Single-cell transcriptomics of multifocal glioblastomas characterized properties of natural tumor evolution, as well as tumor migration and immunoenvironment remodeling.
Abstract
Isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM) has a dismal prognosis. A better understanding of tumor evolution holds the key to developing more effective treatment. Here we study GBM's natural evolutionary trajectory by using rare multifocal samples. We sequenced 61,062 single cells from eight multifocal IDH wild-type primary GBMs and defined a natural evolution signature (NES) of the tumor. We show that the NES significantly associates with the activation of transcription factors that regulate brain development, including MYBL2 and FOSL2. Hypoxia is involved in inducing NES transition potentially via activation of the HIF1A–FOSL2 axis. High-NES tumor cells could recruit and polarize bone marrow–derived macrophages through activation of the FOSL2–ANXA1–FPR1/3 axis. These polarized macrophages can efficiently suppress T-cell activity and accelerate NES transition in tumor cells. Moreover, the polarized macrophages could upregulate CCL2 to induce tumor cell migration.
Significance:
GBM progression could be induced by hypoxia via the HIF1A–FOSL2 axis. Tumor-derived ANXA1 is associated with recruitment and polarization of bone marrow–derived macrophages to suppress the immunoenvironment. The polarized macrophages promote tumor cell NES transition and migration.
This article is highlighted in the In This Issue feature, p. 2711
INTRODUCTION
Isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM) is the most common and aggressive type of brain tumor in adults. Due to the evolution of malignancy in the tumor, improving GBM prognosis remains challenging (1). Standard of care, debulking surgery followed by radiation combined with temozolomide, results in a median survival of around 15 months (2, 3). Most patients experience recurrence within 10 months of primary treatment (4). Currently, immune-checkpoint inhibitors improve the prognosis of many cancers (5, 6), but they have not yet delivered substantive clinical benefits to patients with GBM, likely due to the significant immunosuppressive tumor environment in GBM (7).
Communication between tumor cells and immune cells plays a pivotal role in tumor progression (8). In particular, the infiltrating macrophages promote tumor angiogenesis, growth, and drug resistance (9) via various cytokine and chemokine release. Thus, it is crucial to systematically characterize the natural progression of GBM and unravel the potential mechanism of immune microenvironment remodeling during disease progression.
Currently, several models have been constructed to explore GBM initiation and progression. For instance, Ozawa and colleagues (10) analyzed human GBM data and found that chromosome alterations may be an initiating factor for GBM. Additional studies (11–15) focused on longitudinal sample models to explore the spatiotemporal progression of GBM. The disease trajectory was projected from paired primary and recurrent samples and showed a genomic GBM landscape driven by treatment. Though these studies significantly improved our understanding of tumor evolution and therapy failure (11–13, 15), they mainly focused on genomic changes after treatment. How tumor cells interact with the tumor microenvironment (TME) during natural disease progression remains unclear due to a lack of appropriate models and technology (16). Multifocal GBMs are defined as synchronously occurring lesions in a patient. Although rare, these tumors are considered to be an excellent model for studying GBM natural evolution (17), because they may represent various phases of tumor progression. Increasing evidence shows that most multifocal GBMs descend from a common tumor precursor cell clone in an early stage and undergo long-term parallel genetic evolution (17, 18). Here, we investigate tumor progression by applying single-cell RNA sequencing (scRNA-seq) to multifocal IDH wild-type primary GBMs, allowing unprecedented high-resolution characterization of tumor natural evolutionary processes.
RESULTS
A Landscape of Primary Multifocal GBMs Based on scRNA-seq Data
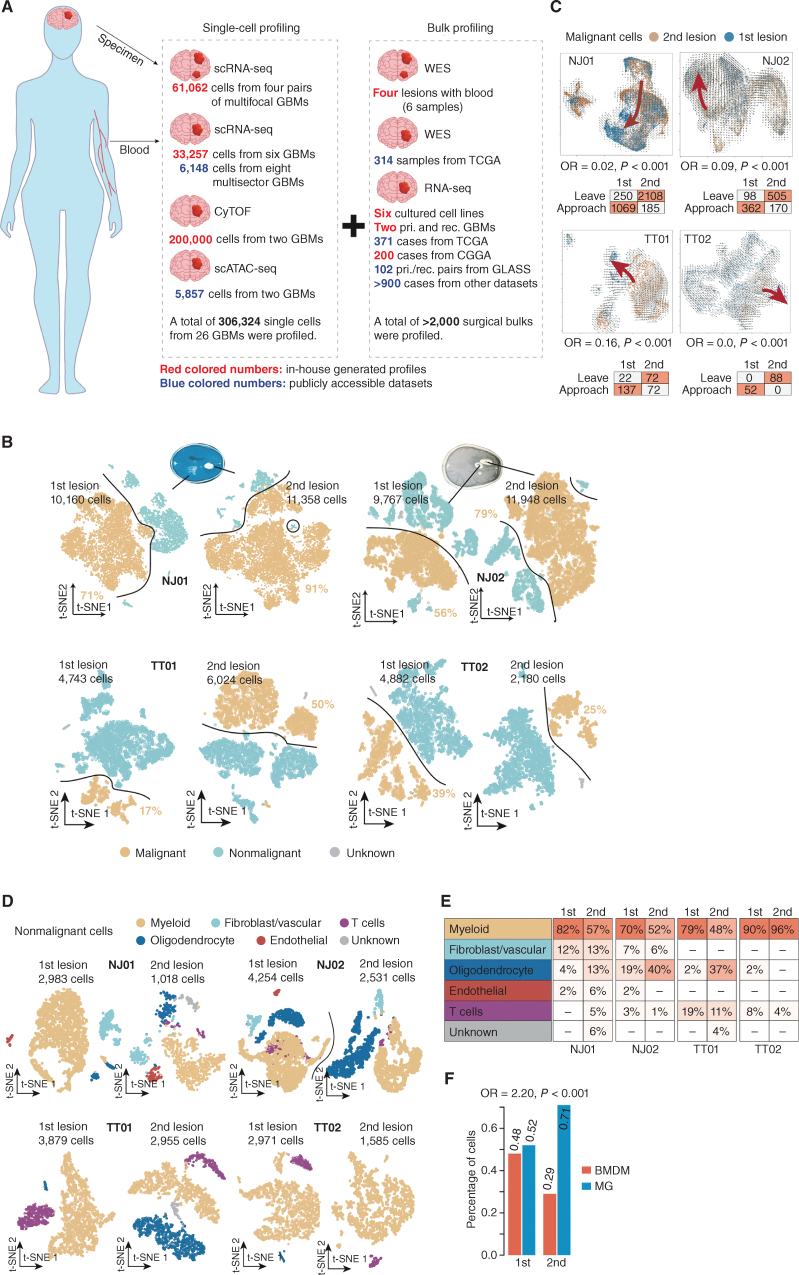
We integrated 306,324 single cells, involving transcriptome, epigenome, and proteome data, to investigate the relationships between tumor cells and the microenvironment under the natural evolution of primary GBMs (Fig. 1A). Specifically, we first enrolled four patients (i.e., NJ01, NJ02, TT01, and TT02) diagnosed with multifocal GBMs (two primary IDH wild-type lesions per case), of which the specimens were subjected to scRNA-seq (NJ01: 21,518 cells; NJ02: 21,715; TT01: 10,767 cells; TT02: 7,062 cells). Using the K-means clustering algorithm on scRNA-seq profiles, we classified the cells in each lesion into distinct clusters, which was followed by tumor cell inferring (see Supplementary Methods; Fig. 1B; Supplementary Fig. S1A–S1D). We next explored the genomic features of these cases by inferring chromosome copy numbers for each tumor cell based on the single-cell mRNA expression profile using inferCNV (19). The resulting copy-number profiles revealed that two lesions in each of the four cases derived from the same ancestor reflecting shared copy-number alterations commonly found in GBM (refs. 20, 21; Supplementary Fig. S2A–S2D). A temporal order was implied for NJ01 and NJ02, in which one lesion harbored copy-number changes (e.g., chromosome 3 and 12q amplification, and chromosome 15 deletion in NJ01; chromosome 6 deletion in NJ02) not detected in the other lesion. This suggested that one lesion may have seeded the other. Whole-exome sequencing of tumor tissue corroborated the findings and was in accordance with those derived from scRNA-seq (Supplementary Fig. S2A and S2B; see Supplementary Methods). Besides, both lesions shared a high percentage of somatic mutations (clonal mutations, first lesion: 63%, second lesion: 55% for NJ01; first lesion: 36%, second lesion: 26% for NJ02; Supplementary Fig. S2E), which further confirmed that the two lesions shared a common ancestor. To further substantiate the potential evolution sequence order between two lesions for each case, we performed RNA velocity analysis on tumor cells from the two lesions derived from the same patient (Fig. 1C; Supplementary Fig. S2F). The approach aims to predict the time derivative of gene abundance in cells by comparing the ratio of spliced to unspliced transcripts per gene. By enumerating the number of cells transiting to each group (the second or first lesion) based on the transition trajectory predicted by RNA velocity analysis, we performed a Fisher exact test analysis to evaluate such a transition tendency (see Supplementary Methods). Notably, we found that the four tumors all shared a potential transition tendency from the second to the first lesions. Moreover, we applied the stem-like, differentiated-like, and proliferation stem-like gene sets from our earlier study (22) to tumor cells using the single-sample gene set enrichment analysis (ssGSEA) algorithm. By enumerating the tumor cells belonging to each phenotype according to the enrichment score, we found the tumor cells with stem-associated features (combining stem-like and proliferating stem-like) were enriched in the younger lesions (one-tailed Student paired t test, P = 0.036). Of note, the tumor cells with both stem-associated and differentiated-like features showed a higher proportion in the older lesions (P = 0.045; Supplementary Fig. S2G). Therefore, these results implied that tumor cells in the second lesion might act as new “bamboo shoots,” which may finally develop into the phenotype of tumor cells in the first lesion, suggesting that the first lesion may be older than the second lesion.
Figure 1.
