Abstract
Genes encoding the Mycoplasma arthritidis surface-exposed lipoprotein MAA1 were cloned and sequenced from MAA1-expressing strains 158p10p9 and PG6, from a low-adherence (LA) variant derived from 158p10p9 that expresses a truncated version of MAA1 (MAA1Δ) and from two MAA1-negative strains, 158 and H39. The deduced amino acid sequences of maa1 from 158p10p9 and PG6 predicted, respectively, 86.5- and 86.4-kDa basic, largely hydrophilic lipoproteins with 29-amino-acid signal peptides and predicted cleavage sites for signal peptidase II (Ala-Ala-Ala↓Cys). The truncation in the LA variant resulted from a G→T substitution at nucleotide 695, which created a premature stop codon. This, in turn, generated a predicted 26.6-kDa prolipoprotein (23.6 kDa after processing), consistent with an Mr of ∼24,000 calculated for MAA1Δ. Similarly, absence of MAA1 expression in H39 and 158 resulted from C→A substitutions at nucleotide 208, generating premature stop codons at that site in both strains.
Mycoplasma arthritidis causes an acute, self-limited, septic arthritis of rats under both natural and experimental conditions (4). As with most infectious diseases, adherence to host tissues is likely to play an important role. We have identified two surface proteins, MAA1 and MAA2, that may be involved in this process (14). Both MAA1 and MAA2 are lipid modified (17). MAA2 has been cloned and sequenced and shown to be phase variable due to changes in the length of a poly(T) tract in the promoter region (17).
The ability of monoclonal antibodies (MAbs) against these two proteins to inhibit attachment of M. arthritidis to rat cells in vitro (14) led to their original characterization as putative adhesins. Although definitive evidence for their role in cytadherence is still lacking, an additional indication for the involvement of MAA1 came with our identification of a spontaneously appearing mutant from M. arthritidis 158p10p9, which attached to rat cells in culture very poorly compared to the parent strain (14). We originally designated the low-adherence (LA) mutant LC1; here we refer to it as the LA variant. The only discernible difference between LA and the parent strain is that the LA variant expresses a highly truncated version of MAA1 (MAA1Δ, Mr ∼24,000 [versus 90,000 for the parent strain]). Although MAA1Δ seems to be properly inserted into the membrane and is exposed on the cell surface, it is apparently nonfunctional, at least with regard to cytadherence (14).
MAA2 is a marker for a group of M. arthritidis strains related to strain 158p10p9, while MAA1 is not strain specific (15). MAA1 is expressed by 14 of 20 strains tested (15), although all 20 attach to rat cells in culture (unpublished observation). Therefore, while MAA1 may be an important adhesin for the strains that express it, there are no doubt alternative adhesins in other strains; this is not uncommon among pathogenic bacteria (6). Although MAA1 does not appear to be a marker for any particular set of strains, it does contain important antigenic epitopes, because loss of approximately two-thirds of the molecule significantly reduced the ability of the LA variant to adsorb antibody from a strain-specific anti-M. arthritidis antiserum (15).
The possible involvement of MAA1 in adherence was intriguing, but a more fundamental question was whether it was in any way relevant to the disease process. We found that the LA variant induced slightly but significantly less severe arthritis in rats than did the wild type (14). In addition, MAA1 induced a strong antibody response in rats with M. arthritidis-induced arthritis, and rats passively immunized with an anti-MAA1 MAb were nearly completely protected against a challenge injection of virulent M. arthritidis (16). Rats actively immunized with MAA1 or passively immunized with a polyclonal monospecific antiserum against MAA1 were also protected, although to a lesser extent. These data provided at least indirect evidence that MAA1 plays a role in pathogenesis, possibly in mediating adherence to joint tissues early in the infectious process.
Evidence for involvement of MAA1 in both cytadherence and pathogenesis prompted a more detailed characterization of this protein. In addition to lipid modification, preliminary studies showed that MAA1 was neither size nor phase variable and that the epitope recognized by the protective, attachment-inhibiting MAb was exposed on the surface of the mycoplasmal cell and removed by treatment of the intact cells with carboxypeptidase Y (17). Here we report the cloning and sequencing of the maa1 gene from MAA1-expressing strains 158p10p9 and PG6, from the LA variant derived from strain 158p10p9, which expresses a product about one-third the size of the wild-type protein (MAA1Δ), and from strains 158 and H39, which do not express MAA1.
