Abstract
We identified resistance mechanisms to abiraterone acetate/prednisone (AA/P) in patients with metastatic castration-resistant prostate cancer (mCRPC) in the Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) study.
We analyzed whole-exome sequencing (WES) and RNA-sequencing data from 83 patients with metastatic biopsies before (V1) and after 12 weeks of AA/P treatment (V2). Resistance was determined by time to treatment change (TTTC).
At V2, 18 and 11 of 58 patients had either short-term (median 3.6 months; range 1.4–4.5) or long-term (median 29 months; range 23.5–41.7) responses, respectively. Nonresponders had low expression of TGFBR3 and increased activation of the Wnt pathway, cell cycle, upregulation of AR variants, both pre- and posttreatment, with further deletion of AR inhibitor CDK11B posttreatment. Deletion of androgen processing genes, HSD17B11, CYP19A1 were observed in nonresponders posttreatment. Genes involved in cell cycle, DNA repair, Wnt-signaling, and Aurora kinase pathways were differentially expressed between the responder and non-responder at V2. Activation of Wnt signaling in nonresponder and deactivation of MYC or its target genes in responders was detected via SCN loss, somatic mutations, and transcriptomics. Upregulation of genes in the AURKA pathway are consistent with the activation of MYC regulated genes in nonresponders. Several genes in the AKT1 axis had increased mutation rate in nonresponders. We also found evidence of resistance via PDCD1 overexpression in responders.
Implications:
Finally, we identified candidates drugs to reverse AA/P resistance: topoisomerase inhibitors and drugs targeting the cell cycle via the MYC/AURKA/AURKB/TOP2A and/or PI3K_AKT_MTOR pathways.
Introduction
The management of metastatic prostate cancer is changing rapidly, with the inclusion of several novel drugs and drug combinations in the treatment of hormone-sensitive and castration-resistant disease (1). However, despite considerable progress, progression of prostate cancer to castration-resistant prostate cancer (CRPC) remains a lethal development as the majority of patients will inevitably experience progression and death, with 29,430 deaths attributed to prostate cancer in the United States in 2018 (2). Although several drug choices are available to control disease progression after the development of CRPC, predictive biomarkers for drug resistance and sensitivity remain mostly unknown. Biomarkers based on the stage-specific landscape of genomic alterations in prostate cancer are under investigation (3) but are not yet incorporated into clinical practice for CRPC-stage disease. Abiraterone acetate, a CYP17A1 inhibitor that is a standard treatment option for patients with metastatic CRPC (mCRPC; refs. 5, 6) has no well-defined predictive genomic biomarkers. Recently, we reported that increased expression of genes in the Wnt pathway and cell-cycle proliferation in pretreatment metastases were associated with 12 week-primary resistance to abiraterone acetate/prednisone (AA/P) in patients with mCRPC (4). As the next step in our analysis of this prospective clinical trial (https://clinicaltrials.gov/ identifier NCT No. 01953640), we have now evaluated biomarkers of AA/P efficacy by analyzing the posttreatment genomic landscape of metastatic biopsies in these same patients with mCRPC to identify mechanisms of acquired resistance and, equally important, molecular signatures for AA/P exposure by analyzing the genomic and transcriptomic evolution of metastatic biopsies before and after AA/P treatment.
Materials and Methods
The Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) study, initiated in May 2013 after approval by the Mayo Clinic Institutional Review Board (IRB), enrolled patients with metastatic castration-resistant prostate cancer (mCRPC) after the failure of androgen deprivation therapy. All patients provided written informed consent to undergo two serial metastatic tissue or bone biopsies (5), with the first biopsy performed prior to the initiation of AA/P treatment (visit 1: pretreatment) and the second after 12 weeks of treatment (visit 2: posttreatment). The eligibility criteria and the study protocol have been reported previously (4). The primary goal of the study was to determine genomic alterations associated with pre-chemotherapy AA/P treatment resistance, and those results have been reported previously (4). We now report a secondary aim of the study, which was to identify biomarkers for acquired resistance to AA/P in the posttreatment metastases. Clinical data for all patients is reported in Supplementary Table S1.
Sequencing and genomic aberration analysis
Sequencing of all pre-AA/P treatment biopsy specimens, visit 1 (V1), was reported previously (4). For the analysis of the visit 2 (V2) post-AA/P specimens, similar sequencing methods for whole-exome sequencing (WES) were performed on Illumina HiSeq 2500, and whole transcriptome sequencing (RNA-seq) was performed on Illumina HiSeq 2000 instruments, respectively. Methods have been reported previously (4) and are also described in Supplementary Materials and Methods accompanied by quality control (QC) information for samples in this study in Supplementary Tables S2 to S4 and Supplementary Figs. S1 to S9.
Somatic mutations pathogenicity analysis
Pathogenicity of selected variants were estimated by two primary bioinformatics tools, the cancer-specific OncoKB database (6) and the Variant Effect Scoring Tool (VEST; refs. 7, 8). The VEST statistics and other annotations were obtained via the CRAVAT server (9).
Statistical analysis
Medical records of all patients were collected after enrollment for long-term follow-up and for determination of time to treatment change (TTTC), defined as the time from enrollment until the change of AA/P treatment due to progressive disease (4). The date of the last follow-up is the last patient contact date recorded in the electronic medical records as of October 2017. The lower and upper quartiles of TTTC for the entire cohort were used to define nonresponders and responders, respectively. Statistical tests performed are described in the Supplementary Materials and Methods. Gene sets scoring for RNA-seq was performed by transforming the gene expression data into sample scores per gene set using GSVA (10), followed by association with TTTC using logistic regression or survival Cox-model.
Functional analyses
We performed in silico functional analyses using up- and downregulated genes. For posttreatment results, we selected genes with FDR ≤ 0.05 and fold changes ≥2 or ≤0.5.
Upstream regulator analyses
Upstream transcription factors likely driving the gene expression at V2 were identified with X2Kweb (11). This tool also identified likely signaling kinases responsible for the regulation of the transcription factors driving the differential gene expression. The function of the signaling genes was then further studied using DAVID by clustering all annotations (12, 13).
Secondary drug candidate analysis
The L1000CDS2 (14) tool and L1000FWD (15) were used to select candidate treatments that produced a gene expression signature negatively correlated with the differential gene expression of responders versus nonresponders. These tools can suggest secondary drug treatments that can rescue nonresponders and identify target genes that are potential drivers of the nonresponse. We report signatures found to be significant by requiring a P value to be below the Bonferroni-adjusted threshold.
Data availability
The anonymized raw data (bam files for DNA and fastqs for RNA-seq) will be available in dbGAP (phs001141.v2.p1). All results in the Supplementary Tables refer to the anonymized dbGAP subject ids.
