bhandari D.R., Römpp A., Spengler B. High-resolution MALDI mass spectrometry imaging of gallotannins and monoterpene glucosides in the root of Paeonia lactiflora. Sci. Rep. 2016;6 doi: 10.1038/srep36074. 36074-36074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Y., Liu J.a., Han J., Guan M., Yang H., Lin Y., Xiong S., Zhao Z. Electrospray deposition device used to precisely control the matrix crystal to improve the performance of MALDI MSI. Sci. Rep. 2016;6(1):37903. doi: 10.1038/srep37903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wang W., Liu Y., Jiang S., Huang G., Ye L. Near-infrared spectroscopy as a process analytical technology tool for monitoring the parching process of traditional Chinese medicine based on two kinds of chemical indicators. Phcog. Mag. 2017;13(50):332–337. doi: 10.4103/pm.pm_416_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang X., Han L., Jia L., Liu E., Li Z., Yu H., Wang Y., Gao X., Yang W. Integration of multicomponent characterization, untargeted metabolomics and mass spectrometry imaging to unveil the holistic chemical transformations and key markers associated with wine steaming of Ligustri Lucidi Fructus. J. Chromatogr. A. 2020;1624:461228. doi: 10.1016/j.chroma.2020.461228. [DOI] [PubMed] [Google Scholar]
- Li S., Zhu N., Tang C., Duan H., Wang Y., Zhao G., Liu J., Ye Y. Differential distribution of characteristic constituents in root, stem and leaf tissues of Salvia miltiorrhiza using MALDI mass spectrometry imaging. Fitoterapia. 2020;146:104679. doi: 10.1016/j.fitote.2020.104679. [DOI] [PubMed] [Google Scholar]
- Li B., Ge J., Liu W., Hu D., Li P. Unveiling spatial metabolome of Paeonia Suffruticosa and Paeonia Lactiflora roots using MALDI MS imaging. New Phytol. 2021;231(2):892–902. doi: 10.1111/nph.17393. [DOI] [PubMed] [Google Scholar]
- Liao Y., Fu X., Zhou H., Rao W., Zeng L., Yang Z. Visualized analysis of within-tissue spatial distribution of specialized metabolites in tea (Camellia sinensis) using desorption electrospray ionization imaging mass spectrometry. Food Chem. 2019;292:204–210. doi: 10.1016/j.foodchem.2019.04.055. [DOI] [PubMed] [Google Scholar]
- Liu C., Chen S., Xiao X., Zhang T., Hou W., Liao M. A new concept on quality marker of Chinese materia medica: quality control for Chinese medicinal products. Chin. Tradit. Herb. Drugs. 2016;47(9):1443–1457. doi: 10.7501/j.issn.0253-2670.2016.09.001. [DOI] [Google Scholar]
- Liu C., Guo D.-a., Liu L. Quality transitivity and traceability system of herbal medicine products based on quality markers. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2018;44:247–257. doi: 10.1016/j.phymed.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhou Y., Wang J., Xiong C., Xue J., Zhan L., Nie Z. N-Phenyl-2-naphthylamine as a novel MALDI matrix for analysis and in situ imaging of small molecules. Anal. Chem. 2018;90(1):729–736. doi: 10.1021/acs.analchem.7b02710. [DOI] [PubMed] [Google Scholar]
- Meng X., Fu W., Huo M., Liu Y., Zhang Z., Wei J., Wang Z., Abliz Z. In situ label-free visualization of tissue distributions of salidroside in multiple mouse organs by MALDI-MS imaging. Int. J. Mass Spectrom. 2020;453:116347. doi: 10.1016/j.ijms.2020.116347. [DOI] [Google Scholar]
- Merdas M., Lagarrigue M., Vanbellingen Q., Umbdenstock T., Da Violante G., Pineau C. On-tissue chemical derivatization reagents for matrix-assisted laser desorption/ionization mass spectrometry imaging. J. Mass Spectrom. 2021 doi: 10.1002/jms.4731. JMS, e4731. [DOI] [PubMed] [Google Scholar]
- Morikawa-Ichinose T., Fujimura Y., Murayama F., Yamazaki Y., Yamamoto T., Wariishi H., Miura D. Improvement of sensitivity and reproducibility for imaging of endogenous metabolites by matrix-assisted laser desorption/ionization-mass spectrometry. J. Am. Soc. Mass Spectrom. 2019;30(8):1512–1520. doi: 10.1007/s13361-019-02221-7. [DOI] [PubMed] [Google Scholar]
