Abstract
Atherosclerosis (AS) is an important cause of common vascular diseases. The present study aimed to investigate whether Krüppel like transcription factor 2 (KLF2) could protect against endothelial cell injury and promote cholesterol excretion from foam cells through autophagy. An in vitro AS model was established by the induction of oxidized low-density lipoprotein (ox-LDL) for human umbilical vein endothelial cells (HUVECs). Phorbol-12-myristate-13-acetate (PMA)-induced THP-1 monocytes were differentiated into macrophages which were transformed to foam cells by ox-LDL incubation. The expression of KLF2, adhesion factors, cholesterol efflux regulatory proteins and autophagy-associated proteins in HUVECs or/and THP-1 monocytes was detected by reverse transcription-quantitative PCR and western blot analysis. HUVECs viability, levels of inflammatory factors, formation of foam cells and cholesterol efflux were respectively analyzed by CCK-8 assay, ELISA and Oil Red O staining. KLF2 expression was decreased in ox-LDL-induced HUVECs. KLF2 overexpression attenuated ox-LDL-induced endothelial cell injury, as evidenced by increased cell viability and decreased levels of TNF-α, IL-6, IL-1β, intercellular adhesion molecule 1, vascular cell adhesion molecule-1 and E-selectin. In addition, KLF2 overexpression inhibited the formation of THP-1 macrophage-derived foam cells and promoted lipid efflux. ox-LDL induced decreased KLF2 expression in THP-1 macrophage derived foam cells and KLF2 overexpression activated Nrf2 expression and enhanced autophagy. In conclusion, KLF2 alleviated endothelial cell injury and inhibited the formation of THP-1 macrophage-derived foam cells by activating Nrf2 and enhancing autophagy.
Keywords: Krüppel like transcription factor 2, endothelial cell injury, lipid efflux, autophagy, THP-1 macrophage-derived foam cells
Introduction
Atherosclerosis (AS) is an inflammatory disease of the large and middle arteries characterized by abnormal aggregation of subcutaneous lipids, especially oxidized low-density lipoprotein (ox-LDL), and macrophage-derived foam cells and is a major cause of death from cardiovascular disease (1). Ox-LDL can act as the main trigger of aseptic inflammation by recruiting and attaching to a variety of inflammatory cells, mainly monocytes and neutrophils and aggravating the inflammatory response that damages blood vessels (2). Increased levels of intercellular adhesion molecule-1 (ICAM-1), vascular cell adhesion molecule-1 (VCAM-1) and E-selectin are observed in atherosclerotic vascular endothelial cells (3,4). Ox-LDL induced increased adhesion activity of endothelial cells, which is mainly manifested by significantly increased cell adhesion molecules expressed by endothelial cells (5), to contribute to endothelia cell damage.
Currently, statins are the first-line drugs in clinical treatment of AS. Statins play an anti-AS role mainly by improving vascular endothelial function (6), inhibiting proliferation and migration of vascular smooth muscle cells (7) and inhibiting formation of foam cells (8). Statin can significantly induce the expression of Krüppel like transcription factor 2 (KLF2), a member of the zinc finger subfamily of transcription factors and a member of the SPL/Krüppel like transcription factor family (9,10). KLF2 is evolutionarily conserved between rodents and humans and maintains endothelial homeostasis through anti-inflammatory, anti-thrombosis, anti-oxidation and anti-proliferation effects in endothelial cells (11,12). However, the protective role of KLF2 in the process of endothelial cell injury has not been fully elucidated. Further exploration of the regulatory mechanism of KLF2 on endothelial injury may provide a new target for the clinical treatment of AS.
KLF2 has also been shown to be involved in the differentiation of monocytes. KLF2 expression is reduced during the differentiation of monocytes to macrophages (13), which negatively regulates the pro-inflammatory activation of monocytes/macrophages and is also an important regulator of innate immune response (13,14). Myeloid cell-specific knockout of KLF2 enhances the adhesion of macrophages to endothelial cells and induces the uptake of ox-LDL by macrophages (10,15), thereby increasing the formation of macrophage-derived foam cells. The appearance of foam cells is considered to be one of the early manifestations of AS and the potential extracellular and intracellular lipid deposition is also a key trigger factor for the progression of AS lesions (16,17). Therefore, it is important to enhance the outflow of cholesterol from macrophages in the treatment process of AS, which can effectively reduce the formation of macrophage-derived foam cells and contribute to plaque stability.
Autophagy may play an active role in cholesterol effluent from macrophages. A recent study showed that interfering with KLF2 inhibited Beclin and LC3 levels in abdominal aortic aneurysms, suggesting that KLF2 activates autophagy-related genes in smooth muscle cells (18). Simvastatin maintains microvascular function by inhibiting RAC1, thereby releasing RAB7, activating autophagy and increasing the expression of KLF2, which in turn further promotes the activation of autophagy (19). However, it remains to be elucidated whether KLF2 plays a role in the process of macrophage foaming, and whether KLF2 can participate in the process of cholesterol effusion in macrophages by regulating autophagy is worth exploring.
Therefore, the present study aimed to investigate whether KLF2 could protect against endothelial cell injury and promote cholesterol excretion from foam cells through autophagy.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) and human acute monocytic leukemia cells (THP-1 monocytes) were purchased from CoBioer Biosciences Co., Ltd. HUVECs were cultured in DMEM medium (Gibco; Thermo Fisher Scientific Inc.) supplemented with 10% FBS in a humidified atmosphere containing 5% CO2 at 37˚C. An in vitro AS model was established by the induction of 100 µg/ml ox-LDL (MilliporeSigma) for HUVECs for 24 h. For ox-LDL treatment, the cells were exposed to 100 µg/ml of ox-LDL for 24 h after transfection.
THP-1 monocytes were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific Inc.) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) in a humidified atmosphere containing 5% CO2 at 37˚C. THP-1 monocytes were treated with 50 nM phorbol-12-myristate-13-acetate (PMA) for 48 h to induce their differentiation into macrophages. Then, THP-1 derived macrophages were continuously treated with 50 µg/ml ox-LDL for 48 h to form THP-1 macrophage-derived foam cells.
Cell transfection
HUVECs or THP-1 derived macrophages were seeded into 6-well plates at the density of 1x106 cells/well. The sequence of KLF2 (5'-CAACAGCGTGCTGGACTTCA-3') was cloned into a pcDNA3.1 vector to generate overexpressing plasmid (pcDNA3.1-KLF2) by GenePharma Co., Ltd. and the empty pcDNA3.1 vector (pcDNA3.1-NC) was used as a negative control. The accession no. of this KLF2 was NC_000019. Transfection of 20 µg pcDNA3.1 plasmids into HUVECs or THP-1 derived macrophages was performed at 37˚C for 6 h by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the kit instructions. At 48 h post-transfection, reverse transcription-quantitative (RT-q) PCR and western blot analysis were used to examine gene expression levels.
