The authors show that subcapsular sinus macrophages promote aggressiveness of metastatic melanoma cells by releasing IL1α, which upregulates STAT3 signaling in the tumor. Blocking IL1α could synergize with STAT3 inhibitors in treating metastatic melanoma.
Abstract
During melanoma metastasis, tumor cells originating in the skin migrate via lymphatic vessels to the sentinel lymph node (sLN). This process facilitates tumor cell spread across the body. Here, we characterized the innate inflammatory response to melanoma in the metastatic microenvironment of the sLN. We found that macrophages located in the subcapsular sinus (SS) produced protumoral IL1α after recognition of tumoral antigens. Moreover, we confirmed that the elimination of LN macrophages or the administration of an IL1α-specific blocking antibody reduced metastatic spread. To understand the mechanism of action of IL1α in the context of the sLN microenvironment, we applied single-cell RNA sequencing to microdissected metastases obtained from animals treated with the IL1α-specific blocking antibody. Among the different pathways affected, we identified STAT3 as one of the main targets of IL1α signaling in metastatic tumor cells. Moreover, we found that the antitumoral effect of the anti-IL1α was not mediated by lymphocytes because Il1r1 knockout mice did not show significant differences in metastasis growth. Finally, we found a synergistic antimetastatic effect of the combination of IL1α blockade and STAT3 inhibition with stattic, highlighting a new immunotherapy approach to preventing melanoma metastasis.
Introduction
Melanoma is the most lethal form of skin cancer and a serious threat to public health. In recent years, the incidence of this type of cancer has progressively increased, and it is currently one of the most common malignancies in both adult and young individuals (1). During melanoma progression, malignant cells in the skin acquire additional genetic mutations that direct them toward the lymphatic vessels, which serve as a transportation system (2). Once in the lymphatics, the metastatic cells initiate an active migration that leads them toward the sentinel lymph node (sLN; ref. 3). The presence of melanoma metastasis in this organ is indicative of a poor prognosis and drastically decreases the survival rate of patients (4).
Upon breaching the LN capsule, metastatic cells access the LN sinuses via the afferent lymphatics, following chemokine gradients generated by lymphatic endothelial cells (5). The invasion of sLNs initiates in the subcapsular sinus (SS) area (6) and progressively spreads toward the inner structures (7). This process facilitates the access of the metastatic cells to the bloodstream via high-endothelial venules and their consequent spread to other organs (8, 9).
The LN sinuses are populated by resident phagocytic cells, including three distinct macrophage subsets termed SS macrophages (SSM), medullary macrophages (MM), and medullary cord macrophages (MCM), according to the area in which they reside (10). Strategically positioned along the SS area, SSMs are the first immune cells to encounter lymph-transported antigens and pathogens, preventing their systemic dissemination (11). They play a critical role in the initiation of immune responses against immune complexes and viruses (12–14), as well as in the promotion of humoral immunity (15, 16). Despite the role of macrophages against infectious pathogens having been largely demonstrated, their involvement in the response against tumors remains somewhat controversial (17–20). This issue is mainly due to the ability of these cells to activate either anti- or protumoral responses, which allow them to be traditionally classified as M1 or M2 macrophages (21). For instance, some authors have described a protective function of SSMs associated with the capture of cell-derived antigens originating from dying tumor cells (18) and their cross-presentation to CD8+ T cells (17). Tacconi and colleagues have reported a protective role of CD169+ LN macrophages in breast cancer metastasis that was dependent on the presence of B cells (20). Conversely, other studies have revealed a protumoral effect of these cells, mainly linked with their capacity to trigger and maintain the inflammatory response both in peripheral and lymphoid tissues (6, 22, 23).
The inflammatory response plays a fundamental role in the behavior of cancer cells. Some cancers, including melanoma, are able to grow in chronically inflamed conditions and take advantage of inflammation (24). One of the mechanisms by which innate inflammation supports tumor growth is associated with the IL1 family of cytokines (25). IL1β, the best-characterized molecule of this family, promotes the growth of several tumor types, mainly by mediating immune suppression and activating endothelial cells (26, 27), and evidence suggests that blocking IL1R signaling might prolong the survival time of patients with different tumors (28–32). In addition, IL1β antagonism can synergize with immune-checkpoint inhibitor therapy (27). However, the inflammatory mechanisms responsible for tumor promotion might vary between the primary tumor and the metastatic areas, including the sLN (33). Understanding these differences will influence the design of specific immunotherapies intended to control tumor dissemination in both locations (34) and in different types of tumors, including melanoma (35).
In this study, we characterized the innate immune response of the sLN to melanoma metastasis invasion. Furthermore, we identified a mechanism that associates the inflammatory reaction, initiated by SSMs, with the progression of the metastatic melanoma cells. These results will affect the generation of new therapies and have the potential to improve the efficiency of current immunotherapies against metastatic melanoma by modifying the protumoral role of the SSMs.
Materials and Methods
Cell culture and lentiviral transduction
B16-F1, B16-F10, and A375 cell lines were provided, respectively, by F. Grassi [Institute for Research in Biomedicine (IRB), Bellinzona], G. Guarda (IRB, Bellinzona), and C. Catapano [Institute of Oncology Research (IOR), Bellinzona] in 2018, 2017, and 2020. E0771 cell lines were acquired from Ch3 BioSystems in 2018. None of the cell lines were reauthenticated in the past year. All cell lines were expanded for 3–4 passages before freezing several aliquots. At need, aliquots were thawed and used for in vivo or in vitro experiments within 5 passages from thawing. All cells were maintained in a complete RPMI medium composed of RPMI-1640 (Gibco, 31870-025), 1% Hepes (Gibco, 15630-056), 10% heat-inactivated FBS (Gibco, 10270-106), 1% glutamax (Gibco, 35050-038), 1% sodium pyruvate (Gibco, 11360–039), 1% nonessential amino acids (Gibco, 11140-035), 50 units/mL penicillin + 50 μg/mL streptomycin (Gibco, 15070-063), and 50 μmol/L β-mercaptoethanol (Gibco, 31350-010). Cells were regularly tested for Mycoplasma infection (MycoAlert Mycoplasma Detection kit, Lonza, LT07-418). B16-F1–mCherry and B16-F1–Azurite cell lines were generated by lentiviral transduction. Briefly, lentiviral plasmids pSicoR-Ef1a-mCh (Addgene, 31847) or pLV-Azurite (Addgene, 36086) were purified using a Plasmid Maxi Kit (QIAGEN, 12162). Next, we transfected HEK293T cells, acquired from ATCC (CRL-3216) in 2017 and maintained in culture according to the manufacturer's instructions for less than 10 passages, using pMD2G and psPAX (Addgene, 12260 and 12259) as packaging vectors to generate viral particles. After concentration by centrifugation, the virus was collected and used for B16-F1 transduction. Cells were incubated with different serial dilutions of the virus for 48 hours, before checking the transduction efficiency by flow cytometry. The condition presenting the highest percentage of transduced fluorescent cells was then selected to purify the fluorescent population by live-cell sorting using a BD FACSAria Sorter.