- Morisasa M., Sato T., Kimura K., Mori T., Goto-Inoue N. Application of matrix-assisted laser desorption/ionization mass spectrometry imaging for food analysis. Foods. 2019;8(12) doi: 10.3390/foods8120633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukrimin M., Conrad A.O., Kovalchuk A., Julkunen-Tiitto R., Bonello P., Asiegbu F.O. Fourier-transform infrared (FT-IR) spectroscopy analysis discriminates asymptomatic and symptomatic Norway spruce trees. Plant Sci. : an international journal of experimental plant biology. 2019;289:110247. doi: 10.1016/j.plantsci.2019.110247. [DOI] [PubMed] [Google Scholar]
- Nilsson A., Forngren B., Bjurström S., Goodwin R.J., Basmaci E., Gustafsson I., Annas A., Hellgren D., Svanhagen A., Andrén P.E., Lindberg J. In situ mass spectrometry imaging and ex vivo characterization of renal crystalline deposits induced in multiple preclinical drug toxicology studies. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimesh S., Mohottalage S., Vincent R., Kumarathasan P. Current status and future perspectives of mass spectrometry imaging. Int. J. Mol. Sci. 2013;14(6):11277–11301. doi: 10.3390/ijms140611277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer S.R., Drexler D.M. Tissue analysis by imaging MS. Bioanalysis. 2011;4(1):95–112. doi: 10.4155/bio.11.282. [DOI] [PubMed] [Google Scholar]
- Parrot D., Papazian S., Foil D., Tasdemir D. Imaging the unimaginable: desorption electrospray ionization - imaging mass spectrometry (DESI-IMS) in natural product research. Planta Med. 2018;84(9–10):584–593. doi: 10.1055/s-0044-100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta Siegel T., Hamm G., Bunch J., Cappell J., Fletcher J.S., Schwamborn K. Mass spectrometry imaging and integration with other imaging modalities for greater molecular understanding of biological tissues. Mol. Imag. Biol. 2018;20(6):888–901. doi: 10.1007/s11307-018-1267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prideaux B., Stoeckli M. Mass spectrometry imaging for drug distribution studies. J Proteomics. 2012;75(16):4999–5013. doi: 10.1016/j.jprot.2012.07.028. [DOI] [PubMed] [Google Scholar]
- Qin L., Zhang Y., Liu Y., He H., Han M., Li Y., Zeng M., Wang X. Recent advances in matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI-MSI) for in situ analysis of endogenous molecules in plants. Phytochem. Anal. 2018;29(4):351–364. doi: 10.1002/pca.2759. [DOI] [PubMed] [Google Scholar]
- Ràfols P., Heijs B., Del Castillo E., Yanes O., McDonnell L.A., Brezmes J., Pérez-Taboada I., Vallejo M., García-Altares M., Correig X. rMSIproc: an R package for mass spectrometry imaging data processing. Bioinformatics. 2020;36(11):3618–3619. doi: 10.1093/bioinformatics/btaa142. [DOI] [PubMed] [Google Scholar]
- Ren J.-l., Zhang A.-H., Kong L., Han Y., Yan G.-L., Sun H., Wang X.-J. Analytical strategies for the discovery and validation of quality-markers of traditional Chinese medicine. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2020;67:153165. doi: 10.1016/j.phymed.2019.153165. [DOI] [PubMed] [Google Scholar]
- Römpp A., Spengler B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013;139(6):759–783. doi: 10.1007/s00418-013-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara T., Bhandari D.R., Spengler B., Vollmann J. Spermidine and other functional phytochemicals in soybean seeds: spatial distribution as visualized by mass spectrometry imaging. Food Sci. Nutr. 2019;8(1):675–682. doi: 10.1002/fsn3.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Watanabe Y., Shirakawa M., Matsushita Y., Imai T., Koike T., Sano Y., Funada R., Fukazawa K., Fukushima K. Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant J. 2012;69(3):542–552. doi: 10.1111/j.1365-313X.2011.04811.x. [DOI] [PubMed] [Google Scholar]
- Schulz S., Becker M., Groseclose M.R., Schadt S., Hopf C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr. Opin. Biotechnol. 2019;55:51–59. doi: 10.1016/j.copbio.2018.08.003. [DOI] [PubMed] [Google Scholar]