Comparative characterization of multifocal GBMs and the TME by scRNA-seq. A, Schematic workflow for the construction of single-cell multifocal GBM transcriptomes. CGGA, Chinese Glioma Genome Atlas; cyTOF, cytometry by time of flight; pri., primary; rec., recurrent; scATAC-seq, single-cell sequencing assay for transposase-accessible chromatin; TCGA, The Cancer Genome Atlas; WES, whole-exome sequencing. B, T-distributed stochastic neighbor embedding (t-SNE) plots of malignant (orange dots) and nonmalignant (blue dots) cells in the first and second lesions derived from the indicated cases. C, Top, RNA velocity illustrates the direction of tumor cell transition in the t-SNE plots between two lesions across the indicated cases. Bottom, the table shows the number of cells with a putative transition tendency to the center of each lesion. OR, odds ratio.D, t-SNE plot of nonmalignant cells in the first and second lesions derived from the indicated cases. E, Comparison of the percentages of different nonmalignant cell types between the first and second lesions across the indicated cases. F, Comparison of percentages of cells identified as BMDMs and MGs in the first and second lesions across the four cases (NJ01, NJ02, TT01, and TT02).
We further clustered nontumor cells based on scRNA-seq data. The cell type for each cluster was confirmed using cell-specific markers, including FCGR1A (myeloid-like), COL1A1 (fibroblast-like), CD3D (T-cell-like), CDH5 (endothelial-like), and MAG (oligodendrocyte-like; Fig. 1D and E). Notably, we observed that more than 45% of nontumor cells were myeloid-like cells (mainly macrophages; Fig. 1E), which was consistent with our previous findings (23) that myeloid cells were the major nonneoplastic cell population in GBM. In GBM, tumor-associated macrophages (TAM) may originate from either peripheral bone-marrow–derived macrophage (BMDM) or resident microglia (MG). By applying the signatures provided by Müller and colleagues (24), we identified BMDM and MG cells in scRNA-seq data across the four cases (see Supplementary Methods). We found that the ratio of BMDMs to MGs was higher in the first lesion than in the second lesion (Fisher exact test, odds ratio = 2.20, P < 0.001; Fig. 1F).
A Natural Evolution Signature Is Associated with Poor GBM Prognosis
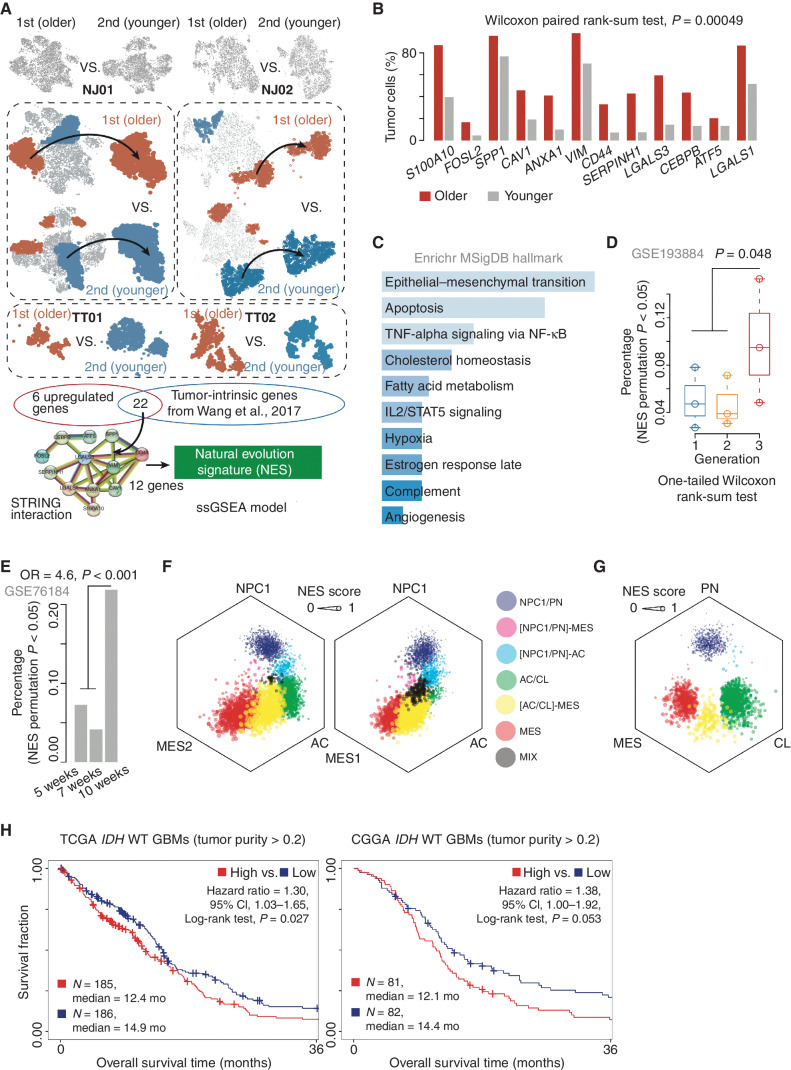
Because we had confirmed the putative evolution order between two lesions (i.e., the first lesion was older than the second lesion) in four cases (i.e., NJ01, NJ02, TT01, and TT02), we asked what kind of transcriptional signatures could characterize such an evolution. To solve this problem, we first focused on NJ01 and NJ02 to determine differentially expressed genes (DEG) between the tumor cells in the first and second lesions (i.e., the highly expressed genes derived from first vs. second and vice versa; see Supplementary Methods; Fig. 2A). We found the percentage of DEGs expressed in the first lesion was significantly higher than in the second lesion, suggesting that GBM natural evolution may be accompanied by broad gene activation. We next iterated over all the tumor clusters to select ones expressing the top or bottom proportion of these DEGs (designated as the older and younger clusters; Fig. 2A). The top 20% of the older clusters in the first lesion and the bottom 20% of the younger clusters in the second lesion were further analyzed for differential gene expression. To ensure that the older GBMs ubiquitously expressed these genes, we applied a similar strategy as above to separately analyze TT01 and TT02 (see Supplementary Methods). Twenty-eight genes were retained through the overlap of DEGs in NJ01, NJ02, TT01, and TT02. These genes were further filtered for a signature of 11,529 genes intrinsically expressed in GBM (23) and subjected to STRING protein–protein interaction to obtain a module (Supplementary Fig. S3A) consisting of 12 genes, including CD44, FOSL2, and ANXA1 (Supplementary Table S1). We labeled the genes in this module as a natural evolution signature (NES) and hypothesized that higher NES reflected a longer evolutionary history. As shown in Fig. 2B, the older lesions consist of a higher percentage of tumor cells expressing NES genes than the younger lesions (Wilcoxon paired rank-sum test, P < 0.001). All NES genes showed a higher expression level in the tumor compared with nontumor samples across The Cancer Genome Atlas (TCGA) GBM dataset (Wilcoxon rank-sum test, P < 0.01; Supplementary Fig. S3B). Moreover, we explored the single-cell expression profile consisting of eight GBMs (6,148 cells in total) derived from Yu and colleagues (25). We enumerated the number of NES genes expressed in each cell and found that, on average, the tumor cells expressed more NES genes than the nontumor cells (Wilcoxon paired rank-sum test, P = 0.039; Supplementary Fig. S3C). Notably, the tumor cells accounted for an average of 58% of all cells expressing NES genes, which was 2-fold higher than that of macrophages (23%; Supplementary Fig. S3D), suggesting that the tumor cells were the major source for expressing NES genes. We next used the ssGSEA algorithm (26) to construct a scoring algorithm based on NES genes (see Supplementary Methods). Trajectory analysis (see Supplementary Methods) showed that tumor cells with high NES (hNES) were enriched near trajectory branch points, indicating hNES may be a GBM evolution endpoint (Supplementary Fig. S3E). Functional enrichment analysis using Enrichr (27) showed that these signature genes were associated with pathways including apoptosis, angiogenesis, and hypoxia (Fig. 2C).
Figure 2.
The NES characterizes tumor progression. A, Schematic diagram of the identification of the GBM NES. B, Bar plot demonstrates the percentage of tumor cells expressing the indicated genes between the first and second lesions across four multifocal GBMs. C, Functional enrichment analysis of the 12 NES genes. MSigDB, Molecular Signatures Database. D, Comparison of the percentage of NES-high tumor cells between the indicated groups. E, A bar plot demonstrates the distribution of NES-high tumor cells across the samples after treatment for 2, 5, and 7 weeks. OR, odds ratio. F and G, Hexagonal plots depict different cellular state or subtype signature scores for malignant cells in the first and second lesions. Each data point corresponds to a single cell and is positioned along three axes according to its relative scores for the indicated cellular states. The size of the data point reflects the NES score of the cell. AC, astrocyte-like; CL, classic-like; MES, mesenchymal-like; NPC, neural progenitor–like; PN, proneural-like. H, Survival analysis of IDH wild-type GBMs (tumor purity >0.2) between NES-high and NES-low groups across the indicated datasets. CI, confidence interval.
To evaluate the performance of our algorithm, we first collected two sectors (i.e., P1 and P2) from one patient tumor (TT03) and performed scRNA-seq for each sector (Supplementary Fig. S3F–S3H). We found that the two sectors shared copy-number alterations (CNA; inferred by inferCNV), including amplification of chromosome 7/18q and deletion of chromosome 6p/10p. Furthermore, P2 harbored private CNAs, including amplification of chromosome 3p and deletion of chromosome 13q/19q (Supplementary Fig. S3I), suggesting that P2 was a younger sector compared to P1. By performing the ssGSEA algorithm combined with a permutation analysis, we found that P1 showed a significantly higher percentage (8.8%) of NES-high tumor cells than P2 (3.7%; Fisher exact test, odds ratio = 2.53, P < 0.001; Supplementary Fig. S3J). A similar result was also observed by analyzing the dataset derived from a GBM (i.e., GS5) with two sectors (i.e., P1 and P3), provided by Yu and colleagues (ref. 25; see Supplementary Methods; Supplementary Fig. S3K–S3M). However, for those multisector GBMs without definitive genetic evolutionary order, the NES showed no significant difference between the two sectors (Supplementary Fig. S3N). We next applied the NES algorithm to analyze a dataset derived from patient-derived explants (PDE; ref. 28). We found that the samples from the third generation have a high percentage of NES-high tumor cells (one-tailed Wilcoxon rank-sum test, P = 0.048; Fig. 2D; Supplementary Fig. S4A for dataset of patient-derived GBM organoid). We also explored a dataset of the RCAS-TVA glioma mouse model (GSE76184) and found the percentage of NES-high tumor cells in the sample collected after radiation for 7 weeks was more than 4-fold higher than that after 2 or 5 weeks (odds ratio = 4.6, P < 0.001; Fig. 2E). Finally, we collected two specimens from a patient diagnosed as primary and recurrent GBM (occurred from the primary site) only treated with surgical resection due to the medical treatment limitation. We found 10 of 12 (83.3%) NES genes showed a higher expression in the recurrent sample (permutation, P = 0.030; Supplementary Fig. S4B).