(The results of this study were presented in part at the 99th General Meeting of the American Society for Microbiology, Chicago, Ill., May 30 to June 3, 1999.)
MATERIALS AND METHODS
Mycoplasma strains and culture conditions.
M. arthritidis culture and storage conditions were as described previously (15). Stock cultures were stored frozen at −70°C; each 1-ml aliquot contained approximately 3 × 1010 to 4 × 1010 CFU. Most of the work in this study was done with strains 158p10p9 (3), PG6 (9), 158 (5), and H39 (8). Additional strains used in Southern hybridization are described in detail elsewhere (15).
Isolation and N-terminal sequencing of MAA1 and design of a degenerate oligonucleotide probe.
MAA1 was sliced from a sodium dodecyl sulfate (SDS)-polyacrylamide gel, extracted, reelectrophoresed, and transferred to a polyvinylidine difluoride membrane as described previously for MAA2 (17). N-terminal sequencing was performed by the Mayo Protein Core Facility, Department of Biochemistry and Molecular Biology, Mayo Medical School, Rochester, Minn. From this amino acid sequence (X-Asp-Asp-Thr-Lys-Lys-Lys-Pro-Glu-Glu-Pro-Lys-Lys-Glu-Asp-Pro-Lys-Gln-Lys-Pro), a degenerate 23-base oligonucleotide was designed for use in screening an M. arthritidis 158p10p9 DNA library and in Southern hybridization experiments. The sequence of the probe was 5′-AAAGAAGATCCWAAACAAAAACC-3′, where W represents either A or T. Oligonucleotides and primers used in this study were synthesized by GIBCO BRL Life Technologies (Grand Island, N.Y.) or by the Iowa State University DNA Synthesis and Sequencing Facility (Ames, Iowa).
Mycoplasmal chromosomal and recombinant DNA preparation and Southern hybridization.
Mycoplasmal chromosomal DNA was prepared from 1-ml stock cultures as described previously (13, 17).
For routine screening, recombinant plasmids were isolated from Escherichia coli by boiling (1) or by modified alkaline lysis (pRSET Xpress Kit manual; Invitrogen Corp., Carlsbad, Calif.). For DNA sequencing, plasmid preparations were purified chromatographically by using Qiagen (Chatsworth, Calif.) columns according to the manufacturer's instructions.
For Southern hybridization, 9 μg of mycoplasmal chromosomal DNA or 0.25 to 0.5 μg of recombinant plasmid DNA was digested with the appropriate restriction endonucleases, electrophoresed on agarose gels according to standard procedures (1), and transferred by vacuum blotting to nylon membranes (Hybond-N+; Amersham Life Sciences, Arlington Heights, Ill.). Blots were exposed to the degenerate oligonucleotide probe designed from the N-terminal amino acid sequence of MAA1. The oligonucleotide was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase as instructed by the supplier (Promega, Madison, Wis.).
Molecular cloning of chromosomal fragments hybridizing with the MAA1-derived probe.
Samples (9 μg) of chromosomal DNA from strains 158p10p9 and PG6 were digested with EcoRI and ligated into the EcoRI-digested, dephosphorylated (1) vectors pSPORT 1 (GIBCO BRL) and pBluescript SK(+) (Stratagene, La Jolla, Calif.), respectively, by using T4 DNA ligase (Promega). Recombinant plasmids were introduced into E. coli XL1-Blue MRF′ by electroporation (Cell-Porator; Bethesda Research Laboratories Life Technologies, Inc., Bethesda, Md.), and colonies containing recombinant DNA were screened with the 32P-labeled MAA1-derived oligonucleotide (1). Positive colonies were expanded, and plasmid DNA was digested with EcoRI and hybridized with the probe. Then, 7-kb inserts containing maa1-like sequences were recovered from both strains. A 500-bp internal HindIII/PstI fragment was subcloned from the 158p10p9 insert into pSPORT 1 for DNA sequencing. Sequence analysis revealed a portion of an open reading frame (ORF) containing a sequence identical to the probe. The remainder of this ORF plus some flanking DNA were sequenced from the original clone by primer walking. Initial sequencing of the PG6 insert was done with a primer designed from the 5′ end of the 158p10p9 maa1 gene, and the rest of the gene was sequenced by primer walking.
