Abstract
Background
The proliferation and increasing maturity of biometric monitoring technologies allow clinical investigators to measure the health status of trial participants in a more holistic manner, especially outside of traditional clinical settings. This includes capturing meaningful aspects of health in daily living and a more granular and objective manner compared to traditional tools in clinical settings.
Summary
Within multidisciplinary teams, statisticians and data scientists are increasingly involved in clinical trials that incorporate digital clinical measures. They are called upon to provide input into trial planning, generation of evidence on the clinical validity of novel clinical measures, and evaluation of the adequacy of existing evidence. Analysis objectives related to demonstrating clinical validity of novel clinical measures differ from typical objectives related to demonstrating safety and efficacy of therapeutic interventions using established measures which statisticians are most familiar with.
Key Messages
This paper discusses key considerations for generating evidence for clinical validity through the lens of the type and intended use of a clinical measure. This paper also briefly discusses the regulatory pathways through which clinical validity evidence may be reviewed and highlights challenges that investigators may encounter while dealing with data from biometric monitoring technologies.
Keywords: Digital medicine, Digital health, Biometric monitoring technologies, Clinical trials, Statistics, Clinical validation
Introduction
The emergence of digital medicine in clinical trials has been propelled in recent years due to a surge of health-related technologies that enable holistic health assessments and data production for trial participants, sponsors, and healthcare systems, especially outside of traditional clinical settings [1]. Rigorous processes for verification, analytical and clinical validation, and evaluation of usability are needed to develop fit-for-purpose digital medicine tools for medical data generation in today's clinical research, care, and decision-making [2, 3].
This paper uses the previously coined term biometric monitoring technologies (BioMeTs) [2] to refer to “connected digital medicine products that process data captured by mobile sensors, using algorithms to generate measures of behavioral and/or physiological function and level of functional activity that may ultimately result in the identification and deployment of clinical digital measures.” As in any rapidly evolving field, multiple terms referring to the same kind of tools can be found in the literature and regulatory documents, including Digital Health Technologies, which have a broader scope. The focus of this paper is on the BioMeTs used to enable reliable and comprehensive clinical evaluations of trial participants and to add flexibility outside of the traditional clinical trial assessment setting to yield data for the development of investigational medical products (IMPs), i.e., drugs, biologics, and vaccines. This paper will not address clinical validation in support of clearance for a medical device, software as a medical device, or a digital behavioral intervention and the corresponding digital endpoints.
There are significant advantages to use of BioMeTs in clinical trials, including enhancing the quality and efficiency of clinical investigations [4] and facilitating the development of personalized medicine by expanding the pool of potential predictive biomarkers and granular participant-monitoring methods [5]. BioMeTs can make health-related data more representative of the composition of contemporary societies by extending access to clinical trials to diverse populations and measuring health-related outcomes that truly matter to trial participants, reflecting their health and functioning in daily living and real-world settings [6].
As evidenced by a growing list of Digital Medicine Society (DiMe) crowdsourced digital endpoints in industry-sponsored clinical investigations [7], there is a need for standardized methodologies to validate and analyze health-related digital measures so that they become scientifically accepted in clinical research [2, 3]. This paper provides a viewpoint on the scientific framework for demonstrating the clinical validity of novel digital measures and highlights key considerations for using some statistical methods. Future work will delve into providing further technical guidance on the analytical methods to achieve the analysis objectives. This paper also discusses key aspects of the regulatory framework guiding BioMeT-derived evidence generation and review, challenges specific to clinical trial data sets, and perspectives on future directions.
Background
A foundational approach for developing health-related digital measures in clinical trials follows a well-accepted framework of outcomes research where measures are defined in terms of (i) a meaningful aspect of health (MAH); (ii) a concept of interest (COI); (iii) and a clinical measure [8, 9, 10]. The MAH refers to elements of a disease that the trial participant wants to prevent, improve, or avoid making worse. From the MAH, a measurable COI can be identified. In turn, a specific clinical measure can be derived from the COI. Digital clinical measures are health outcomes or physiological characteristics of an individual's health, wellness, and/or condition that are collected digitally and include both technology and measurement considerations [2]. The above framework applies both to direct and indirect measures of health-related outcomes, which are referred to as Clinical Outcome Assessments (COA) and biomarkers, respectively, with several subcategories defined for each [11].
A COA directly measures what matters to trial participants and reflects how they feel, function, or survive. When a COA is collected using electronic or sensor technologies, it is referred to as an electronic COA (eCOA). All types of COAs (i.e., patient-reported outcome [PRO], observer-reported outcome, clinician-reported outcome, and performance outcome) are commonly digitalized and used in remote activities in modern clinical trials. However, not all eCOAs are collected using BioMeTs; e.g., electronic PROs use technology to capture survey data. This paper focuses on eCOAs that are collected using BioMeTs.
Biomarkers are characteristics that measure indicators of a normal biological process, pathogenic process, or responses to an exposure or a medical intervention, including therapeutic interventions [12]. When a biomarker is collected using digital technologies, it is referred to as a digital biomarker [13]. Following the BEST framework, biomarkers can be of seven types: pharmacodynamic (PD)/response, monitoring, safety, susceptibility/risk, prognostic, diagnostic, and predictive [12].
Both COAs and biomarkers may serve as the basis for a definition of an endpoint to be used in clinical trials − a precisely defined, statistically analyzed health-related variable to demonstrate a clinical benefit of an experimental medical intervention. The evaluation of the fit-for-purpose of BioMeTs for collecting digital clinical measures comprises the verification, analytical validation, and clinical validation phases as described in the V3 framework [2]. This paper will address only the aspects of clinical validation.
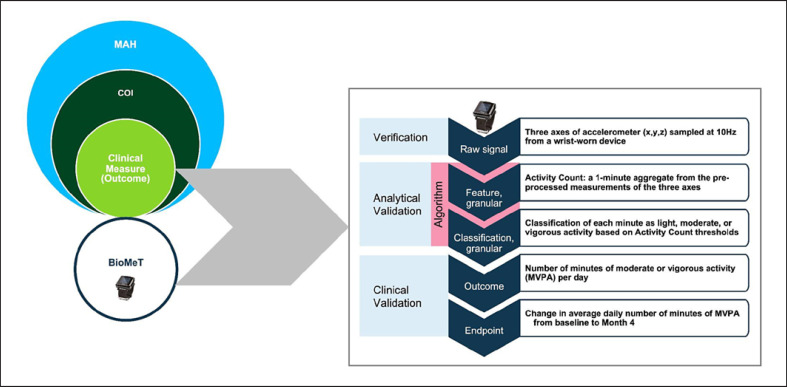
As an illustration of the above concepts, consider a MAH in trial participants with pulmonary arterial hypertension (PAH) that reflects physical functioning in daily living. A measurable COI related to this aspect of health could be the physical capacity to perform activities of daily living [14]. A clinical measure that reflects this COI could be the number of minutes spent daily in moderate or vigorous physical activity (MVPA) categories, which include the type of daily activities that matter to patients. It can be measured using a BioMeT containing an accelerometer sensor. An endpoint for use in clinical trials could be defined as the change in the average daily number of minutes of MVPA from baseline to month four after initiation of treatment [15, 16].
Figure 1 provides an example of a measurement process that may be employed to arrive at an endpoint value from a raw BioMeT-derived signal. Verification is expected to provide assurance that the raw data harnessed from an accelerometer sensor is an accurate physical measurement of acceleration associated with the movement of a wrist. Analytical validation is intended to focus on the next two steps that represent a data-processing algorithm aiming at transforming sensor signal into classification of the physical activity intensity level per minute when motion is being monitored and digitally collected. This validation phase could be carried out by assessing classification accuracy compared to another method, e.g., direct human observation, measurement of metabolic equivalent units, and labeling of various types of movement. Given an assurance of acceptable algorithm accuracy, a specific clinical measure (outcome) can be defined using the output of the classification algorithm, e.g., the number of minutes of MVPA aggregated at the day level. This outcome can then serve to define an endpoint. Clinical validation is expected to provide evidence that the BioMeT-derived outcome and endpoint are clinically meaningful, e.g., they can capture a clinically significant effect of a medical intervention on the corresponding COI that is meaningful for the target population. This granular view of the measurement process highlights the fact that changes at any of its steps (e.g., change of sensor that generates raw data or its body positioning or change in the classification algorithm) may necessitate revalidation of the endpoint or showing some degree of equivalence or superiority of modified components compared to the initial ones.