A previous study (29) demonstrated that GBM mainly consists of four cellular states, including neural progenitor–like (NPC), oligodendrocyte progenitor–like (OPC), astrocyte-like (AC), and mesenchymal-like (MES) cells and that the cellular state exhibits plasticity. Therefore, we explored the NES change among the cellular states at the scRNA-seq level in the first and second lesions from NJ01. The gene signatures for each cellular state were derived from Neftel and colleagues (29) and were used to construct a prediction model with the ssGSEA algorithm (26). We focused on tumor cells with at least one significant cellular state (permutation, P < 0.01; see Supplementary Methods). By mapping cells into a hexagonal plot according to cellular state signature scores, we observed a definite cellular state transition along the NPC/OPC–AC–MES axis with a proportional stepwise NES increase (Fig. 2F; Supplementary Fig. S4C), consistent with the transition direction reported by Neftel and colleagues (29). A similar trend was observed in the relationship between the NES and GBM subtypes reported in our previous work [e.g., proneural-like (PN), classic-like (CL), and MES; ref. 23], in which NES showed a stepwise increase and transition direction from the PN-to-MES subtype (Fig. 2G). Analysis of tumor samples from a different independent dataset (e.g., TCGA) also showed that the MES subtype had higher NES scores than CL, which increased relative to the PN subtype (MES vs. CL, P < 0.001; MES vs. PN, P < 0.001; Supplementary Fig. S4D). We found that NES, to some extent, showed an association with MES-like features. However, when performing the hypergeometric distribution analysis (30) on NES- and MES-state genes, we found that there was no significant enrichment association between NES and MES states (NES–MES1: P = 0.38; NES–MES2: P = 1.00), suggesting that NES acts with distinct biological functions from MES states.
We next explored the relationship between NES and clinical outcomes in GBM IDH wild-type patients. We categorized samples with tumor purity >0.2 (predicted by ESTIMATE; ref. 31) from TCGA into low NES (lNES) and hNES groups (classified by median NES, the samples whose NES score was greater than the median NES score were labeled as hNES, otherwise lNES) and compared their overall survival. The lNES group showed longer overall survival than hNES [high vs. low NES, hazard ratio = 1.30; 95% confidence interval (CI), 1.03–1.65; log-rank test, P = 0.027; Fig. 2H; Supplementary Fig. S4E for all IDH wild-type GBMs]. We further validated this result using data from the Chinese Glioma Genome Atlas (CGGA; ref. 32; hazard ratio = 1.38; 95% CI, 1.00–1.92; log-rank test, P = 0.053; Fig. 2H; Supplementary Fig. S4E). Multivariate analysis controlling for age suggested that NES was an independent prognostic factor (based on TCGA dataset, hazard ratio = 1.2; 95% CI, 0.98–2.3; log-rank test, P = 0.071; Supplementary Fig. S4F; Supplementary Table S1).
Hypoxia Involves in NES Transition Potentially by the HIF1A–FOSL2 Axis
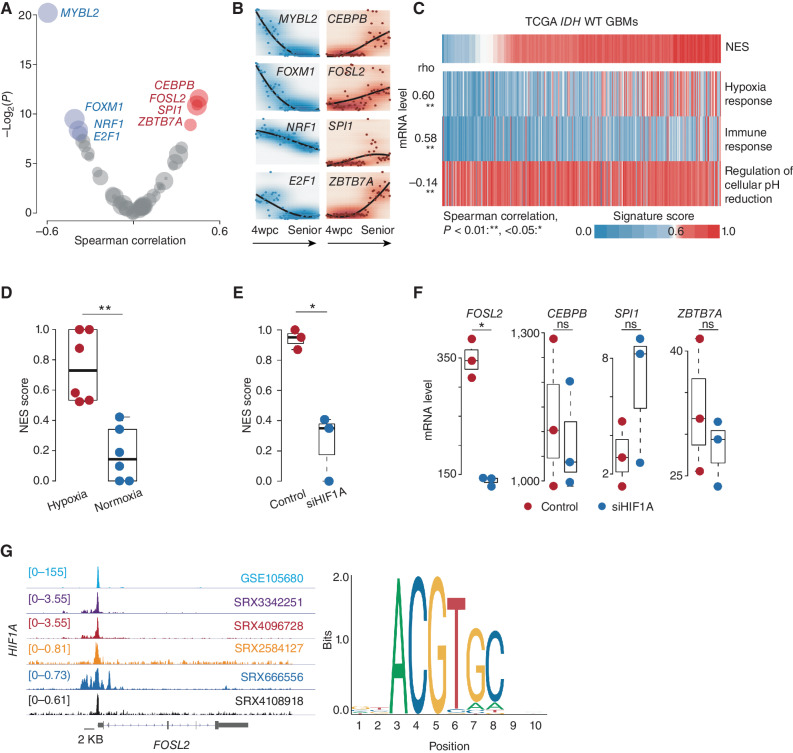
We next investigated the potential transcriptional regulation during NES transition. Using the RABIT algorithm (33) to search for tumor-associated transcription factors (TF) across TCGA IDH wild-type GBMs, we found four TFs whose regulatory activity scores were significantly positively correlated with increased NES (designated as hNES-related), including CEBPB, FOSL2, SPI1, and ZBTB7A (Spearman correlation test, adjusted P < 0.2; Fig. 3A). The NES genes CEBPB and FOSL2 have been reported as critical regulators in promoting tumor progression (34). MYBL2, FOXM1, NRF, and E2F1 TFs were significantly activated in the samples with low NES (Fig. 3A). Among these TFs, both E2F1 and MYBL2 participate in the regulation of cell proliferation and growth (35, 36). These TFs also showed a distinct expression pattern in brain development (Fig. 3B), in which hNES-related TFs were more likely to express in the later developmental stages, which was different from the observation in the development of other tissue types (Supplementary Fig. S5). Additionally, we analyzed GBM single-nuclei sequencing assay for transposase-accessible chromatin (snATAC-seq) and scRNA-seq datasets provided by Wang and colleagues (ref. 37; see Supplementary Methods). By performing motif enrichment analysis for the open regions using Homer (38), we observed that the open chromatin regions associated with TFs such as FOSL2 were prone to enrich in hNES tumor clusters (Supplementary Fig. S6), which was consistent with the findings above.
Figure 3.
Microenvironment remodeling is associated with NES transition. A, Scatter plot of the Spearman correlation between NES score and regulatory activity across TCGA IDH wild-type GBMs. The x-axis and y-axis represent the correlation coefficient (rho) and log-transformed P value, respectively. TFs whose regulatory activity is positively and negatively correlated (adjusted P < 0.2) with NES score (red and blue, respectively). B, mRNA expression of indicated TFs across different stages of brain development from 4 weeks post-conception (wpc) to advanced age. C, Correlation between the NES and indicated pathways across TCGA IDH wild-type (WT) GBMs. Spearman correlation; **, P < 0.01; *, <0.05. D, Comparison of NES scores between tumor cells under hypoxic and normoxic conditions. Wilcoxon rank-sum test; **, P < 0.01; *, <0.05 (P values here also apply to E and F). E, Comparison of NES scores between tumor cells in the indicated groups. F, Comparison of the mRNA level of the indicated gene between tumor cells in the indicated groups. ns, not significant. G, Left, ChIP-seq analysis of HIF1A across various cancer types. The representative Integrative Genomics Viewer tracks at the FOSL2 locus show the distribution of peaks upstream of the transcription start site (<5 KB). Right, binding motif of HIF1A (from JASPAR).
Many studies demonstrated that tumor evolution is orchestrated by various factors, including the microenvironment (39). To identify the potential factors promoting NES state transition, we first tested the correlation between NES and a set of microenvironment-related pathways, including hypoxia response, immune response, and pH reduction [gene set collected from GSEA Molecular Signatures Database (MSigDB); Fig. 3C]. The hypoxia response showed a significant association with NES (Spearman correlation test, rho = 0.60, P < 0.001). By comparing the change of NES between glioma cells cultured with different oxygen levels (i.e., 5% and 20%), we found that tumor cells under hypoxia exhibited a higher NES (20% vs. 5%, Wilcoxon rank-sum test, P = 0.0049; Fig. 3D). Knocking down hypoxia-inducible factor (e.g., HIF1A) on the tumor cells under hypoxia condition showed a significant decrease in NES (P < 0.05; Fig. 3E), indicating that hypoxia played an important role in accelerating the NES transition. Moreover, the tumor cells treated with siHIF1A dramatically reduced the FOSL2 mRNA level (P < 0.05; Fig. 3F). Chromatin immunoprecipitation sequencing (ChIP-seq) analysis based on ChIP-Atlas (https://chip-atlas.org) and ENCODE (https://www.encodeproject.org) showed a sharp HIF1A peak in the upstream region (chr2:28,618,500–28,618,900) of the FOSL2 promoter across various cancer types (Fig. 3G). A similar finding was also observed in our recent study (22), in which FOSL2 binding sites demonstrated increased DNA methylation disorder under stress conditions (e.g., hypoxia), which may be associated with response to stress stimuli. Taken together, these findings suggested that hypoxia may be involved in NES transition potentially through the HIF1A–FOSL2 axis.