- Schwamborn K., Caprioli R.M. Molecular imaging by mass spectrometry — looking beyond classical histology. Nat. Rev. Cancer. 2010;10(9):639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- Shi J., Cao B., Wang X.W., Aa J.Y., Duan J.A., Zhu X.X., Wang G.J., Liu C.X. Metabolomics and its application to the evaluation of the efficacy and toxicity of traditional Chinese herb medicines. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2016;1026:204–216. doi: 10.1016/j.jchromb.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Shi R., Dai X., Li W., Lu F., Liu Y., Qu H., Li H., Chen Q., Tian H., Wu E., Wang Y., Zhou R., Lee S.-T., Lifshitz Y., Kang Z., Liu J. Hydroxyl-group-dominated graphite dots reshape laser desorption/ionization mass spectrometry for small biomolecular analysis and imaging. ACS Nano. 2017;11(9):9500–9513. doi: 10.1021/acsnano.7b05328. [DOI] [PubMed] [Google Scholar]
- Song X., Luo Z., Li X., Li T., Wang Z., Sun C., Huang L., Xie P., Liu X., He J., Abliz Z. In situ hydrogel conditioning of tissue samples to enhance the drug's sensitivity in ambient mass spectrometry imaging. Anal. Chem. 2017;89(12):6318–6323. doi: 10.1021/acs.analchem.7b00091. [DOI] [PubMed] [Google Scholar]
- Sturtevant D., Lee Y.-J., Chapman K.D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 2016;37:53–60. doi: 10.1016/j.copbio.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Sun C., Liu W., Ma S., Zhang M., Geng Y., Wang X. Development of a high-coverage matrix-assisted laser desorption/ionization mass spectrometry imaging method for visualizing the spatial dynamics of functional metabolites in Salvia miltiorrhiza Bge. J. Chromatogr. A. 2020;1614:460704. doi: 10.1016/j.chroma.2019.460704. [DOI] [PubMed] [Google Scholar]
- Sun H., Zhang A., Wang X. Potential role of metabolomic approaches for Chinese medicine syndromes and herbal medicine. Phytother Res. : PT. 2012;26(10):1466–1471. doi: 10.1002/ptr.4613. [DOI] [PubMed] [Google Scholar]
- Taira S., Ikeda R., Yokota N., Osaka I., Sakamoto M., Kato M., Sahashi Y. Mass spectrometric imaging of ginsenosides localization in Panax ginseng root. Am. J. Chin. Med. 2010;38(3):485–493. doi: 10.1142/s0192415x10008007. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Kozuka T., Anegawa A., Nagatani A., Mimura T. Development and application of a high-resolution imaging mass spectrometer for the study of plant tissues. Plant Cell Physiol. 2015;56(7):1329–1338. doi: 10.1093/pcp/pcv083. [DOI] [PubMed] [Google Scholar]
- Takáts Z., Wiseman J.M., Gologan B., Cooks R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Talaty N., Takáts Z., Cooks R.G. Rapid in situ detection of alkaloids in plant tissue under ambient conditions using desorption electrospray ionization. Analyst. 2005;130(12):1624–1633. doi: 10.1039/b511161g. [DOI] [PubMed] [Google Scholar]
- Thunig J., Hansen S.H., Janfelt C. Analysis of secondary plant metabolites by indirect desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 2011;83(9):3256–3259. doi: 10.1021/ac2004967. [DOI] [PubMed] [Google Scholar]
- Tian F., Liu R., Fan C., Sun Y., Huang X., Nie Z., Zhao X., Pu X. Effects of Thymoquinone on small-molecule metabolites in a rat model of cerebral ischemia reperfusion injury assessed using MALDI-MSI. Metabolites. 2020;10(1):27. doi: 10.3390/metabo10010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Xie B., Zou Z., Jiao Y., Lin L.-E., Chen C.-L., Hsu C.-C., Peng J., Yang Z. Multimodal imaging of amyloid plaques: fusion of the single-probe mass spectrometry image and fluorescence microscopy image. Anal. Chem. 2019;91(20):12882–12889. doi: 10.1021/acs.analchem.9b02792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillner J., Wu V., Jones E.A., Pringle S.D., Karancsi T., Dannhorn A., Veselkov K., McKenzie J.S., Takats Z. Faster, more reproducible DESI-MS for biological tissue imaging. J. Am. Soc. Mass Spectrom. 2017;28(10):2090–2098. doi: 10.1007/s13361-017-1714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trim P.J., Henson C.M., Avery J.L., McEwen A., Snel M.F., Claude E., Marshall P.S., West A., Princivalle A.P., Clench M.R. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 2008;80(22):8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- Van de Plas R., Yang J., Spraggins J., Caprioli R.M. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods. 2015;12(4):366–372. doi: 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Végvári Á. Drug localizations in tissue by mass spectrometry imaging. Biomarkers Med. 2015;9(9):869–876. doi: 10.2217/bmm.15.64. [DOI] [PubMed] [Google Scholar]