Results
Patient characteristics, samples for sequencing, analysis, and QCs
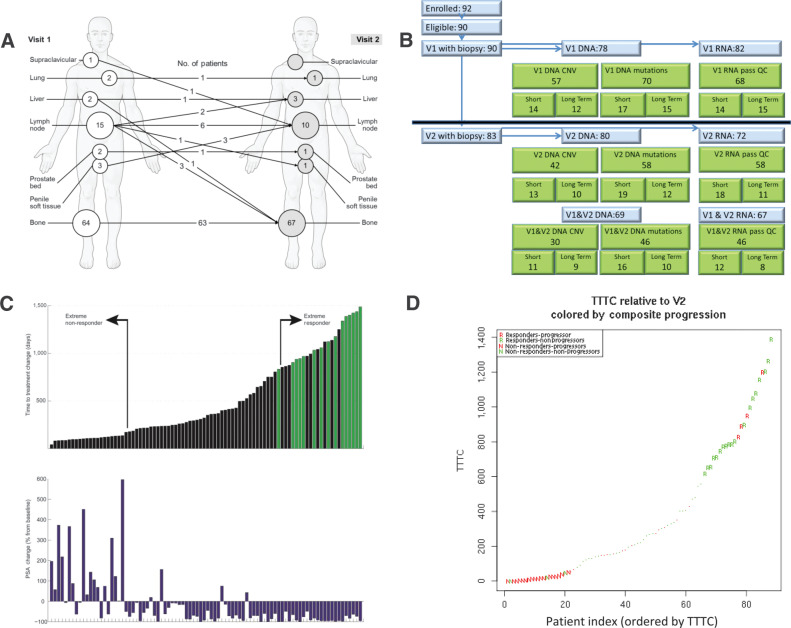
Between May 2013 and September 2015, 83 of the 92 patients enrolled in the PROMOTE study successfully underwent metastatic biopsies during both the before (V1) and after (V2) AA/P treatment visits. The biopsy sites obtained at V1 and V2 are shown in Fig. 1A and B, shows the subsets of these 83 patients that passed QC standards for RNA expression, somatic DNA mutations, and somatic copy number (SCN) that were included in the V2-only or V1 and V2 analyses. Indicator variables in the clinical data Supplementary Table S1 indicate which genomic data passed QC (QC metrics in Supplementary Tables S2–S4). Supplementary Table S5 presents the demographic characteristics of the patient subsets with biopsies that passed the RNA-seq QC. 83 of 92 patients came for a second visit, with a median TTTC for those 83 patients of 303 days. We defined nonresponders as patients in the first quartile of TTTC (≤147 days) and responders as patients in the upper quartile of TTTC (≥667 days; Fig. 1C). All responders had negative PSA changes at 12 weeks posttreatment, whereas most (16/21) nonresponders had increase in PSA after 12 weeks (Fig. 1C). More than half (13/22) of the responders remained on therapy as of the last follow-up date in the medical records as of October 2017. There was good agreement between the two different response phenotypes: the “Composite Progression” phenotype at 12 weeks posttreatment (from ref. 4) and the long-term TTTC phenotype (Fig. 1D) in responders (18/22) and nonresponders (18/21). To avoid mortality bias, the time to treatment change (TTTC) relative to V2 for acquired resistance was used as a readout for outcomes in V2.
Figure 1.
Basic information of the study. A, Biopsy sites pre- and posttreatment. B, Consort flow diagram and samples passing QC per data type (C) PSA change versus TTTC (or last-follow-up—green bar). Nonresponders (low TTTC) and responders (high TTTC) show positive and negative PSA changes respectively (D) V2 TTTC versus composite progression criteria at 12 weeks from V1 manuscript.
Somatic mutations
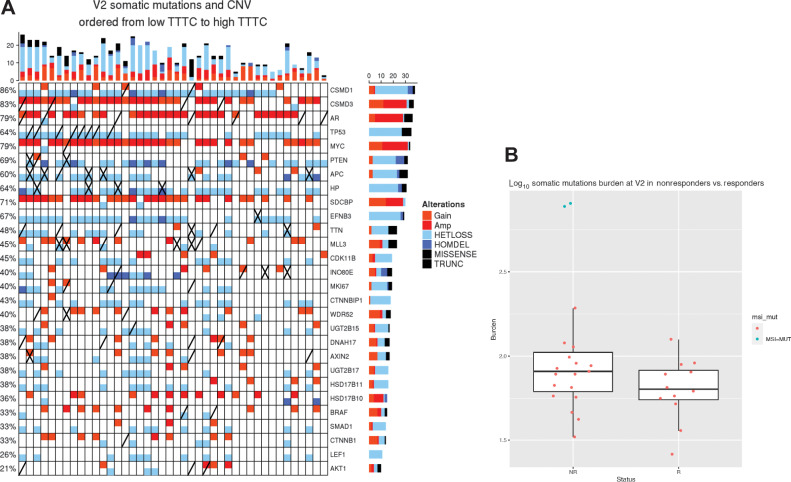
The frequency of somatic mutations in pretreatment samples was comparable with other studies (Supplementary Table S6), as we reported previously (4). Two samples had mutations in DNA mismatch repair (MMR). One sample had missense mutations in MLH3 and APOBEC2B; another sample had truncating mutations in MLL3, APC, and PMS2, as well as missense mutations in ATM. Both samples were nonresponders and were hypermutator (outliers) in the burden plot on (Fig. 2B). Eighty-six genes were found to be frequently mutated in four or more post AA/P treatment samples, including MUC2, TP53, MLL3, MUC16, AR, NOTCH2NL, APC, TTN, PSPH, COL11A1, DNAH12, BRCA2, RYR2, CSMD3, SPOP, FGFR3, AXIN2, PTEN, and FOXA1 (Supplementary Table S6). Two genes (PLPPR3 and TTN) have somatic mutation counts associated with response (Supplementary Table S7). Eighteen genes were significantly differentially mutated between pre- and posttreatment samples (Supplementary Table S7). For four of those genes, the mutation count was different between paired pre- and posttreatment samples (DYNC2H1, ZFC3H1 gained mutations and FBRSL1, ZFPM1 lost mutations).
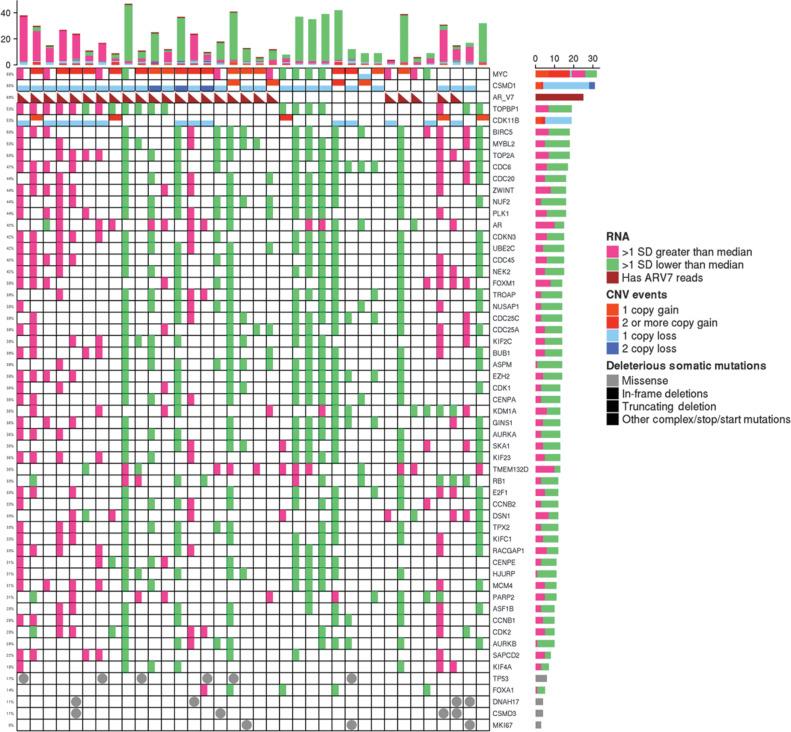
Figure 2.