Fig. 1.
Measurement process example for a BioMeT.
Digital clinical measures collected by BioMeTs can be used in clinical trials in many ways, for instance, as prognostic biomarkers for stratification purposes during trial enrollment [17]; for collection of additional evidence on the trial participants' quality of life, e.g., via passive monitoring of physical activity or sleep [8]; as monitoring biomarkers for the safety and efficacy [9, 18], as surrogate endpoints, e.g., in early phase trials for “go/no-go” decisions to accelerate clinical development programs [2]; and, as predictive biomarkers in the framework of precision medicine. Given the novelty and complexity of BioMeTs, to use digital clinical measures to support regulatory decisions (e.g., labeling), early regulatory interactions are needed to discuss the validation process [18].
Novel digital clinical measures are expected to be most beneficial when no assessment tools exist for the concept of interest or existing tools have important limitations. In such cases, validation of novel digital measures cannot proceed by simply showing sufficient equivalence to existing measurement methods. Although working with data from BioMeTs entails many unique challenges, general principles of development and validation of fit-for-purpose drug development tools apply, and the existing regulatory and scientific guidelines should be followed. The next section will focus on key considerations when using some statistical analyses to demonstrate the clinical validity of novel digital measures depending on their type.
Key Considerations for Statistical Analysis Planning to Support Clinical Validation
A body of clinical validation evidence should provide insight into the reliability, accuracy, sensitivity, and generalizability of a novel clinical outcome measurement method [9]. Validation evidence should be transparent, comprehensive, and traceable across different analysis stages, which can be broadly divided into two categories − analyses of exploratory versus confirmatory nature.
Exploratory analyses may first be carried out to suggest a definition of a promising clinical measure or its components that may be strongly associated with the COI. These analyses have a hypothesis generation purpose and may employ predictive modeling to assess the strength of association between one or more digital parameters and disease-related clinical outcomes [8]. A precise definition of a clinical measure can be proposed based on both qualitative input from patients, caregivers, and investigators as well as quantitative input from data-driven exploratory analyses, although the relative influence of these two factors may vary. In qualitatively driven use cases (more likely for eCOAs), exploratory analyses may be helpful to fine-tune digital features that would represent the COI most accurately [19], e.g., to suggest thresholds on “activity counts” (summaries of accelerometer measurements over time intervals) that best delineate activities of daily living that are important to trial participants. More exploratory data-driven modeling may first be conducted to evaluate the feasibility and face validity of the approach, assess the maturity of BioMeTs to be deployed into clinical development, and identify the features that are most correlated with relevant clinical outcomes when it is less clear which digital parameters are most related to the COI. This is often done as part of early-phase trials during a clinical development program or in separate studies designed specifically to explore several candidate BioMeTs and digital clinical measures. For example, the utility and accuracy of multiple digital technologies have been assessed as potential diagnostic tools for unipolar depression, as well as digital biomarkers that may complement existing clinically validated psychometric questionnaires in depression [20]. Another example is the study of participants with elevated blood glucose or prediabetes [21] where the feasibility of using noninvasive and widely accessible methods, including smartwatches and food logs, was assessed for continuous detection of personalized glucose deviations and prediction of interstitial glucose value in real-time.
Once one or more candidate digital clinical measure definitions are proposed, confirmatory analysis is conducted to demonstrate desirable measurement properties and all aspects of clinical validity relevant to the type of measure and its intended context of use [8]. Several candidate definitions may undergo these types of validation analyses to determine which one is the most reliable and sensitive in the target population. The latter is especially pertinent when high-resolution longitudinal data are collected by BioMeTs, which need to be summarized into high-level summary measures representative of COI. All aspects of clinical validity should be demonstrated in the target population for which the clinical measure is intended to be used. A digital clinical measure previously validated in a different patient population generally requires revalidation for use in a new population, although revalidation requirements may differ between COA and biomarkers and depending on clinical characteristics of the two populations. For example, the FDA guidance on development of COA [22] mentions that “Additional qualitative research may be recommended if the instrument will be used in a significantly different patient population (e.g., a different disease or age-group), and sufficient evidence is not available to support content relevance to the target population. Additional analyses may be recommended to evaluate the instrument's measurement properties within the new population.”
The rest of this section discusses the types of analysis objectives that would need to be pursued to demonstrate various aspects of clinical validity. While the requirements for validation of eCOAs and digital biomarkers are somewhat different, there are also many commonalities. For biomarkers, requirements also depend on the biomarker type as defined earlier, and reliability as well as sensitivity to change (also referred to as responsiveness) needs to be demonstrated [19].
In general, responsiveness is defined as the ability of an instrument to accurately detect change when it has occurred. Although this definition is very simple, responsiveness is not a static attribute of an instrument but rather depends on the context in which it is used. The concept of “change” itself can represent many different distinct states, either within or across individuals, concurrently or over time, and possibly in relation to other measures. Clarity on the aspects of change that are relevant for a specific use case is crucial for choosing appropriate clinical trial designs and methods for assessing and interpreting responsiveness. The taxonomy of responsiveness introduced by Beaton et al. [23] is very useful in this respect. Essential aspects are briefly summarized and expanded here.
The three axes of Beaton et al.'s [23] taxonomy of responsiveness are as follows:
1. Individual-level (change in an individual patient) versus group-level (average amount of change for a group) interpretation. A smaller amount of change may sometimes be considered “important” at a group level compared to the individual level.
2. Between-individual differences at one point in time versus within-person change over time versus a hybrid of both. The first category may contrast trial participants with different disease severities at diagnosis, the second − evolution of the disease over time within the same participant, and the third may target between-participant differences in their individual changes over time.
3. Minimum change (potentially) detectable by the instrument versus observed change in a population at two different occasions versus estimated change in a population deemed to have changed based on an external (reference) criterion. The first category reflects mainly the measurement error of the instrument. The observed change is likely to be most relevant for eCOAs, where sensitivity to a change typically primarily focuses on the effect size (mean-to-standard-deviation ratio) of decline/improvement, i.e., change in eCOA values over time during which the disease is expected to progress or improve relative to inherent population variability. The estimated change would most often be relevant for biomarkers because, unlike eCOA, they are indirect measures. Clinically meaningful changes in biomarker values are primarily established by correlating and anchoring them to clinical outcomes and/or a response to interventions known to induce change in the relevant health aspect.
The three axes are orthogonal, although not all possible combinations may be relevant. With respect to the third axis, especially for the estimated change, there is additional granularity that determines with respect to what (reference) the responsiveness is determined. The details will often depend on whether the validation is for an eCOA or a specific type of digital biomarker, as outlined in Table 1. Cells marked by “X” in Table 1 indicate which objective may apply to the various types of digital clinical measures.
Table 1.
Key analysis objectives to demonstrate clinical validity of eCOA and digital biomarkers
| Demonstrate that a digital measure can: | eCOA | Digital biomarker |
||||||
|---|---|---|---|---|---|---|---|---|
| PD | monitoring | safety | risk | prognostic | diagnostic | predictive | ||
| Reliably measure a COI within and across individuals in stable disease states within a range of environmental conditions | X | X | X | X | X | X | X | X |
|
| ||||||||
| Differentiate between healthy individuals and those with disease | X | X | X | X | X | |||
|
| ||||||||
| Differentiate between concurrent disease/symptoms severity categories; correlate with other concurrent clinical outcomes | X | X | X | X | X | |||
|
| ||||||||
| Detect disease progression or a clinical event of interest | X | X | X | X | X | |||
|
| ||||||||
| Predict future outcomes (short or long term) | X | X | X | X | ||||
|
| ||||||||
| Accurately detect functional states or activities of interest | X | X | X | X | X | |||
|
| ||||||||
| Capture response to an intervention | X | X | X | X | ||||
|
| ||||||||
| Predict response to an intervention | X | |||||||
Considerations for the Assessment of Reliability
Assessment of (test-retest) reliability aims at evaluating the degree to which the results obtained by a measurement procedure can be replicated, or in other words, the variability of repeated measurements under the same conditions. This analysis is relevant to all types of clinical measures and is a prerequisite for all subsequent analyses and interpretations of meaningful differences and changes, i.e., what constitutes a signal versus noise.