Tumor Cells with hNES Associate with BMDM Infiltration
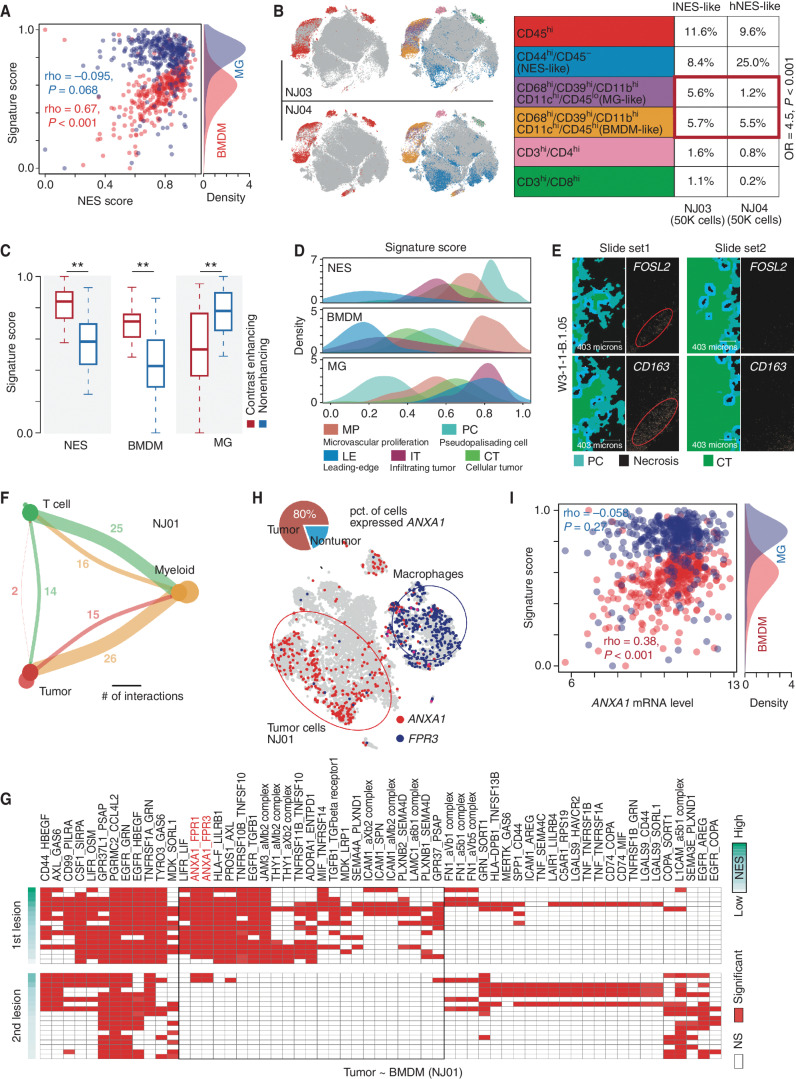
Our single-cell analysis showed that BMDMs were enriched in the older (the first) lesion. To confirm such a finding, we associated NES with the BMDM and MG signatures in TCGA IDH wild-type GBMs using ssGSEA. We found that the BMDM signature (24) was positively correlated with NES (Spearman correlation test, rho = 0.67, P < 0.001), whereas the MG signature showed no correlation with NES (rho = −0.095, P = 0.068; Fig. 4A). Cytometry by time of flight (CyTOF) analysis (see Supplementary Methods and Supplementary Table S2 for the mass cytometry antibodies) based on two GBM biopsies (NJ03 and NJ04) also showed that hNES-like GBM (NJ04, with a high percentage of CD44+CD45− cells) had a higher ratio of number of BMDMs to MGs (Fisher exact test, odds ratio = 4.5, P < 0.001; Fig. 4B).
Figure 4.
Associations between NES and immune microenvironment. A, Correlation between NES score (x-axis) and signature scores (y-axis) of MGs (blue points) and BMDMs (red points) in TCGA IDH wild-type GBMs. The density plot on the right represents the distribution of MG and BMDM signature scores. B, Left, distribution of CD45 overlaid on the 2D t-distributed stochastic neighbor embedding (t-SNE) plot of the GBM sample derived from NJ03 (top) and NJ04 (bottom) patients. Distribution of combination of gene markers overlaid on the 2D t-SNE plot of GBM samples derived from NJ03 (top) and NJ04 (bottom) patients. Right, comparison of percentages of indicated cell types between two samples (NJ03 and NJ04). OR, odds ratio. C, Comparison of indicated signature scores between samples in contrast-enhancing and nonenhancing regions. Wilcoxon rank-sum test; **, P < 0.01. D, The distribution of indicated signatures (top: NES; middle: BMDM; bottom: MG) among different GBM regions. E, Image of in situ hybridization and tumor feature annotation (derived from IVY GAP, W3-1-1-B.1.05) for FOSL2 and CD163 expression among different regions (top: PC; bottom: CT). F, Interaction among different cell types. The width of links represents the number of significant ligand–receptor interactions between the indicated cell types. G, A heat map for ligand–receptor interactions between indicated cell types across the first and second lesions of NJ01. NS, not significant. H, Distribution of the indicated genes (ANXA1 and FPR3) overlaid on the 2D t-SNE plot of the first lesion of NJ01. The pie plot demonstrates the percentage of the indicated cells highly expressing ANXA1. I, Correlation between ANXA1 mRNA expression (x-axis) and MG (blue) and BMDM (red) signature scores across TCGA IDH wild-type GBMs.
To further investigate the potential relationship between BMDM/MG and NES, we analyzed GBM mRNA expression profiles of MRI-localized biopsies (dataset from GSE59612). We observed that BMDMs and NES both showed higher enrichment scores in the contrast-enhancing region (contrast-enhancing vs. nonenhancing, Wilcoxon rank-sum test, P < 0.001; Fig. 4C), suggesting that BMDM and hNES tumor cells were prone to enrich in the core region of tumor tissues. Furthermore, we focused on the analysis of the dataset derived from IVY GAP (ref. 40; see Supplementary Methods). In this dataset, biopsies were collected from various regions of GBMs, including the cellular tumor region (CT), the infiltrating tumor region (IT), the leading-edge region (LE), the microvascular proliferation region (MP), and the pseudopalisading cell region (PC; a region associated with tumor necrosis and located in the center of the tumor). We found that hNES samples were significantly enriched (PC vs. other regions; Wilcoxon rank-sum test, P < 0.001) in the PC region (Fig. 4D). BMDMs and MGs were enriched mainly in the MP and LE regions, respectively (Wilcoxon rank-sum test, P < 0.001; Fig. 4D). Intriguingly, BMDMs and NES both showed a similar enrichment tendency in MP and PC regions (Fig. 4D). Such a finding was partially confirmed by an image of in situ hybridization derived from IVY GAP (W3-1-1-B.1.05), in which FOSL2 and CD163 were observed colocalizing in the PC region (Fig. 4E). Taken together, these findings suggested that BMDM infiltration may associate with hNES tumor cells.
We also integrated and reclustered all the myeloid cells from four cases (eight lesions, 16,051 cells) to further dissect the myeloid subclusters potentially associated with hNES tumor cells. Specifically, we identified 17 subclusters and named these clusters (e.g., mac_CD69; Supplementary Fig. S7A and S7B; Supplementary Methods). We used the ssGSEA algorithm to calculate the M2-like and BMDM-like signature scores for each subcluster. We found that mac_PLIN2, mac_IL32, and mac_FCN1 showed a high proportion of cells significantly enriched with BMDM- and M2-like features. When analyzing the IVY GAP dataset, we found that the BMDM-associated subclusters were prone to be enriched in the MP and PC regions (Supplementary Fig. S7C). Furthermore, we analyzed links between the molecule markers of those BMDM-associated subclusters (e.g., mac_FCN1, mac_IL32, mac_PLIN2, mac_F13A1, mac_CCNB2, and mac_MANF) and NES based on the mRNA profile of TCGA IDH wild-type GBMs. We found that 83% of the markers were positively correlated with the NES score across TCGA IDH wild-type GBMs (Supplementary Fig. S7D).
By exploring cell–cell communication using CellphoneDB (41) based on scRNA-seq data from the second lesion of NJ01 (see Supplementary Methods), we found that the “tumor cell–myeloid–T cell” communication axis was a significant part of the cellular communication network (Fig. 4F; Supplementary Fig. S8A for other cases). Notably, myeloid cells (mainly macrophages) were likely to act as intermediaries between tumor cells and T cells, suggesting an essential role in promoting immune microenvironment remodeling alongside tumor progression. To unravel the potential interaction axis between BMDMs and hNES tumor cells, we analyzed cell–cell communication between tumor clusters and BMDMs based on the ligand–receptor interactions predicted by CellphoneDB (41). In general, the older (the first) lesion showed more ligand–receptor interactions than the younger (the second) lesion (Fig. 4G; Supplementary Fig. S8B for NJ02, TT01, and TT02). Specifically, we identified 59, 45, 35, and 78 pairs of ligand–receptor interactions in NJ01, NJ02, TT01, and TT02, respectively (Supplementary Fig. S8C). Furthermore, we took the intersection among the ligand/receptors mainly on tumor cells in the older lesion (older lesion: ≥50% of tumor clusters; younger lesion: <50%) derived from four samples and observed that only ANXA1 was shared by four samples (Supplementary Fig. S8C). The ANXA1 functions as a ligand in tumor clusters that interact with FPR1/3 receptors on BMDMs, mainly in the first lesion (Fig. 4G and H; Supplementary Fig. S8B and S8D). Such ligand–receptor interactions were also observed in the integration analysis of the other four scRNA-seq GBMs derived from Tiantan Hospital [Supplementary Fig. S8E; Supplementary Fig. S8F and S8G for the t-distributed stochastic neighbor embedding (t-SNE) plot of four integrated GBMs]. Especially, we observed that the frequency of ANXA1–FPR3 interaction in tumor–BMDM communication was increased compared with tumor–MG communication, in which the percentage of BMDMs expressing FPR3 was 5-fold higher than that in MGs (Supplementary Fig. S9A and S9B). A similar expression bias was also observed in nontumor samples (reference) or combined samples (reference coupled with IDH wild-type glioma) from datasets provided by Brain TIME (https://joycelab.shinyapps.io/braintime/; Supplementary Fig. S9C). Finally, by exploring mRNA expression of TCGA IDH wild-type GBMs, we found ANXA1 expression was significantly correlated with the BMDM signature (24) score (Spearman correlation test, rho = 0.38, P < 0.001; Fig. 4I) but not the MG signature score, suggesting that ANXA1 may associate with BMDM infiltration.