Mice
The IRB hosted animal experiments in facilities defined as specific pathogen-free facilities, according to the Federation of European Laboratory Animal Science Associations guidelines. Experiments involving Il1a KO mice were conducted at the Ben-Gurion University animal facility. In both facilities, mice were housed in Individually Ventilated Cages with controlled light–dark cycle (12:12), room temperature (20–24°C) and relative humidity (30%–70%). Animal caretakers, researchers, and veterinarians provided mice with a daily check of general health conditions. All animal experiments were conducted in accordance with the Swiss Federal Veterinary Service guidelines and the Israel Animal Welfare Act. All mouse procedures were authorized by the relevant institutional committees (Commissione Cantonale per gli Esperimenti sugli Animali) of the Cantonal Veterinary Office and the Israeli Council for Animal Experimentation of the Ministry of Health, with licensing numbers TI 25/2017, TI 24/2018, TI 55/2018, and TI 30/2020. Charles River Laboratories, F. Sallusto (IRB, Bellinzona), and R. Apte [Ben-Gurion University (BGU), Be'er Sheva] provided C57BL/6J, B6.129S7-Il1r1tm1Imx/J (Il1r1 KO/KO, Jackson code 003245) and Il1a KO mice, respectively (36). These mice were then bred in-house. B6.129P2(Cg)-Cx3cr1tm1Litt/J (CX3CR1GFP/wt) mice were originally acquired from Jackson Laboratories (cat. 005582) and bred in-house. The genotype of all mice was confirmed as previously described (37, 38). Mice of ages from 6 to 12 weeks, showing good health conditions and no abnormal clinical signs were used in experiments. Equal numbers of males and females were assigned to experimental groups through a statistical randomization process. Power calculation per group size determination, performed by using R software (R: A Language and Environment for Statistical Computing, R Core Team, R Foundation for Statistical Computing, Vienna, Austria), estimated 10 animals per group to obtain > 99% statistical power.
Allograft model
To implant tumors, 106 syngeneic B16-F1, B16-F10, and E0771 cells were resuspended in 10 μL sterile PBS and injected subcutaneously in the right footpad of mice anesthetized with isoflurane (5% for induction, 3% for maintenance, FiO2 = 1 L/min). After cell injection and anesthesia recovery, mice were monitored to check for the absence of pain or impaired movement. Mouse body weight and tumor size were measured every one or two days. Tumor volume was calculated with the formula V = (length × width2) / 2 and mice were euthanized when tumors reached 250 mm3. Euthanasia was performed by isoflurane overdose followed by cervical dislocation and organs were collected immediately. We excluded from experiments mice that did not develop tumors [V = 0 mm3 at day 20 post tumor implantation (p.t.i.)] or that developed tumors in the popliteal fossa, which impeded the collection of the popliteal lymph node. In some experiments, we injected 15 μL of B16-F1 tumor cell lysate originating from 5 × 105 cells and generated by sonication at constant cycles of 30 seconds.
In vivo treatments
To maximize the specific effect of treatments on LN metastases and to minimize the effect on tumor engraftment and primary tumor growth, all treatments were administered when the primary tumor reached a size of 40 mm3, which corresponds to the time of arrival of the first metastatic cells to the LN. Additionally, all local treatments were injected in the calf, to minimize their distribution to the primary tumor. All treatments were resuspended in a maximum volume of 10 μL in calcium- and magnesium-free PBS (PBS−). Injections in the subcutis were performed under anesthesia as described above, and mice were allowed to recover and monitored for the absence of any sign of pain in the foot. Carrier-free recombinant mouse IL1α (BioLegend, 575006) was locally administered at a dose of 1 μg / 10 μL per day. The anti-mouse IL1α (InVivoMAb anti-mouse IL1α, clone ALF-161, Bio X Cell) was administered to block IL1α at a dose of 200 μg i.v. plus 60 μg locally, as previously reported (16). Blockade was then maintained with a daily local injection of 60 μg. STAT3 was inhibited by local injection of stattic (SelleckChem, S7024-50MG) 3.75 mg/kg every two days. Stattic was reconstituted, according to the manufacturer's instructions, in 5% DMSO (VWR, N182-5×10ML), 40% PEG300 (MedChem Express, HY-Y0873), 5% Tween 80 (Sigma-Aldrich, P1754), and 50% distilled water. For macrophage depletion, mice received locally 10 μL of clodronate- or PBS-containing liposomes (Liposoma, CP-010-010), followed by a second dose 2 days later.
In vivo imaging
To monitor primary tumor growth and mCherry expression of fluorescent cancer cells, we used the IVIS Spectrum Imaging System (PerkinElmer). Mice were anesthetized with isoflurane as above described to measure epifluorescence. Immediately after image acquisition, animals were allowed to recover from anesthesia. Images were later analyzed using Living Image Software 4.2 (Caliper LifeSciences).
Immunofluorescence and IHC
For mouse microscopy experiments, organs were fixed immediately after collection in 4% paraformaldehyde (PFA; Merck-Millipore) for 12 hours at 4°C, then washed in PBS− and embedded in 4% Low Gelling Temperature Agarose (Sigma-Aldrich). Sections (50 μm) were cut with a vibratome (VT1200S, Leica). Slices were stained in a blocking buffer composed of TritonX100 (VWR) 0.1–0.3%, BSA 5% (VWR), and fluorescently labeled antibodies at the appropriate concentration, all diluted in PBS supplemented with calcium and magnesium (PBS+). After 72 hours of incubation at 4°C, samples were washed in 0.05% Tween 20 (Sigma-Aldrich), fixed with PFA 4%, washed in PBS− and mounted on glass slides. Confocal images were acquired using a Leica TCS SP5 microscope with a 20 × 0.7 oil objective. To quantify the rate of invasion of melanoma in each sLN region, we first identified metastatic mass on the mCherry channel with an automatic Otsu threshold, after noise filtering with ImageJ Despeckle plugin and size filtering for regions bigger than 30 μm2. LN regions were manually identified based on CX3CR1 and CD21/35 expression. Next, we quantified the total tumor area and the percentage of overlap of metastasis with each other LN region. Sample sizes were distributed as follows: n = 21, 7, and 11 for week 1, 2, and 3 p.t.i., respectively. To quantify the expression of CD169, the LN regions were manually identified as described above and the mean fluorescence intensity of CD169 in each region was calculated. For human studies, we collected metastatic sLNs from 7 patients (6 males and 1 female). Patients were selected based on a previous diagnosis of melanoma and IHC confirmation of the presence of MelanA+ lesions in the sLN. Samples were collected immediately after routine surgery, formalin-fixed, and stored embedded in paraffin. All patients signed a declaration of consent for the use of samples for research purposes. The study was approved by the Ethics Commission of the Canton of Bern, and all procedures were in accordance with the standards of the Helsinki Declaration of 1975, as revised in 1983. To stain for IL1α, samples were stained using the BOND-III fully automated IHC/ISH stainer (BOND Polymer Refine Red Detection, Leica Biosystems, DS9390, and BOND Polymer Refine Detection, Leica Biosystems, DS9800) according to the manufacturer's instructions. To stain STAT3 and pSTAT3, primary antibodies (mouse anti-Stat3, clone 124H6, and mouse anti-Phospho-Stat3, Tyr705, clone M9C6, Cell Signaling Technology) were incubated overnight at 4°C and the MACH 4 Universal HRP-Polymer Detection System (Biocare Medical, BRI4012L) was applied according to the manufacturer's protocol. 3D cell reconstruction was performed using Imaris 9.7.2 Cell Imaging Software (Oxford Instruments).