The measurement error is typically viewed as having two components: (1) random error of the measurement tool and/or measurement process and (2) natural variability in the measured COI. The first component is likely to be evaluated, to a large extent, during the verification and analytical validation. However, those validation stages may not always cover all conditions that are relevant for a specific use case. The second component is especially likely to necessitate evaluation during the clinical validation stage as it examines both the within-individual sources of variability (including the individual's physiology and fluctuations in behavioral patterns and environmental factors while the individual remains in a stable disease state) and the between-individual sources of variability.
Reliability of a digital measure can be assessed based on data from a repeated-measure design, where measurements are collected from each participant multiple times under various conditions that would reflect both error components. For example, for a measure of sedentary behavior, measurements should be obtained over two or more weeks, to reflect wear compliance and day-to-day variability. Multiple measurements should be taken over periods of time where the participant's disease status is stable, e.g., during screening/baseline, maintenance treatment period, or follow-up. At the same time, participants with different disease severities should be included in the study and/or participants should be assessed during multiple periods when their disease can be confirmed as deteriorating or improving based on established clinical criteria. This would enable assessment of whether the measurement error depends on the disease state.
Analyses of reliability typically rely on random (or mixed) effect models, from which one of the key reliability indices, the intra-class correlation, is estimated [24]. For categorical clinical measures, reliability is often characterized using Cohen's kappa, prevalence-adjusted and bias-adjusted kappa, and average positive and negative agreement indices [25].
Assessments of reliability lead to the estimate of the standard error of measurement and contribute to the determination of minimal detectable change and Minimal Clinically Important Change (MCIC). Determination of MCIC requires additional considerations, including qualitative clinical arguments and/or analyses of responsiveness (sensitivity to change).
In cases where BioMeTs are used in a free-living environment, outside of the controlled clinical center setting, and over long periods of time, digital clinical measures may be affected by multiple sources of variability, and it is critical to thoroughly evaluate its magnitude and opportunities to improve reliability.
Assessment of Ability to Differentiate between Healthy Individuals and Those with Disease
This is a key objective for diagnostic biomarkers but may also be relevant for eCOAs, monitoring biomarkers, and for some PD/response and safety biomarkers. Validation may be based on a clinical trial that includes participants with and without a disease as determined by a reference diagnostic method. For example, a novel method for detecting the presence or absence of PAH based on the number of minutes of MVPA as discussed in Figure 1 with a wrist-worn device over extended periods of time in daily living would need to be evaluated in terms of accuracy against one or more reference standard measuring methods. A new digital PAH measure can then be used as a diagnostic biomarker as well as a monitoring biomarker to determine treatment success.
Analysis typically relies on classification techniques and performance measures, including sensitivity (recall), specificity, precision, and related measures of classification accuracy. Care must be taken to select appropriate metrics as different measures may be biased or misleading depending on the base rates of target categories. One of the challenges is the selection and validation of cutoff levels for classification, which should ideally be done based on data that were not used in biomarker discovery/development.
Assessment of Ability to Differentiate between Concurrent Disease/Symptoms Severity Categories and Correlations with Other Concurrent Clinical Outcomes
For eCOAs and several types of digital biomarkers, a digital clinical measure may need to differentiate between disease or symptom severity categories as determined by other clinical assessments concurrently. For example, a novel digital clinical measure of physical activity intended for monitoring trial participants with heart failure may need to distinguish between New York Heart Association (NYHA) classes of heart failure and/or Kansas City Cardiomyopathy Questionnaire (KCCQ) overall summary score categories [26].
This aspect also covers more general objectives of investigating how the digital clinical measure is associated with other related clinical outcomes. For example, for a digital measure of physical activity in trial participants with heart disease, an association with the concurrent assessments using the KCCQ physical limitation score [27] may be of interest. Although it should be noted that sometimes lack of correlation may not necessarily be evidence of invalidity: e.g., objective BioMeTs-derived measures and PROs may be measuring different and complementary aspects of the same MAH.
Analysis methods used for this objective may include both classification and regression models that focus on classification accuracy and significance of the digital clinical measure effect on the reference outcome or vice versa. At a minimum, a strong association would need to be shown or that the distribution of digital measurements within each category of reference outcome are sufficiently distinct. There is no universal rule as to what constitutes a strong association, as the interpretation depends on the statistical method used to characterize the association as well as on the context of use (e.g., a stronger association may be required for a PD biomarker intended to be used for go/no-go decisions during a clinical development program, while a weaker association may be acceptable for a monitoring biomarker intended to be used as a stratification factor in a randomized treatment assignment).
One of the goals of these analyses may also be to evaluate responsiveness and support the determination of Minimal Clinically Important Difference (MCID). The discussion of Beaton et al.'s [28] taxonomy of responsiveness above highlighted that this is a multifaceted aspect, and it should always be clearly defined what type of responsiveness is being assessed and relative to what. Determination of MCID may, at least in part, rely on anchor-based methods that explore the magnitude of change in a novel clinical measure versus a reference (anchor) measure [22, 28, 29]. The MCID is not an intrinsic characteristic of a clinical measure, but rather may vary, at both the group and individual level, depending on the target population, clinical context, the patient's baseline, and whether improvement or deterioration is being measured. In addition to quantitative evidence, qualitative clinical considerations may often be included in the determination of MCID.
Assessment of Ability to Detect Disease Progression or a Clinical Event of Interest
A somewhat different objective for eCOAs and several types of digital biomarkers may be to demonstrate their ability to detect a clinically meaningful disease progression or a critical clinical event of interest, i.e., a concurrent (and sometimes abrupt) health status change. Statistical methods and performance metrics, in this case would be similar to those mentioned above for differentiating between healthy and diseased individuals. A study that could support this evaluation would ideally include participants who are monitored for an event of interest over a period, with one group using a BioMeT and a control group not using it. The importance of thorough clinical validation in the context of a specific use case and careful evaluation of both false negative and false positive detection rates and their implications cannot be overstated here, as the target events may have a big impact both on the individuals' health and the healthcare system. An example of this is a clinical validation study in participants who presented for a cardiovascular evaluation at a clinic after an abnormal pulse was detected by Apple Watch [30]. The device and its algorithm were previously cleared by the FDA for an optical abnormal pulse and ECG features detection to opportunistically reveal a notification of possible atrial fibrillation in over-the-counter use [31, 32]. The Wyatt et al. [30] study concluded that only 11% of participants who received an alert received a clinically actionable diagnosis and that the false positive alerts may lead to overutilization of healthcare resources, which highlights the importance of use case-specific clinical validation.
For these interventions to be fit-for-purpose, successful deployment of BioMeTs needs to account for different demographics that reflect the clinically relevant populations with regards to age, gender, sex, race, and ethnicity [33, 34]. Additionally, there can be significant differences in physiologic and activity parameters measured via BioMeTs across sex, race, ethnicity, and clinical conditions [35]. For example, the measurement of blood oxygen saturation via optical sensors may need to be validated in some specific populations [21, 33, 36, 37]. Furthermore, technology utilization may vary across different demographics [38, 39].
Assessment of Ability to Predict Future Outcomes (Short- or Long-Term)
Unlike in previous cases, here the goal is to demonstrate the ability of a digital clinical measure to predict future clinical outcomes and events. This is most relevant for risk and prognostic biomarkers. For example, gait speed may be a susceptibility/risk biomarker in patients with HIV as an early indicator of future decline in mobility [40]. This aspect may also be relevant for some safety and monitoring biomarkers. For example, deterioration in the number of minutes of MVPA, as discussed in Figure 1 in participants with heart disease, may be used to predict the likelihood of major cardiovascular events in the future.
To support such an evaluation, data from a longitudinal clinical trial would be necessary, where values of the digital clinical measure are collected at trial entry and possibly over time with the information on the outcomes or events of interest. Longitudinal studies require more resources compared to cross-sectional studies. Consequently, the objectives of evaluating the association between the novel digital measure and future outcomes (or the clinical events of interest as discussed in the previous subsection) are often embedded as secondary or exploratory objectives in interventional trials rather than being pursued as the primary objectives in studies designed specifically for clinical validation. Statistical methods useful for this objective will typically include classification and regression models with an evaluation of the significance of the biomarker effect on the outcome of interest after adjusting for other relevant covariates [41].