RT-qPCR
HUVECs or THP-1 monocytes in each group (24-well plates at a density of 2x105 cells per well) were treated with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) to obtain the total RNA according to the manufacturer's protocols. RNA purity was assessed by spectrophotometric analysis (CWBIO) wherein the A260/280 ratios were between 1.8 and 2.2. Equal RNA was used for cDNA generation by using a PrimeScript RT reagent kit with a gDNA eraser (Takara Bio, Inc.) according to the manufacturer's protocols. Subsequently, SYBR Premix Ex Taq II (Takara Bio, Inc.) was then used to amplify cDNA through qPCR according to the manufacturer's protocols using an ABI Prism 7500 Fast Real-time PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The amplification efficiencies of the qPCRs were 90-110%. The thermocycling conditions were: Initial denaturation at 95˚C for 10 min; followed by 40 cycles of 95˚C for 1 min and 60˚C for 1 min. The sequences of oligonucleotide primers used for qPCR were: KLF2 forward, 5'-AGACCTACACCAAGAGTTCGCATC-3' and reverse, 5'-ATCGCACAGATGGCACTGGAATG-3'; ICAM-1 forward, 5'-GCCCGAGCTCAAGTGTCTAA-3' and reverse, 5'-GGAGAGCACATTCACGGCA-3'; VCAM-1 forward, 5'-AATTTATGTGTGTGAAGGAG-3' and reverse, 5'-GCATGTCATATTCACAGAA-3'; E-selectin forward, 5'-TGTGGGTCTGGGTAGGAACC-3' and reverse, 5'-AGCTGTGTAGCATAGGGCAAG-3'; ATP-binding cassette transporters A1 (ABCA1) forward, 5'-GCTGGTGTGGACCCTTACTC-3' and reverse, 5'-GCAGCTTCATATGGCAGCAC-3'; ATP-binding cassette transporter G1 (ABCG1) forward, 5'-TGTCTGATGGCCGCTTTCTC-3' and reverse, 5'-GGACCCATAATGGCCACCAA-3'; scavenger receptor BI (SR-BI) forward, 5'-ATGACTCCTGAGTCCTCGCT-3' and reverse, 5'-GGTCAGCGTTGAGGAAGTGA-3'; peroxisome proliferator-activated receptor-γ (PPARγ) forward, 5'-CCAGAAGCCTGCATTTCTGC-3' and reverse, 5'-CACGGAGCTGATCCCAAAGT-3'; liver X receptor (LXR) α forward, 5'-TCTGGACAGGAAACTGCACC-3' and reverse, 5'-AAGAATCCCTTGCAGCCCTC-3'; GAPDH forward, 5'-CCTCAAGATCATCAGCAATG-3' and reverse, 5'-CCATCC ACAGTCTTCTGGGT-3'. The 2-ΔΔCq method was used to analyze the relative expression of KLF2, ICAM-1, VCAM-1 and E-selectin (20), which were normalized to GAPDH. The experiment was repeated three times.
Western blotting
For the detection of nuclear factor erythroid 2-related factor 2 (Nrf2), nucleoprotein and cytoplasmic protein extraction kit obtained from Nanjing KeyGen Biotech Co., Ltd. was used to extract the nucleoprotein and cytoplasmic proteins from HUVECs or THP-1 monocytes. For the detection of other proteins, HUVECs or THP-1 monocytes in each group were homogenized in RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS; Beyotime institute of Biotechnology], which was centrifuged at 4˚C at 850 x g for 15 min to collect the total protein. The protein concentration was determined by bicinchoninic acid (BCA) method. The equivalent amount of 50 µg protein was separated by 12% SDS-PAGE and then transferred to a PVDF membranes (MilliporeSigma) which were then blocked with 5% non-fat milk for 2 h at room temperature. Subsequently, the membranes were incubated with primary antibodies as KLF2 (cat. no. ab236507; 1:1,000; Abcam), ICAM-1 (cat. no. ab282575; 1:1,000; Abcam), VCAM-1 (cat. no. ab174279; 1:1,000; Abcam), E-selectin (cat. no. 20894-1-AP; 1:800; ProteinTech Group, Inc.), ABCA1 (cat. no. 96292S; 1:1,000; Cell Signaling Technology, Inc.), ABCG1 (cat. no. ab52617; 1:1,000; Abcam), SR-BI (cat. no. ab217318; 1:1,000; Abcam), PPARγ (cat. no. 16643-1-AP; 1:1,000; ProteinTech Group, Inc.), LXRα (cat. no. 14351-1-AP; 1:1,000; ProteinTech Group, Inc.), Nrf2 (cat. no. ab62352; 1:1,000; Abcam), Lamin B (cat. no. 12987-1-AP; 1:1,000; ProteinTech Group, Inc.), LC3II (cat. no. 4108S; 1:1,000; Cell Signaling Technology, Inc.), Beclin1 (cat. no. 3495T; 1:1,000; Cell Signaling Technology, Inc.), p62 (cat. no. 18420-1-AP; 1:1,000; ProteinTech Group, Inc.) and GAPDH (cat. no. 5174T; 1:1,000; Cell Signaling Technology, Inc.) at 4˚C overnight. Following incubation with the horseradish peroxidase-conjugated anti-rabbit secondary antibody (cat. no. 7074P2, 1:3,000; Cell Signaling Technology, Inc.) for 1 h at room temperature, protein bands were visualized by a chemiluminescent detection system (ECL; Cytiva) using an ECL reagent (Thermo Fisher Scientific, Inc.). Bands grey values were semi-quantified analyzed using ImageJ software (v1.46; National Institutes of Health). Protein expression was normalized to the internal reference gene GAPDH or Lamin B.
CCK-8 assay
HUVECs (5x103 cells per well) were seeded into 96-well plates and transfection performed for 48 h. Then, the transfected HUVECs were treated with 100 µg/ml of ox-LDL for 24 h. After PBS washing, each well was added with 10 µl CCK-8 solution and the 96-well plates were incubated at 37˚C for 2 h. The absorbance of each well was detected by a microplate reader (Bio-Rad Laboratories, Inc.) at 450 nm.
ELISA
HUVECs were cultured in 96-well plates at a density of 1x104 cells/ml. HUVECs were transfected for 48 h and then exposed to ox-LDL (100 µg/ml) for 24 h, then the medium was obtained and centrifuged at 4˚C at 850 x g for 10 min to collect the culture supernatant. The levels of TNF-α, IL-6 and IL-1β in the supernatant was assessed using a TNF-α ELISA Kit (cat. no. PT518), IL-1β ELISA Kit (cat. no. PI305) and IL-6 ELISA Kit (cat. no. PI330) all from Beyotime Institute of Biotechnology.