Flow cytometry
LNs were collected, disrupted with tweezers, and enzymatically digested for 10 minutes at 37°C. DNase I (0.28 mg/mL, VWR, A3778.0100), dispase (1 U/mL, Corning, 354235), and collagenase P (0.5 mg/mL, Roche, 11213857001) were resuspended in PBS−. Digestion was stopped using a solution of 2 mmol/L EDTA (Sigma-Aldrich, A3145) and 2% heat-inactivated filter-sterilized fetal bovine serum (Gibco, 10270-106) diluted in PBS−. We blocked Fc receptors (αCD16/32, BioLegend, 101302), stained dead cells (Zombie Aqua Fixable Viability Kit, BioLegend, 423101), and performed surface staining (see Antibodies). For IL1α detection, intracellular staining was performed after surface staining with a dedicated kit (eBioscience, 88/8824/00) according to the manufacturer's instructions. Stained cells were run through an LSRFortessa or FACSymphony (BD Biosciences) and data were analyzed using FlowJo 10.7.1 software (FlowJo LLC).
To measure cytokine and chemokine expression in the LN, LEGENDPlex assays (Mouse Proinflammatory Chemokine Panel and Mouse Inflammation Panel, BioLegend, 740150) were used according to the manufacturer's instructions. Briefly, pLNs were collected and carefully disrupted in 75 μL ice-cold phosphate buffer, minimizing cell rupture. The suspension was centrifuged at 100 rcf for 5 minutes, and the supernatant was collected. 25 μL supernatant was used for cytokine and chemokine detection. Samples were analyzed by flow cytometry on an LSRFortessa or FACSymphony (BD Biosciences) and data were analyzed using the LEGENDplex Data Analysis Software Suite (BioLegend).
Antibodies
We used the following antibodies for flow cytometry experiments: anti-CD45 (clone 30-F11, BioLegend, 1 μg/mL), anti-CD3 (clone 17A2, BioLegend, 600 ng/mL), anti-B220 (CD45R, clone RA3–6B2, BioLegend, 600 ng/mL), anti-Gr-1 (clone RB6-8C5, BioLegend, 600 ng/mL), anti-NK1.1 (clone PK136, BioLegend, 800 ng/mL), anti-MHC II (I-A/I-E, clone M5/114.15.2, BioLegend, 200 ng/mL), anti-CD11b (clone M1/70, BioLegend, 600 ng/mL), anti-CD11c (clone N418, BioLegend, 600 ng/mL), anti-F4/80 (clone BM8, BioLegend, 800 ng/mL), anti-CD169 (Siglec-1, clone 3D6.112, 800 ng/mL), anti-IL1R1 (clone FAB7712N, R&D Systems, 1 μg/mL), anti-IL1α (clone ALF-161, BioLegend; clone REA288, Miltenyi Biotec, 2 μg/mL), anti-CD4 (clone RM4-5, BioLegend, 600 ng/mL), anti-CD8a (clone 53-6.7, Invitrogen, 600 ng/mL), anti-CD25 (clone PC61, BioLegend, 600 ng/mL), anti-CD80 (clone 16-10A1, BioLegend, 600 ng/mL), anti-CD86 (clone GL-1, BioLegend, 600 ng/mL), anti-CD68 (clone FA-11, BioLegend, 600 ng/mL), anti-CD27 (clone LG.7F9, Thermo Fisher Scientific, 600 ng/mL), anti-NKG2D (clone CX5, BioLegend, 800 ng/mL), anti-NKG2A (clone 18d3, BioLegend, 800 ng/mL), and anti-CD69 (clone H1.2F3, BioLegend, 800 ng/mL).
For confocal experiments, we used the following antibodies, all at 2 μg/mL concentration: anti-CD21/35 (CR1/CR2, clone 7E9, BioLegend), antipodoplanin (clone eBio8.1.1, Invitrogen), anti-NK1.1 (clone PK136, BioLegend), anti-F4/80 (clone BM8, BioLegend), anti-CD169 (Siglec-1, clone 3D6.112), and anti-IL1α (clone ALF-161, BioLegend; clone REA288, Miltenyi Biotec).
Human samples were stained with anti-IL1α (clone OTI2F8, Novus Biologicals), anti-CD68 (clone PG-M1, Dako), anti-Stat3 (clone 124H6, Cell Signaling Technology), and anti-Phospho-Stat3 (Tyr705, clone M9C6, Cell Signaling) antibodies.
Single-cell RNA sequencing
Metastatic LNs were obtained from four PBS-injected mice, six tumor-bearing mice and four tumor-bearing mice treated with anti-IL1α as described above. Metastases from tumor-bearing mice were microsurgically dissected using sterile microsurgical tools. SS, interfolicular (IF) and follicular (F) regions were dissected in negative controls. After dissection, samples were disrupted into single-cell suspension as described for flow cytometry, using sterile nuclease-free tools. Single cells were barcoded using the 10X Chromium Single-Cell platform and cDNA libraries were prepared according to the manufacturer's protocol (Chromium Next GEM Single-Cell 3′ GEM, Library and Gel Bead Kit v3.1, 4 rxns, 10X Genomics, 1000128). In brief, cell suspensions, reverse transcription master mix, and partitioning oil were loaded on a single-cell chip and then run on the Chromium Controller. Reverse transcription was performed within the droplets at 53°C for 45 minutes. cDNA was amplified for 12 cycles on a Biometra thermocycler. cDNA size selection was performed using beads (CleanNGS, CleanNA, CNGS-0050) and a ratio of SpriSelect reagent volume to sample volume of 0.6. cDNA was analyzed on a DNA chip (Agilent High Sensitivity DNA Kit, Agilent 2100 Bioanalyzer, 5067-4626) for qualitative control purposes. cDNA was fragmented using the proprietary fragmentation enzyme blend for 5 minutes at 32°C, followed by end repair and A-tailing at 65°C for 30 minutes. cDNA was double-sided size selected using SpriSelect beads. Sequencing adaptors were ligated to the cDNA at 20°C for 15 minutes. cDNA was amplified using a sample-specific index oligo as a primer, followed by another round of double-sided size selection using SpriSelect beads. Final libraries were analyzed on an Agilent Bioanalyzer High Sensitivity DNA chip for quality control. cDNA libraries were sequenced on a NextSeq500 Illumina platform aiming for 50,000 reads per cell. Base calls were converted to reads with the software Cell Ranger 3.1 (10X Genomics).
Quality control, processing, annotation, and differential gene-expression analysis of single-cell RNA sequencing data
We used the cellranger pipeline (39) to generate gene-expression count matrices from the raw data. For each sample, a gene-by-cell counts matrix was used to create a Seurat object using Seurat (40). We filtered cell barcodes with < 500 unique molecular identifiers and > 5% mitochondrial contents. Each individual sample was then normalized by a factor of 10,000 and log transformed (NormalizeData). The top 2,000 most variable genes were then identified within each sample using the FindVariableFeatures method (40). We then integrated the cells from all samples together using FindIntegrationAnchors and IntegrateData (ref. 40; 2,000 genes). The integrated gene-expression matrix obtained by applying the filtering steps above was then used to perform principal component analysis (RunPCA), preliminary clustering analysis, including nearest neighbor graph (FindNeighbors) and unbiased clustering (FindClusters; ref. 40), and cell type annotation. Uniform Manifold Approximation and Projection (UMAP) was then used to visualize the integrated expression data. We identified gene-expression markers for each cluster using FindAllMarkers from Seurat with default settings, including the Wilcoxon test and Bonferroni P value correction (40). Differential gene expression between specified clusters (or subclusters) was performed using FindMarkers (Wilcoxon rank sum test) with Benjamini–Hochberg false discovery rate (FDR) correction, average log fold change (logFC) and detection/expression percentage rate (pct). Genes were considered (significantly) differentially expressed if FDR < 0.05, logFC > 0.2, and pct > 20% within the cells in a given group.