Assessment of Accuracy of Detection of Functional States or Activities of Interest
Clinical measures can also be developed to detect various functional states or activities of interest and to quantify associated parameters. Examples include detecting sleep versus wake state, walking versus other types of locomotion, scratching behavior, hand movements associated with smoking or eating, etc.
Evaluating the accuracy of such detection would typically require data collected by a reference method, e.g., polysomnography for sleep or video recording for walking or smoking. Some related digital parameters may be evaluated as part of the analytical validation. However, during clinical validation, additional evaluation, e.g., over more extended periods of time, in specific environments, or for specific aggregate measures, may need to be performed. It is important to clearly set and justify acceptable accuracy targets, taking into account a clinically meaningful difference for the population of interest, including whether they are most relevant on an individual or group level, as well as for cross-sectional assessments versus changes in response to treatment. For example, the accuracy of actigraphy-based measures of sleep varies across different sleep parameters and sleep disorders, and acceptable levels of accuracy also depend on whether the measure is intended to be used for individual clinical care decisions or for the assessment of treatment-related changes [42].
Assessment of Ability to Capture Intervention Effect
If eCOAs or digital biomarkers (PD/response, monitoring, or safety) are planned to be used in clinical trials as endpoints, it is necessary to show that the measure is sensitive to intervention-induced changes. For a biomarker, this would typically be shown by assessing the correlation between changes in the biomarker values from pre- to post-intervention and the corresponding changes in some other clinical measurements. Analyses done to address some of the questions listed above lay a foundation for this analysis, e.g., determining minimal detectable change and MCID is necessary to interpret meaningful differences between treatment groups. Statistical approaches may include anchor-based analyses with anchors such as Patient Global Assessment of (disease) Severity (PGI-S) or Patient Global Assessment of Change (PGI-C) to determine or confirm clinically meaningful differences between treatment groups for the digital measure [43].
Assessment of Ability to Predict Response to an Intervention
Predictive biomarkers measure patient characteristics prior to the start of an intervention and are used to predict a future response to a treatment or its magnitude, with the latter often measured by a different clinical measure (either efficacy or safety). Predictive biomarkers are a cornerstone of precision/personalized medicine, which aims at selecting the right treatment for the right patient when there is considerable treatment response heterogeneity that can be explained by a measurable patient or disease characteristics. There is a growing recognition that in many diseases, it would be difficult to identify a single predictive biomarker capturing complex effects of the disease and treatment, and pursuing multivariate biomarker signatures may hold more promise to capture the interplay between genomic, demographic, physiological, behavioral, and environmental factors [44]. Digital biomarkers may be particularly suited for measuring some of these characteristics. Digital predictive biomarkers are also likely to play an essential role in digital or digitally enhanced therapeutics [45].
Over the past two decades, the number of methods in the literature has been growing for a data-driven identification of predictive biomarkers, capable of considering a large number of candidate biomarkers and identifying a few with the most robust predictive properties [46, 47, 48]. Predictive biomarker identification is laden with methodological difficulties and pitfalls, including multiplicity and treatment effect estimation “optimism bias” (also referred to as data resubstitution bias). Ideally, data-driven predictive biomarker identification should be considered as an exploratory, hypothesis generation stage carried out on one dataset. In contrast, a confirmatory analysis would be carried out on a separate dataset with the objective of estimating and testing the significance of the treatment effect in a biomarker positive subgroup and its complement. In cases where data are limited, and both analyses have to be done using the same dataset, naive data resubstitution should be avoided in favor of more principled methods, examples of which can be found in [49, 50].
Assessment of Biomarkers for Use as Surrogate Endpoints
Biomarkers may be used in clinical trials as surrogate endpoints, if there is a “clear mechanistic rationale and clinical data providing strong evidence that an effect on the surrogate endpoint predicts a specific clinical benefit” [11]. A well-known example is hemoglobin A1c which is a validated surrogate endpoint for microvascular complications associated with diabetes mellitus. Surrogate endpoints are used in cases where a clinical endpoint requires a very long follow-up or invasive assessment procedures.
To validate a surrogate endpoint, it must be shown that (1) the biomarker is prognostic with respect to the clinical outcome and (2) treatment effects on the surrogate endpoint reliably predict treatment effects on the clinical outcome [51]. The second condition is especially challenging to demonstrate because of the possibility of confounding factors that may lie on a causal path and may be difficult to account for even in randomized clinical trials. Multiple statistical approaches have been suggested, including meta-analyses and model-based estimations of direct and indirect effects. For the body of validation evidence to be considered robust, data are required from multiple clinical trials, which may be a mix of randomized and observational studies, as well as prospective and retrospective. Demonstration of surrogacy typically requires the highest level of evidence for regulatory acceptance among all biomarker types. It is expected to be based on a sound scientific hypothesis about the biomarker's role in the target disease and treatment's mechanism of action.
Recent research also suggests that in some disease areas, a combination of several biomarkers (composite biomarkers) have stronger surrogate properties compared to any single surrogate parameter [52]. In this respect, digital biomarkers may play an even more critical role, measuring aspects of health not well captured by other means.
Handling of Missing BioMeT Data
It is paramount to identify statistical approaches to deal with missing data to ensure that derived endpoints are valid, accurate, and reliable [53, 54]. The taxonomy for classifying missing data mechanisms is based on the likelihood of being missed: MCAR (missing completely at random), MAR (missing at random), and MNAR (missing not at random) [53]. Currently, derived values of digital clinical measures are often left missing when some underlying measurements are not available. Emerging statistical approaches for addressing missing BioMeT data include within-patient imputations across standard periods, functional data analysis, deep learning methods, imputation approaches, and robust modeling [53].
Investigators should anticipate the impact of the missing data on trial results and the types of missing data that are likely to occur [1] and implement strategies to optimize data quality starting from the study design [55, 56]. Additionally, the authors should discuss the study's findings, including the handling of missing data and any technical BioMeT problems that impacted the study results in publications and study reports [57, 58].
Key Regulatory Considerations
Regulatory Pathways for Acceptance, Qualification, and Approval of Novel BioMeTs and Their Corresponding Digital Endpoints
The individual regulatory review division within the FDA that may accept a novel clinical measure for a given digital endpoint may differ depending on the disease area in which a specific type of IMP will be evaluated for safety and efficacy in a clinical trial program relative to drugs, biological products, and vaccines [1]. For example, different types of BioMeTs and their corresponding digital endpoints may be used in clinical programs (phase 1–3 trials) to evaluate different experimental IMPs under different investigational new drug applications (INDs). Once the clinical evaluations have been completed, these IMPs are reviewed for US market approval under a new drug application (NDA for drugs) or a biologics license application (BLA for biologics and vaccines).
The regulatory interactions between a sponsor using a BioMeT and the FDA should occur early during the IMP development process to discuss the appropriateness of a specific technology to measure a given digital endpoint. The FDA's feedback on a BioMeT to test the safety and efficacy of an IMP within a specific clinical program may be obtained as part of a pre-IND meeting within the IND pathway [59].
BioMeTs are used to measure a clinical measure do not need to be qualified to be included in a clinical program. However, sponsors may choose to qualify an eCOA or digital biomarker and its corresponding digital endpoint if they determine that they can be used in multiple clinical programs to test different IMPs (e.g., a digital accelerometer may be used in clinical trials related to heart disease, Parkinson's disease, obesity, and other conditions in which mobility may be an important symptom). The regulatory pathway for qualification is described in FDA's Drug Development Tool (DDT) Qualification Programs [60].
Like the FDA, the EMA recommends early discussions for the qualification of BioMeT-derived novel clinical measures. An iterative qualification process is often recommended for applicants [61]. The qualification submission has to provide evidence that the methodology to be qualified is reliable, accurate, precise, generalizable, clinically relevant, and applicable [61]. A successful example of close and continuous collaboration between investigators and the regulatory agencies is the qualification of a digital clinical measure of stride velocity 95th centile as a measure of functional ability in daily life in patients with Duchenne muscular dystrophy [62].
Regulatory agencies require the quality, integrity, reliability, and robustness of data generated in clinical trials for which the sponsors have the ultimate responsibility, even if they delegate all or part of trial activities [19, 63]. To ensure the authenticity, integrity, and confidentiality of data, sponsors should develop a data management plan that depicts the flow of data from creation to final storage and corresponding electronic systems.