A, OncoPrint of samples with both somatic mutations and SCN posttreatment (V2). Mutations worse than missense (truncating, frame-shift, early stop) denoted with an “X,” nonsynonymous mutations denoted by a diagonal line. Samples are ordered by TTTC from nonresponders (left) to responders (right). Second and third TTTC quartile patients are included as well. B, Somatic burden higher in nonresponders. Two outliers, in nonresponders, were found to have mutations in MSI genes.
Somatic mutations in MYC target genes are associated with the TTTC in the posttreatment tumors
We next determined the mutation profile in posttreatment samples associated with outcome measures, including both the binary responders/nonresponders and the continuous TTTC. Four hundred and ninety mutated genes identified in posttreatment samples were associated with TTTC in the survival analysis with Cox-model P values ≤0.05 (Supplementary Table S8). However, most significant gens only involved two samples (such as BRAF—deleterious K601E and VUS R239Q). Six genes CSMD3, DNAH17, WDR52, INO80E, HP, and MKI67 had mutations in at least three samples. Mutations in these genes was associated with poor response (left of Fig. 2A).
We then performed a gene set association with outcome in posttreatment samples. We identified 29/424 gene sets with an excess of mutations in nonresponders (Fisher exact test; Supplementary Table S9), with 22 that were also significantly differentiated between responders and nonresponders. Driving this gene set signal, were several genes with at least two more mutations in nonresponders compared with responders: APC (four nonresponders with mutation vs. 0 mutations in responders), CSMD3 (4 vs. 0), AXIN2 (3 vs. 0), and TP53 (4 vs. 1). Only EFNB3 (0 vs. 2) had more mutations in responders, but only one of these mutations is consistently predicted to be pathogenic, and the other one has inconsistent pathogenicity predictions in dbNSFP4.0 (16–18) 8 of these 29 gene sets had 10 or more genes that were more frequently mutated in nonresponders than responders.
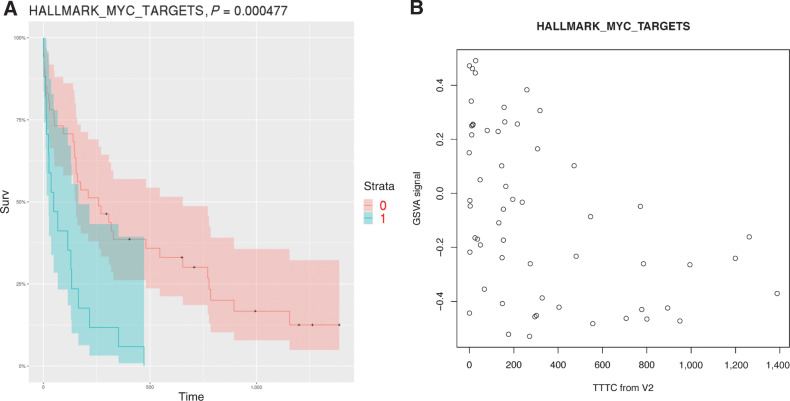
The survival data analysis of the most significant gene set (for MYC target genes) is shown in Fig. 3A. Figure 3B shows gene set mutation scores (the value from the GSVA analysis) in MYC target genes as a function of TTTC. We also found that some genes in the Wnt-pathway (CTNNB1, APC, AXIN2), AKT1 pathways (AKT1, BRAF, PTEN), and PI3K_AKT_MTOR signaling gene sets have a higher mutation rate in nonresponders in the posttreatment samples (84% of these mutations are judged pathogenic by VEST (Supplementary Table S10). For Wnt, mutations of these genes are associated with constitutive activation of the Wnt-signaling pathway (19).
Figure 3.
A, Kaplan–Meyer plot of mutation load for posttreatment samples in MYC targets gene set (version 1 in MSIGDB) vs. TTTC. Samples with high mutation load (above median) are in green band. The P value calculated using a Cox model. B, Gene set mutation load (GSVA value) in MYC targets genes as a function of TTTC.
Somatic mutations changes from V1 to V2 associated with response
We also analyzed genesets with changes from V1 to V2 associated with response (Supplementary Table S11), and found 10 genesets showing an somatic changes associated with TTTC. One of those gene set was significant in a nonresponder only subset analysis (Supplementary Table S12). This geneset associated with a PDCD1 signature had many more mutations at V2 in nonresponders with additional mutations gained in MAP3K8, MAP4, TOR2A, among others.
SCN alterations frequently observed in androgen biosynthesis-metabolism genes in posttreatment responders
We evaluated SCN alterations in post-AA/P treatment samples using WES data in 99 samples with a median tumor purity of 40% (Supplementary Tables S13–S21). Twenty-four regions were identified with significantly different frequencies between responders and nonresponders in the posttreatment tumors (Fig. 4; Supplementary Table S14). The nonresponders showed increased SCN deletions for both CTNNBIP1, a negative regulator of Wnt, and CDK11B, an inhibitor of AR (20). Another segment on chromosome 8, containing the CSMD1 gene, was more frequently deleted in nonresponders whereas another part of chromosome 8 containing SDCBP (aka Synthenin/mda-9) was more amplified in nonresponders. Five nonresponders had a heterozygous deletion in a small chromosome 4 region containing HSD17B11/HSD17B13, which are components of the androgen biosynthesis-metabolism pathway.
Figure 4.
Whole genome representation of copy-number data at Visit2: median copy number gains (alternating red- and orange-shaded regions) and median copy-number deletions (alternating green and blue shaded regions). Regions with significant association with TTTC are shown by black bands along the center axis (under horizontal axis if there were more deletions in nonresponders or above the axis if gains were more frequent). A subset of genes are shown for each band (in black), focusing with genes that were present in the significant genesets, genes that were known targets of AR, genes frequently altered in prostate cancer, or genes with published relevance to prostate cancer. Other genes plotted in red are either at peaks of focal amplification/deletions (PTEN/AR/TP53) or were significant in this paper (e.g., MYC, WNT3A). The focal deletions that are not annotated are in regions with many related genes, zinc fingers, miRNA, or long noncoding RNAs.