Gene relevance analysis of single-cell RNA sequencing data
To determine gene relevance across the single-cell RNA sequencing (scRNA-seq) data, we used a network science approach. To study nodes’ relevance, we applied Graph Theory rules (41) using mathematical and social network analysis concepts. We restricted the analysis to protein–protein interactions and to the pathways in which the gene Il1a is involved. Relevant pathways (cytokine–cytokine receptor interaction, necroptosis, hematopoietic cell lineage, type I diabetes mellitus, pertussis, leishmaniasis, tuberculosis, and inflammatory bowel disease) and related were selected using the Kyoto Encyclopedia of Genes and Genomes (KEGG; ref. 42). Then we measured the expression of these genes in our scRNA-seq data set and we used their expression values as input of STRING (43). The resulting graph was used for the network analysis. We implemented a previously described comprehensive algorithm for evaluating node influences in social networks (44). This algorithm is based on three centrality measures: eigenvector centrality, current-flow betweenness centrality, and reachability. Eigenvector centrality computes the centrality for a node based on the centrality of its neighbors. Current-flow betweenness centrality starts from an electrical current model describing the spreading pattern, to which betweenness centrality, which uses shortest paths, is applied. Finally, reachability refers to the local reaching centrality of a node in a directed graph as the proportion of other nodes reachable from that node. In addition, gene average expressions per single-cell (count per million) are taken as weights of the nodes. Based on these parameters, the algorithm ranked node influences by analyzing preference relations and performing random walk. In the first step, a partial preference graph was derived from the analysis of the preference relation between every node pair for each measure. Later, the comprehensive preference graph (CPG) originated from the combination of preference relations and the three previously indicated measures. Finally, a random walk was executed on CPG to determine the node effect. By applying this implementation to the scRNA-seq data, it was possible to obtain a list of genes related to il1a pathways, according to their importance in our data set.
NanoString analysis
mRNA expression profiling was performed for all samples, including control nonmetastatic LNs, with the “Immune Exhaustion Panel” by NanoString (NanoString Technologies, XT-M-EXHAUST-12), as per the manufacturer's instructions, according to the “Hybridization Protocol for nCounter XT CodeSet Gene Expression Assays Including Panels,” and starting from 30 ng of total RNA per sample. Raw values were log transformed and normalized using quantile method. The background thresholds have been calculated per samples as the mean of the negative controls plus two times their standard deviation. Genes with lower counts than the estimated background were excluded from the subsequent analysis.
Sample-specific background was calculated by adding the average of all the negative controls to their standard deviation multiplied by two and then subtracting this value from the raw values for each gene. Biological normalization was performed to correct for differences in sample abundances. Quantile normalization was applied to the data set and revealed homogeneous distribution and comparability of our samples. The data were log transformed (base 2) for analysis, and linear models for microarray data analysis (limma, R/Bioconductor (45)) were used with a contrast matrix for the comparisons of interest. A P value < 0.05 was considered statistically significant. We corrected the P value for multiple comparisons using the Benjamini–Hochberg (BH) method (46). Heat maps and volcano plots were visualized using corresponding R packages (47, 48).
Analysis of gene pathways
Gene ontology (WikiPathways 2019 Mouse), KEGG pathways (KEGG 2019 Mouse) analysis of significantly overrepresented genes, and gene set enrichment analysis were performed using the fgsea (49) and enrichR (50) R packages. The C7 immunologic signatures gene set and H hallmark gene set from the Broad institute (https://www.gsea-msigdb.org/gsea/msigdb) were supplied as input to the function.
T-cell classification
Projection of cells onto a reference UMAP was done using the R package ProjecTILs (51), a T-cell reference atlas developed to interpret T-cell states. Each experimental condition was projected individually onto the tumor-infiltrating T lymphocytes atlas. Then, we quantified the fraction of cells residing in each ProjectTILS-defined subset.
Survival analysis TCGA
The reverse-phase protein arrays data set for Skin Cutaneous Melanoma (TCGA-SKCM, study ID phs000178), including data from 349 patients, was downloaded from the TCPA Portal and subsequently scaled into z-scores. Overall survival information along with mutational genes’ status and other clinical information were retrieved for each patient from the Firehose website developed at the Broad Institute (https://gdac.broadinstitute.org/). For the survival analysis, we utilized the survival (v2.41-3) and survminer (v0.4.2) R packages. Hazard ratios were derived using the Cox proportional hazard regression model and are based on the high-versus-low protein expression comparison.
Proliferation (MTT)
To evaluate tumor cell proliferation and response to treatments, B16-F1 and A375 cells were seeded in a 96-well plate. Carrier-free recombinant mouse (BioLegend, 575006) or human (Sino Biological, 10128-HNCH-20UG) IL1α were administered at 10 ng/mL and cells were incubated, respectively, for 24 hours or 72 hours. Anti-mouse IL1α (InVivoMAb anti-mouse IL1α, clone ALF-161, Bio X Cell) was added at the indicated doses 24 hours before data collection. To inhibit STAT3, stattic was added at the indicated dosages. Proliferation was assessed by MTT (Methylthiazolyldiphenyl-tetrazolium bromide) assay according to the manufacturer's recommendations (Sigma-Aldrich, M5655-500MG). Absorbance (OD, 560 nm) was measured in a microplate reader (Cytation 5, BioTek). Sensitivity to single drug treatments was evaluated by IC50 (4-parameter calculation upon log-scaled doses), as previously reported (52). The beneficial effect of the combinations versus the single agents was considered both as synergism according to the Chou–Talalay combination index (53), as previously performed (52), and as potency and efficacy according to the MuSyC algorithm (54).
Quantitative RT-PCR
To measure the expression of Stat3, Myd88, and Gapdh genes, the following sets of primers were designed: Stat3 forward, 5′-CACAAATATTTTTGAGTCGGCGC-3′; Stat3 reverse 5′-AAAGCCCCCGATGAGGTAATTC-3′; Myd88 forward, 5′-CGGCAACTAGAACAGACAGACT-3′; Myd88 reverse, 5′-GCAAACTTGGTCTGGAAGTCAC-3′; Gapdh forward, 5′-ACATCATCCCTGCATCCACT-3′; Gapdh reverse, 5′-AGATCCACGACGGACACATT-3′. To isolate RNA from B16-F1, cells were dispersed into single-cell suspension in PBS−. RNA was isolated using an RNeasy Mini Kit (QIAGEN, 74106). cDNA (2 μg) was synthesized using a cDNA synthesis kit (Applied Biosystems, 4368813) following the manufacturer's recommendations. For the quantitative RT-PCR (qPCR) reaction, an SYBR Master Mix (Applied Biosystems, 4472942) was used, and samples were run as technical duplicates on a QuantStudioTM 3 Real-Time PCR System (Thermo Fisher). mRNA levels of Myd88 and Stat3 were expressed relative to Gapdh expression. The Pfaffl method (55) was used to calculate the relative expression of the transcripts.