Data Standards for the Regulatory Submission of BioMeTs
Data standards for BioMeTs should be fit for purpose and follow the needs of a clinical development program so that the meaning and traceability of the data are clear and transparent to the researchers. It is recommended to involve biostatisticians and data scientists from the protocol design stage to plan data collection in a way that would be appropriate both for an initial proof-of-concept scope and long-term objectives [64].
It is advisable to provide the definition of “sample-level data” and “derived data” in regulatory submissions, as well as details related to the development of algorithms employed to convert the sample-level data into parameters of interest and to disclose the mobile technology specifications and testing (e.g., calibration) and metadata collection [55]. Standardized clinical trial data such as defined by the Clinical Data Interchange Standards Consortium (CDISC) are required, preferred, or endorsed for regulatory submissions of clinical and nonclinical data [65, 66]. Literature highlights that these standards are not designed to collect, organize, and analyze large volumes of BioMeT data, but more suitable common standards are yet to be developed [64, 67, 68].
In this context, an emerging standard for BioMeTs is Open mHealth, which recently became an official IEEE family of standards [69]. A summary of relevant data standards is provided in Table 2.
Table 2.
Data standards for regulatory submissions of BioMeTs
| Standard name | Format | Focus |
|---|---|---|
| CDISC | Tabulation data: | Objective: regulatory submission of clinical trial data |
| − The Study Data Tabulation Model (SDTM) including (SDTMIG-MD for medical devices collected data) | Note: additional metadata collected from medical devices is required with respect to data collected from BioMeTs (e.g., the location where the device was used) (Badawy et al. [64]) | |
| − Standard Exchange for Nonclinical Data (SEND) | Geographic scope: | |
| Analysis data: | − Recommended by FDA and Japan PMDA for regulatory submissions | |
| − Analysis Data Model (ADaM) | − Preferred by China NMPA | |
| − Endorsed by EMA | ||
|
| ||
| Open mHealth | Open Mobile Health | Objective: Mobile health data interoperability standard for patient health generated data |
Conclusions
The novelty and potential of BioMeTs to produce reliable data that give investigators a comprehensive picture of participants' health have encouraged research organizations to explore the feasibility of using these tools in clinical trials. However, this has been done primarily as pilot or exploratory trials that offer limited evidence of their clinical validity. This is partially due to the cost, time, and technical and regulatory complexities of more formal validation approaches.
In the coming years, it is expected that these initial trials will be followed by more structured and sound designs that offer stakeholders the evidence they need to increase BioMeT adoption [8]. One of the fundamental aspects of trial design is sample size which needs to be sufficient to fulfill study objectives. Clinical validation of a novel clinical measure is often positioned as a secondary or tertiary objective in a clinical trial, while the sample size is typically calculated to fulfill the primary objective. In order to ensure robust clinical validation evidence, statisticians should determine the sample size required for analyses planned to support clinical validation. Sample size requirements are also driven by expectations for statistical power and model accuracy which, in turn, depend on the intended context of use of a digital clinical measure and its impact on clinical care and clinical development decision-making. For some analysis methods, e.g., determination of intra-class correlation for reliability assessment, estimation of correlation coefficients, and classical analysis of variance, statistical software, such as SAS, R, and SPSS, provide easy-to-use tools for sample size estimation. For more complex methods that may need to be employed for modeling associations between digital measures and other clinical outcomes, several papers on sample size estimation have emerged in recent years [70, 71, 72, 73, 74], but more emphasis and guidance on this topic would be beneficial.
In general, organizations are likely to face multiple challenges as they move toward the formal validation of novel digital measures. The amount of data generated by continuous-monitoring BioMeTs used in daily living is unprecedented. Integration across different tools, platforms, and technology vendors becomes a complicated task for data streaming, storage, and real-time analysis due to the lack of standards for health-related digital data. The absence of standards for BioMeT-derived data results in inefficiencies, and barriers for many stakeholders and some initiatives are already underway to address this critical issue [74]. Additionally, data quality can be impacted by trial participants' adherence to BioMeTs [75], technology literacy, and access to reliable and fast internet for data transmission [4]. Certain types of biases may be introduced in the data via the use of previously developed algorithms either in their entirety or as pre-trained layers in machine learning models. It is essential to examine the characteristics of the individuals whose data contributed to the development of such algorithms to evaluate whether they will be generalizable to other or broader populations [76]. For example, algorithms developed with data from young individuals may not generalize to elderly populations. Another example would be an AI-powered tool for the recognition of skin conditions which may have poor generalizability if it was developed using training data without good representation of various skin colors [77]. Such factors do not necessarily affect all BioMeTs in the same way. For example, a recent study, where the participants used photoplethysmography devices to track the heart rate, has found that the type of activity being performed by an individual had more of a statistical impact on the BioMeT's error than the skin tone of the participant [76]. Precise and quantitative data collected by BioMeTs may also highlight some of the differences between representative populations for the first time. Nevertheless, hypotheses regarding potential biases should be carefully considered and tested.
For many BioMeTs, the processing that occurs between raw data collection and derived parameters used for the calculation of a clinical measure may be a “black box.” Various types of data smoothing and filtering may be applied before data are run through one or more proprietary algorithms to generate parameters of interest. This aspect challenges the flexibility of the interchangeable use of different BioMeTs designed for a similar purpose and makes it difficult to compare or integrate data across different trials. Additionally, it is often unclear how the algorithms have been modified in newer software versions that may be rolled out in the middle of a clinical trial or a clinical development program and if any of the data generated and processed with the previous software version are reliable, comparable, and transferable.
A device-agnostic clinical validation of a digital clinical measure may be challenging because it rests upon the validity of all components, including hardware, raw signal, data preprocessing methods, and algorithms. The equivalence between certain technologies and components should not be taken for granted, although it may be assumed and demonstrated in some cases.
There are also challenges related to the financial resources, time resources, the knowledge base required to navigate the evolving regulatory pathways toward clinical validation of eCOAs and digital biomarkers, and the technical expertise necessary to collect, integrate, manage, analyze, and interpret BioMeT data. Considerations for key analysis objectives required for clinical validation of an eCOA or a specific type of biomarker were highlighted in Table 1. Relevant methodologies and expertise exist in communities involved in the development of non-digital drug development tools but are often fragmented and not known to those currently engaged in the use of BioMeT.
Given these challenges, we believe that for- and nonprofit stakeholders would benefit from pre-competitive collaborations focusing on three particular areas. First, develop data, hardware, and software standards and regulations to work around and connect the diverse BioMeT ecosystem. Second, improve regulations for data rights, access, privacy, and governance [56], as well as scientific transparency required for fulfilling validation requirements. Third, provide guidance on existing analytical methodologies for validation of novel clinical measures and extend or adapt them for BioMeT data as needed [3]. Overall, it would be expected that these partnerships can accelerate the adoption of novel BioMeTs in clinical trials. For example, DiMe has convened several pharmaceutical companies to help advance nocturnal scratch as a digital endpoint for atopic dermatitis [78].
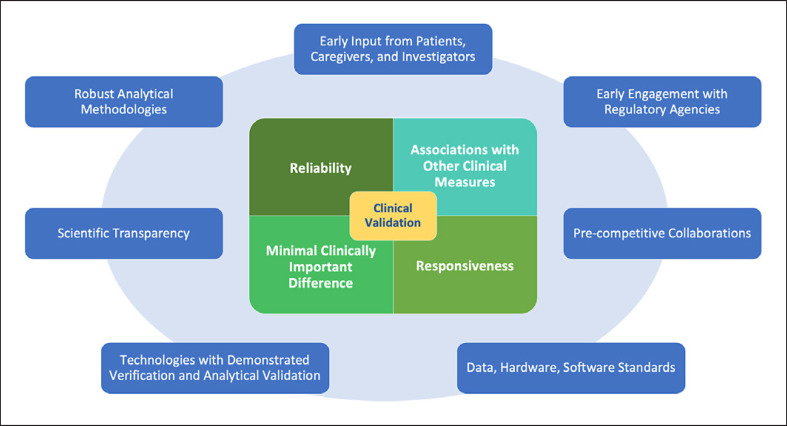
As a first step toward this goal, this paper provides the reader with an overview of the statistical considerations toward clinical validation of eCOAs and digital biomarkers for clinical trial applications. As summarized in Figure 2, we discussed the objectives of statistical analyses that need to be pursued to support the critical elements of clinical validation: reliability, associations between novel digital measures and other relevant clinical measures, responsiveness, and MCID. Factors that play a critical role in enabling a successful validation of novel digital measures are as follows: starting with early input from patients, caregivers, and investigators; engaging early with regulatory agencies; participating in pre-competitive collaborations across the BioMeT ecosystem; adopting data, hardware, and software standards; using technologies that underwent rigorous verification and analytical validation; promoting scientific transparency with regards to the algorithms used to process BioMeT data; and, last but not least, developing, sharing, and using robust statistical methodologies to demonstrate clinical validity. In future publications, statistical approaches that can be used to tackle the key analysis objectives described in this paper will be explored in more detail.