Oil Red O staining
After THP-1 monocytes were induced with 50 nM PMA and/or 50 µg/ml ox-LDL, Oil Red O staining was used to evaluate the formation of THP-1 macrophage-derived foam cells. Following the treatment, cells were fixed with 4% paraformaldehyde (MilliporeSigma) for 30 min and then incubated with filtered 0.5% Oil Red O solution (MilliporeSigma) for 15 min at room temperature. Images were captured under a confocal microscope (Olympus Corporation) and positive Oil Red O areas (staining areas) was used to reflect the total lipid content. The % positive Oil Red O areas was calculated by ImageJ (v.1.8.0; National Institutes of Health).
Liquid scintillation counting
Cholesterol efflux was analyzed as previously described (21). The macrophages and foam cells were cultured and labeled with 0.4 µCi/ml [3H]-cholesterol for 24 h and incubated overnight in RPMI-1640 medium with 0.1% BSA to allow equilibration of [3H]-cholesterol in all cellular pools. Following PBS washing, equilibrated [3H] cholesterol-labeled cells were incubated in RPMI-1640 medium containing 0.1% BSA with or without 15 µg/ml apoA-I for 12 h. A 150 µl sample of efflux medium was obtained at and passed through a 0.45 µm filter to remove any floating cells. Monolayers were washed twice in PBS and cellular lipids were extracted with isopropanol. Medium and cell-associated [3H] cholesterol was then determined by liquid scintillation counting through detecting the α-rays derived from [3H] cholesterol. Percent efflux was evaluated according to the equation: [total media counts/(total cellular counts + total media counts)] x100%.
Statistical analysis
All of the experiments were repeated three times and the data were conformed to normal distribution using Shapiro-Wilk test. Data are presented as means ± standard deviation and analyzed by GraphPad 8.0 (GraphPad Software, Inc.). Two-group comparisons were analyzed with Student's t-test. Comparisons between three or more groups were analyzed by One way analysis of variance, followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
KLF2 overexpression attenuates ox-LDL-induced endothelial cell injury
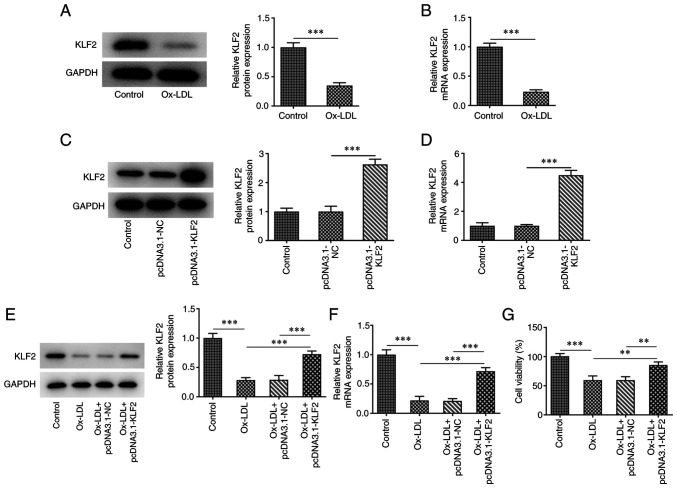
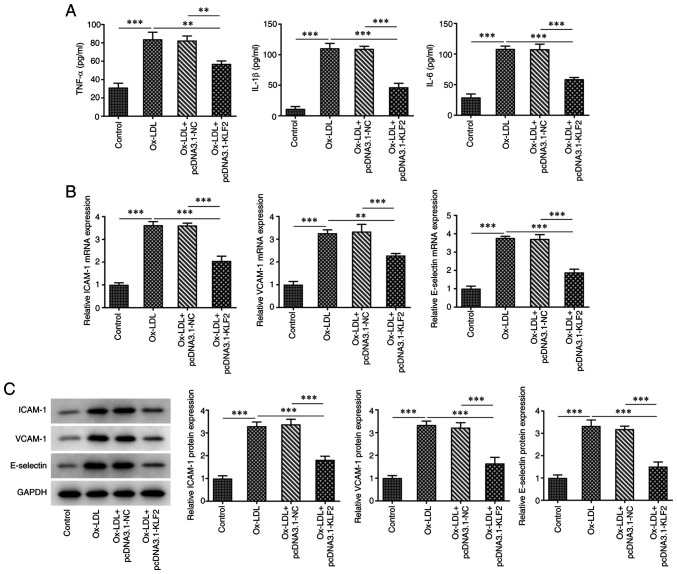
The expression of KLF2 in HUVECs was downregulated after ox-LDL induction compared to the control group (Fig. 1A and B). The expression of KLF2 in HUVECs transfected with pcDNA3.1-KLF2 was upregulated (Fig. 1C and D). When ox-LDL-induced HUVECs were transfected with pcDNA3.1-KLF2, KLF2 protein and mRNA expression was significantly upregulated (Fig. 1E and F). The result of Fig. 1E indicated that ox-LDL suppressed the HUVECs viability while the further KLF2 overexpression improved the decreased viability of ox-LDL-induced HUVECs (Fig. 1G). The levels of TNF-α, IL-1β and IL-6 were increased in ox-LDL-induced HUVECs, which was suppressed by KLF2 overexpression (Fig. 2A). The expression of adhesion factors (ICAM-1, VCAM-1 and E-selectin) was increased in HUVECs treated with ox-LDL and decreased in ox-LDL-induced HUVECs transfected with pcDNA3.1-KLF2 (Fig. 2B and C). These data suggested that KLF2 overexpression attenuated ox-LDL-induced endothelial cell injury.
Figure 1.
KLF2 overexpression increases viability of ox-LDL-induced HUVECs. The (A) protein and (B) mRNA expressions of KLF2 in ox-LDL-induced HUVECs were determined by western blotting and RT-qPCR analysis. The (C) protein and (D) mRNA expressions of KLF2 in HUVECs transfected with pcDNA3.1-KLF2 were determined by western blotting and RT-qPCR analysis. The (E) protein and (F) mRNA expressions of KLF2 in ox-LDL-induced HUVECs transfected with pcDNA3.1-KLF2 were determined by western blotting and RT-qPCR analysis. (G) The viability of ox-LDL-induced HUVECs transfected with pcDNA3.1-KLF2 was detected by CCK-8 assay. **P<0.01 and ***P<0.001. KLF2, Krüppel like transcription factor 2; ox-LDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control.
Figure 2.