ANXA1 Associates with Recruiting and Polarizing BMDMs to Induce Immunosuppression
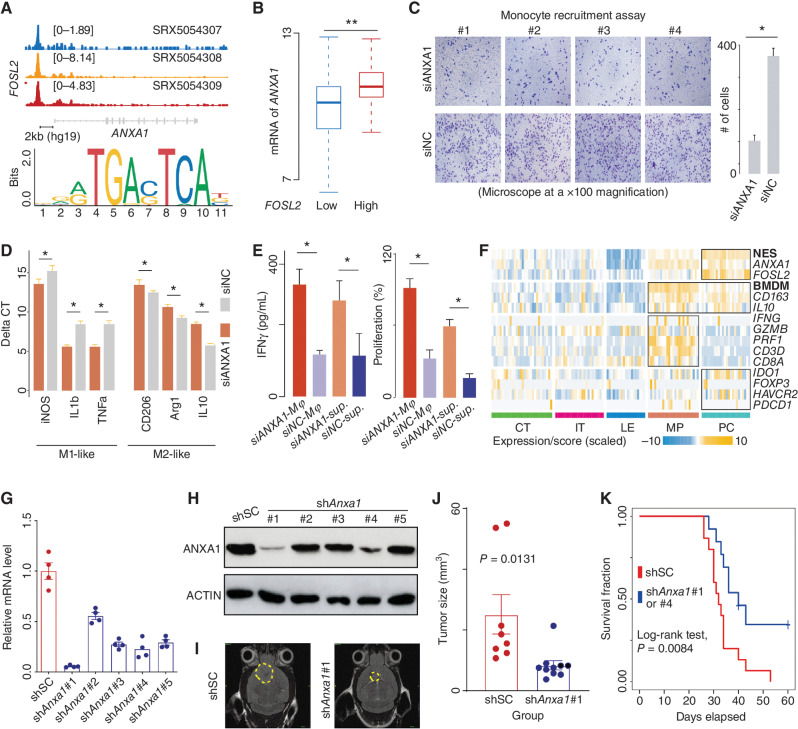
We next searched for potential regulators of ANXA1 using the ChIP-Atlas. We found that the transcript factor FOSL2, one of the NES signatures, showed a sharp peak in the upstream region (chr9:75,764,500–75,764,700) of the ANXA1 promoter across three neural progenitor cells (Fig. 5A). A similar peak was also observed in the upstream region (chr9:75,764,001–75,764,962) of the ANXA1 promoter across GBM cells in an analysis performed on a previously published snATAC-seq dataset (Supplementary Fig. S10A; see Supplementary Methods). The ANXA1 expression was positively correlated with FOSL2 expression across TCGA IDH wild-type GBMs (FOSL2 high vs. low, Wilcoxon rank-sum test, P < 0.001; Fig. 5B).
Figure 5.
ANXA1 is associated with recruiting and polarizing macrophages to suppress CD8+ T cells. A, Top, ChIP-seq analysis of FOSL2 across three neural progenitor cells. The representative Integrative Genomics Viewer (IGV) tracks at the ANXA1 locus show the distribution of peaks upstream of the transcription start site (TSS; <5 KB). Middle, snATAC analysis of GBM cells. The representative IGV tracks at the ANXA1 locus show the location with significant peaks upstream of the TSS (<5 KB). Bottom, binding motif of FOSL2 (from JASPAR). B, Comparison of ANXA1 mRNA expression between TCGA IDH wild-type GBMs classified by the expression of FOSL2. Wilcoxon rank-sum test; **, P < 0.01; *, <0.05. C, Comparison of the migration ability of monocytes between the indicated groups (siANXA1 or siNC) based on transwell assay (microscope at ×100 magnification). D, mRNA level of indicated genes measured by qRT-PCR using the delta-delta Ct method. E, Comparison of IFNγ level (left) and proliferation percentage (right) of CD8+ T cells among indicated groups. F, A heat map for expression of indicated genes and signature scores among different regions. qRT-PCR (G) and immunoblot (H) analysis of Anxa1 mRNA expression in GL261 cells transduced with lentiviral vectors carrying five shRNAs. I, Representative MRI from mice after intracranial injection of GL261 with lentiviral vectors carrying scrambled shRNA or shAnxa1. T2 sequences demonstrate infiltrative tumors in the mouse brain (yellow line). J, Tumor volume was measured by the T2 MRI scan. K, Survival analysis of cases treated with shSC or shAnxa1.
To confirm the influence of ANXA1 derived from tumor cells on monocytes (see Methods), we performed transwell migration assays based on monocytes and U251MG GBM cell lines. In these assays, the tumor cells were treated by knocking down ANXA1 (siANXA1) or not (siNC, control; see Methods; Supplementary Fig. S10B). We observed that the monocytes were more likely to migrate toward tumor cells in the control group (Fig. 5C). After 72 hours, the monocytes in the control group showed characteristics of M2-like macrophages compared with those in the siANXA1 group (Fig. 5D), suggesting that ANXA1 induces monocytes to differentiate into an immunosuppressive state. Therefore, we next compared the activity state of CD8+ T cells cocultured with macrophages derived from the transwell assays (see Methods). Notably, the IFNγ level and proliferation rate of T cells in the control group were significantly lower than those in the other groups (siANXA1 group; Wilcoxon rank-sum test, P < 0.05; see Methods; Fig. 5E). A similar result was observed in the T cells in coculture with macrophages only treated with the supernatant from the transwell assay of the control group (Fig. 5E). These findings were also confirmed in the mRNA expression of IVY GAP, where cytotoxicity-related molecules, such as IFNG, were rarely observed in the NES-enriched regions (Fig. 5F).
Because ANXA1 could profoundly remodel the immune environment of tumor tissue, we next asked whether ANXA1 may help tumor cells take advantage of survival. By comparing mice intracranially injected with GL261 murine GBM cell lines with Anxa1 knockdown versus controls (Fig. 5G and H), we found the mice in the control group had significantly larger tumor sizes than the other group [P = 0.0131; Fig. 5I and J for shAnxa1#1 (short hairpin RNA of ANXA1, or shANXA1); Supplementary Fig. S10C and S10D for shAnxa1#4]. Consistent with this observation, mice in the shAnxa1 group showed a significantly longer median survival time compared with controls (log-rank test, P = 0.0084; Fig. 5K). IHC analysis using the M2-like macrophage marker F4/80/CD163 further showed that M2-like macrophages showed a higher percentage in the control group (shSC) compared with the shAnxa1 group (Student t test, P < 0.05; Supplementary Fig. S10E). Clinical survival analysis based on multiple datasets (e.g., TCGA) also showed that patients with highly expressed ANXA1 showed a shorter survival time than the other patients (hazard ratio = 1.34; 95% CI, 1.12–1.62; log-rank test, P = 0.0014; Supplementary Fig. S10F). Additionally, we explored our mouse transplant model injected with the macrophage-depleting agent clodronate (Supplementary Fig. S10G). We utilized the IVIS Spectrum In Vivo Imaging System to quantify the tumor size (growth) in mice bearing 2-week-old gliomas. Notably, the samples treated with shAnxa1 showed a significantly smaller tumor size than those treated with clodronate alone (P = 0.0016; Supplementary Fig. S10H). Taken together, the findings above suggested that ANXA1 played an important role in suppressing immune activity and promoting tumor progression, which may be a potential target for GBM treatment.
BMDM Infiltration Accelerates Tumor NES Transition and Migration
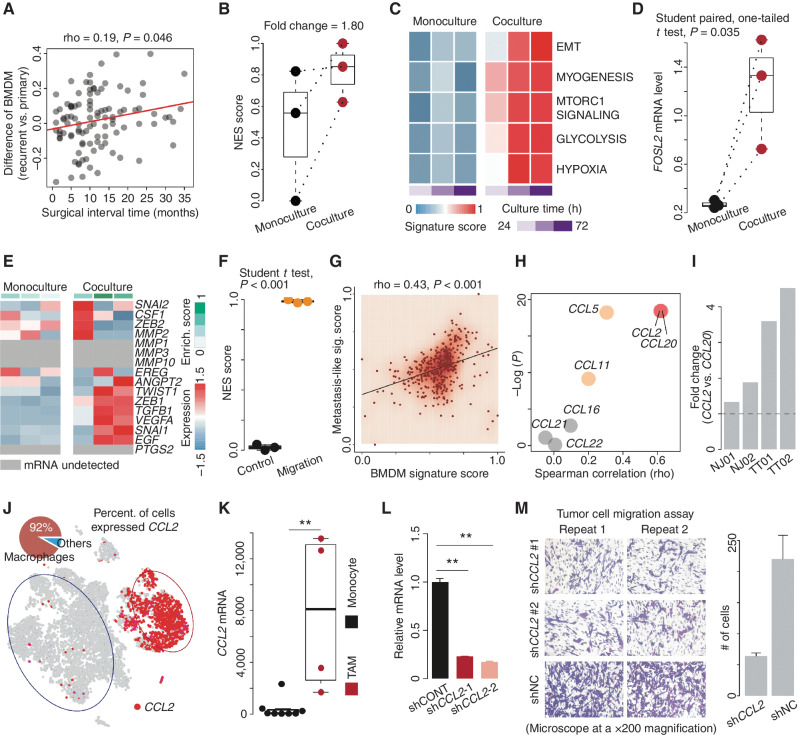
By analyzing the mRNA profile of primary and recurrent GBMs derived from The Glioma Longitudinal AnalySiS (GLASS; 13), we found that the difference in the BMDM-normalized signature score (i.e., BMDM vs. MG signature score calculated by the ssGSEA algorithm) between recurrent and matched primary GBMs was significantly correlated with the increase of surgical interval time (Spearman correlation test, rho = 0.19, P = 0.046; Fig. 6A), which was consistent with the finding reported by Pombo Antunes and colleagues (42), suggesting that BMDM infiltration is associated with tumor progression. Next, we performed RNA-seq on tumor cells cocultured with M2-like macrophages (induced from monocytes) or in monoculture for 24, 48, and 72 hours (see Methods; a total of six samples). Cocultured tumor cells showed a higher NES score than those in the monoculture condition (fold change = 1.80; Fig. 6B), suggesting that BMDMs contribute to accelerating the NES transition. A similar result was further confirmed by comparing the expression profile of GBM stem cells (GSC) derived from the 3D triculture (in culture with astrocytes and neural progenitor cells) and tetra-culture (in culture with astrocytes, neural progenitor cells, and macrophages) GBM tissue models provided by Tang and colleagues (43), in which 75% of NES genes showed a higher expression level in the GSCs cocultured in the tetra-culture model (P = 0.008; Supplementary Fig. S11A). Moreover, enrichment analysis of MSigDB hallmark gene sets via the ssGSEA algorithm showed that the epithelial–mesenchymal transition (EMT) pathway was significantly correlated with NES (Spearman correlation test, rho = 0.94, P = 0.017) and highly upregulated in the coculture condition (one-tailed Student paired t test, P = 0.042), exhibiting a stepwise increase with culture time (Fig. 6C; Supplementary Fig. S11B for 3D culture). It has been reported that FOSL2 is an important TF promoting EMT. The mRNA of FOSL2 showed a higher expression level in tumor cells cocultured with macrophages (one-tailed Student paired t test, P = 0.035; Fig. 6D; Supplementary Fig. S11C). Moreover, the enrichment of EMT also suggested that these tumor cells may acquire invasion capacity after coculture. Previous studies have summarized that tumor metastasis mainly undergoes a cascade, including local invasion and intra-/extravasation, which involves gene sets such as TWIST, ZEB2, COX2, and VEGFs (designated as the metastasis-like gene set; see Supplementary Table S3). Using the ssGSEA algorithm, we found that tumor cells in the coculture condition showed a higher enrichment of these genes (one-tailed Student t test, P = 0.040; Fig. 6E; Supplementary Fig. S11D). Additionally, the analysis based on the data provided by Miyai and colleagues (ref. 44; GSE126725) showed that the diffuse glioma cells with high migration ability showed a higher NES score compared with the control group (Student t test, P < 0.001; Fig. 6F).