EcoRI fragments of 2 and 0.9 kb hybridizing with the MAA1-derived probe were also recovered from strain 158p10p9 and sequenced from vector and internal primers in both directions to determine whether they encoded additional genes similar to maa1.
PCR amplification of maa1 homologs from the LA variant and from MAA1-negative strains 158 and H39.
For amplification of maa1 homologs from the LA variant and from strains 158 and H39, PCR primers PF4 (5′-GATAAAGCGCACTGTTGTGCTC-3′) and PR2 (5′-CATAATTGAAAAAGAATGCC-3′) were designed from sequences flanking the maa1 gene from 158p10p9 (see Fig. 2). The corresponding regions were amplified from 100-ng samples of chromosomal DNA in 50-μl reaction mixes, each containing 2.5 mM MgCl2, 1× reaction buffer (Promega), 10 mM concentrations of deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.), 5 μM concentrations of each primer, and 2.5 U of Taq DNA polymerase (Promega). Thermal cycler settings were as follows: denaturation for 1 min at 94°C, annealing for 1 min at 52°C, and polymerization for 3 min at 72°C for 30 cycles, followed by 1 cycle of 1 min at 94°C, 1 min at 52°C, and 10 min at 72°C. PCR products were ligated into the A-T vector pGEM-T Easy (Promega) for DNA sequencing. They were sequenced by using the PCR primers, primers from internal sequences generated from strains 158p10p9 and 158, and primers from the vector.
FIG. 2.
Nucleotide sequence of maa1 from M. arthritidis 158p10p9 (uppercase letters, first line), PG6, 158, and H39 (lowercase letters, second line). PCR primers PF4 and PR2, used for amplifying this region from LA, 158, and H39, are underlined and italicized. The sequence corresponding to the degenerate oligonucleotide probe is shaded in black. The probe was designed with an A residue instead of the authentic G (unshaded) at the sixth position. A putative ribosome binding site (RBS) and the translation start site are underlined and marked by arrows. A TAG stop codon is underlined (nt 2250), and a putative Rho-independent transcription terminator is boxed. A “-” indicates that the nucleotide sequence is the same as for 158p10p9; an asterisk indicates that nucleotides are missing from those sites in strains PG6, 158, and H39. Sites at which PG6, 158, or H39 differ from each other are annotated. A C→A substitution at nt 204 (underlined) generates premature TAA stop codons in strains 158 and H39. A G→T substitution at nt 695 (underlined) generates a premature TAA stop codon in maa1Δ. Otherwise, the sequence of maa1Δ from the LA variant is identical to that of maa1 from the 158p10p9 parent strain.
DNA sequencing.
DNA sequencing, including primer walking, was performed by the Iowa State University DNA Synthesis and Sequencing Facility, Ames. DNA and deduced amino acid sequence data were analyzed with the MacDNASIS Pro version 3.6 software package (Hitachi Software Engineering Co., Ltd., South San Francisco, Calif.).
Nucleotide sequence accession numbers.
Accession numbers for the sequences described in this report are as follows: maa1 from strain 158p10p9, AF154919; maa1 from strain PG6, AF154920; maa1 from the LA variant, AF154921; maa1 from strain 158, AF154922; and maa1 from strain H39, AF154923.
RESULTS
Hybridization of chromosomal DNA from 20 M. arthritidis strains with the MAA1-derived oligonucleotide probe.