Posttreatment, the APC gene, an inhibitor of Wnt-signaling (Supplementary Table S14), showed more SCN gains in responders whereas deletion events were significantly increased in nonresponders. These results suggested that AA/P nonresponders may have activated Wnt-signaling whereas Wnt was repressed in responders, supporting our findings in our previous analysis for baseline samples (4).
An increased rate of deletion of the HSD17B11/HSD17B13 region in nonresponders was observed after 12 weeks of AA/P treatment compared with baseline (Supplementary Table S15), which might lead to increased androgen levels in the nonresponders because these enzymes catalyze testosterone metabolism through glucuronidation.
Gene set analysis of SCN identified signatures of resistance to treatment
We performed gene set analysis on SCN data in posttreatment samples and identified 20 gene sets that differed significantly between responders and nonresponders (Supplementary Table S17; Supplementary Fig. S11). Among the 20 significant gene sets, we found one gene set indicating more frequent TMPRSS2–ERG fusion events in nonresponders and one gene set involved in TCR signaling that included LEF1, a downstream target of β-catenin.
Three SCN gene sets (200125, 200146, 200185) were significantly associated with both predictive (pretreatment) and acquired (posttreatment) AA/P resistance [Supplementary Tables S16 and S17 (V2) and S18 and S19 (V1); Supplementary Figs. S10 and S11]. In 200125 (E-cadherin signaling in the nascent adherens junction) is driven by KLH20, exclusively gained in five nonresponders, 200146 (IL3-mediated signaling events) does not have a key gene, and 200185(IL2 signaling events mediated by STAT5) is driven by SDCBP (aka Syntenin-1), which is has 10 Gains in nonresponders versus only three in responders. SDCBP has been shown to be a marker of resistance in CRPC that correlates with increased MYC activity (21), suggesting that SDCBP may be a predictive biomarker for AA/P treatment in this gene set.
Supplementary Tables S20 and S21 and Supplementary Fig. S12 shows gene sets with different changes from pre- to posttherapy in responders versus nonresponders. Interestingly, a four-gene signature (HSD17B10, UBE2C, NUSAP1, and ANLN) identified from a previous abiraterone trial, NCT00997198, appeared to show different evolution after 12 weeks of AA/P treatment in nonresponders versus responders.
Genes are differentially expressed between pairs of samples from V1 to V2
Cell cycle, DNA repair, Wnt-signaling, and Aurora kinase signaling pathway genes are differentially expressed between the responder and nonresponder cohorts in AA/P posttreatment tumors
In the posttreatment samples, 819 genes were significantly differentially expressed (FDR ≤ 0.05) in nonresponders compared with responders (Supplementary Table S22), whereas at baseline, only 89 genes were differentially expressed with FDR ≤ 0.05 (Supplementary Table S23). This large increase in the number of differentially expressed genes indicates a transcriptomic change due to AA/P treatment. Five hundred and seventy-two genes for which expression was altered from pre- to post-AA/P treatment showed differences between nonresponders and responders (Supplementary Table S24; FDR ≤ 0.05). Among these 572 genes, 360 overlapped with the differentially expressed genes between responders and nonresponders in the posttreatment samples, whereas six genes overlapped with the differentially expressed genes (FDR ≤ 0.05) in the pretreatment samples. These six genes, including LDHA, CST7, CLEC17A, GIMAP4, WEE1, and GIMAP7 showed the same directionality for differential expression, indicating possible predictive potential.
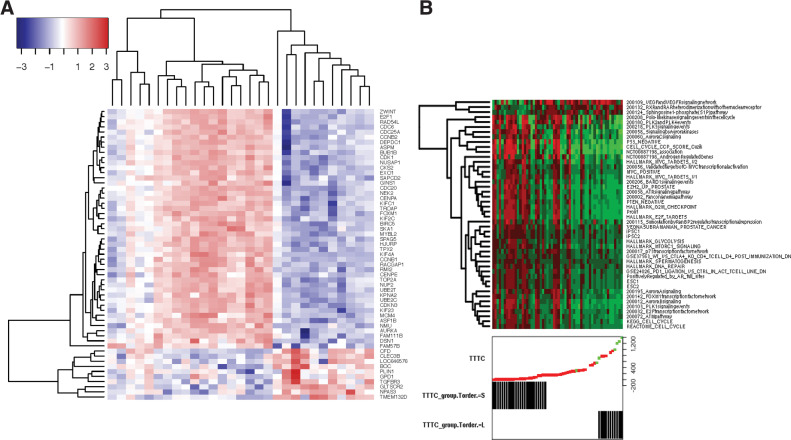
Many of the top differentially expressed genes in the posttreatment samples between responders and nonresponders (Fig. 5A) had known associations with CRPC. We performed a literature search focused on CRPC for the top half of the 93 confidently upregulated genes (FWER ≤ 0.05) and all 10 downregulated genes (FWER ≤ 0.05; see Supplementary Results) and identified genes such as EZH2, E2F1, FOXM1, FOXA1, and genes involved in the cell cycle (kinetochores, spindle, and microtubule related genes), Wnt-signaling, and DNA repair, many of which are regulated by AR, AR-V7, or Aurora kinase. The genes seem to cluster mainly in three groups (two nonresponder groups and one responder groups), except for CFD, CLEC3B, BOC, LOC646576, PLIN, GPD1, GLTSCR2, NPAS3, TMEM132D, and TGFBR3 (bottom of Fig. 5A) who show low expression across all nonresponders at V2.
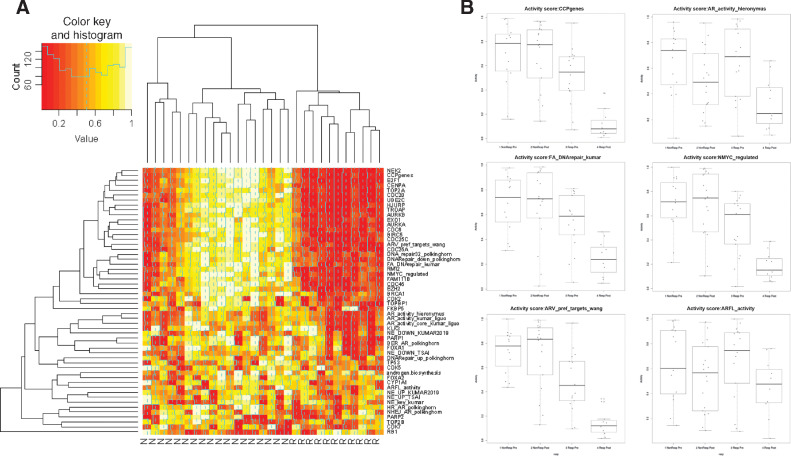
Figure 5.
Gene expression activity. A, Heatmap of most differentially expressed genes from Supplementary Results. B, A heatmap of the most significant gene sets at V2, with samples (columns) ordered by TTTC. Red indicates elevated signature and green indicates low levels of the signature.