Figure 6.
BMDM infiltration is associated with tumor progression. A, Correlation analysis of surgical interval time and the difference in normalized BMDM signature score between primary and recurrent GBMs based on the GLASS dataset. B, Comparison of NES scores between tumor cells in monoculture and coculture with macrophages. C, Comparison of indicated MSigDB hallmark pathways between tumor cells in monoculture and coculture with macrophages for 24, 48, and 72 hours. D, Comparison of the FOSL2 mRNA level between tumor cells in monoculture and coculture with macrophages. E, Comparison of metastasis-related gene expression between tumor cells in monoculture and coculture with macrophages. Top bar colors from white to green represent the enrichment score of metastasis-related genes from low to high. F, Comparison of NES scores between glioma cells with high and low (control) migration ability. Enrich., enrichment. G, Correlation between the NES score and metastasis-like signature (sig.) score across TCGA IDH wild-type GBMs. H, Correlation between the mRNA level of the indicated gene and NES score across TCGA IDH wild-type GBMs. I, The ratio of the number of macrophages expressing CCL2 to that expressing CCL20. J, Distribution of the indicated genes (CCL2 and CCR10) overlaid on the 2D t-SNE plot of the first lesion. The pie plot demonstrates the percentage (percent.) of the macrophages (red) highly expressing CCL2. K, Comparison of the CCL2 mRNA level between TAMs and monocytes. Wilcoxon rank-sum test; **, P < 0.01. L, qRT-PCR analysis of CCL2 mRNA expression in macrophages with lentiviral vectors carrying shRNAs. M, Comparison of migration ability of tumor cells between the indicated groups (shCCL2 or shNC) based on transwell assay (microscope at a ×200 magnification).
We further investigated whether BMDMs interact with GBMs and could promote tumor migration. The analysis based on the TCGA IDH wild-type dataset showed that metastasis-like signatures were significantly correlated with BMDM (Spearman correlation test, rho = 0.43, P < 0.001) and monocyte (rho = 0.40, P < 0.001) signature scores (Fig. 6G for BMDM; Supplementary Fig. S11E for monocyte and MG). An increasing number of studies have demonstrated that TAMs might promote tumor metastasis and progression by secreting cytokines, including CCL2 and CCL20 (45, 46). To further determine which cytokine was associated with such a migration ability during the NES transition, we applied correlation analysis to these cytokines and NES across TCGA IDH wild-type GBMs. Notably, CCL2 (rho = 0.62, P < 0.001) and CCL20 (rho = 0.62, P < 0.001) showed a high correlation with NES score (Fig. 6H). When comparing the percentage of macrophages expressing these two genes across four pairs of multifocal GBMs, we found that CCL2 was prone to express among macrophages broadly (Fig. 6I). Further analysis showed that CCL2 was mainly expressed in macrophages (Fig. 6J; Supplementary Fig. S11F and S11G). By analyzing the data provided by Gabrusiewicz and colleagues (47), we found that CCL2 mRNA levels were higher in tumor-infiltrating macrophages than in monocytes in the periphery (Wilcoxon rank-sum test, P < 0.01; Fig. 6K), suggesting that the upregulation of CCL2 was only the phenotype of the macrophages infiltrating into the tumor. Inspired by these findings, we performed a transwell migration assay in shCCL2 or shNC (control) conditions for 48 hours (Fig. 6L). As expected, the tumor cells in the control group were more likely to migrate toward the lower compartment of the transwell apparatus (where macrophages were; Fig. 6M), confirming that CCL2 could promote tumor cell migration. Moreover, by analyzing TCGA IDH wild-type GBM samples, we found that patients classified as CCL2–high level showed a poor prognosis compared with other patients (hazard ratio = 1.25; 95% CI, 1.08–1.45; log-rank test, P = 0.0033; Supplementary Fig. S11H).
DISCUSSION
Multifocal tumors, especially those lesions that originated from a single ancestor, are an excellent model for investigating natural tumor evolution because the noise of initial genetic alteration and microenvironment are well controlled between different lesions. We characterized GBM evolution through an in-depth analysis of four pairs of multifocal GBMs. We defined an NES to recapitulate molecular changes during untreated tumor evolution. The validity of the NES was corroborated by its significant correlation with tumor development, clinical outcome, and tumor cellular states. These findings collectively reveal that the NES characterized the molecular change of GBM's natural evolution history.
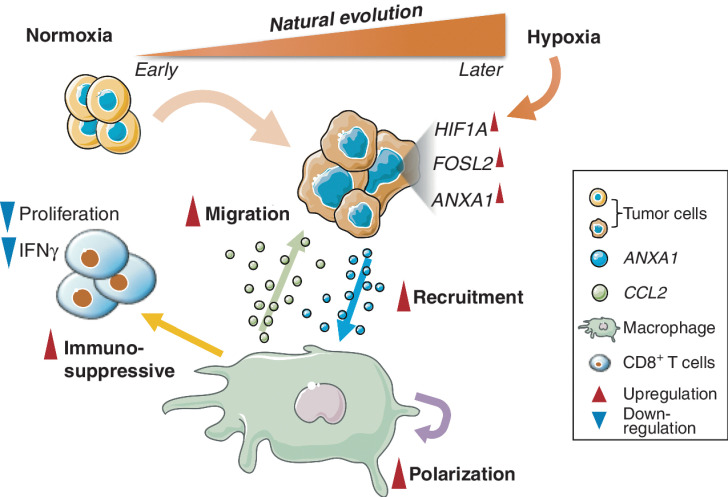
Tumor evolution is considered to be associated with many factors (hypoxia, etc.). Our study integrated scRNA-seq of multifocal GBMs to identify that FOSL2 was one of the signatures highly expressed in the older lesions, suggesting it was potentially involved in tumor evolution. Besides, our study demonstrated that hypoxia could promote NES transition potentially by activating the HIF1A–FOSL2 axis (Fig. 7). FOSL2 regulated ANXA1, which may subsequently recruit and induce monocyte polarization to M2-like macrophages (Fig. 7). These polarized BMDMs showed an immunosuppression phenotype that significantly reduces the proliferation and IFNγ production of CD8+ T cells, posing a challenge to developing immunotherapy for GBM. A similar study was recently reported by Yeo and colleagues (48), in which BMDMs were observed with an increased proportion at the advanced stage of GBMs. Intriguingly, we found that knocking down ANXA1 significantly decreased the suppression effect of macrophages on CD8+ T cells, and tumor progression in GL261-derived syngeneic glioma mouse models, implying that ANXA1 could be a potential target for GBM treatment. Taken together, the “HIF1A–FOSL2–ANXA1” regulatory axis offers us a new perspective to understand natural tumor evolution and its association with immune microenvironment remodeling.
Figure 7.
Schematic representation of tumor natural evolution and interaction with macrophages.
Our findings also show that tumor cells cocultured with the polarized macrophages tend to increase NES, which may be associated with the activation of multiple transcript factors (e.g., FOSL2) and pathways (e.g., EMT). Such progression was also coupled with an enhancement of migration capacity, which is associated with the CCL2 produced by the polarized macrophages (Fig. 7). These findings may together help explain our previous observation, in which macrophage enrichment was significantly correlated with rapid relapse of GBM after treatment (23). Because CCL2 is mainly expressed in macrophages that infiltrate tumor tissues, targeting CCL2 might be a potential therapy for slowing tumor progression.
Increasing evidence has now emerged indicating that GBM does not follow a unidirectional hierarchical stem cell model (49). Our study revealed that the NES-high tumor cells tended to express stem- and differentiating-like signatures simultaneously. This phenotype suggests that NES is associated with high plasticity, which could help tumor cells respond to different stressors in the microenvironment.
While introducing the natural evolution of GBMs, we recognize that the current study has not fully established the causality between the NES transition and microenvironment remodeling. This is limited by the technical challenges to precisely capturing the full stages of GBM evolution. Therefore, additional studies are needed to explore the relationship between tumor cells and the microenvironment during tumor evolution. In addition, this study focused only on the influence of the ligand on the target cells (e.g., ANXA1) rather than cell receptors (e.g., FPR1/3). This is mainly because these ligands showed abnormal expression levels in tumors compared with those receptors, which may be more suitable for targeting. It is also worth pointing out that the mechanism of the NES transition of tumor cells under coculture with macrophages is unclear. The coculture assay based on the cytokine-polarized (e.g., IL4) macrophages is oversimplistic and could not completely reflect the features of macrophages in vivo. Further research should be conducted to systematically evaluate the influence of TAMs on the tumor cells.
In conclusion, our study provides an unprecedented high-resolution characterization of natural tumor evolution followed by immune environment remodeling and tumor migration. The frequent interaction between tumor cells and macrophages may enhance our understanding of GBM progression.
METHODS
Human Tumor and Peripheral Blood Specimens
The scRNA-seq data were derived from 12 samples, including four pairs of multifocal GBMs and four other GBMs. In detail, all patients at the Neurosurgery Department of The First Affiliated Hospital of Nanjing Medical University and the Beijing Tiantan Hospital preoperatively consented to participate in the study. All tumor specimens were collected after written informed consent was obtained from the patients and in accordance with the institutional review board–approved protocols of The First Affiliated Hospital of Nanjing Medical University or Beijing Tiantan Hospital, Capital Medical University. Two pairs of fresh brain tissue samples (NJ01 and NJ02) were preserved using tumor storage solution (MACS, 130-100-008) immediately after surgical resection. The tumors were located in the frontal and parietal lobes. At the same time, paired peripheral blood samples were collected into EDTA anticoagulant tubes. Nine fresh brain tissue samples from six patients (two pairs of multifocal samples, TT01 and TT02; one pair of multisector samples, TT03; and four GBMs) at the Beijing Tiantan Hospital were collected and snap-frozen in liquid nitrogen immediately after surgical resection.