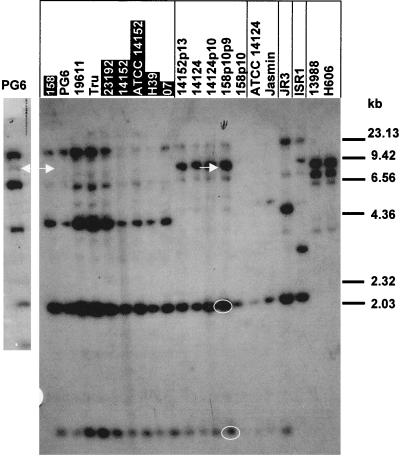
To identify DNA fragments on which maa1 might be located and to determine whether MAA1-negative strains also possessed similar genes, we subjected EcoRI-digested chromosomal DNA from 20 M. arthritidis strains to Southern hybridization with the MAA1-derived oligonucleotide probe (Fig. 1). Two to four fragments from each strain hybridized with the probe, including strains not expressing MAA1 (strains 158, 23192, 14152, ATCC 14152, H39, and O7). The 7-kb EcoRI fragments from strains 158p10p9 and PG6 contained ORFs with characteristics appropriate for maa1 (see below and Fig. 2 and 3); these are marked with arrows. The 7-kb fragment from PG6 can be seen more clearly on the small blot to the left in Fig. 1. The probe also hybridized with EcoRI fragments from the LA variant that were the same size as those from the 158p10p9 parent strain (not shown).
FIG. 1.
Southern hybridization of EcoRI-digested chromosomal DNA from 20 M. arthritidis strains with the degenerate oligonucleotide probe designed from the N-terminal amino acid sequence of MAA1. Each strain contained at least two EcoRI fragments that hybridized with the probe. Strains previously shown not to express MAA1 (15) are shaded in black. Strains that are closely related to each other are boxed (15). The 7-kb fragments from strains 158p10p9 and PG6 that contain genes encoding MAA1 are marked with white arrows. Because the PG6 lane was underloaded on this blot and the 7-kb band was hard to see, a second hybridization was done with another preparation of PG6 (small panel to the left). Additional fragments cloned and sequenced from 158p10p9 are circled in white.
FIG. 3.
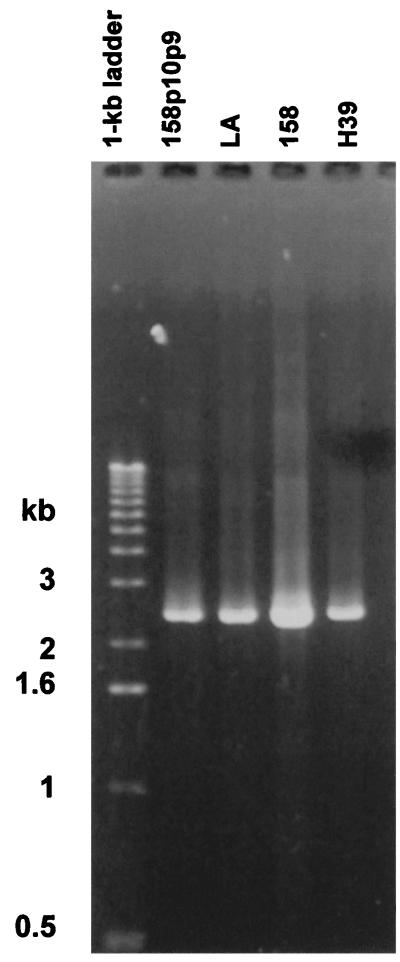
PCR products amplified from chromosomal DNA from strain 158p10p9, the LA variant derived from 158p10p9, strain 158, and strain H39 by using primers PF4 and PR2. Size markers are in the first lane. No product was amplified from controls lacking M. arthritidis template DNA (not shown).
Molecular cloning and sequencing of maa1 from M. arthritidis 158p10p9 and PG6.
The MAA1-derived oligonucleotide probe was used to screen transformants containing EcoRI fragments cloned from 158p10p9 and PG6 chromosomal DNA. Transformants containing 7-kb inserts from both strains hybridized with the probe. The hybridizing region from strain 158p10p9 was localized to a 500-bp internal HindIII/PstI fragment. This fragment was subcloned, sequenced, and found to encode an incomplete ORF with some of the characteristics of a surface protein. This included a sequence at its 5′ end which could encode a putative cleavage site for signal peptidase II (Ala-Ala-Ala↓Cys) (12). The remainder of the ORF and 5′ and 3′ flanking DNA were sequenced by primer walking. The corresponding region from the PG6 clone was sequenced by using a similar strategy. DNA sequences of these regions are shown in Fig. 2.