Fig. 2.
Key elements are supporting clinical validation and factors contributing to a successful development of novel digital clinical measures.
Conflict of Interest Statement
Bohdana Ratitch is a salaried employee of Bayer. Isaac R. Rodriguez-Chavez is a salaried employee of ICON plc. Abhishek Dabral is a salaried employee and stockholder of Amgen. Adriano Fontanari is a salaried employee of PatchAi srl (an Alira Health Company). Francesco Onorati is a salaried employee of Current Health. Benjamin Vandendriessche is a salaried employee of Byteflies. Stuart Morton is a salaried employee of Eli Lilly & Co. Yasaman Damestani is a salaried employee and stockholder of Karyopharm Therapeutics Inc.
Funding Sources
The authors have no funding sources to disclose.
Author Contributions
Bohdana Ratitch, Isaac R. Rodriguez-Chavez, Abhishek Dabral, Adriano Fontanari, Julio Vega, Francesco Onorati, Benjamin Vandendriessche, Stuart Morton, and Yasaman Damestani made substantial contributions to the conception or design of the work. Bohdana Ratitch, Isaac R. Rodriguez-Chavez, Abhishek Dabral, Adriano Fontanari, Julio Vega, Francesco Onorati, Benjamin Vandendriessche, Stuart Morton, and Yasaman Damestani drafted this work and revised it critically for important intellectual content and gave final approval of the version to be published. Bohdana Ratitch, Isaac R. Rodriguez-Chavez, Abhishek Dabral, Adriano Fontanari, Julio Vega, Francesco Onorati, Benjamin Vandendriessche, Stuart Morton, and Yasaman Damestani agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Acknowledgments
This publication is a result of collaborative research performed under the auspices of the Digital Medicine Society (DiMe). DiMe is a 510(c)(3) nonprofit professional society for the digital medicine community and is not considered a Sponsor of this work. All the authors are members of DiMe who volunteered their contributions to this systematic review. DiMe research activities are overseen by a research committee, the members of which were invited to comment on the manuscript prior to submission. The authors acknowledge Elizabeth Kunkoski for sharing regulatory insights, as well as Jennifer C. Goldsack and Ieuan Clay for critically reviewing and editing the manuscript.
Funding Statement
The authors have no funding sources to disclose.
References
- 1.FDA. Digital health technologies for remote data acquisition in clinical investigations U.S. Food and Drug Administration. FDA. 2021. [cited 2022 Jan 8]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/digital-health-technologies-remote-data-acquisition-clinical-investigations.
- 2.Goldsack JC, Coravos A, Bakker JP, Bent B, Dowling AV, Fitzer-Attas C, et al. Verification, analytical validation, and clinical validation (V3): the foundation of determining fit-for-purpose for Biometric Monitoring Technologies (BioMeTs) NPJ Digit Med. 2020 Apr 14;3((1)):55–15. doi: 10.1038/s41746-020-0260-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godfrey A, Vandendriessche B, Bakker JP, Fitzer-Attas C, Gujar N, Hobbs M, et al. Fit-for-purpose biometric monitoring technologies: leveraging the laboratory biomarker experience. Clin Transl Sci. 2021 Jan;14((1)):62–74. doi: 10.1111/cts.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coravos A, Khozin S, Mandl KD. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. NPJ Digit Med. 2019 Mar 11;2((1)):14–15. doi: 10.1038/s41746-019-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schüssler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, et al. A longitudinal big data approach for precision health. Nat Med. 2019 May;25((5)):792–804. doi: 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khozin S, Coravos A. Decentralized trials in the age of real-world evidence and inclusivity in clinical investigations. Clin Pharmacol Ther. 2019 Jul;106((1)):25–27. doi: 10.1002/cpt.1441. [DOI] [PubMed] [Google Scholar]
- 7.Digital Medicine Society (DiMe) Library of digital endpoints. Digital Medicine Society (DiMe) 2021. [cited 2021 Jun 27]. Available from: https://www.dimesociety.org/communication-education/library-of-digital-endpoints/
- 8.Goldsack JC, Dowling AV, Samuelson D, Patrick-Lake B, Clay I. Evaluation, acceptance, and qualification of digital measures: from proof of concept to endpoint. Digit Biomark. 2021 Mar 23;5((1)):53–64. doi: 10.1159/000514730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walton MK, Cappelleri JC, Byrom B, Goldsack JC, Eremenco S, Harris D, et al. Considerations for development of an evidence dossier to support the use of mobile sensor technology for clinical outcome assessments in clinical trials. Contemp Clin Trials. 2020 Apr 1;91:105962. doi: 10.1016/j.cct.2020.105962. [DOI] [PubMed] [Google Scholar]
- 10.Manta C, Patrick-Lake B, Goldsack JC. Digital measures that matter to patients: a framework to guide the selection and development of digital measures of health. Digit Biomark. 2020 Sep–Dec;4((3)):69–77. doi: 10.1159/000509725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FDA Biomarker qualification program. FDA. 2021. [cited 2021 Aug 8]. Available from: https://www.fda.gov/drugs/drug-development-tool-ddt-qualification-programs/biomarker-qualification-program.
- 12.FDA-NIH Biomarker Working Group . BEST (Biomarkers, EndpointS, and other Tools) resource. Silver Spring, MD: Food and Drug Administration (US); 2016. [cited 2021 Aug 10]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK326791/ [PubMed] [Google Scholar]
- 13.De Brouwer W, Patel CJ, Manrai AK, Rodriguez-Chavez IR, Shah NR. Empowering clinical research in a decentralized world. NPJ Digit Med. 2021 Jul 1;4((1)):102–105. doi: 10.1038/s41746-021-00473-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FDA The voice of the patient: pulmonary arterial hypertension. 2014. Dec, [cited 2021 Oct 31]. Available from: https://www.fda.gov/media/90479/
- 15.Nathan SD, Flaherty KR, Glassberg MK, Raghu G, Swigris J, Alvarez R, et al. A randomized, double-blind, Placebo-controlled study of pulsed, inhaled nitric oxide in subjects at risk of pulmonary hypertension associated with pulmonary fibrosis. Chest. 2020 Aug 1;158((2)):637–645. doi: 10.1016/j.chest.2020.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Bellerophon Pulse Technologies A study to assess pulsed inhaled nitric oxide in subjects with pulmonary fibrosis at risk for pulmonary hypertension (REBUILD) 2017. Aug, clinicaltrials.gov [cited 2021 Oct 28]. Report No.: study/ NCT03267108. Available from: https://clinicaltrials.gov/ct2/show/study/NCT03267108.
- 17.Stephenson D, Alexander R, Aggarwal V, Badawy R, Bain L, Bhatnagar R, et al. Precompetitive consensus building to facilitate the use of digital health technologies to support Parkinson disease drug development through regulatory science. Digit Biomark. 2020 Nov 26;4((Suppl 1)):28–49. doi: 10.1159/000512500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coravos A, Goldsack JC, Karlin DR, Nebeker C, Perakslis E, Zimmerman N, et al. Digital medicine: a primer on measurement. Digital Med. 2019;41 doi: 10.1159/000500413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.European Medical Agency (EMA) Questions and answers: qualification of digital technology-based methodologies to support approval of medicinal products. 2020. Available from: https://www.ema.europa.eu/en/documents/other/questions-answers-qualification-digital-technology-based-methodologies-support-approval-medicinal_en.pdf.