The fresh brain tissue samples were washed with prechilled phosphate-buffered saline (PBS; BBI Life Sciences, E607008-0500). Tissue digestion was performed with a Human Tumor Dissociation Kit (MACS, 130-095-929), and the tissue slurry was passed through a 70-μm cell strainer. Red blood cell lysis and debris removal were performed after filtering the single-cell suspension (MACS, 130-094-183 and 130-109-398). Then, the samples were washed and resuspended in PBS with 0.04% bovine serum albumin (BSA; BBI Life Sciences, A600332-0005). Finally, the cell suspensions were filtered using a 40-μm cell strainer.
Single-Cell Library Preparation and Sequencing
Cells from the samples collected from The First Affiliated Hospital of Nanjing Medical University were counted using a Countess instrument (Thermo) and diluted to 700 to 1,200 cells/μL. Cells were processed according to the Chromium single-cell RNA 3′ kit protocol (10X Genomics V2.0 chemistry) to capture 10,000 cells/chip position. All the remaining procedures, including library construction, were performed according to the manufacturer's protocol. Libraries were quantified using an Agilent high-sensitivity DNA chip on a Bioanalyzer 2100 system (Agilent). Single-cell libraries were then sequenced on an Illumina HiSeqXTen instrument (PE150).
The cells collected from Beijing Tiantan Hospital were loaded into Chromium microfluidic chips with 3′ (10X Genomics V2.0) chemistry and barcoded with a 10X Chromium Controller (10X Genomics). RNA from the barcoded cells was subsequently reverse-transcribed, and sequencing libraries were constructed with a Chromium single-cell RNA 3′ v2 reagent kit (10X Genomics). The libraries were sequenced with an Illumina HiSeq 2000 (Illumina).
The single-cell sequencing data were analyzed using Cell Ranger (version 2.2.0, 10X Genomics) with default parameters. The data quality was assessed using the Seurat package (version 2.3.4).
Whole-Exome Sequencing
Single-cell suspensions and paired peripheral blood of NJ01 and NJ02 were processed for whole-exome sequencing. Massively Parallel Sequencing Exome capture was performed using an Agilent SureSelect Human All Exon V6 kit. All exome–genome sequencing was performed on an Illumina NovaSeq 6000 platform. The sequencing data were aligned to the human reference genome build hg19 with bwa (v0.7.15). Aligned reads were sorted by genome coordinate using Picard Tools (v2.18.12) after marking duplicate reads. Alignment quality was further improved by local realignment around insertions and deletions and base quality score recalibration. Somatic single-nucleotide variants were identified using Mutect (v.1.1.17). Then, variants in dbSNP build 138 or whose mutation allele frequency (MAF) was >0.7 were filtered. Allelic copy numbers in exome sequencing data were estimated using Sequenza with the default options. MAF was adjusted by tumor purity estimated from scRNA-seq (NJ01: first lesion: 0.7, second lesion: 0.9; NJ02: first lesion: 0.6, second lesion: 0.8). The scaled MAF was calculated by integration analysis of MAFs from two lesions.
Sample Collection and Processing for CyTOF
Tumor specimens were obtained from patients (NJ03 and NJ04) diagnosed with GBM at the Department of Neurosurgery of The First Affiliated Hospital of Nanjing Medical University. The fresh tumor tissue was mechanically cut into small pieces and enzymatically digested with RPMI 1640 containing 2 mg/mL collagenase, 250 μg/mL hyaluronase, and 20 μg/mL DNase I.
Antibodies were purchased, purified, and conjugated in-house using MaxPar X8 Polymer kits (Fluidigm). The mass cytometry antibodies used are shown in Supplementary Table S2. First, 1–3 × 106 cells from each sample were washed with protein-free PBS, stained with 1 μmol/L cisplatin for 5 minutes at room temperature, and stained for cell-surface markers in staining media (PBS containing 0.5% BSA and 0.02% NaN3) for 30 minutes at 4°C. Then, cells were fixed and stained with DNA Intercalator-Ir overnight. Finally, cells were stained with a Foxp3/TF staining buffer (eBioscience) for 30 minutes at 4°C and washed. Cells were stored at 4°C until analysis.
Cells were washed twice with deionized water before adding EQ normalization beads containing Ce140, Eu151, Eu153, Ho165, and Lu175 (Fluidigm). Data were acquired with a Helios instrument. After normalizing and randomizing values near zero using Helios software, FCS files were obtained. Data generated from different batches were normalized using the bead normalization method. To get accurate immune subset information, we analyzed all samples with the FLOWSOM algorithm. All cell events in each sample were pooled and included in this analysis. t-SNE visualization was based on 50,000 CD45+ cells randomly selected from all cells.
Transwell Coculture Assay for Examining Macrophage Migration and Polarization
According to the manufacturer's instructions, peripheral blood (20 mL) was collected from healthy individuals, and peripheral blood mononuclear cells were extracted using the Ficoll separation method (GE Healthcare Life Sciences). Mononuclear cells were obtained by CD14 magnetic bead separation according to the manufacturer's instructions (Life Technologies) and cultured in 20% FBS RPMI 1640 medium (GM-CSF; 50 ng/mL, PeproTech) for 5 days to induce M0-like macrophages. T cells were obtained by CD3 magnetic bead separation according to the manufacturer's instructions (Life Technologies) and cultured in 10% FBS RPMI 1640 medium (IL2; 10 ng/mL, PeproTech).
Human glioma cells U251MG (RRID:CVCL_0021) were obtained from the Cell Bank of Shanghai Institutes of the Chinese Academy of Sciences and cultured in a 10% FBS DMEM high-glucose medium. siRNA (e.g., siANXA1; Supplementary Table S4) was transfected with Lipofectamine RNAIMAX transfection reagent according to the manufacturer's instructions (Life Technologies). After 48 hours, the cells were collected for protein extraction. The interference efficiency was detected by Western blotting.
Transwell cells (Millipore) were placed in a 24-well plate. A total of 105 U251MG cells (RRID: CVCL_0021) with different treatments (siANXA1 or siNC) were added to the lower chamber and 500 μL of 10% FBS DMEM high-glucose medium. M0 macrophages (100 μL) were added to the upper compartment and 10% FBS RPMI 1640 medium. After 48 to 72 hours of coculture, the upper chamber was fixed with methanol, and the cells inside the membrane were wiped off with a cotton swab (Beyotime). The cells outside the membrane were stained with crystal violet (Beyotime) and photographed under a microscope.
To examine the phenotype of macrophages under different culture conditions, total RNA was extracted from the macrophages using the TRIzol method according to the manufacturer's instructions (Takara) and then subjected to reverse transcription [HiScript II First-Strand cDNA Synthesis Kit (+ gDNA wiper), Vazyme] and an SYBR Real-Time PCR assay (AceQ qPCR SYBR Green Master Mix, Vazyme; Supplementary Table S5 for primers).
Examination of Immune Activity of T Cells
To examine the influence of cocultured macrophages on the activity of T cells, 104 T cells and 1 mL of 10% FBS RPMI 1640 medium were added to the bottom of 12-well plates coated with 1 μg/mL anti-CD3 and 5 μg/mL soluble anti-CD28 (BioLegend). After 48 hours, the macrophages or the supernatant from the coculture in the above transwell experiment was also added. After 48 hours of culture, 20 ng/mL PMA and 1 M ionomycin (Beyotime) were added. After 72 hours of culture, the supernatant was collected and tested using the IFNR assay kit (see kit instructions for details). T cells in the ELISA experiment were collected, and 103 T cells were added to each well of the 96-well plate, followed by 100 μL of 10% FBS RPMI 1640 medium and 10 μL CCK8 solution (CCK8 kit, Beyotime). The culture was continued for 2 hours, and the absorbance was measured at 450 nm.
Intracranial Xenograft Tumor Models and MRI
All mouse experiments were performed under the guidelines of and were approved by the Animal Care and Use Committee of Nanjing Medical University. Female and male C57BL/6J mice (RRID:IMSR_JAX:000664; 6–8 weeks of age) were purchased from The Jackson Laboratory. The intracranial xenograft tumor models were established as previously described (50), and 1 × 105 cells in 5 μL PBS of the murine GBM cell line GL261 (RRID:CVCL_Y003) with shRNA scramble control (shSC) or ANXA1 knockdown (shANXA1) were injected. To make the GL261 lines (RRID:CVCL_Y003) bearing shSC or shANXA1, lentivirus containing respective sequences were used to infect the cells that were subjected to puromycin selection for 1 week, followed by confirmation with real-time PCR (ANXA1 primer: Qiagen, cat. # QT00145915) and western blotting (Anti-Annexin A1 Ab, Abcam, cat. # ab214484). For lentivirus production, the pLKO.1 target gene set for the mouse ANXA1 gene was purchased from Sigma-Aldrich. The packaging system from Invitrogen including plasmids pCMVR8.74 (RRID:Addgene_22036), pMD2.G (RRID:Addgene_12259), and pRSV-Rev (RRID:Addgene_12253) was used to generate lentiviruses in HEK293T cells (KCB, cat. # KCB 200744YJ, RRID:CVCL_0063).
The MRI of mice was performed at Rangos Research Center Animal Imaging Core at Children's Hospital of UPMC of Pittsburgh. The tumor size of animals from MRI imaging was analyzed with ITK-SNAP.
Macrophage Depletion
Lentiviral model-derived mouse glioblastoma cells (GL261; RRID:CVCL_Y003; 2 × 105 cells) were suspended in 5 μL PBS and stereotactically injected into the right striatum of C57 female animals (6 weeks). The mice were randomly separated into two groups for macrophage depletion. Two thousand microliters of liposome–clodronate (5 mg/mL) or liposome–PBS were given to the animals intravenously on day 4, followed by treatment with a reduced dosing of 100 μL per mouse on days 11 and 18. Tumor growth was monitored by bioluminescence imaging on an IVIS Spectrum In Vivo Imaging System (PerkinElmer) weekly and quantified with Live Image v.4.0 software (Living Image; PerkinElmer).