On the basis of RNA-seq data for the posttreatment samples, 71 gene sets were significantly (P ≤ 0.05) differentially expressed between nonresponders and responder, with 46 being significant at the FDR ≤ 0.05 level (Supplementary Table S25). This stands in stark contrast with baseline, which showed no gene sets with FDR ≤ 0.05 (Supplementary Table S26). The significant gene sets included many common prostate cancer gene sets, E2F family targets genes, EZH2, cell cycle, MYC targets, TP53 targets, AR-regulated gene sets, DNA repair, Aurora kinase signaling, MTOR signaling, iPS cell signatures, FOXM1, TOP2A, sphingosine pathway, and retinoic acid receptors (RARs) hetero-dimerization (Fig. 5B). Most of those gene sets were elevated in nonresponders, but three were elevated in responders posttherapy (but not pretherapy) the VEGFA and KDR signaling network, the Sphingosine1 phosphate pathway, and the RXR and RAR gene sets (top 3 gene sets in Fig. 5B). For the latter, the most significant genes were VDR and RXRG that were upregulated in posttreatment samples of responders (FDR = 0.043; Supplementary Table S22; Supplementary Fig. S13). Notably, the RXRG gene is elevated in responders and has been shown to form dimers with AR and reduce binding to AR target genes in the presence of RXRG ligands (22). At baseline, these two genes did not show significant differences between responders and nonresponders. The VDR gene is also important for MYC downregulation in nonresponders as it promotes MYC turnover (23, 24)
Of those 46 posttherapy gene sets strongly associated with response (FDR < 0.05; Supplementary Table S25), 13 were also significantly (P ≤ 0.05) associated with response at baseline (Supplementary Table S26). These 13 gene sets with predictive potential were all elevated in nonresponders at baseline and included signatures for proliferation, PTEN_NEGATIVE, KEGG_CELL_CYCLE, networks for Aurora kinases A&B, PLK1, FOXM1, E2F signaling, glycolysis, and validated targets of MYC transcriptional activation. In Supplementary Table S27, are five genesets with a statistically significant correlation between the scores from V1 and V2. One geneset 200090 (“Role of Calcineurin-dependent NFAT signaling in lymphocytes,” involving MAPK13, MAPK3, and CASP3) was highly associated with response at both visits and is a good candidate for prognosis. The other four genesets were highly associated at V2, but did not reach significance at V1 despite the score correlation (STEROID_BIOSYNTHETIC_ PROCESS, 200197 (“Insulin-mediated glucose transport”), ESC1, ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_ DN, STEROID_BIOSYNTHETIC_PROCESS).
The gene expression difference between post-therapy and pre-therapy was associated (P ≤ 0.05) with TTTC for 3 gene sets related to PDCD1, Wnt-signaling, and cell polarity (Supplementary Table S28). We were able to support our results from our paper on the V1 data with the composite progression criteria. Supplementary Table S29 shows that the majority of the cell cycle genes identified in our visit 1 manuscript were significantly upregulated at V1 and even more significant at V2. PLK1 was also significant downregulated in non-responders. Most Wnt Inhibitor activity genes have consistent differential expression, but were not significant at V1 (half were at V2).
In Supplementary Table S30, we show genes that are differentially expressed between V1 and V2 (columns A–H), the remaining present the P values related to TTTC. We see that genes KIF188, SPC24, UBE2T, and were slightly associated with TTTC at V1, but had strong decrease of expression in responders from V1 to V2 and then became even more strongly associated with TTTC at V2, whereas TEF was not even significantly associated with TTTC at V1 and became associated at V2 (increase in responders), suggesting a response to treatment. In general, the gene expression is more strongly associates with response at V2 and this is not a factor of the purity of the biopsies (Supplementary Fig. S3), which is lower, but not statistically significantly.
Finally, we collected a set of published signatures to verify the important biological pathways we found by various analyses (see Supplementary Materials and Methods for details). Figure 6A contrasts the RNA-seq signature values (or key genes) between responders and nonresponders for the top activity signatures in the posttreatment samples. Notable is that nonresponders cluster in two groups. One group (rightmost nonresponder group) has strong associations with elevated cell cycle, DNA repair, MYC, androgen biosynthesis, AR, and aurora kinase signaling in nonresponders. The second group (leftmost nonresponder group) was very similar to responders except for elevated PARP2/TOP2B expression and two signatures for DNA repair genes (HR and NHEJ) that are target of AR (note that the HR and NHEJ gene sets that is not limited to AR targets is not elevated). The two nonresponding samples on the left have elevated neuroendocrine signatures seemed to have inactivated DNA repair (the second sample has a truncating MLL3 mutation R2609* that support this, while the first sample is at the limit of somatic mutation detection (7% cellularity) so that we cannot rule out mutations in DNA repair genes). Responders were associated with high levels of RB1 at V2 (one of the 10 confidently downregulated genes in responders from Supplementary Table S22).
Figure 6.
A, Heatmap of gene expression activity signatures and driver/key genes. B, Top activity signatures in gene expression activity heatmap.
PD-1 gene elevated at V2 in nonresponders
The PDCD1 gene (aka PD-1) showed no prognosis at V1, but it is significantly elevated in responders and relative to V2 (P = 0.03), whereas the expression dropped slightly in nonresponders at V2 relative to V1. Only responders showed that effect as PDCD1 was slightly, but not significantly elevated from V1 to V2 in the paired analysis (Supplementary Table S30). The PDL1 gene (CD274) was slightly elevated at V2 (more in nonresponders), but not statistically significantly.
Upstream regulators of pathways disrupted in nonresponders versus responders
After Bonferroni correction, the significantly differently expressed genes were highly enriched in targets for transcription factors E2F4, FOXM1, AR, and SIN3A and that 22 kinase genes being activated (Supplementary Table S31). Functional annotation clustering of Biocarta, KEGG, and Reactome pathways (Supplementary Table S32) revealed four major clusters of function: (i) AKT1, PRKACA, MAPK14, MAPK8, CHUK signaling; (2) cell cycle and TP53 checkpoint (CDK2, CDK4, CHEK1, ATM, CDK1); (3) pathways involving CDK1, MAPK1, and MAPK3 signaling; and (4) pathways involving CSNK2A1, MAPK3, and MAPK8 signaling.
AR_V(7, 8) and ARV7 isoforms are significant in posttreatment nonresponders
We counted reads supporting specific AR isoform splice junctions, with splice junction AR_V(1, 2, 3, 4) common to four isoforms (V1, V2, V3, V4). Using a Fisher exact test and a Wilcox rank test, we found AR_V(1, 2, 3, 4), AR_V8, AR_V9, and AR_V23 occurred significantly more frequently in nonresponders at both time points. AR_V3, AR_V(3, 4) and AR-45 were only significant at baseline while AR_V(7, 8) and ARV7 were most significant in the posttreatment samples (Supplementary Table S33; Supplementary Fig. S14). Interestingly, 22 of 26 genes that were exclusively upregulated by AR-V7 but not by AR-full length (25) were significantly different (FDR ≤ 0.05) between responders and non-responders after AA/P treatment, but none were at baseline. (annotation column Q in Supplementary Table S22 and Supplementary Table S23).