The 2-kb and 0.9-kb EcoRI fragments from 158p10p9 circled in white on Fig. 1 were also cloned and sequenced. Both contained sequences with some similarity to the MAA1-derived oligonucleotide probe. However, the 2-kb fragment contained no obvious functional ORFs, while the 0.9-kb fragment contained what appeared to be a small ORF with a premature stop codon near the 5′ end and little similarity to maa1 other than the probe sequence.
PCR amplification and sequencing of maa1 homologs from the LA variant and MAA1-negative strains 158 and H39.
If maa1 genes were absent from the non-MAA1-expressing strains or if maa1 had undergone significant deletion or recombination in the LA variant, we would expect these changes to manifest as restriction fragment polymorphisms on Southern hybridization. However, this was not the case (Fig. 1, e.g., strains 158 and PG6). Instead, the restriction fragment length polymorphisms seen in the figure corresponded to previously identified strain relationships (15) and were independent of MAA1 expression. Our designation of “MAA1-negative” was based on Western immunoblotting with an MAA1-specific MAb that recognized a surface epitope from strain 158p10p9 (14, 15). The DNA hybridization results could be explained if the so-called “MAA1-negative” strains produced proteins similar to MAA1, with surface-exposed domains that were antigenically variable but with membrane-embedded N termini that were more highly conserved. In this case, the probe designed from the N-terminal sequence of MAA1 might not distinguish among them. However, another explanation is required for the absence of restriction fragment length polymorphisms in the LA variant. This next set of experiments was designed (i) to determine whether maa1-like genes were present in MAA1-negative strains, and if so, (ii) to determine whether their deduced amino acid sequences were consistent with expression of a set of antigenically variable MAA1-like proteins, and finally (iii) to identify the nature of the mutation in the LA variant.
PCR primers PF4 and PR2 (Fig. 2) were used to amplify the regions encoding maa1 homologs from MAA1-negative strains 158 and H39 and from the LA variant (Fig. 3). These primers were designed to amplify 2.4-kb products from strains expressing MAA1 (e.g., 158p10p9). However, 2.4-kb products were also amplified from MAA1-negative strains 158 and H39, as well as from the LA variant, which expresses a version of MAA1 that is less than one-third the size of the wild-type protein (14). This is consistent with the Southern hybridization results described above (Fig. 1).
The 2.4-kb products amplified from two preparations of chromosomal DNA from the LA variant and from two preparations each from the MAA1-negative strains 158 and H39 were cloned into the A-T vector pGEM-T Easy and sequenced. Comparison of all six sequences with each other and with those of 158p10p9 and PG6 allowed PCR-induced errors to be identified and corrected. Since the 158 and H39 sequences were almost identical to each other, we treated inconsistencies that appeared in only one of the four samples, but not in the other three, as PCR induced. Similarly, except as described below, sequences from the two LA preparations were identical to the sequence from the parent strain; inconsistencies that appeared in only one, but not in both, were also considered PCR induced.
Comparison of nucleotide and deduced amino acid sequences of maa1 from 158p10p9, PG6, the LA variant, 158, and H39.
DNA sequences flanking the 5′ ends of all five genes were highly conserved, as were the 5′ ends of the coding regions (Fig. 2). Putative ribosome binding sites were located just upstream of the ATG start site, although no obvious promoter sequences were seen. In addition, putative Rho-independent transcription terminators (ΔG = −16.2) were found just downstream of the TAG stop codons. There was a 2-nucleotide (nt) mismatch between the probe sequence and the authentic sequence in strains PG6, 158, and H39 but only a single mismatch with the corresponding sequence from strain 158p10p9, which may explain why the 7-kb EcoRI fragments containing maa1 from strains PG6, 158, and H39 hybridized only weakly with the MAA1-derived oligonucleotide probe on the blot in Fig. 1.