Transwell Coculture Assay for Examining Tumor Cell Migration
The shCCL2 sequence was obtained from Sigma-Aldrich (Supplementary Table S6). Two packaging vector plasmids, VSV-G (RRID:Addgene_138479) and psPAX2 (RRID:Addgene_12260), were added to each test tube (concentration × volume, concentration: ng/μL, mass: 5 μg, 5:5, or other ratios), and mixed with 1 mL HBS and 70 μL CaCl2. After 10 to 15 minutes, DMEM was changed to make HEK293T cells (KCB, cat. # KCB 200744YJ, RRID:CVCL_0063) fully nourished. The reagent was mixed 3 to 5 times and absorbed away. After 12 to 16 hours, HEK293T cells (KCB, cat. # KCB 200744YJ, RRID:CVCL_0063) were added to 10 mL of neurobasal medium for 48 hours. Lentiviruses were collected after the liquid exchange, and titers were measured. The lentiviruses were stored at −80°C.
The prepared macrophages were cultured with 2 mL of Accutase and incubated for 3 to 5 minutes, and then 3 mL of medium was added. All the supernatants were absorbed for centrifugation. The supernatant was removed, and 3 mL of the medium was added for resuspension. A total of 106 cells were counted in each dish and cultured with 3 mL of medium. The lentiviruses were incubated at 37°C and centrifuged at 3,000 rpm for 5 minutes. Then, 4 to 5 mL of the virus supernatant was added to the macrophages. The medium was changed after 12 hours of infection. After 48 hours, 1 μg/mL of puromycin was added to screen the cells. The remaining cells were collected after 2 days.
The purine-screened macrophages were digested, counted with a counting plate adjusted to 2 × 104 cells, and placed into a transwell chamber. Media (100–200 μL) without FBS were added to the upper chamber. Media (600 μL) containing 10% FBS were added to the bottom of the transwell chamber. The transwell chamber was then placed in an incubator at 37°C and 5% CO2 for 48 hours. The upper chamber was fixed with methanol, and the cells inside the membrane were wiped off with a cotton swab. The cells outside the membrane were stained with crystal violet (Beyotime) and photographed under a microscope.
RNA-seq of Tumor Cells in Coculture or Monoculture Condition
The normal peripheral blood lymphocytes were obtained from healthy donors. In brief, peripheral blood mononuclear cells were separated by Ficoll-Hypaque (Sigma-Aldrich) density gradient centrifugation. Monocytes were isolated using anti-CD14–coated microbeads (Miltenyi Biotec) and cultured for 7 to 10 days in RPMI 1640 medium supplemented with 10% FBS, 1% P/S, 4 mmol/L L-glutamine, and 50 U/mL GM-CSF (R&D Systems). Following differentiation, the cells were treated with 20 ng/mL IL4 (Sigma-Aldrich) for 2 days, resulting in M2-like TAMs. The U251MG cells (RRID:CVCL_0021) were cultured in a serum-free medium containing DMEM/F12 medium (Gibco) supplemented with 20 ng/mL EGF (Gibco), 20 ng/mL bFGF (Gibco), 2% B27 (Gibco), 1 × N2 supplement (Gibco), 1 × MEM NEAA (Gibco), and 1:100 antibiotic–antimycotic (Gibco). For coculturing experiments, coculture inserts (0.4-μm pores; BD Biosciences) were placed in 6-well tissue culture plates. The TAMs were added to the tissue culture wells (1 × 106 cells per well), and U251MG cells (RRID:CVCL_0021; 1 × 105) were added to the inserts. The two types of cells in the transwell chambers were cocultured for 24, 48, and 72 hours. The U251MG cells (RRID:CVCL_0021) and TAMs were harvested at each time point and washed with cold PBS for RNA-seq. Total RNA was extracted using TRIzol Reagent (Invitrogen) and reverse-transcribed using HiScript II Q RT SuperMix (Vazyme). RT-qPCR was performed using the SYBR Green Master Mix (Vazyme). A total of 500 ng of total RNA per group were selected to build the cDNA library using the Smart-seq2 protocol. RNA-seq was performed using a PE150 strategy (HiSeq X Ten, Illumina). The RNA-seq data were subjected to mRNA analysis (https://docs.gdc.cancer.gov) for mRNA quantification.
Data and Code Availability
scRNA-seq, whole-exome sequencing, and bulk RNA-seq data supporting this study are available from the China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn; GSA: HRA002914 and HRA002913). Custom codes used for this study are accessible at https://github.com/woolingxiang/NES.
Supplementary Material
Acknowledgments
The authors thank Zhe Lin for assistance with the transwell assay analysis of examining macrophage migration and polarization. This research is supported by grants from the National Natural Science Foundation of China (91959113, 81972358), Key Research and Development Program of Jiangsu Province (BE2017733), Basic Research Program of Jiangsu Province (BK20180036), Jiangsu Province's Science and Technology Foundation (BE2018724), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
R.G.W. Verhaak reports personal fees from NeuroTrials, Inc. and Stellanova, Inc. outside the submitted work. Q. Wang reports nonfinancial support from Oncocare Life Technology (Suzhou) Co., Ltd. during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
L. Wu: Investigation, writing–original draft, project administration. W. Wu: Investigation, writing–original draft. J. Zhang: Data curation. Z. Zhao: Investigation, visualization. L. Li: Visualization, methodology. M. Zhu: Visualization, methodology. M. Wu: Methodology. F. Wu: Validation, visualization. F. Zhou: Validation, visualization. Y. Du: Validation, visualization. R.-C. Chai: Validation, visualization. W. Zhang: Data curation. X. Qiu: Data curation. Q. Liu: Validation. Z. Wang: Formal analysis, visualization. J. Li: Software. K. Li: Formal analysis, methodology. A. Chen: Validation. Y. Jiang: Validation. X. Xiao: Validation. H. Zou: Validation. R. Srivastava: Validation. T. Zhang: Visualization, methodology. Y. Cai: Formal analysis, methodology. Y. Liang: Software. B. Huang: Visualization, methodology. R. Zhang: Validation. F. Lin: Validation. L. Hu: Validation. X. Wang: Validation. X. Qian: Validation. S. Lv: Validation, methodology. B. Hu: Validation. S. Zheng: Methodology. Z. Hu: Resources. H. Shen: Resources. Y. You: Conceptualization, resources. R.G.W. Verhaak: Conceptualization, resources, writing–review and editing. T. Jiang: Conceptualization, resources, writing–review and editing. Q. Wang: Conceptualization, resources, supervision, writing–review and editing.
References
- 1. Gatenby RA, Brown JS. Integrating evolutionary dynamics into cancer therapy. Nat Rev Clin Oncol 2020;17:675–86. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. [DOI] [PubMed] [Google Scholar]
- 3. Jiang T, Nam DH, Ram Z, Poon WS, Wang J, Boldbaatar D, et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 2021;499:60–72. [DOI] [PubMed] [Google Scholar]
- 4. Mallick S, Benson R, Hakim A, Rath GK. Management of glioblastoma after recurrence: a changing paradigm. J Egypt Natl Canc Inst 2016;28:199–210. [DOI] [PubMed] [Google Scholar]
- 5. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oiseth SJ, Aziz MS. Cancer immunotherapy: a brief review of the history, possibilities, and challenges ahead. J Cancer Metastasis Treat 2017;3:250–61. [Google Scholar]
- 7. McGranahan T, Therkelsen KE, Ahmad S, Nagpal S. Current state of immunotherapy for treatment of glioblastoma. Curr Treat Options Oncol 2019;20:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol 2012;2012:948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 2014;26:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 2015;28:318–28. [DOI] [PubMed] [Google Scholar]
- 12. Wang JG, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48:768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019;576:112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res 2015;25:316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578: 122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McDonald TO, Chakrabarti S, Michor F. Currently available bulk sequencing data do not necessarily support a model of neutral tumor evolution. Nat Genet 2018;50:1620–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abou-El-Ardat K, Seifert M, Becker K, Eisenreich S, Lehmann M, Hackmann K, et al. Comprehensive molecular characterization of multifocal glioblastoma proves its monoclonal origin and reveals novel insights into clonal evolution and heterogeneity of glioblastomas. Neuro Oncol 2017;19:546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JK, Wang J, Sa JK, Ladewig E, Lee HO, Lee IH, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 2017;49:594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tickle T, GC TI, Brown M, Haas B. inferCNV of the Trinity CTAT Project [software]. Klarman Cell Observatory, Broad Institute of MIT and Harvard. 2019. Available from:https://github.com/broadinstitute/infercnv. [Google Scholar]
- 20. Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson KC, Anderson KJ, Courtois ET, Gujar AD, Barthel FP, Varn FS, et al. Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat Genet 2021;53:1456–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017;32:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Müller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol 2017;18:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu K, Hu Y, Wu F, Guo Q, Qian Z, Hu W, et al. Surveying brain tumor heterogeneity by single-cell RNA-sequencing of multi-sector biopsies. Natl Sci Rev 2020;7:1306–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–U22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with enrichr. Curr Protoc 2021; 1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LeBlanc VG, Trinh DL, Aslanpour S, Hughes M, Livingstone D, Jin D, et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell 2022;40:379–92. [DOI] [PubMed] [Google Scholar]
- 29. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, et al. Chinese Glioma Genome Atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics 2021;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang P, Freedman ML, Liu JS, Liu XS. Inference of transcriptional regulation in cancers. Proc Natl Acad Sci U S A 2015;112:7731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 2011;25:2594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 2017;8:e2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle 2009;8:532–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang L, Babikir H, Muller S, Yagnik G, Shamardani K, Catalan F, et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov 2019;9:1708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol 2017;27:863–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon JG, et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018;360:660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 42. Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci 2021;24:595–610. [DOI] [PubMed] [Google Scholar]
- 43. Tang M, Xie Q, Gimple RC, Zhong Z, Tam T, Tian J, et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res 2020;30:833–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miyai M, Kanayama T, Hyodo F, Kinoshita T, Ishihara T, Okada H, et al. Glucose transporter Glut1 controls diffuse invasion phenotype with perineuronal satellitosis in diffuse glioma microenvironment. Neurooncol Adv 2021;3:vdaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou J, Tang Z, Gao S, Li C, Feng Y, Zhou X. Tumor-associated macrophages: recent insights and therapies. Front Oncol 2020;10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016;1:e85841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol 2022;23:971–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun 2019;10:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen A, Jiang Y, Li Z, Wu L, Santiago U, Zou H, et al. Chitinase-3-like 1 protein complexes modulate macrophage-mediated immune suppression in glioblastoma. J Clin Invest 2021;131:e147552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq, whole-exome sequencing, and bulk RNA-seq data supporting this study are available from the China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn; GSA: HRA002914 and HRA002913). Custom codes used for this study are accessible at https://github.com/woolingxiang/NES.