KLF2 overexpression downregulates the levels of inflammatory factors and adhesion factors in ox-LDL-induced HUVECs. (A) The levels of inflammatory factors in ox-LDL-induced HUVECs transfected with pcDNA3.1-KLF2 were determined by ELISA kits. The (B) protein and (C) mRNA expressions of adhesion factors in ox-LDL-induced HUVECs transfected with pcDNA3.1-KLF2 were determined by western blotting and reverse transcription-quantitative PCR. **P<0.01 and ***P<0.001. KLF2, Krüppel like transcription factor 2; ox-LDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; NC, negative control.
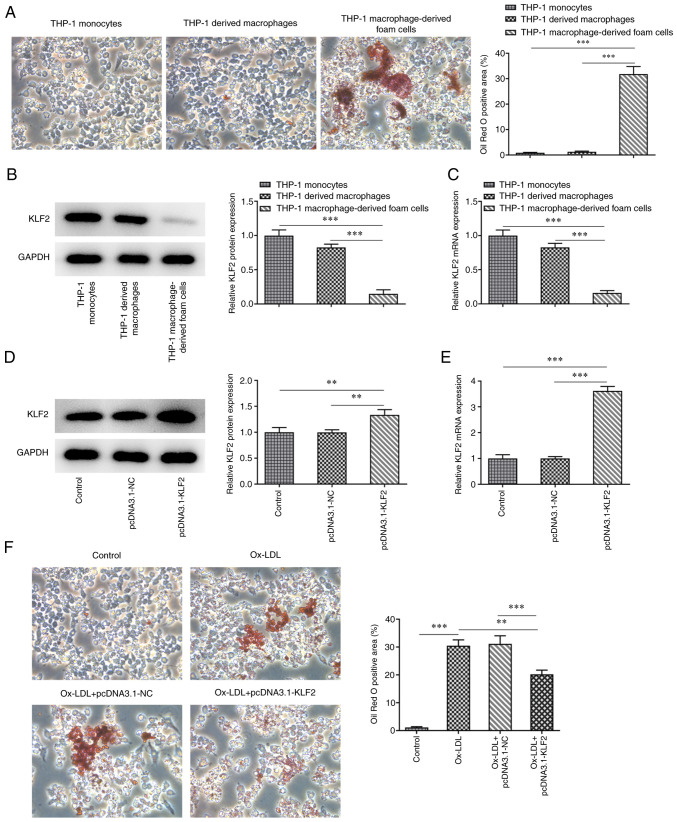
KLF2 overexpression inhibits the formation of THP-1 macrophage-derived foam cells
After PMA induced THP-1 monocytes for 48 h, the cells were fusiform in shape with extended pseudopods to differentiate into macrophages. The mature macrophages were differentiated. The mature differentiated macrophages were incubated with ox-LDL for 48 h and the cells were stained with oil red O. Under the light microscope, there were a large number of red lipid particles in the cells and the cell volume increased (Fig. 3A). The expression of KLF2 in THP-1 macrophage-derived foam cells was decreased compared with that in THP-1 monocytes and THP-1 derived macrophages (Fig. 3B and C). After THP-1 derived macrophages (control) were transfected with pcDNA3.1-KLF2, KLF2 expression was upregulated (Fig. 3D and E). A large number of red lipid particles were observed in the THP-1 derived macrophages induced by ox-LDL, which were decreased by KLF2 overexpression (Fig. 3F). On the whole, KLF2 overexpression inhibited the formation of THP-1 macrophage-derived foam cells.
Figure 3.
KLF2 overexpression inhibits the formation of THP-1 macrophage-derived foam cells. (A) The formation of THP-1 macrophage-derived foam cells was confirmed by Oil Red O staining. Magnification, x200. The (B) protein and (C) mRNA expressions of adhesion factors in THP-1 monocytes, THP-1 derived macrophages and THP-1 macrophage-derived foam cells were determined by western blotting and RT-qPCR analysis. The (D) protein and (E) mRNA expressions of adhesion factors in THP-1 derived macrophages transfected with pcDNA3.1-KLF2 were determined by western blotting and RT-qPCR analysis. (F) The formation of THP-1 macrophage-derived foam cells affected by KLF2 overexpression was confirmed by Oil Red O staining. Magnification, x200. **P<0.01 and ***P<0.001. KLF2, Krüppel like transcription factor 2; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control.
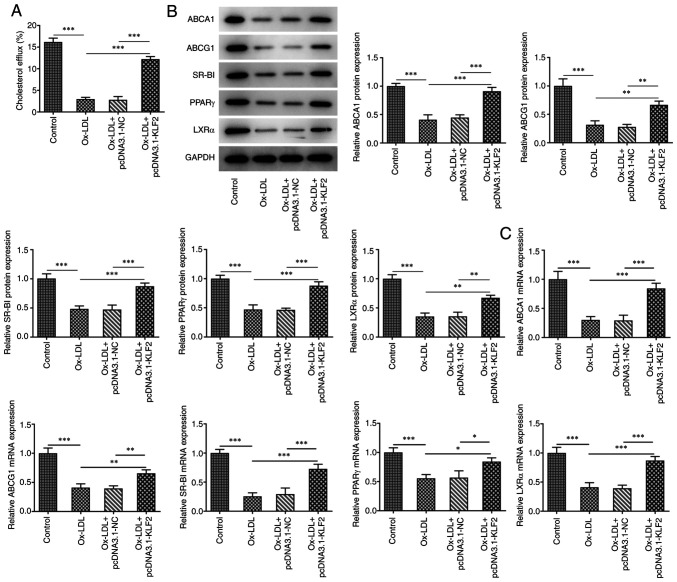
KLF2 overexpression promotes lipid efflux in THP-1 macrophage-derived foam cells
As shown in Fig. 4A, Ox-LDL reduced the cholesterol efflux in THP-1 derived macrophages, which was promoted by KLF2 overexpression. Ox-LDL suppressed the expression of cholesterol efflux regulatory proteins (ABCA1, ABCG1, SR-BI, PPARγ and LXRα) in THP-1 derived macrophages and KLF2 overexpression could reverse the above changes (Fig. 4B). Consistently, the mRNA expression levels of ABCA1, ABCG1, SR-BI, PPARγ and LXRα showed the same changing tendency as their protein expression levels (Fig. 4C). In brief, KLF2 overexpression promoted lipid efflux in THP-1 macrophage-derived foam cells.
Figure 4.