- 20.Sverdlov O, Curcic J, Hannesdottir K, Gou L, De Luca V, Ambrosetti F, et al. A study of novel exploratory tools, digital technologies, and central nervous system biomarkers to characterize unipolar depression. Front Psychiatry. 2021 May 6;12:640741. doi: 10.3389/fpsyt.2021.640741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bent B, Cho PJ, Henriquez M, Wittmann A, Thacker C, Feinglos M, et al. Engineering digital biomarkers of interstitial glucose from noninvasive smartwatches. NPJ Digit Med. 2021 Jun 2;4((1)):89–11. doi: 10.1038/s41746-021-00465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.FDA Methods to identify what is important to patients and select, develop or modify fit-for-purpose clinical outcomes assessments. 2018. Oct, [cited 2021 Nov 3]. Available from: https://www.fda.gov/media/116277/download.
- 23.Beaton DE, Bombardier C, Katz JN, Wright JG. A taxonomy for responsiveness. J Clinical Epidemiol. 2001 Dec 1;54((12)):1204–1217. doi: 10.1016/s0895-4356(01)00407-3. [DOI] [PubMed] [Google Scholar]
- 24.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016 Jun;15((2)):155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looney SW, editor. Biostatistical methods. Totowa, NJ: Humana Press; 2002. Statistical methods for assessing biomarkers; pp. p. 81–109. [cited 2021 Nov 2]. Methods in Molecular Biology™. Available from: [DOI] [PubMed] [Google Scholar]
- 26.Alpert CM, Smith MA, Hummel SL, Hummel EK. Symptom burden in heart failure: assessment, impact on outcomes, and management. Heart Fail Rev. 2017 Jan;22((1)):25–39. doi: 10.1007/s10741-016-9581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green CP, Porter CB, Bresnahan DR, Spertus JA. Development and evaluation of the Kansas City cardiomyopathy questionnaire: a new health status measure for heart failure. J Am Coll Cardiol. 2000 Apr 1;35((5)):1245–1255. doi: 10.1016/s0735-1097(00)00531-3. [DOI] [PubMed] [Google Scholar]
- 28.Beaton DE, Boers M, Wells GA. Many faces of the minimal clinically important difference (MCID): a literature review and directions for future research. Curr Opin Rheumatol. 2002 Mar;14((2)):109–114. doi: 10.1097/00002281-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 29.King MT. A point of minimal important difference (MID): a critique of terminology and methods. Expert Rev Pharmacoecon Outcomes Res. 2011 Apr;11((2)):171–184. doi: 10.1586/erp.11.9. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt KD, Poole LR, Mullan AF, Kopecky SL, Heaton HA. Clinical evaluation and diagnostic yield following evaluation of abnormal pulse detected using Apple Watch. J Am Med Inform Assoc. 2020 Sep 1;27((9)):1359–1363. doi: 10.1093/jamia/ocaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FDA Review of de novo request for classification of the ECG app. 2018. [cited 2021 Sep 6]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf18/den180044.pdf.
- 32.FDA Review of de novo request for classification of the irregular rhythm notification feature. 2018. [cited 2021 Sep 6]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf18/DEN180042.pdf.
- 33.Cho PJ, Yi JJ, Ho E, Dinh YH, Shandhi MH, Patil A, et al. Demographic imbalances resulting from bring-your-own-device study design. JMIR Mhealth Uhealth. 2022 Apr 8; doi: 10.2196/29510. [cited 2022 Mar 10]; 10(4): e29510. http://preprints.jmir.org/preprint/29510/accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.FDA. Enhancing the diversity of clinical trial populations: eligibility criteria, enrollment practices, and trial designs guidance for industry U.S. Food and Drug Administration. FDA. 2020. [cited 2022 Mar 20]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/enhancing-diversity-clinical-trial-populations-eligibility-criteria-enrollment-practices-and-trial.
- 35.Golbus JR, Pescatore NA, Nallamothu BK, Shah N, Kheterpal S. Wearable device signals and home blood pressure data across age, sex, race, ethnicity, and clinical phenotypes in the Michigan Predictive Activity & Clinical Trajectories in Health (MIPACT) study: a prospective, community-based observational study. Lancet Digit Health. 2021 Nov;3((11)):e707–15. doi: 10.1016/S2589-7500(21)00138-2. [DOI] [PubMed] [Google Scholar]
- 36.Sjoding MW, Dickson RP, Iwashyna TJ, Gay SE, Valley TS. Racial bias in pulse oximetry measurement. N Engl J Med. 2020 Dec 17;383((25)):2477–2478. doi: 10.1056/NEJMc2029240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaisdell CJ, Goodman S, Clark K, Casella JF, Loughlin GM. Pulse oximetry is a poor predictor of hypoxemia in stable children with sickle cell disease. Arch Pediatr Adolesc Med. 2000 Sep 1;154((9)):900–903. doi: 10.1001/archpedi.154.9.900. [DOI] [PubMed] [Google Scholar]
- 38.Hilty DM, Armstrong CM, Edwards-Stewart A, Gentry MT, Luxton DD, Krupinski EA. Sensor, wearable, and remote patient monitoring competencies for clinical care and training: scoping review. J Technol Behav Sci. 2021 Jun;6((2)):252–277. doi: 10.1007/s41347-020-00190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nouri SS, Adler-Milstein J, Thao C, Acharya P, Barr-Walker J, Sarkar U, et al. Patient characteristics associated with objective measures of digital health tool use in the United States: a literature review. J Am Med Inform Assoc. 2020 May 1;27((5)):834–841. doi: 10.1093/jamia/ocaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrack JA, Althoff KN, Jacobson LP, Erlandson KM, Jamieson BD, Koletar SL, et al. Accelerated longitudinal gait speed decline in HIV-infected older men. J Acquir Immune Defic Syndr. 2015 Dec 1;70((4)):370–376. doi: 10.1097/QAI.0000000000000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.FDA. Adjusting for covariates in randomized clinical trials for drugs and biological products U.S. Food and Drug Administration. FDA. 2022. [cited 2022 Mar 20]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/adjusting-covariates-randomized-clinical-trials-drugs-and-biological-products.
- 42.Smith MT, McCrae CS, Cheung J, Martin JL, Harrod CG, Heald JL, et al. Use of actigraphy for the evaluation of sleep disorders and circadian rhythm sleep-wake disorders: an American Academy of Sleep Medicine systematic review, meta-analysis, and GRADE assessment. J Clin Sleep Med. 2018 Jul 15;14((7)):1209–1230. doi: 10.5664/jcsm.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guyatt GH, Osoba D, Wu AW, Wyrwich KW, Norman GR, Clinical Significance Consensus Meeting Group Methods to explain the clinical significance of health status measures. Mayo Clin Proc. 2002 Apr 1;77((4)):371–383. doi: 10.4065/77.4.371. [DOI] [PubMed] [Google Scholar]
- 44.Fröhlich H, Balling R, Beerenwinkel N, Kohlbacher O, Kumar S, Lengauer T, et al. From hype to reality: data science enabling personalized medicine. BMC Med. 2018 Aug 27;16((1)):150. doi: 10.1186/s12916-018-1122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guthrie NL, Carpenter J, Edwards KL, Appelbaum KJ, Dey S, Eisenberg DM, et al. Emergence of digital biomarkers to predict and modify treatment efficacy: machine learning study. BMJ Open. 2019 Jul 1;9((7)):e030710. doi: 10.1136/bmjopen-2019-030710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alemayehu D, Chen Y, Markatou M. A comparative study of subgroup identification methods for differential treatment effect: performance metrics and recommendations. Stat Methods Med Res. 2018 Dec 1;27((12)):3658–3678. doi: 10.1177/0962280217710570. [DOI] [PubMed] [Google Scholar]
- 47.Lipkovich I, Dmitrienko A, D'Agostino RB., Sr Tutorial in biostatistics: data-driven subgroup identification and analysis in clinical trials. Stat Med. 2017;36((1)):136–196. doi: 10.1002/sim.7064. [DOI] [PubMed] [Google Scholar]
- 48.Loh WY, Cao L, Zhou P. Subgroup identification for precision medicine: a comparative review of 13 methods. Wiley Interdiscip Rev Data Min Knowl Discov. 2019;9((5)):e1326. [Google Scholar]
- 49.Guo X, He X. Inference on selected subgroups in clinical trials. J Am Stat Assoc. 2021 Jul 3;116((535)):1498–506. [Google Scholar]
- 50.Thomas M, Bornkamp B. Comparing approaches to treatment effect estimation for subgroups in clinical trials. Stat Biopharm Res. 2017 Apr 3;9((2)):160–171. [Google Scholar]
- 51.Buyse M, Molenberghs G, Paoletti X, Oba K, Alonso A, Van der Elst W, et al. Statistical evaluation of surrogate endpoints with examples from cancer clinical trials: statistical evaluation of surrogate endpoints. Biom J. 2016 Jan;58((1)):104–132. doi: 10.1002/bimj.201400049. [DOI] [PubMed] [Google Scholar]
- 52.Van der Elst W, Alonso AA, Geys H, Meyvisch P, Bijnens L, Sengupta R, et al. Univariate versus multivariate surrogates in the single-trial setting. Stat Biopharm Res. 2019 Jul 3;11((3)):301–310. [Google Scholar]
- 53.Sunny JS, Patro CPK, Karnani K, Pingle SC, Lin F, Anekoji M, et al. Anomaly detection framework for wearables data: a perspective review on data concepts, data analysis algorithms and prospects. Sensors. 2022 Jan 19;22((3)):756. doi: 10.3390/s22030756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di J, Demanuele C, Kettermann A, Karahanoglu FI, Cappelleri JC, Potter A, et al. Considerations to address missing data when deriving clinical trial endpoints from digital health technologies. Contemp Clin Trials. 2022 Feb;113:106661. doi: 10.1016/j.cct.2021.106661. [DOI] [PubMed] [Google Scholar]
- 55.CTTI Recommendations: advancing the use of mobile technologies for data capture & improved clinical trials. 2020. Available from: https://www.ctti-clinicaltrials.org/sites/www.ctti-clinicaltrials.org/files/mobile-devices-recommendations.pdf#Pg7Ln5.