maa1 from the LA variant differed from that of the parent strain at a single site, a G→T substitution at nt 695, which generated an inframe, premature TAA stop codon (Fig. 2). maa1 from strains 158 and H39 differed from each other by only a single nucleotide and were identical to PG6 except at positions 208, 456, 458, 2056, and 2031. There were three fewer nucleotides in these three strains between sites 2086 and 2102 than in strain 158p10p9 (Fig. 2), so that the reading frame was disrupted and then restored. Of greater interest was the C→A substitution at nt 208 in 158 and H39, which generated premature TAA stop codons much closer to the 5′ end of the gene than in the LA variant. While these results do not support our hypothesized set of antigenically variable MAA1-like proteins with conserved N termini, they do provide an explanation for the absence of MAA1 from these strains. The predicted products may be too small for proper processing or more highly susceptible to proteolysis.
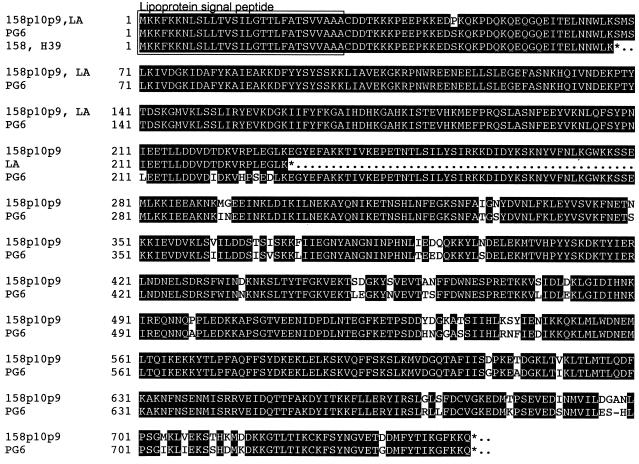
Deduced amino acid sequences of these proteins are shown in Fig. 4. Sequences from strains 158p10p9 and PG6 predicted 747- and 746-amino-acid, 86.5- and 86.4-kDa proteins, respectively, with identical 29-amino-acid lipoprotein signal peptides with predicted cleavage sites for signal peptidase II (Ala-Ala-Ala↓Cys [12]). Both were basic (pI = 8.14 and 8.37, respectively) and largely hydrophilic. The two proteins were highly conserved, especially at the N termini. Short nonidentical regions were clustered in the middle and toward the C terminus. A GenBank search revealed no significant similarity to other known proteins.
FIG. 4.
Deduced amino acid sequence of MAA1 from strains 158p10p9, PG6, 158, and H39 and of MAA1Δ from the LA variant. Identical regions are shaded in black. A putative lipoprotein signal peptide is boxed.
The deduced amino acid sequence of the corresponding gene from the LA variant predicted a 231-amino-acid, 26.6-kDa lipoprotein that was otherwise identical to the N-terminal 231 amino acids of MAA1 from 158p10p9. The mature lipoprotein, after cleavage of the signal peptide, would be 23.6 kDa in size, in agreement with our earlier estimation of an Mr of 24,000 for MAA1Δ (14).
DISCUSSION
In this report we describe the isolation and sequencing of maa1 and maa1 homologs from two M. arthritidis strains expressing MAA1 (158p10p9 and PG6), an LA variant expressing a truncated version of the protein, and two nonexpressing strains (158 and H39). We were unable to express the genes encoding MAA1 from strains 158p10p9 and PG6 in E. coli because they each contained six TGA tryptophan codons. However, we believe that they do, in fact, encode MAA1, based upon (i) the predicted molecular masses from deduced amino acid sequences, which coincided roughly with the Mr of 90,000 calculated for MAA1 from SDS-polyacrylamide gels (14); (ii) the predicted hydrophilicity and the presence of putative lipoprotein signal peptides, compared with our earlier observation that MAA1 was surface exposed and lipid modified (14, 17); (iii) the premature stop codon in maa1 from the LA variant, generating a predicted 23.6-kDa product after processing of the signal peptide, in comparison with an estimated Mr of 24,000 for MAA1Δ; and (iv) the very early truncation of the maa1 genes in MAA1-negative strains 158 and H39, explaining why no products were detected.