KLF2 overexpression promotes lipid efflux from THP-1 macrophage-derived foam cells. (A) The cholesterol efflux in ox-LDL induced THP-1 derived macrophages transfected with pcDNA3.1-KLF2 was analyzed by liquid scintillation counting. (B) The expression of cholesterol efflux related proteins in ox-LDL induced THP-1 derived macrophages transfected with pcDNA3.1-KLF2 was determined by western blotting. (C) The mRNA expression of ABCA1, ABCG1, SR-BI, PPARγ and LXRα in ox-LDL induced THP-1 derived macrophages transfected with pcDNA3.1-KLF2 was tested with RT-qPCR. *P<0.05, **P<0.01 and ***P<0.001. KLF2, Krüppel like transcription factor 2; ox-LDL, oxidized low-density lipoprotein; NC, negative control.
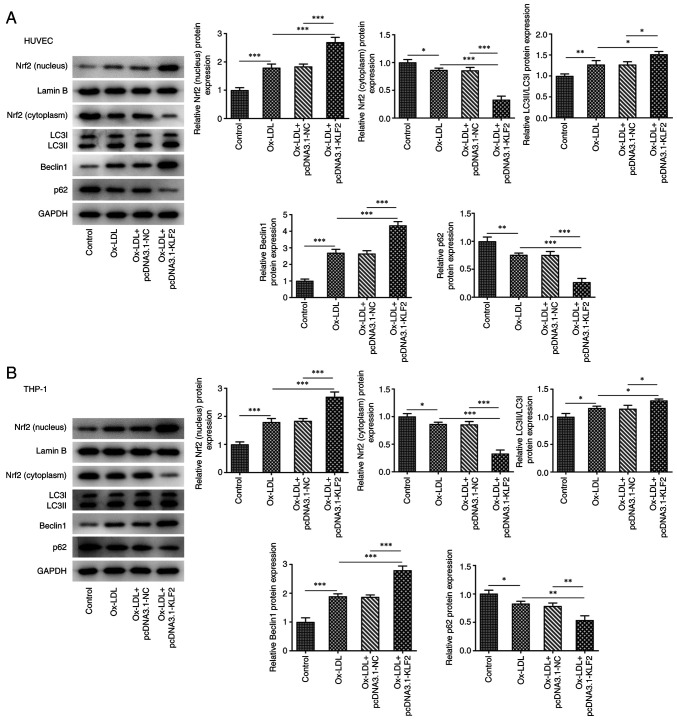
KLF2 overexpression activities Nrf2 expression and enhances autophagy in ox-LDL-induced HUVECs and THP-1 macrophage derived foam cells
In HUVECs, ox-LDL promoted the expression of Nrf2 (nucleus), LC3II/LC3I and Beclin1 while suppressing the expression of Nrf2 (cytoplasm) and p62. KLF2 overexpression further enhanced the expression of Nrf2 (nucleus), LC3II/LC3I and Beclin1 while downregulating the expression of Nrf2 (cytoplasm) and p62 in ox-LDL induced HUVECs (Fig. 5A). As shown in Fig. 5B, the expression of Nrf2 (nucleus), LC3II/LC3I and Beclin1 was increased while the expression of Nrf2 (cytoplasm) and p62 was decreased in THP-1 derived macrophages induced by ox-LDL and the expression of Nrf2 (nucleus), LC3II/LC3I and Beclin1 was further increased while the expression of Nrf2 (cytoplasm) and p62 was further decreased by KLF2 overexpression in THP-1 derived macrophages induced by ox-LDL. These observations revealed that KLF2 overexpression activates Nrf2 expression and enhances autophagy in ox-LDL-induced HUVECs and THP-1 macrophage-derived foam cells.
Figure 5.
KLF2 overexpression activities Nrf2 expression and enhances autophagy in ox-LDL-induced HUVECs and THP-1 macrophage derived foam cells. The expression of Nrf2 and autophagy-related proteins in (A) ox-LDL induced HUVECs and (B) THP-1 macrophages transfected with pcDNA3.1-KLF2 was analyzed by western blotting. *P<0.05, **P<0.01 and ***P<0.001. KLF2, Krüppel like transcription factor 2; ox-LDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; NC, negative control.
Discussion
The expression of KLF2 can be significantly induced by statins in the clinical treatment of AS. The present study indicated that KLF2 expression was decreased in ox-LDL induced HUVECs. In vivo animal experiments have shown that the specific loss of KLF2 in endothelial cells is prone to the occurrence of AS (22). In addition, KLF2 can also reduce endothelial permeability and enhance endothelial barrier function by activating endothelial RAP guanine nucleotide exchange factor 3(23). Fork head transcription factor 1, regulated by KLF2, directly inhibits the activation of endothelial inflammasome and delays the formation of AS lesions (24). In the present study, KLF2 overexpression improved viability and alleviated inflammation of ox-LDL-induced HUVECs, which indicated that KLF2 could protect against injury in HUVECs induced by ox-LDL.
Aggregation of macrophage-derived foam cells in the vascular wall is the main pathological feature of AS (25,26). Macrophages phagocytize excess ox-LDL, resulting in intracellular lipid metabolism disorder and excessive lipid accumulation in macrophages formed foam cells. The formation of foam cells is closely associated with excessive lipid uptake by cells and intracellular bile sterol outflow disorder (27). It has been shown that inhibiting cholesterol outflow can lead to cholesterol accumulation in macrophages and accelerate the formation of foam cells (25). Therefore, the key to inhibiting the formation of foam cells depends on the cholesterol outflow pathway. There are three vectors for cholesterol outflow: ABCA1, ABCG1 and SR-BI (28). PPARγ is expressed in a variety of vascular cells, including monocytes/macrophages (29), endothelial cells (30) and smooth muscle cells (31), which are involved in the AS process. As members of the nuclear receptor superfamily, PPARα and LXRα bind corresponding ligands and then form active heterodimer with retinoid X receptor, which is mainly involved in lipid metabolism and promotes cholesterol excretion (32). The present study demonstrated that KLF2 overexpression suppressed the formation of macrophage-derived foam cells by increasing cholesterol outflow through the promotion of the expression of ABCA1, ABCG1, SR-BI, PPARγ and LXRα.