- 56.DiMe Society The playbook: digital clinical measures. 2021. [cited 2021 Jun 27]. Available from: https://playbook.dimesociety.org/
- 57.Manta C, Mahadevan N, Bakker J, Ozen Irmak S, Izmailova E, Park S, et al. EVIDENCE publication checklist for studies evaluating connected sensor technologies: explanation and elaboration. Digit Biomark. 2021 May–Aug;5((2)):127–147. doi: 10.1159/000515835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clinical Trials Transformation Initiative − CTTI CTTI recommendations: advancing the use of mobile technologies for data capture & improved clinical trials. 2021 [Google Scholar]
- 59.FDA The FDA's drug review process: ensuring drugs are safe and effective. FDA. 2017. [cited 2021 Sep 23]. Available from: https://www.fda.gov/drugs/information-consumers-and-patients-drugs/fdas-drug-review-process-ensuring-drugs-are-safe-and-effective. [PubMed]
- 60.FDA Drug Development Tool (DDT) qualification programs. FDA. 2021. [cited 2021 Sep 22]. Available from: https://www.fda.gov/drugs/development-approval-process-drugs/drug-development-tool-ddt-qualification-programs.
- 61.European Medical Agency (EMA) Notice to sponsors on validation and qualification of computerised systems used in clinical trials. 2020 [Google Scholar]
- 62.Servais L, Camino E, Clement A, McDonald CM, Lukawy J, Lowes LP, et al. First regulatory qualification of a novel digital endpoint in Duchenne muscular dystrophy: a multi-stakeholder perspective on the impact for patients and for drug development in neuromuscular diseases. Digit Biomark. 2021;5((2)):183–190. doi: 10.1159/000517411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Office of the Federal Register U.S. 21 code of federal regulations part 312.3: investigational new drug application. 2021. [cited 2021 Oct 31]. Available from: https://www.ecfr.gov/current/title-21/chapter-I/subchapter-D/part-312.
- 64.Badawy R, Hameed F, Bataille L, Little MA, Claes K, Saria S, et al. Metadata concepts for advancing the use of digital health technologies in clinical research. Digit Biomark. 2019;3((3)):116–132. doi: 10.1159/000502951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.CDISC CDISC foundational standards. 2021. [cited 2021 Nov 17]. Available from: https://www.cdisc.org/standards/foundational.
- 66.European Medical Agency (EMA) Final advice to the European Medicines Agency from the clinical trial advisory group on clinical trial data formats. 2013. Available from: https://www.ema.europa.eu/en/documents/other/ctag2-advice-european-medicines-agency-clinical-trial-advisory-group-clinical-trial-data-formats_en-4.pdf.
- 67.Bent B, Wang K, Grzesiak E, Jiang C, Qi Y, Jiang Y, et al. The digital biomarker discovery pipeline: an open-source software platform for the development of digital biomarkers using mHealth and wearables data. J Clin Trans Sci. 2020;5((1)):e19. doi: 10.1017/cts.2020.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalali A, Richerson S, Ouzunova E, Westphal R, Miller B. Handbook of behavioral neuroscience. Elsevier; 2019. Digital biomarkers in clinical drug development; pp. p. 229–238. [cited 2021 Jul 15] [Google Scholar]
- 69.IEEE Standard Association P1752.2: standard for mobile health data: representation of cardiovascular, respiratory, and metabolic measures. 2021. [cited 2021 Sep 23]. Available from: https://standards.ieee.org/project/1752_2.html.
- 70.Riley RD, Snell KIE, Ensor J, Burke DL, Harrell FE, Jr, Moons KGM, et al. Minimum sample size for developing a multivariable prediction model: part I − continuous outcomes. Stat Med. 2019;38((7)):1262–1275. doi: 10.1002/sim.7993. [DOI] [PubMed] [Google Scholar]
- 71.Riley RD, Snell KI, Ensor J, Burke DL, Harrell FE, Jr, Moons KG, et al. Minimum sample size for developing a multivariable prediction model: part II − binary and time-to-event outcomes. Stat Med. 2019;38((7)):1276–1296. doi: 10.1002/sim.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snell KIE, Archer L, Ensor J, Bonnett LJ, Debray TPA, Phillips B, et al. External validation of clinical prediction models: simulation-based sample size calculations were more reliable than rules-of-thumb. J Clin Epidemiol. 2021 Jul 1;135:79–89. doi: 10.1016/j.jclinepi.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stark M, Zapf A. Sample size calculation and re-estimation based on the prevalence in a single-arm confirmatory diagnostic accuracy study. Stat Methods Med Res. 2020 Oct 1;29((10)):2958–2971. doi: 10.1177/0962280220913588. [DOI] [PubMed] [Google Scholar]
- 74.Clay I, Angelopoulos C, Bailey AL, Blocker A, Carini S, Carvajal R, et al. Sensor data integration: a new cross-industry collaboration to articulate value, define needs, and advance a framework for best practices. J Med Internet Res. 2021 Nov 9;23((11)):e34493. doi: 10.2196/34493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olaye IM, Belovsky MP, Bataille L, Cheng R, Ciger A, Fortuna KL, et al. Recommendations for defining and reporting adherence measured by Biometric Monitoring Technologies (BioMeTs): a systematic review. J Med Internet Res. 2022 Apr 14;24((4)):e33537. doi: 10.2196/33537. http://preprints.jmir.org/preprint/33537/accepted . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bent B, Goldstein BA, Kibbe WA, Dunn JP. Investigating sources of inaccuracy in wearable optical heart rate sensors. NPJ Digit Med. 2020 Feb 10;3((1)):18–19. doi: 10.1038/s41746-020-0226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen D, Khan SM, Xu AJ, Ibrahim H, Smith L, Caballero J, et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit Health. 2022 Jan; doi: 10.1016/S2589-7500(21)00252-1. [cited 2021 Dec 19]; 4(1): e64-74. https://www.thelancet.com/journals/landig/article/PIIS2589-7500(21)00252-1/fulltext. [DOI] [PubMed] [Google Scholar]
- 78.DiMe Society Digital Medicine Society convenes pharmaceutical leaders to collaborate on new digital endpoint. 2021. [cited 2021 Nov 17]. Available from: https://www.prnewswire.com/news-releases/digital-medicine-society-convenes-pharmaceutical-leaders-to-collaborate-on-new-digital-endpoint-301413561.html.