- Verbeeck N., Spraggins J.M., Murphy M.J.M., Wang H.D., Deutch A.Y., Caprioli R.M., Van de Plas R. Connecting imaging mass spectrometry and magnetic resonance imaging-based anatomical atlases for automated anatomical interpretation and differential analysis. Biochim. Biophys. Acta Protein Proteonomics. 2017;1865(7):967–977. doi: 10.1016/j.bbapap.2017.02.016. [DOI] [PubMed] [Google Scholar]
- Vestal M., Vestal C., Li S., Parker K. The seven S criteria for evaluating the performance of a MALDI mass spectrometer for MSI. J. Am. Soc. Mass Spectrom. 2020;31(12):2521–2530. doi: 10.1021/jasms.0c00216. [DOI] [PubMed] [Google Scholar]
- Wang J., van der Heijden R., Spijksma G., Reijmers T., Wang M., Xu G., Hankemeier T., van der Greef J. Alkaloid profiling of the Chinese herbal medicine Fuzi by combination of matrix-assisted laser desorption ionization mass spectrometry with liquid chromatography–mass spectrometry. J. Chromatogr. A. 2009;1216(11):2169–2178. doi: 10.1016/j.chroma.2008.11.077. [DOI] [PubMed] [Google Scholar]
- Wang X., Han J., Pan J., Borchers C.H. Comprehensive imaging of porcine adrenal gland lipids by MALDI-FTMS using quercetin as a matrix. Anal. Chem. 2014;86(1):638–646. doi: 10.1021/ac404044k. [DOI] [PubMed] [Google Scholar]
- Wang X., Han J., Yang J., Pan J., Borchers C.H. Matrix coating assisted by an electric field (MCAEF) for enhanced tissue imaging by MALDI-MS. Chem. Sci. 2015;6(1):729–738. doi: 10.1039/c4sc01850h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Bai H., Cai Z., Gao D., Jiang Y., Liu J., Liu H. MALDI imaging for the localization of saponins in root tissues and rapid differentiation of three Panax herbs. Electrophoresis. 2016;37(13):1956–1966. doi: 10.1002/elps.201600027. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cai Y., Wang Y., Zhou X., Zhang Y., Lu H. Improved MALDI imaging MS analysis of phospholipids using graphene oxide as new matrix. Sci. Rep. 2017;7:44466. doi: 10.1038/srep44466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Hao R., Liu Y., Wang Y., Man S., Gao W. Tissue distribution, metabolism and absorption of Rhizoma Paridis Saponins in the rats. J. Ethnopharmacol. 2021;273:114038. doi: 10.1016/j.jep.2021.114038. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhu T., Xu H.-B., Pu X.-P., Zhao X., Tian F., Ding T., Sun G.-B., Sun X.-B. Effects of notoginseng leaf triterpenes on small molecule metabolism after cerebral ischemia/reperfusion injury assessed using MALDI-MS imaging. Ann. Transl. Med. 2021;9(3) doi: 10.21037/atm-20-4898. 246-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Lee H.K., Yue G.G.L., Chung A.C.K., Lau C.B.S., Cai Z. A novel binary matrix consisting of graphene oxide and caffeic acid for the analysis of scutellarin and its metabolites in mouse kidney by MALDI imaging. Analyst. 2021;146(1):289–295. doi: 10.1039/d0an01539c. [DOI] [PubMed] [Google Scholar]
- Wang T., Liu J., Luo X., Hu L., Lu H. Functional metabolomics innovates therapeutic discovery of traditional Chinese medicine derived functional compounds. Pharmacol. Ther. 2021;224:107824. doi: 10.1016/j.pharmthera.2021.107824. [DOI] [PubMed] [Google Scholar]
- Watson B., Urbanczyk-Wochniak E., Lei Z. Joint Annual Meeting of the American-Fern-Society-American-Society-of-Plant-Biologists/American-Society-of-Plant-Taxonomists/Botanical-Society-of-America; 2007. Spatially Resolved Metabolomics and Proteomics of Medicago Truncatula Border Cells and Root Tips. [Google Scholar]