Under basal conditions, autophagy may help maintain cell balance. As a stress response to hunger or oxidative, autophagy can be activated as an adaptive process (33). Evidence suggests that dysregulation of autophagy could result in AS and that autophagy is implicated in endothelial cells response to pathophysiological stimuli during AS (34). Autophagy participates in the defense mechanism against oxidative stress and inflammation, thereby preventing vascular cell death (35). Ox-LDL is reported to be an inducer of autophagy in HUVECs (36,37), which is in agreement with the present study, as evidenced by increased LC3II/LC3I, Beclin1 expression and decreased p62 expression in HUVECs. Additionally, a previous study shows that enhanced autophagy can improve age-related phenotypes, reduce age-related heart and kidney disease and improve the health status of mice (38). It can also protect the body from stress injury and delay the development of AS (39). Macrophage autophagy serves an important role in the occurrence and development of AS by promoting the efflux of intracellular cholesterol (40). Defective autophagy of macrophages in AS impairs cholesterol metabolism and inflammatory body activation (41,42). Ox-LDL exposure has been shown to promote the expression of autophagosome marker proteins in cultures, possibly by increasing oxidative stress and inflammation (43,44). However, there is currently disagreement regarding the reported autophagic activity in ox-LDL-treated macrophages, as some reports suggest that autophagic flux is inhibited in ox-LDL-stimulated macrophages (45,46) and others insisted that that ox-LDL could lead to activation of autophagy (47,48). The present study also suggested that autophagy was induced after ox-LDL exposure in THP-1 derived macrophages. It has been reported that metformin could inhibit foam cell formation by activating KLF2-mediated autophagy (49). KLF2 possibly contributes to regulation of autophagy in a model of acute liver injury (19). KLF-autophagy pathway modulates the life span of nematodes and regulates mammalian age-associated vascular dysfunction (50). A recent study showed that interfering with KLF2 inhibits Beclin and LC3 levels in abdominal aortic aneurysms (18). The present study demonstrated that overexpression of KLF2 increased the autophagy in ox-LDL-induced HUVECs and THP-1 macrophage derived foam cells.
Nrf2 is a transcription factor involved in cellular redox homeostasis. Under oxidative stress, Nrf2 separates from its inhibitor KEAP-1 and translocates into the nucleus, leading to transcriptional activation of cellular defense genes (51,52). Nrf2 activation can protect human coronary endothelial cells from oxidative stress and knockdown of Nrf2 expression promotes hydrogen peroxide-induced apoptosis (53). Hu et al (54) found that activation of Nrf2 signaling pathway reduced the level of reactive oxygen species in vascular endothelial cells and inhibited NLRP3 dependent endothelial cell pyrolysis. Induction of Nrf2/HO-1 activation can upregulate the expression of ABCA1 and ABCG1 and enhance cholesterol excretion (55,56). Wang et al (57) found that Nrf2 induces the formation of cytolysosome and enhances autophagy activity, thereby inhibiting tumor cell apoptosis. It is noteworthy that Nrf2 inducer upregulates the gene and protein expression of autophagy-related molecules and also enhances autophagic flux in diabetic mouse aorta, suggesting that Nrf2 could activate autophagy in AS (58). Notably, KLF2 can stimulate Nrf2 to enter into the nucleus and initiate activation of the Nrf2 pathway (59) and artesunate enhances nuclear translocation of Nrf2 in vascular smooth muscle cells by upregulating KLF2 expression (60). The present study indicated that KLF2 overexpression stimulated Nrf2 to transport into the nucleus and enhanced autophagy by increasing the expression of LC3II/LC3I and Beclin1 and decreasing the p62 expression in ox-LDL induced HUVECs and THP-1 macrophage-derived foam cells. The addition of some siRNA-Nrf2 or some inhibitor of autophagy might provide stronger evidence for the present findings, which is a limitation of the present study and will be conducted in the next experiments.
In conclusion, KLF2 expression was decreased in ox-LDL induced HUVECs and THP-1 macrophage-derived foam cells. In addition, KLF2 alleviated endothelial cell injury and promotes lipid outflow by inhibiting the formation of THP-1 macrophage-derived foam cells through enhancing autophagy mediated by Nrf2. The findings provided a promising target for the treatment of AS. Some limitations that need to be addressed later. The addition of some siRNA-Nrf2 or some inhibitor of autophagy might provide stronger evidence for the present findings. Whether the changes in steady-state nuclear and cytoplasmic levels of Nrf2 are transcriptional, due to changes in the stability of Nrf2, or release of Nrf2 from KEAP1 in the cytoplasm needs to be explored. The use of commercial fluorescent sensors (LC3B-RFP and LC3B-GFP) and electron microscopy to measure the process of autophagy will be considered in future study.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZT and XD conceived and designed the experiments. HR, YL, HY and QL performed the experiments. ZT, HR and XD analyzed the data and wrote the manuscript. ZT and YL confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang Y, Kou J. Research progress of oxLDL in atherosclerotic thrombosis. Chin J Cardiovasc Rehabil Med. 2021;30:344–347. [Google Scholar]
- 2.Trpkovic A, Resanovic I, Stanimirovic J, Radak D, Mousa SA, Cenic-Milosevic D, Jevremovic D, Isenovic ER. Oxidized low-density lipoprotein as a biomarker of cardiovascular diseases. Crit Rev Clin Lab Sci. 2015;52:70–85. doi: 10.3109/10408363.2014.992063. [DOI] [PubMed] [Google Scholar]
- 3.Wojakowski W, Gminski J. Soluble ICAM-1, VCAM-1 and E-selectin in children from families with high risk of atherosclerosis. Int J Mol Med. 2001;7:181–185. doi: 10.3892/ijmm.7.2.181. [DOI] [PubMed] [Google Scholar]
- 4.Davies MJ, Gordon JL, Gearing AJ, Pigott R, Woolf N, Katz D, Kyriakopoulos A. The expression of the adhesion molecules ICAM-1, VCAM-1, PECAM, and E-selectin in human atherosclerosis. J Pathol. 1993;171:223–229. doi: 10.1002/path.1711710311. [DOI] [PubMed] [Google Scholar]
- 5.Zhong XJ, Chen TW, Chen YH, Chen FH, Zhou-Xue LI, Liu LL, Liu MQ, Huang QR. Effects of ox-LDL on the proaggregation and proadhesion-related molecules expression of vascular endothelial cells. Chin J Arterioscler. 2014;9(e89877) [Google Scholar]