The other two EcoRI fragments from 158p10p9 that hybridized strongly with the MAA1-derived probe (circled in white in Fig. 1) did not contain maa1-like sequences other than those with limited homologies to the probe itself, suggesting that maa1 was present in only a single copy in this, and probably related, strains.
Similarities among the nucleotide sequences of maa1 from the four strains examined were consistent with previously identified strain relationships. In an earlier study, we showed by serologic and DNA restriction analysis that strains PG6, H39, and 158 were more closely related to each other than they were to 158p10p9 (15). Here we show that maa1 sequences from PG6, H39, and 158 were 95% identical to maa1 from 158p10p9, while maa1 sequences from H39 and 158 were >99% identical to maa1 from PG6. maa1 from H39 and 158 differed from each other by only a single nucleotide.
The deduced amino acid sequences of MAA1 from 158p10p9 and PG6 predicted basic, largely hydrophilic, prolipoproteins that differed in size by a single amino acid. Their 29-amino-acid signal peptides were identical to each other and shared 79% identity with the 29-amino-acid signal peptide of the M. arthritidis variable surface lipoprotein MAA2 (17). The N-terminal amino acid sequence obtained from the gel-purified protein matched the deduced amino acid sequence of MAA1 from 158p10p9 immediately adjacent to the predicted signal peptidase II cleavage site (Fig. 2), confirming that the signal peptide was removed prior to insertion of the protein into the membrane. The molecular mass of processed MAA1 estimated from the deduced amino acid sequence was less than the 90 kDa predicted by SDS-polyacrylamide gel electrophoresis (PAGE) (14). However, acylation and other posttranslational modifications are known to affect electrophoretic migration (2, 10, 11, 18, 19), and MAA2 also migrated more slowly by SDS-PAGE than predicted from its amino acid sequence (17).
The mutation in the LA variant that resulted in truncation of MAA1Δ proved to be a G→T substitution at nt 695, which generated a TAA stop codon at that site. The resulting product, after processing, would be about 23.6 kDa in size, corresponding to the 24-kDa product identified earlier by Western immunoblotting (14). These observations are consistent with our earlier suggestion that the loss of adherence in the LA variant was probably due to truncation of MAA1 (14), although definitive evidence will require the introduction of specific mutations in maa1 from the parent strain. It is interesting that in spite of the absence of more than two-thirds of the molecule, the epitope recognized by the attachment-inhibiting MAb remained intact (14). It is possible that this epitope was not directly involved in adherence and that the MAb's effect on attachment was due to steric hindrance. Alternatively, generation of the actual binding site may require precise conformation of the larger molecule. In that regard, there were two Cys residues near the C terminus of the molecule that may generate some secondary structure in that region.
A similar substitution, much closer to the 5′ end of the coding region, was responsible for the absence of detectable products in strains such as 158 and H39 that do not express MAA1. Such products would be only 7.6 kDa in size (4.5 kDa after processing) and may be rapidly degraded or not properly processed. In a similar vein, Henrich et al. showed that truncation of protein P50, a variant of the adherence-related molecule Vaa, in certain Mycoplasma hominis isolates were due to the generation of premature stop codons deeper within the coding region (7). As for MAA1, these were not reversible. Results of the present study do not support the hypothesis that M. arthritidis strains produce a family of MAA1-like proteins that are conserved at the membrane-embedded N termini but with structurally variable surface-exposed domains. However, the absence of MAA1 from these strains did not render them incapable of cytadherence (unpublished observation), indicating that they possess other adhesins. Further efforts to identify and characterize these alternative adhesins are under way in our laboratory.
ACKNOWLEDGMENTS
We thank Richard Duman for assistance in preparation of many of the media and reagents used in this study.