Immunoblotting
To evaluate protein expression in tumor cells, B16-F1 and A375 cells were treated using carrier-free recombinant mouse or human IL1α, at 100 ng/mL. To block IL1α, anti-mouse IL1α was added at the dose of 100 ng/mL. All treatments were added either for 24 hours or 72 hours. Cells were harvested and lysed by boiling samples in 2× Laemmli sample buffer (Bio-Rad, 1610737), supplemented with β-mercaptoethanol (Sigma-Aldrich, M6250), for 10 minutes. Lysates (30–50 μg) were resolved according to molecular weight by electrophoresis using Mini-PROTEAN TGX Precast gels 4%–20% gradient (Bio-Rad, 4561096). Next, proteins were blotted onto nitrocellulose membrane (Bio-Rad, 1620115) by electric transfer, and the membranes were blocked in TBST (20 mmol/L Tris-HCl [pH 7.5], 150 mmol/L NaCl, 0.1% Tween 20) with 5% nonfat dry milk (Bio-Rad, 1706404) for 1 hour at room temperature. The following primary antibodies were used in TBST 5% BSA buffer: mouse monoclonal, anti-Stat3 (clone 124H6, 9139, Cell Signaling Technology) and rabbit monoclonal, anti-p(Y705)Stat3 (9131, Cell Signaling Technology). Mouse monoclonal anti-GAPDH (clone FF26A/F9, CNIO) was used in TBST with 5% nonfat dry milk. The secondary antibodies used were: ECL anti-mouse IgG horseradish peroxidase–linked species-specific whole antibody and ECL anti-rabbit IgG horseradish peroxidase-linked species-specific whole antibody (Amersham, NA931 and NA934, respectively). Membranes were treated with Westar ηC 2.0 chemiluminescent substrate (Cyanagen, XLS142) and signals were detected using digital imaging with Fusion Solo (Vilber Lourmat).
Statistical analyses
All raw data were analyzed, processed, and presented using GraphPad Prism 8.2.1 (GraphPad Software). First, we applied the Shapiro–Wilk normality test to analyze the distribution of data. Then we compared means among groups using one-way ANOVA or unpaired t test for data with normal distribution, and the nonparametric Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
Data availability
The data generated in this study are available within the article and its supplementary data files or from the corresponding author upon reasonable request. scRNA-seq and NanoString data generated in this study are publicly available in Gene-Expression Omnibus (GEO) at GSE212227 and GSE208247, respectively. Data used for the survival analysis were obtained from the TCPA Portal at TCGA-SKCM-L3-S64.
Results
Development of a murine model of melanoma metastasis to the popliteal lymph node
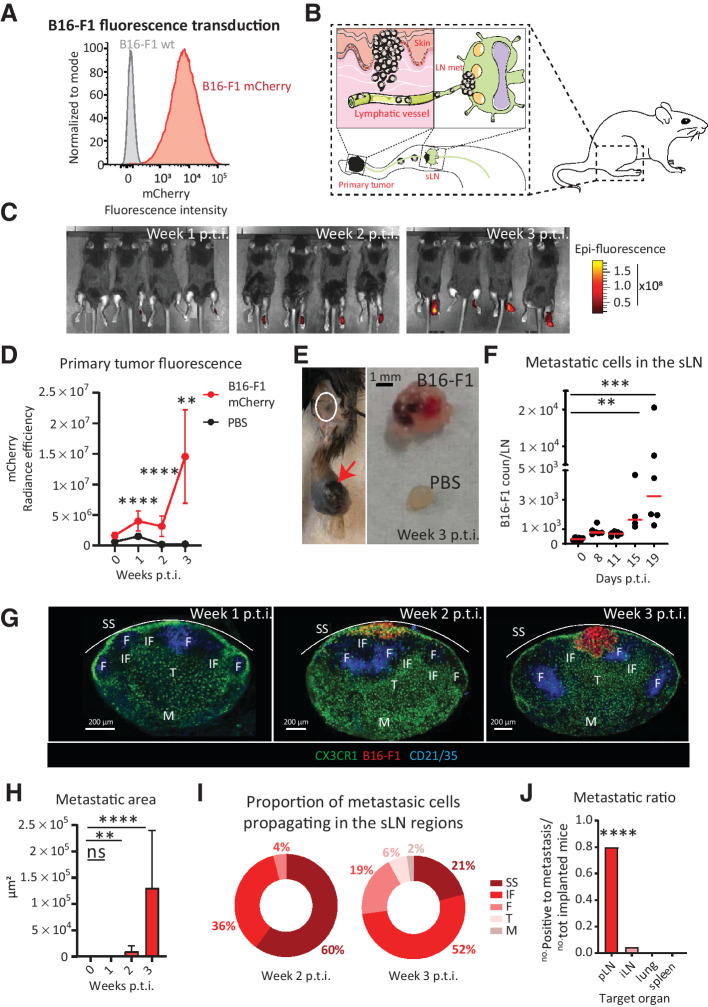
To study the metastatic process in the sentinel popliteal lymph node, we transduced the melanoma cell line B16-F1 with a lentiviral vector codifying for mCherry and characterized the expression of this fluorescent protein by flow cytometry and microscopy (Fig. 1A and Supplementary Fig. S1A, respectively). The primary tumor was induced by subcutaneous injection of the cancer cells in the mouse footpad, similar to what was previously reported (Fig. 1B; ref. 56). Next, the formation of the primary tumor was monitored by measuring tumor volume (Supplementary Fig. S1B and S1C), and tumor fluorescence was quantified using in vivo imaging (IVIS, PerkinElmer; Fig. 1C and D). Following this approach, we observed a significant engraftment starting from week 1 p.t.i (Fig. 1D). Macrometastases were detectable in the sLNs three weeks p.t.i. (Fig. 1E).
Figure 1.
Growth of mouse melanoma metastases in the sLNs. We used B16-F1 melanoma cells transduced with a lentiviral vector codifying for mCherry to develop a mouse model of melanoma metastasis to the popliteal lymph nodes, which we characterized. A, Histogram of flow cytometry data showing fluorescent expression of mCherry in representative samples of the transduced B16-F1 mCherry cells (red) in comparison with wild-type melanoma cells (gray). B, Schematic representation of the tumor model, including primary tumor engraftment (left) and migration of cells to the sLN (right). C, Representative images and (D) quantification of IVIS time course showing increasing primary tumor fluorescence (red; n = 4). E, Representative pictures of (left) primary tumor (red arrow), draining sentinel LN (white circle) and (E, right) comparison between metastatic and healthy LN at week 3 p.t.i. F, Time course of metastatic cell invasion of the sLN quantified by flow cytometry (n > 4 for all groups, dots indicate individual values). G, Representative confocal micrographs of sLNs at weeks 1, 2, and 3 p.t.i., showing the position of B16 melanoma (red) with respect to CD21/35+ (blue) follicular dendritic cells and CX3CR1+ (green) myeloid cells. SS, IF, F, T, and M stand for subcapsular sinus, interfollicular, follicular, T cells, and medullary areas, respectively. H, Quantification of total metastatic area in the sLN, measured by confocal microscopy (n > 7 for all groups). I, Quantification of confocal microscopy images showing tumor cell fluorescence in the different compartments of the LN at weeks 2 and 3 p.t.i. (n > 7 for all groups). J, Metastatic ratio, defined as the number of mice with metastases in the target organ divided by the total number of implanted mice, at week 3 p.t.i. iLN stands for inguinal LN (n > 7 for all groups). All data indicate mean values for each group. Lines in D and H indicate standard deviation. All results have been confirmed by at least three independent experiments. Data have been analyzed with one-way ANOVA or unpaired t test in case of normal distribution, and Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
To study in more detail the progression of melanoma cells toward the sLNs, we used flow cytometry, observing a significant increase of the metastatic cells at day 15 p.t.i. compared with control samples (Fig. 1F). In addition, to characterize the metastasis in detail, we used confocal microscopy analysis of the sLNs at different time points following tumor induction (Fig. 1G), and we measured a significant increase in the metastatic area beginning at 2 weeks p.t.i. (Fig. 1H). The morphology of sLN metastases was in concordance with previous work in humans (5), with metastatic cells initially observed to invade the SS area (Fig. 1G and I). At later time points, the metastasis progressively expanded through the interfollicular area (IF), invading the transverse sinus (Fig. 1I; Supplementary Fig. S1D). Conversely, we did not observe the presence of metastatic cells in distant organs, such as the spleen or lungs, at equivalent time points (Fig. 1J), confirming the lymphatic dissemination of the tumor at the evaluated time points.