- 6.Sen-Banerjee S, Mir S, Lin Z, Hamik A, Atkins GB, Das H, Banerjee P, Kumar A, Jain MK. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 7.Chandrasekar B, Mummidi S, Mahimainathan L, Patel DN, Bailey SR, Imam SZ, Greene WC, Valente AJ. Interleukin-18-induced human coronary artery smooth muscle cell migration is dependent on NF-kappaB- and AP-1-mediated matrix metalloproteinase-9 expression and is inhibited by atorvastatin. J Biol Chem. 2006;281:15099–15109. doi: 10.1074/jbc.M600200200. [DOI] [PubMed] [Google Scholar]
- 8.Takemoto M, Liao JK. Pleiotropic effects of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Arterioscler Thromb Vasc Biol. 2001;21:1712–1719. doi: 10.1161/hq1101.098486. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, Wang S. Role of kruppel-like transcription factors in adipogenesis. Dev Biol. 2013;373:235–243. doi: 10.1016/j.ydbio.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 10.Atkins GB, Wang Y, Mahabeleshwar GH, Shi H, Gao H, Kawanami D, Natesan V, Lin Z, Simon DI, Jain MK. Hemizygous deficiency of Krüppel-like factor 2 augments experimental atherosclerosis. Circ Res. 2008;103:690–693. doi: 10.1161/CIRCRESAHA.108.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novodvorsky P, Chico TJ. The role of the transcription factor KLF2 in vascular development and disease. Prog Mol Biol Transl Sci. 2014;124:155–188. doi: 10.1016/B978-0-12-386930-2.00007-0. [DOI] [PubMed] [Google Scholar]
- 12.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA Jr, García-Cardeña G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci USA. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity. 2011;34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingrel JB, Pilcher-Roberts R, Basford JE, Manoharan P, Neumann J, Konaniah ES, Srinivasan R, Bogdanov VY, Hui DY. Myeloid-specific Krüppel-like factor 2 inactivation increases macrophage and neutrophil adhesion and promotes atherosclerosis. Circ Res. 2012;110:1294–1302. doi: 10.1161/CIRCRESAHA.112.267310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frostegård J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013;11(117) doi: 10.1186/1741-7015-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summerhill VI, Grechko AV, Yet SF, Sobenin IA, Orekhov AN. The atherogenic role of circulating modified lipids in atherosclerosis. Int J Mol Sci. 2019;20(3561) doi: 10.3390/ijms20143561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmon M, Spinosa M, Zehner ZE, Upchurch GR, Ailawadi G. Klf4, Klf2, and Zfp148 activate autophagy-related genes in smooth muscle cells during aortic aneurysm formation. Physiol Rep. 2019;7(e14058) doi: 10.14814/phy2.14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guixé-Muntet S, de Mesquita FC, Vila S, Hernández-Gea V, Peralta C, García-Pagán JC, Bosch J, Gracia-Sancho J. Cross-talk between autophagy and KLF2 determines endothelial cell phenotype and microvascular function in acute liver injury. J Hepatol. 2017;66:86–94. doi: 10.1016/j.jhep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Hu YW, Wang Q, Ma X, Li XX, Liu XH, Xiao J, Liao DF, Xiang J, Tang CK. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells. J Atheroscler Thromb. 2010;17:493–502. doi: 10.5551/jat.3152. [DOI] [PubMed] [Google Scholar]
- 22.Jain MK, Sangwung P, Hamik A. Regulation of an inflammatory disease: Krüppel-like factors and atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:499–508. doi: 10.1161/ATVBAHA.113.301925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang RT, Wu D, Meliton A, Oh MJ, Krause M, Lloyd JA, Nigdelioglu R, Hamanaka RB, Jain MK, Birukova A, et al. Experimental lung injury reduces Krüppel-like factor 2 to increase endothelial permeability via regulation of RAPGEF3-Rac1 signaling. Am J Respir Crit Care Med. 2017;195:639–651. doi: 10.1164/rccm.201604-0668OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang T, Liu J, Chen X, Zhang L, Pi J, Sun H, Li L, Bauer R, Wang H, Yu Z, et al. Endothelial Foxp1 suppresses atherosclerosis via modulation of Nlrp3 inflammasome activation. Circ Res. 2019;125:590–605. doi: 10.1161/CIRCRESAHA.118.314402. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Li L, Xie W, Wu JF, Yao F, Tan YL, Xia XD, Liu XY, Liu D, Lan G, et al. Apolipoprotein A-1 binding protein promotes macrophage cholesterol efflux by facilitating apolipoprotein A-1 binding to ABCA1 and preventing ABCA1 degradation. Atherosclerosis. 2016;248:149–159. doi: 10.1016/j.atherosclerosis.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Chistiakov DA, Bobryshev YV, Orekhov AN. Macrophage-mediated cholesterol handling in atherosclerosis. J Cell Mol Med. 2016;20:17–28. doi: 10.1111/jcmm.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher EA. Regression of atherosclerosis: The journey from the liver to the plaque and back. Arterioscler Thromb Vasc Biol. 2016;36:226–235. doi: 10.1161/ATVBAHA.115.301926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu XH, Fu YC, Zhang DW, Yin K, Tang CK. Foam cells in atherosclerosis. Clin Chim Acta. 2013;424:245–252. doi: 10.1016/j.cca.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Castrillo A, Tontonoz P. PPARs in atherosclerosis: The clot thickens. J Clin Invest. 2004;114:1538–1540. doi: 10.1172/JCI23705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vattulainen-Collanus S, Akinrinade O, Li M, Koskenvuo M, Li CG, Rao SP, de Jesus Perez V, Yuan K, Sawada H, Koskenvuo JW, et al. Loss of PPARγ in endothelial cells leads to impaired angiogenesis. J Cell Sci. 2016;129:693–705. doi: 10.1242/jcs.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruemmer D, Blaschke F, Law RE. New targets for PPARgamma in the vessel wall: Implications for restenosis. Int J Obes (Lond) 2005;29 (Suppl 1):S26–S30. doi: 10.1038/sj.ijo.0802910. [DOI] [PubMed] [Google Scholar]
- 32.Ge CX, Yu R, Xu MX, Li PQ, Fan CY, Li JM, Kong LD. Betaine prevented fructose-induced NAFLD by regulating LXRα/PPARα pathway and alleviating ER stress in rats. Eur J Pharmacol. 2016;770:154–164. doi: 10.1016/j.ejphar.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Guo C, Chen Z, Zhang P, Li J, Li Y. Vitexin alleviates ox-LDL-mediated endothelial injury by inducing autophagy via AMPK signaling activation. Mol Immunol. 2017;85:214–221. doi: 10.1016/j.molimm.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Zhou J, Dou Y, Shi Y, Wang Y, Hong J, Zhao J, Zhang J, Yuan Y, Zhou M, Wei X. The protective effects of angelica organic acid against ox-LDL-induced autophagy dysfunction of HUVECs. BMC Complement Med Ther. 2020;20(164) doi: 10.1186/s12906-020-02968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Z, Liu S, Wang X, Khaidakov M, Dai Y, Mehta JL. Oxidant stress in mitochondrial DNA damage, autophagy and inflammation in atherosclerosis. Sci Rep. 2013;3(1077) doi: 10.1038/srep01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin HH. In vitro and in vivo atheroprotective effects of gossypetin against endothelial cell injury by induction of autophagy. Chem Res Toxicol. 2015;28:202–215. doi: 10.1021/tx5003518. [DOI] [PubMed] [Google Scholar]