TOP2A, aurora kinase, and CDK inhibitors may overcome acquired resistance to AA/P treatment
The L1000CDS and L1000FWD tools identified candidate drugs to turn nonresponders into responders. Supplementary Table S34 lists the top candidate drugs many of which target similar or complementary systems. The top candidates, palbociblib and PHA-793887, are CDK inhibitors. PHA-793887 targets CDK2, CDK5, and CDK7. Neither CDK5 nor CDK7 were differentially expressed between responders and nonresponders, but CDK2 (P = 0.0029) and its downstream genes CDC6 (P = 2e−6) and CDC45 (P = 3e−9) were significantly elevated in nonresponders in posttreatment samples. On the other hand palbociclib, selectively targets CDK4 and CDK6. Although a few nonresponders had CDK4 highly expressed in their posttreatment tumors, overall CDK4 and CDK6 were not differentially expressed in the nonresponders. Moreover, most of the cell-cycle genes downstream of CDK4 were not consistently differentially expressed. Three drugs were topoisomerase inhibitors targeting TOP2A, including mitoxantrone, an FDA approved treatment for patients with CRPC. This was consistent with the finding that TOP2A and TOPBP1 were elevated in the nonresponders in the posttreatment samples. Figure 6B supports the use of a topoisomerase inhibitor for mCRPC abiraterone resistant patients; it shows elevated levels of cell-cycle signatures and of cell-cycle genes such as TOP2A, TOP2B, AURKA, AURKB, and PARP2, in the second cluster of nonresponders.
Another cell-cycle regulation target axis is the PI3K_AKT_MTOR inhibition. Three drug candidates inhibit MTOR (torin-2 and NVP-BEZ235, PP-110 (which is also a PI3KCA inhibitor)), four target PI3K (wortmannin, GSK-2126458,GDC-0941, and GDC-0980), two inhibit AKT1 (MK-2206, canertinib), and two more affect the AKT1 axis: IGF1 inhibitor (BMS-536924) and PRKCA agonist (Ingenol 3,20 dibensotate) (26, 27). Of the remaining candidates two are MEK inhibitors (BED-K57080016 and selumetinib) and two are EGFR inhibitors (dovitinib and canartinib, with foretinib being a multikinase inhibitor including PDGFR, KDR, and MAPK). There are still a few other genes/pathways being targeted by other drugs, but the aforementioned drugs were consistently targeting related pathways.
Discussion
Our earlier report showed that activation of the Wnt/β-catenin pathway and increased cell cycle-driven proliferation, were associated with primary resistance to AA/P therapy of patients with mCRPC prior to AA/P treatment in the prospective PROMOTE clinical trial. This study identified genomic and transcriptomic alterations associated with acquired resistance after 12 weeks of AA/P therapy, based on the sequencing data for the metastatic samples after treatment. Once again, Wnt pathway and increased cell-cycle activity were found to play an important role in acquired resistance to AA/P in posttreatment metastases, with the Proliferation gene set being the most significantly associated with acquired resistance. A number of pathways are seen in multiple types of genomic alterations (Supplementary Table S35), including MYC, Wnt-signaling, RXRG-related pathways, and UV response.
Moreover, AR splicing variant AR-V7, AR-V8, AR-V9, AR_V23, AR-V45 were significantly more frequently present in nonresponders, both pretreatment and posttreatment samples. These results suggest that Wnt, cell-cycle signaling, and AR variants might all be predictive biomarkers for AA/P resistance regardless of abiraterone exposure. In posttreatment samples, the presence of AR splicing variants and MYC amplifications were associated with resistance (Fig. 7). Modulation of cell-cycle activity was so critical for resistance that associated alterations were seen in somatic mutation, SCN, and RNA-seq (Fig. 7).
Figure 7.
RNA-seq, CNV, and somatic mutations at V2 of significant genes related to cell-cycle progression, the aurora kinase pathway (including EZH2 and MYC) together with AR expression and AR V7 isoform presence (triangle). Samples ordered by TTTC (low to high TTTC from left to right).
We identified novel genomic biomarkers for acquired resistance gained after AA/P treatment. Compared with pre-AA/P treatment, analysis of the upstream regulators of the top differentially expressed genes between responders and nonresponders after treatment identified several genes regulating or regulated by E2F4, FOXM1, AR, and SIN3A. In addition, EZH2 was also significant (FDR ≤ 0.05), but did not reach Bonferroni significance (Supplementary Table S31). The annotation clustering (Supplementary Table S32) indicates that TP53 checkpointing (supported by our high rate of TP53 mutations in nonresponders) works in concert with cell-cycle regulation. This clustering also shows that AKT1 (found in many of our analyses) also works in concert with several MAPK (MAPK3, MAPK1, MAPK8, MAPK14) as well as PRKACA.
The resistance is mediated by multiple genes for each pathway
AR was activated both by altering genes that modulated AR activity and by an increase in levels of constitutively active AR variants in nonresponders. We found that HSD17B11(P = 0.019), and CYP19A1(P = 0.089), which encode enzymes that metabolize androgens, showed increased SCN deletions in nonresponders after treatment, indicating another modality—other than AR– by which androgen response could be modulated. Another AR modulating gene was RXRG, which had elevated expression in responders and can inhibit AR binding. CDK11B, an inhibitor of AR, was frequently deleted in nonresponders. AR activity was also detected in significant gene sets in Fig. 5B (positively regulated by AR full sites and androgen regulated genes). We also found that the DNA repair pathways that were activated, were limited to genes regulated by AR (Fig. 6A). Figure 6A also shows that AR activity signatures are elevated in a subset of nonresponders.
We detected Wnt involvement in resistance several ways. We found significant RNA-seq gene sets, but also SCN deletions for CTNNBIP1, a negative regulator of Wnt, in nonresponders whereas responders showed SCN gains in APC, a regulator of Wnt. Somatic mutations in CTNNB1, APC, AXIN2 in nonresponders are associated with constitutive activation of Wnt.
We found several lines of evidence that inactivating the MYC pathway was associated with good response and its activation consistent with poor response. The gene set analysis of somatic mutations indicated increased mutations in MYC downstream targets in responders after AA/P treatment. The gene signatures of genes regulated by MYC were significantly downregulated in nonresponders. MYC transcripts were not significantly differently expressed in our data even though it was downregulated by a factor of two in nonresponders (Supplementary Table S22). It is likely that the MYC activity is regulated at the protein level as it is known that the MYC protein is tightly regulated via phosphorylation driven degradation (28) and via cofactors. The MYCBP2 protein, which binds MYC and negatively regulates its activity (29), was lower in nonresponders (FDR = 0.056). At the SCN level, we found gains of SDCBP in non-responders which has been shown to regulate MYC (30). The VDR gene has increased gains in responders (Supplementary Fig. S13A) and this gene was found to increase MYC turnover (31). We also found elevated levels of TOP2A/AURKA/RAD21 in nonresponders and these genes interact with MYC (32) to regulate cell cycle. AURKA both is a target of MYC (33) and promotes elevated levels of MYC (34). Recent analysis of ChIP-seq data found that MYC binds to the promoter of TOP2A, TOP2B, TOP3A, TOP3B genes and that silencing of MYC downregulates these genes (35), consistent with our findings on TOP2B. Supporting this MYC/AURKA/TOP2A axis hypothesis, we find several drugs targeting the AURKA and TOP2A as candidates for second line therapy for nonresponders.