IL1α promotes melanoma growth in sLNs
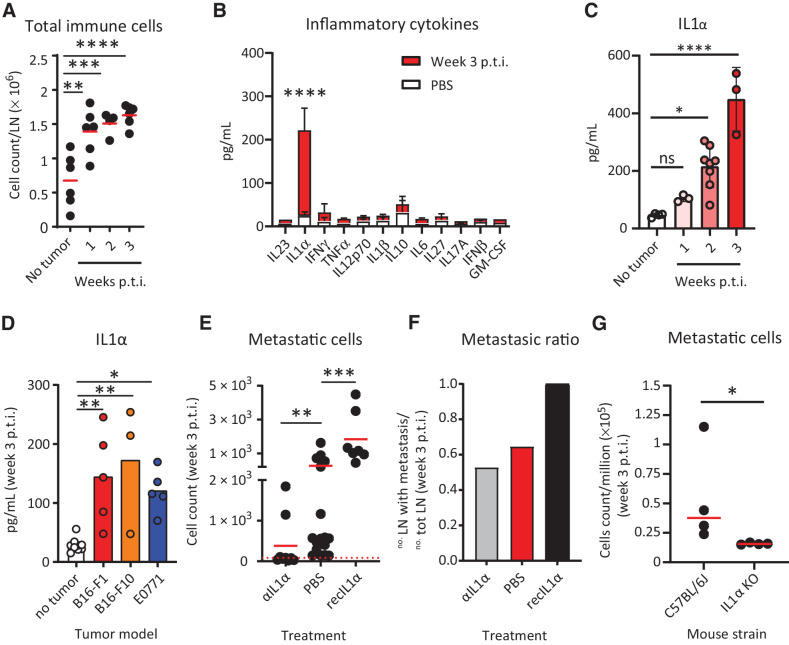
To characterize the inflammatory reaction induced by B16-F1 metastasis, we quantified the total number of immune cells infiltrating the sLNs by flow cytometry, observing a significant 2-fold increase starting from the first week p.t.i. (Fig. 2A). Studying the infiltrated immune cells types in more detail (Supplementary Fig. S2A), we detected significant increases in dendritic cells (MHC II+CD11chighCD11b+ and MHC II+CD11chighCD11b– subsets), NK cells (CD3–NK1.1+), neutrophils (MHC II–GR1high), monocytes (MHC II–GR1int), and macrophages (MHC II+CD11cint/lowCD11b+; Supplementary Fig. S2B), as well as B (B220+) and T cells (CD4+, CD8+, and Treg subtypes, Supplementary Fig. S2C). Among the T-cell subtypes present in the metastasis, Treg increased in a higher fold change than CD4+ and CD8+ T cells (Supplementary Fig. S2D). Moreover, CD8+ T cells exhibited significantly higher numbers of PD1+ cells and higher levels of PD1 expression in comparison with controls, compatibly with a picture of T-cell exhaustion (Supplementary Fig. S2E and S2F, respectively).
Figure 2.
Protumoral release of IL1α in the metastatic sLN. We observed release of IL1α in metastatic sLNs and described the prometastatic role of this cytokine. A, Progressive increase in the size of the sLN correlated with an increase in the total number of CD45+ immune cells, measured by flow cytometry (n = 6). B, Quantification of inflammatory cytokines in the supernatant of metastatic (red) and nonmetastatic (white) LN using LEGENDPlex assays (n > 3 for all groups). C, Time-course kinetics showing IL1α release in the sLNs during the first three weeks p.t.i. (n > 3 for all groups). D, Quantification of IL1α in the sLN at 3 weeks p.t.i. of different cancer models, melanoma B16-F1 and B16-F10, and breast cancer (E0771; n > 3 for all groups). E, Flow-cytometric quantification of mCherry+ LN metastatic cells in tumor-bearing animals treated with IL1α-blocking antibody or recombinant IL1α in comparison with untreated (n > 7 for all groups). F, LN metastatic ratio in mice untreated or treated with IL1α-blocking antibody or recombinant IL1α at week 3 p.t.i. (n > 7 for all groups). G, Metastatic cells in the sLN of wild-type and IL1α KO mice 3 weeks p.t.i. (n = 4). Data in A–E, and G indicate mean values for each group, with individual values represented as dots in A, C, D, E, and G. Lines in B and C indicate standard deviation. The red dashed line in E indicates the background, calculated on the number of events in nontumor-bearing mice. All results have been confirmed by at least 3 independent experiments. Data have been analyzed with one-way ANOVA or unpaired t test in case of normal distribution, and Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
To further characterize the recruitment of immune cells to the metastasized sLNs we used a multiplexed approach to quantify the concentration of different inflammatory chemokines, including CXCL13, CXCL9, CCL22, CCL5, and CCL2, in the sLN supernatant, observing a significant increase of all the tested molecules at week three p.t.i. (Supplementary Fig. S2G). Additionally, we measured the concentration of 12 inflammatory cytokines. Among all the molecules analyzed, only IL1α exhibited a significant increase at week 3 p.t.i., compared with the control group (Fig. 2B). A further study of the dynamics of IL1α release highlighted that upregulation began at week 2 p.t.i. (Fig. 2C). To evaluate whether IL1α secretion was also present in other tumor models, we measured the level of this cytokine in sLNs metastasized with the melanoma cell line B16-F10 or with the breast cancer cell line E0771, observing similar levels of IL1α in both models at 3 weeks p.t.i. (Fig. 2D).
To explore the hypothesis that IL1α might have protumoral properties, we treated mice with a daily subcutaneous injection of either IL1α-blocking antibody or recombinant IL1α. Interestingly, blocking the IL1α pathway by administering the neutralizing antibody significantly decreased the metastasis growth in the sLNs, as indicated by a reduction in the number of metastatic cells measured by flow-cytometric analysis at week three p.t.i. (Fig. 2E). Conversely, the number of melanoma cells significantly increased in the sLNs treated with recombinant IL1α at equivalent time points (Fig. 2E). Moreover, the metastatic ratio, defined as the number of mice developing sLN metastasis at week 3 p.t.i. divided by the total number of mice showing primary tumor engraftment, was higher in the animals treated with recombinant IL1α and it was reduced following IL1α blockade (Fig. 2F). Nevertheless, the observed variation in the metastasis size after treatment could be associated with a reduction in primary tumor size. Therefore, to address that possibility, we normalized the number of metastatic cells to the primary tumor volume, confirming the results previously observed in Fig. 2E (Supplementary Fig. S2H). Additionally, the previously described treatments did not have a significant effect on the growth of the primary tumor in comparison with the untreated control group (Supplementary Fig. S2I). However, we observed that Il1a KO mice showed a reduction not only in the number of metastatic cells at week 3 p.t.i. (Fig. 2G), but also in the primary tumor volume starting from week 4 p.t.i. (Supplementary Fig. S2J). This discrepancy could be partially explained by the mode of administration of the blocking antibody, which promotes the transport toward the draining lymphatics, or by the time of administration of the blocking antibody in comparison with the constant absence of IL1α in the tumor microenvironment in KO mice.