- 37.Che J, Liang B, Zhang Y, Wang Y, Tang J, Shi G. Kaempferol alleviates ox-LDL-induced apoptosis by up-regulation of autophagy via inhibiting PI3K/Akt/mTOR pathway in human endothelial cells. Cardiovasc Pathol. 2017;31:57–62. doi: 10.1016/j.carpath.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Fernández ÁF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitada M, Ogura Y, Koya D. The protective role of Sirt1 in vascular tissue: Its relationship to vascular aging and atherosclerosis. Aging (Albany NY) 2016;8:2290–2307. doi: 10.18632/aging.101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao X, Bandler RC, Rahman K, Fisher EA, Rayner KJ, et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37:1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grootaert MO, da Costa Martins PA, Bitsch N, Pintelon I, De Meyer GR, Martinet W, Schrijvers DM. Defective autophagy in vascular smooth muscle cells accelerates senescence and promotes neointima formation and atherogenesis. Autophagy. 2015;11:2014–2032. doi: 10.1080/15548627.2015.1096485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao H, Jia Q, Yan L, Chen C, Xing S, Shen D. Quercetin suppresses the progression of atherosclerosis by regulating MST1-mediated autophagy in ox-LDL-induced RAW264.7 macrophage foam cells. Int J Mol Sci. 2019;20(6093) doi: 10.3390/ijms20236093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollace V, Gliozzi M, Musolino V, Carresi C, Muscoli S, Mollace R, Tavernese A, Gratteri S, Palma E, Morabito C, et al. Oxidized LDL attenuates protective autophagy and induces apoptotic cell death of endothelial cells: Role of oxidative stress and LOX-1 receptor expression. Int J Cardiol. 2015;184:152–158. doi: 10.1016/j.ijcard.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Fan X, Wang J, Hou J, Lin C, Bensoussan A, Chang D, Liu J, Wang B. Berberine alleviates ox-LDL induced inflammatory factors by up-regulation of autophagy via AMPK/mTOR signaling pathway. J Transl Med. 2015;13(92) doi: 10.1186/s12967-015-0450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu HF, Li HZ, Tang YL, Tang XQ, Zheng XL, Liao DF. Nicotinate-curcumin impedes foam cell formation from THP-1 cells through restoring autophagy flux. PLoS One. 2016;11(e0154820) doi: 10.1371/journal.pone.0154820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li G, Peng J, Liu Y, Li X, Yang Q, Li Y, Tang Z, Wang Z, Jiang Z, Wei D. Oxidized low-density lipoprotein inhibits THP-1-derived macrophage autophagy via TET2 down-regulation. Lipids. 2015;50:177–183. doi: 10.1007/s11745-014-3977-5. [DOI] [PubMed] [Google Scholar]
- 47.Huang B, Jin M, Yan H, Cheng Y, Huang D, Ying S, Zhang L. Simvastatin enhances oxidized-low density lipoprotein-induced macrophage autophagy and attenuates lipid aggregation. Mol Med Rep. 2015;11:1093–1098. doi: 10.3892/mmr.2014.2790. [DOI] [PubMed] [Google Scholar]
- 48.Zhang BC, Zhang CW, Wang C, Pan DF, Xu TD, Li DY. Luteolin attenuates foam cell formation and apoptosis in Ox-LDL-stimulated macrophages by enhancing autophagy. Cell Physiol Biochem. 2016;39:2065–2076. doi: 10.1159/000447902. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, Feng K, Zhang C, Zhang H, Zhang J, Hua Y, Dong Z, Zhu Y, Yang S, Ma C. Metformin attenuates atherosclerosis and plaque vulnerability by upregulating KLF2-mediated autophagy in apoE-/- mice. Biochem Biophys Res Commun. 2021;557:334–341. doi: 10.1016/j.bbrc.2021.04.029. [DOI] [PubMed] [Google Scholar]
- 50.Hsieh PN, Zhou G, Yuan Y, Zhang R, Prosdocimo DA, Sangwung P, Borton AH, Boriushkin E, Hamik A, Fujioka H, et al. A conserved KLF-autophagy pathway modulates nematode lifespan and mammalian age-associated vascular dysfunction. Nat Commun. 2017;8(914) doi: 10.1038/s41467-017-00899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donovan EL, McCord JM, Reuland DJ, Miller BF, Hamilton KL. Phytochemical activation of Nrf2 protects human coronary artery endothelial cells against an oxidative challenge. Oxid Med Cell Longev. 2012;2012(132931) doi: 10.1155/2012/132931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Q, Zhang T, Yi L, Zhou X, Mi M. Dihydromyricetin inhibits NLRP3 inflammasome-dependent pyroptosis by activating the Nrf2 signaling pathway in vascular endothelial cells. Biofactors. 2018;44:123–136. doi: 10.1002/biof.1395. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Wang J, Huang E, Gao S, Li H, Lu J, Tian K, Little PJ, Shen X, Xu S, Liu P. Tanshinone IIA suppresses cholesterol accumulation in human macrophages: Role of heme oxygenase-1. J Lipid Res. 2014;55:201–213. doi: 10.1194/jlr.M040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q, Tang SL, Liu XY, Zhao GJ, Ouyang XP, Lv YC, He PP, Yao F, Chen WJ, Tang YY, et al. Tertiary-butylhydroquinone upregulates expression of ATP-binding cassette transporter A1 via nuclear factor E2-related factor 2/heme oxygenase-1 signaling in THP-1 macrophage-derived foam cells. Circ J. 2013;77:2399–2408. doi: 10.1253/circj.cj-12-1616. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Liu Z, Hu T, Han L, Yu S, Yao Y, Ruan Z, Tian T, Huang T, Wang M, et al. Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle. 2017;16:1053–1062. doi: 10.1080/15384101.2017.1312224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazaro I, Lopez-Sanz L, Bernal S, Oguiza A, Recio C, Melgar A, Jimenez-Castilla L, Egido J, Madrigal-Matute J, Gomez-Guerrero C. Nrf2 activation provides atheroprotection in diabetic mice through concerted upregulation of antioxidant, anti-inflammatory, and autophagy mechanisms. Front Pharmacol. 2018;9(819) doi: 10.3389/fphar.2018.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fledderus JO, Boon RA, Volger OL, Hurttila H, Ylä-Herttuala S, Pannekoek H, Levonen AL, Horrevoets AJ. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:1339–1346. doi: 10.1161/ATVBAHA.108.165811. [DOI] [PubMed] [Google Scholar]
- 60.He LH, Gao JH, Yu XH, Wen FJ, Luo JJ, Qin YS, Chen MX, Zhang DW, Wang ZB, Tang CK. Artesunate inhibits atherosclerosis by upregulating vascular smooth muscle cells-derived LPL expression via the KLF2/NRF2/TCF7L2 pathway. Eur J Pharmacol. 2020;884(173408) doi: 10.1016/j.ejphar.2020.173408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.