- Wiseman J.M., Ifa D.R., Song Q., Cooks R.G. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. 2006;45(43):7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- Wu H., Liu X., Gao Z.-Y., Dai Z.-F., Lin M., Tian F., Zhao X., Sun Y., Pu X.-P. Anti-myocardial infarction effects of Radix Aconiti Lateralis Preparata extracts and their influence on small molecules in the heart using matrix-assisted laser desorption/ionization-mass spectrometry imaging. Int. J. Mol. Sci. 2019;20(19):4837. doi: 10.3390/ijms20194837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Liang Z., Zhao Z., Cai Z. Direct analysis of alkaloid profiling in plant tissue by using matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 2007;42(1):58–69. doi: 10.1002/jms.1138. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Takahashi K., Caputi L., Mizuno H., Rodriguez-Lopez C.E., Iwasaki T., Ishizaki K., Fukaki H., Ohnishi M., Yamazaki M., Masujima T., O'Connor S.E., Mimura T. The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics. New Phytol. 2019;224(2):848–859. doi: 10.1111/nph.16138. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Takahashi K., Mizuno H., Anegawa A., Ishizaki K., Fukaki H., Ohnishi M., Yamazaki M., Masujima T., Mimura T. Cell-specific localization of alkaloids in Catharanthus roseus stem tissue measured with Imaging MS and Single-cell MS. Proc. Natl. Acad. Sci. U. S. A. 2016;113(14):3891–3896. doi: 10.1073/pnas.1521959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.-T., Li S.-D., Li C., Xiong Y.-X., Lu X.-H., Zhou X.-F., Yang L.-Q., Pu L.-J., Luo H.-Y. Panax notoginsenoside saponins Rb1 regulates the expressions of Akt/mTOR/PTEN signals in the hippocampus after focal cerebral ischemia in rats. Behav. Brain Res. 2018;345:83–92. doi: 10.1016/j.bbr.2018.02.037. [DOI] [PubMed] [Google Scholar]
- Yang W., Zhang Y., Wu W., Huang L., Guo D., Liu C. Approaches to establish Q-markers for the quality standards of traditional Chinese medicines. Acta Pharm. Sin. B. 2017;7(4):439–446. doi: 10.1016/j.apsb.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang Y., Qiu H., Ju Z., Shi Y., Wang Z., Yang L. Localization of constituents for determining the age and parts of ginseng through ultraperfomance liquid chromatography quadrupole/time of flight-mass spectrometry combined with desorption electrospray ionization mass spectrometry imaging. J. Pharmaceut. Biomed. Anal. 2021;193:113722. doi: 10.1016/j.jpba.2020.113722. [DOI] [PubMed] [Google Scholar]
- Yoon S., Lee T.G. Biological tissue sample preparation for time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging. Nano Converg. 2018;5(1) doi: 10.1186/s40580-018-0157-y. 24-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Bai G., Han Y., Xu J., Gong S., Li Y., Zhang H., Liu C. The method of quality marker research and quality evaluation of traditional Chinese medicine based on drug properties and effect characteristics. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2018;44:204–211. doi: 10.1016/j.phymed.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Zhang X.J., Qiu J.F., Guo L.P., Wang Y., Li P., Yang F.Q., Su H., Wan J.B. Discrimination of multi-origin Chinese herbal medicines using gas chromatography-mass spectrometry-based fatty acid profiling. Molecules. 2013;18(12):15329–15343. doi: 10.3390/molecules181215329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Li B., He D., Zhao C., Shi Z., Dong B., Pan D., Patil R.R., Yan Z., Guo Z. Chemical characteristic and bioactivity of hemicellulose-based polysaccharides isolated from Salvia miltiorrhiza. Int. J. Biol. Macromol. 2020;165(Pt B):2475–2483. doi: 10.1016/j.ijbiomac.2020.10.113. [DOI] [PubMed] [Google Scholar]
- Zhu T., Wang L., Tian F., Zhao X., Pu X.-P., Sun G.-B., Sun X.-B. Anti-ischemia/reperfusion injury effects of notoginsenoside R1 on small molecule metabolism in rat brain after ischemic stroke as visualized by MALDI–MS imaging. Biomed. Pharmacother. 2020;129:110470. doi: 10.1016/j.biopha.2020.110470. [DOI] [PubMed] [Google Scholar]