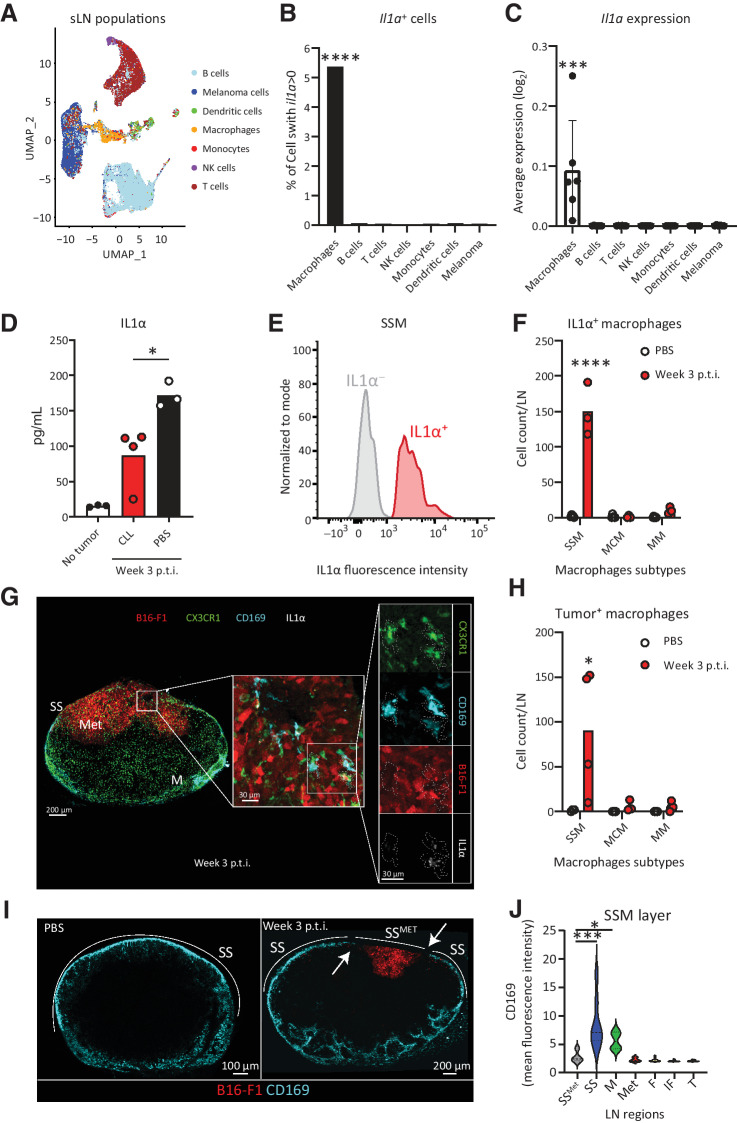
SSMs are the main source of protumoral IL1α and disappear after tumor phagocytosis
In a previous study, we determined that LN macrophages were the main producers of IL1α in the LNs following influenza vaccination (16). To elucidate the main source of this cytokine in the model of melanoma metastasis, we microsurgically dissected the metastatic regions of the sLNs and analyzed them by scRNA-seq (Fig. 3A). These analyses showed that LN macrophages were the main producers of IL1α (Fig. 3B and C). The depletion of macrophages, through injection with clodronate liposomes (CLL), significantly reduced the levels of IL1α in the LN supernatant (Fig. 3D). Depletion of macrophages also abrogated the growth of the metastatic melanoma cells in the LNs, confirming the protumoral nature of the macrophages (Supplementary Fig. S3A and S3B). However, the local administration of CLL in the sLNs did not affect the volume of the primary tumor (Supplementary Fig. S3C). Moreover, we observed that the presence of metastatic cells in the sLNs was accompanied by the upregulation of multiple genes indicative of an immunosuppressive and protumoral environment, such as Il10rb, Nras, Smarca4, Mif, Vegfa, and Lgals9. At the same time, genes associated with activation of the immune system, including Ctsw, Sh2d1a, Hla-dqb2, Jund, Ptk2b, Batf, Ptpn7, Tnfaip3, Pik3cd, Tcf7, Akt3, Malt1, Bcl, Trim33, Gata3, and Pik3r1, were downregulated (Supplementary Fig. S3D). Despite their protumoral function, macrophages did not play a crucial role in general immune regulation in the LN, as their depletion by CLL did not modify the expression of any of the previously mentioned genes or any other gene related to immune suppression and exhaustion among the 785 that we quantified (Supplementary Fig. S3D).
Figure 3.
SSMs are the main sources of IL1α. We demonstrated that IL1α in the sLN is released by SSMs, following tumor phagocytosis and the disappearance of the SSM layer. A, UMAP plot of cell populations, identified by scRNA-seq, in the metastasized sLN of a representative mouse 3 weeks p.t.i. (n > 4 for all groups). B, Percentage of cells expressing Il1a and (C) average Il1a expression in the cells of the sLN 3 weeks p.t.i., measured by scRNA-seq (n > 4 for all groups). D, IL1α quantification in metastasized sLN supernatant of mice depleted for macrophages by clodronate liposome (CLL) injection in comparison with untreated metastasized and nonmetastasized LN, measured by LEGENDPlex assay (n > 3 for all groups). E, Flow-cytometric histograms showing presence, 3 weeks p.t.i., of IL1α+ (red) and IL1α– (gray) SSM in a representative metastatic sLN. F, Flow-cytometric quantification of the number of IL1α+ cells among the three major subtypes of macrophages in the sLN 3 weeks p.t.i. in comparison with negative controls (n = 3). SSM, MCM, and MM stand for subcapsular sinus macrophages (CD169+F4/80–), medullary cord macrophages (CD169–F4/80+) and medullary macrophages (CD169+F4/80+), respectively.G, Confocal micrograph showing the whole sLN (left) and magnifications of the metastatic region (center and right) indicating IL1α and tumor vesicles in CX3CR1+CD169+ macrophages in a representative metastatic sLN. Colors indicate CX3XR1+ cells (green), mCherry+ melanoma (red), CD169+ macrophages (cyan), and IL1α (white). H, Flow-cytometric quantification of the number of each subtype of tumor+ macrophages (n = 4). I, Confocal representative images of CD169+ macrophage distribution in the sLN 3 weeks p.t.i. (right) in comparison with negative controls (left). J, Quantification of CD169 fluorescence in the main regions of the LN 3 weeks p.t.i., indicating disruption of CD169 layer (white arrows) in the SS overlying the metastatic area (SSMET; n = 7). Data represented in C, D, F and H indicate mean values for each group, with individual values represented as dots. Dots in A indicate single cells. Lines in C indicate standard deviation. The violin plot in J shows mean and standard deviation. All results have been confirmed by at least 3 independent experiments. Data have been analyzed with one-way ANOVA or unpaired t test in case of normal distribution, and Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
To identify the specific subset of macrophages responsible for the production of IL1α, we used flow cytometry (Fig. 3E and F) and confocal microscopy (Fig. 3G), both of which supported the premise that SSMs (CD169+F4/80–) were the main source of IL1α in the metastatic region. As seen in other studies (18), we also found that tumor-infiltrating SSMs phagocyted melanoma cells (Fig. 3G and H; Supplementary Fig. S3E, and Supplementary Movie S1). To demonstrate the clinical relevance of these findings, we performed IHC staining of melanoma metastatic LNs from patients, confirming that the local production of IL1α was associated with CD68+ tumor-infiltrating macrophages in the SS region (Supplementary Fig. S3F).