This work was supported in part by grants from the University of South Dakota School of Medicine Parson's Endowment Fund, Public Health Service grant RO1 37924 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Science Foundation EPSCoR Program under grant OSR-9108773, and the South Dakota Future Fund.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 2.Bhugra B, Voelker L L, Zou N, Yu H, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 3.Cole B C, Ward J R, Jones R S, Cahill J F. Chronic proliferative arthritis of mice induced by Mycoplasma arthritidis. I. Induction of disease and histopathological characteristics. Infect Immun. 1971;4:344–355. doi: 10.1128/iai.4.4.344-355.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole B C, Washburn L R, Taylor-Robinson D. Mycoplasma-induced arthritis. In: Razin S, Barile M F, editors. The mycoplasmas. IV. Mycoplasma pathogenicity. New York, N.Y: Academic Press; 1985. pp. 108–160. [Google Scholar]
- 5.Edward D G, Freundt E A. The classification and nomenclature of organisms of the pleuropneumonia group. J Gen Microbiol. 1956;14:197–207. doi: 10.1099/00221287-14-1-197. [DOI] [PubMed] [Google Scholar]
- 6.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrich B, Lang K, Kitzerow A, MacKenzie C, Hadding U. Truncation as a novel form of variation of the p50 gene in Mycoplasma hominis. Microbiology. 1998;144:2979–2985. doi: 10.1099/00221287-144-11-2979. [DOI] [PubMed] [Google Scholar]
- 8.Leberman P R, Smith P F, Morton H E. The susceptibility of pleuropneumonia-like organisms to the in vitro action of antibiotics: aureomycin, chloramphenicol, dihydrostreptomycin, streptomycin, and sodium penicillin G. J Urol. 1950;64:167–173. doi: 10.1016/S0022-5347(17)68616-6. [DOI] [PubMed] [Google Scholar]
- 9.Preston W S. Arthritis in rats caused by pleuropneumonia-like micro-organisms and the relationship of similar organisms to human rheumatism. J Infect Dis. 1942;70:180–184. [Google Scholar]
- 10.Proft T, Hilberg H, Plagens H, Herrmann R. The P200 protein of Mycoplasma pneumoniae shows common features with the cytadherence-associated proteins HMW1 and HMW3. Gene. 1996;171:79–82. doi: 10.1016/0378-1119(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 11.Simmons W L, Zuhua C, Glass J I, Simecka J W, Cassell G H, Watson H L. Sequence analysis of the chromosomal region around and within the V-1-encoding gene of Mycoplasma pulmonis: evidence for DNA inversion as a mechanism for V-1 variation. Infect Immun. 1996;64:472–479. doi: 10.1128/iai.64.2.472-479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Appl Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voelker L L, Weaver K E, Ehle L J, Washburn L R. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect Immun. 1995;63:4016–4023. doi: 10.1128/iai.63.10.4016-4023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn L R, Hirsch S, Voelker L L. Mechanisms of attachment of Mycoplasma arthritidis to host cells in vitro. Infect Immun. 1993;61:2670–2680. doi: 10.1128/iai.61.6.2670-2680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washburn L R, Voelker L L, Ehle L J, Hirsch S, Dutenhofer C, Olson K, Beck B. Comparison of Mycoplasma arthritidis strains by enzyme-linked immunosorbent assay, immunoblotting, and DNA restriction analysis. J Clin Microbiol. 1995;33:2271–2279. doi: 10.1128/jcm.33.9.2271-2279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn L R, Weaver E J. Protection of rats against Mycoplasma arthritidis-induced arthritis by active and passive immunizations with two surface antigens. Clin Diagn Lab Immunol. 1997;4:321–327. doi: 10.1128/cdli.4.3.321-327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washburn L R, Weaver K E, Weaver E J, Donelan W, Al-Sheboul S. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect Immun. 1998;66:2576–2586. doi: 10.1128/iai.66.6.2576-2586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yogev D, Watson-McKown R, Rosengarten R, Im J, Wise K S. Increased structural and combinatorial diversity in an extended family of genes encoding Vlp surface proteins of Mycoplasma hyorhinis. J Bacteriol. 1995;177:5636–5643. doi: 10.1128/jb.177.19.5636-5643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, Teng L-J, Watson H L, Glass J I, Blanchard A, Cassell G H. Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect Immun. 1995;63:891–898. doi: 10.1128/iai.63.3.891-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]