Somatic mutations were increased in nonresponders in the AKT1 axis: AKT1, BRAF, PTEN, PI3K_AKT_MTOR gene sets, as well as TCR signaling gene sets. The AKT1 axis is a potential point of pharmacologic intervention (36). The upstream regulator analysis and the drug signature analysis were very consistent and also identified the PI3K_AKT_MTOR axis and cell cycle as important targets.
Responder biomarkers
Responders were characterized by elevated levels of RB1, increased mutations in MYC downstream targets, increase SCN or expression gains in AR controlling genes (see above), and increase SCN gains in APC, an inhibitor of Wnt-signaling. Three gene sets had elevated expression posttherapy (Fig. 5B), the RXR and RAR heterodimers gene set, the VEGFA and KDR signaling network, and the Sphingosine1 phosphate pathway. We also found that changes in genes in the androgen biosynthesis and elimination pathway, Wnt signaling inhibition, VEGFA/KDR(VEGFR), RXRA, and RAR heterodimers binding to AR, and Sphingosine1 phosphatase play roles in positive response to therapy.
Our in silico analysis finds that cell-cycle inhibitors, topoisomerase inhibitors, MTOR inhibitors, and even a number of inhibitors in the PI3K_AKT_MTOR axis might be additional therapeutic options for patients with mCRPC who do not have a long term response to abiraterone. PI3KCA inhibitors combined with AA trials had difficulty achieving sufficient response without doses causing intolerable side effects (37). However, targeting AKT1, which is downstream in this axis, along with an AA regimen (38) showed promise. These suggest that targeting MTOR and or AURKA, which are downstream of PI3KCA/AKT1 (39) might be targets that are even more robust against the development of resistance. Our data supports that the simultaneous targeting of the MYC/TOP2A/AURKA axis of cell-cycle control would increase effectiveness of a multitarget regimen which includes AA/P. This hypothesis would require additional validation.
We also found increased expression of the PDCD1 gene (PD1) in responders suggesting a developing resistance in responders. We also found increased somatic mutations in genes involved in differential expression of PDCD1 at Visit 2 in nonresponders, suggesting that investigation of combination therapy of abiraterone acetate and PDCD1 inhibitors may be warranted, especially in light of the MYC and AURKA both of which were found to be associated with PDCD1 expression in TNBC (40).
Nonresponder biomarkers
The nonresponders show universal low expression of a few genes, most notably TGFBR3 (Fig. 6A). Low expression of TGFBR3 has been associated with prostate cancer resistance and it was shown that TGFBR3 knockdown led to the enhanced expression of PROM1(CD133) (41), a marker found in cancer stem cells. The majority of nonresponders appear to be AR-driven (Fig. 6A) with activation of all the pathways of resistance discussed at the beginning of the discussion section. A second nonresponder group often has gene expression similar to responders. This second group is not uniform, with different genes or signatures being activated per patients (including PARP2/TOP2B).
Analysis of patients with mCRPC treated with enzalutamide (42) found an association with neuro-endocrine features in nonresponders. In Fig. 5A, three signatures of neuro-endocrine (NE) are elevated in nonresponders, but mainly in the subgroup of patients with elevated AR activity and furthermore the NE signatures are not as discriminating as other we highlighted.
In summary, we find that several patients do exceptionally well on abiraterone treatment, and our study has identified not only critical genomic alterations in response to AA/P for both responders and nonresponders, but also defined a potential strategy that might help us to overcome resistance and prolong survival. Further studies will be needed to test these drug treatments to overcome AA/P resistance and further define subgroups of nonresponders.
Supplementary Material
Acknowledgments
This work was supported by the Mayo Clinic Center for Individualized Medicine (MC1351 to M. Kohli and L. Wang); Minnesota Partnership for Biotechnology and Medical Genomics (MNP#14.37 to M. Kohli and S.M. Dehm); Department of Defense (W81XWH-15-1-0634 to S.M. Dehm and M. Kohli); NIH-NCI, R01 CA174777 to S.M. Dehm AT Suharya and Ghan DH; Joseph and Gail Gassner; and Mayo Clinic Schulze Cancer for Novel Therapeutics in Cancer Research (to M. Kohli and L. Wang). Other contributing groups include the Mayo Clinic Cancer Center (no grant numbers apply), the Pharmacogenomics Research Network (PGRN; no grant numbers apply), and the Janssen Research & Development, LLC (no grant numbers apply), which provided partial drug support on study from April 2014 and funding support for bioinformatics analysis and delivery of the study data (ending in 2018).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
H. Sicotte reports other support from Jannsen during the conduct of the study. S.M. Dehm reports grants from US Department of Defense Prostate Cancer Research Program, grants from US NCI, and grants from Minnesota Partnership for Biotechnology and Medical Genomics during the conduct of the study; personal fees from Bristol Myers Squibb/Celgene and Oncternal Therapeutics; grants from Pfizer/Astellas/Medivation; and grants and personal fees from Janssen R&D, LLC outside the submitted work; also has a patent for Androgen receptor variant inhibitors and methods of using issued. V. Bhargava reports employment with Janssen Research and Development, LLC; also reports ownership of Johnson and Johnson stock. M. Gormley reports other support from Janssen Research and Development outside the submitted work. A.H. Bryce reports other support from Janssen during the conduct of the study; personal fees from Astra Zeneca, Merck, Gilead, Advanced Accelerator Applications, and Bayer; and personal fees and other support from Janssen outside the submitted work. R.M. Weinshilboum reports other support from OneOme LLC outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
H. Sicotte: Data curation, software, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. K.R. Kalari: Conceptualization, resources, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Qin: Writing–review and editing. S.M. Dehm: Supervision, funding acquisition, investigation, writing–review and editing. V. Bhargava: Software, formal analysis, investigation, writing–review and editing. M. Gormley: Software, formal analysis, investigation, writing–review and editing. W. Tan: Software, formal analysis, investigation, writing–review and editing. J.P. Sinnwell: Data curation, software, formal analysis, investigation, writing–review and editing. D.W. Hillman: Data curation, software, investigation, writing–review and editing. Y. Li: Data curation, software, formal analysis, writing–review and editing. P.T. Vedell: Data curation, software, writing–review and editing. R.E. Carlson: Data curation, software, writing–review and editing. A.H. Bryce: Data curation, software, investigation, writing–review and editing. R.E. Jimenez: Data curation, investigation, writing–review and editing. R.M. Weinshilboum: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Kohli: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, investigation. L. Wang: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, writing–review and editing.
References
- 1. Cornford P, Bellmunt J, Bolla M, Briers E, De Santis M, Gross T.et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: Treatment of relapsing, metastatic, and Castration-resistant prostate cancer. Eur Urol 2017;71:630–42. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 3. Sanhueza C, Kohli M. Clinical and novel biomarkers in the management of prostate cancer. Curr Treat Options Oncol 2018;19:8. [DOI] [PubMed] [Google Scholar]
- 4. Wang L, Dehm SM, Hillman DW, Sicotte H, Tan W, Gormley M.et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann Oncol 2018;29:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jimenez RE, Atwell TD, Sicotte H, Eckloff B, Wang L, Barman P.et al. A prospective correlation of tissue histopathology with nucleic acid yield in metastatic castration-resistant prostate cancer biopsy specimens. Mayo Clin Proc Innov Qual Outcomes 2019;3:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J.et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics 2013;14:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Douville C, Masica DL, Stenson PD, Cooper DN, Gygax DM, Kim R.et al. Assessing the pathogenicity of insertion and deletion variants with the variant effect scoring tool (VEST-Indel). Hum Mutat 2016;37:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Douville C, Carter H, Kim R, Niknafs N, Diekhans M, Stenson PD.et al. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics 2013;29:647–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma'ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 2012;28:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 13. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duan Q, Reid SP, Clark NR, Wang Z, Fernandez NF, Rouillard ADet al. L1000CDS(2): LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl 2016;2:16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Z, Lachmann A, Keenan AB, Ma'ayan A. L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics 2018;34:2150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 2011;32:894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 2013;34:E2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 2016;37:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sunaga N, Kohno T, Kolligs FT, Fearon ER, Saito R, Yokota J. Constitutive activation of the Wnt signaling pathway by CTNNB1 (β-catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer 2001;30:316–21. [DOI] [PubMed] [Google Scholar]
- 20. Zong H, Chi Y, Wang Y, Yang Y, Zhang L, Chen H.et al. Cyclin D3/CDK11p58 complex is involved in the repression of androgen receptor. Mol Cell Biol 2007;27:7125–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talukdar S, Das SK, Pradhan AK, Emdad L, Windle JJ, Sarkar D.et al. MDA-9/Syntenin (SDCBP) is a critical regulator of chemoresistance, survival and stemness in prostate cancer stem cells. Cancers (Basel) 2019;12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chuang KH, Lee YF, Lin WJ, Chu CY, Altuwaijri S, Wan YJ.et al. 9-cis-retinoic acid inhibits androgen receptor activity through activation of retinoid X receptor. Mol Endocrinol 2005;19:1200–12. [DOI] [PubMed] [Google Scholar]
- 23. Salehi-Tabar R, Nguyen-Yamamoto L, Tavera-Mendoza LE, Quail T, Dimitrov V, An BS. et al. Vitamin D receptor as a master regulator of the c-MYC/MXD1 network. Proc Natl Acad Sci U S A 2012;109:18827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salehi-Tabar R, Memari B, Wong H, Dimitrov V, Rochel N, White JH. The tumor suppressor FBW7 and the Vitamin D receptor are mutual cofactors in protein turnover and transcriptional regulation. Mol Cancer Res 2019;17:709–19. [DOI] [PubMed] [Google Scholar]
- 25. Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C. et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012;72:3457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng SSW, Tsao M-S, Chow S, Hedley DW. Inhibition of phosphatidylinositide 3-kinase enhances Gemcitabine-induced apoptosis in human pancreatic cancer cells. Cancer Res 2000;60:5451–5. [PubMed] [Google Scholar]
- 27. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer 2019;18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 2004;3:1133–7. [PubMed] [Google Scholar]
- 29. Ge Z, Guo X, Li J, Hartman M, Kawasawa YI, Dovat S. et al. Clinical significance of high c-MYC and low MYCBP2 expression and their association with Ikaros dysfunction in adult acute lymphoblastic leukemia. Oncotarget 2015;6:42300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Talukdar S, Das SK, Pradhan AK, Emdad L, Shen XN, Windle JJ. et al. Novel function of MDA-9/Syntenin (SDCBP) as a regulator of survival and stemness in glioma stem cells. Oncotarget 2016;7:54102–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tabar RS. The hormone-bound vitamin D receptor regulates turnover of target proteins of the SCFFBW7 E3 ligase. In Department of Experimental Medicine. McGill University; 2017:223. https://escholarship.mcgill.ca/downloads/c821gn670?locale=en. [Google Scholar]
- 32. Buchel G, Carstensen A, Mak KY, Roeschert I, Leen E, Sumara O. et al. Association with Aurora-A controls N-MYC-dependent promoter escape and pause release of RNA Polymerase II during the cell cycle. Cell Rep 2017;21:3483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A. et al. Aurora kinases A and B are upregulated by Myc and are essential for maintenance of the malignant state. Blood 2010;116:1498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget 2017;8:23937–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen J, Li W, Cui K, Ji K, Xu S, Xu Y. Artemisitene suppresses tumorigenesis by inducing DNA damage through deregulating c-Myc-topoisomerase pathway. Oncogene 2018;37:5079–87. [DOI] [PubMed] [Google Scholar]
- 36. Nitulescu GM, Van De Venter M, Nitulescu G, Ungurianu A, Juzenas P, Peng Q. et al. The Akt pathway in oncology therapy and beyond (Review). Int J Oncol 2018;53:2319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Massard C, Chi KN, Castellano D, de Bono J, Gravis G, Dirix L. et al. Phase Ib dose-finding study of abiraterone acetate plus buparlisib (BKM120) or dactolisib (BEZ235) in patients with castration-resistant prostate cancer. Eur J Cancer 2017;76:36–44. [DOI] [PubMed] [Google Scholar]
- 38. de Bono JS, De Giorgi U, Rodrigues DN, Massard C, Bracarda S, Font A. et al. Randomized phase II study evaluating Akt blockade with Ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss. Clin Cancer Res 2019;25:928–36. [DOI] [PubMed] [Google Scholar]
- 39. Osher E, Macaulay VM. Therapeutic targeting of the IGF axis. Cells 2019;8:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun S, Zhou W, Li X, Peng F, Yan M, Zhan Yet al. Nuclear Aurora kinase A triggers programmed death-ligand 1-mediated immune suppression by activating MYC transcription in triple-negative breast cancer. Cancer Commun (Lond) 2021;41:851–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharifi N, Hurt EM, Kawasaki BT, Farrar WL. TGFBR3 loss and consequences in prostate cancer. Prostate 2007;67:301–11. [DOI] [PubMed] [Google Scholar]
- 42. Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E. et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci U S A 2020;117:12315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The anonymized raw data (bam files for DNA and fastqs for RNA-seq) will be available in dbGAP (phs001141.v2.p1). All results in the Supplementary Tables refer to the anonymized dbGAP subject ids.