To investigate the mechanism of release of IL1α by SSMs, we first quantified cell numbers by flow cytometry, observing that the total number of SSMs in the sLN remained constant during the first 3 weeks p.t.i. (Supplementary Fig. S3G), whereas their frequency among all macrophages in the sLN decreased (Supplementary Fig. S3H). This finding was in contrast with a significant increase in the number of total macrophages observed in the sLN at equivalent time points (Supplementary Fig. S2B). Therefore, to investigate why SSM numbers do not increase during metastasis growth, we quantified the expression of the macrophage marker CD169 in different regions of the metastatic sLNs by confocal microscopy. We observed that the layer of CD169+ cells in the SS was absent in the proximity of the metastatic area (Fig. 3I). In further detail, CD169 in the SS surrounding the metastatic region was expressed significantly less than in the nonmetastasized SS (Fig. 3J), suggesting that SSMs in direct contact with melanoma undergo cell death. Conversely, myeloid CX3CR1+ cells and NK cells infiltrated the tumor and did not disappear during metastasis (Supplementary Fig. S3I and S3J, respectively). To confirm that the phagocytosis of tumor cell debris was able to induce SSM disappearance, we injected B16-F1 lysate in the mouse footpad and performed flow-cytometric analysis at 12 and 24 hours following injection. We observed that the percentage of SSMs significantly decreased compared with noninjected controls (Supplementary Fig. S3K).
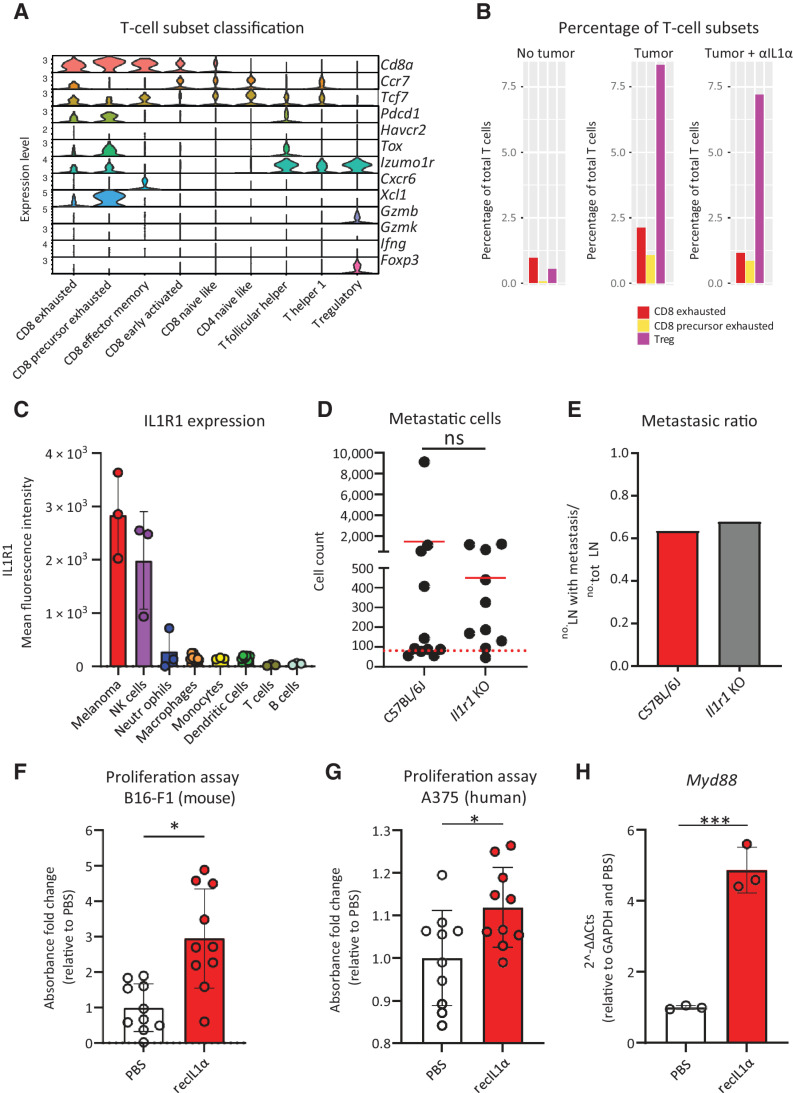
SSM-derived IL1α induces melanoma proliferation
In previous studies, we have demonstrated the involvement of IL1α in the inflammatory reaction in LNs (16, 57). However, here we did not observe any significant effect on the abundance of the major immune cell types in the metastatic sLNs following treatment with anti-IL1α (Supplementary Fig. S4A and S4B). Moreover, IL1α blockade did not significantly affect the percentage of regulatory and exhausted T cells in the metastatic lesion (Supplementary Fig. S4C; Fig. 4A and B). To further characterize the protumoral mechanism of IL1α, we measured the expression of IL1R1, the only known receptor involved in the signaling of this cytokine (26), in the cells residing in the metastatic sLNs. Among the evaluated cells, melanoma and NK cells displayed the highest level of IL1R1 expression (Fig. 4C). To clarify the role of immune cells in mediating the protumoral function of IL1α, we induced melanoma in Il1r1 KO mice. In these mice, the tumor-infiltrating cells cannot be involved in IL1 signaling, only implanted cancer cells express this receptor. The absence of IL1R1 in the immune compartment did not significantly affect either the number of metastatic cells in the sLNs (Fig. 4D) or the metastatic ratio (Fig. 4E), demonstrating that the protumoral effect of IL1 signaling was not associated with the cell response mediated by immune cells. Next, after confirming the expression of IL1R1 in cultured melanoma cells (Supplementary Fig. S4D), we measured their proliferation rate following exposure to IL1α. We discovered that IL1α significantly promoted the proliferation of murine and human melanoma cell lines (Fig. 4F and G, respectively). To further characterize the activation of IL1R1 signaling in B16-F1 melanoma cells, we measured by qPCR the expression of the Myd88 gene, which codifies the main mediator of Interleukin-1 receptor–associated kinase (IRAK) signaling activated by IL1R1 (26). The results confirmed that melanoma cells treated with recombinant IL1α significantly upregulated the Myd88 gene (Fig. 4H).
Figure 4.
Direct effect of protumoral IL1α on metastatic cells. We demonstrated that IL1α in the sLN does not revert immune exhaustion while promoting metastatic cell proliferation via IL1R1 signaling. A, Classification criteria of T-cell subsets using the scRNA-seq data of a representative metastatic sLN. B, Percentage of CD8 exhausted T cells (red), CD8 precursor exhausted T cells (yellow), and Treg (violet) among the total T cells in LN without tumor (left) in comparison with metastatic LN from untreated mice (center) and mice treated with IL1α blockade (right; n > 4 for all groups). C, Flow cytometry quantification of mean fluorescence intensity (MFI) indicating IL1R1 expression in the sLN cell populations (n = 3); gating as in Supplementary Fig. S2A. D, Flow-cytometric quantification of metastatic cells and (E) metastatic ratio in wild-type and Il1r1 knockout mice, 3 weeks p.t.i. (n > 10 for all groups). F, Proliferation assay (MTT) of B16-F1 treated with recombinant IL1α for 24 hours in comparison with untreated cells (n = 10). G, Proliferation assay (MTT) of human melanoma A375 treated with human recombinant IL1α for 72 hours, in comparison with untreated cells (n = 10). H, qPCR quantification of the Myd88 gene in B16-F1 stimulated with recombinant IL1α in comparison with unstimulated (n = 3). Data in all the graphs indicate mean. Lines and dots, where present, represent standard deviation and individual values, respectively. The red dashed line in B indicates the background, calculated on the number of events in nontumor-bearing mice. All results have been confirmed by at least three independent experiments. Data have been analyzed with one-way ANOVA or unpaired t test in case of normal distribution, and Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
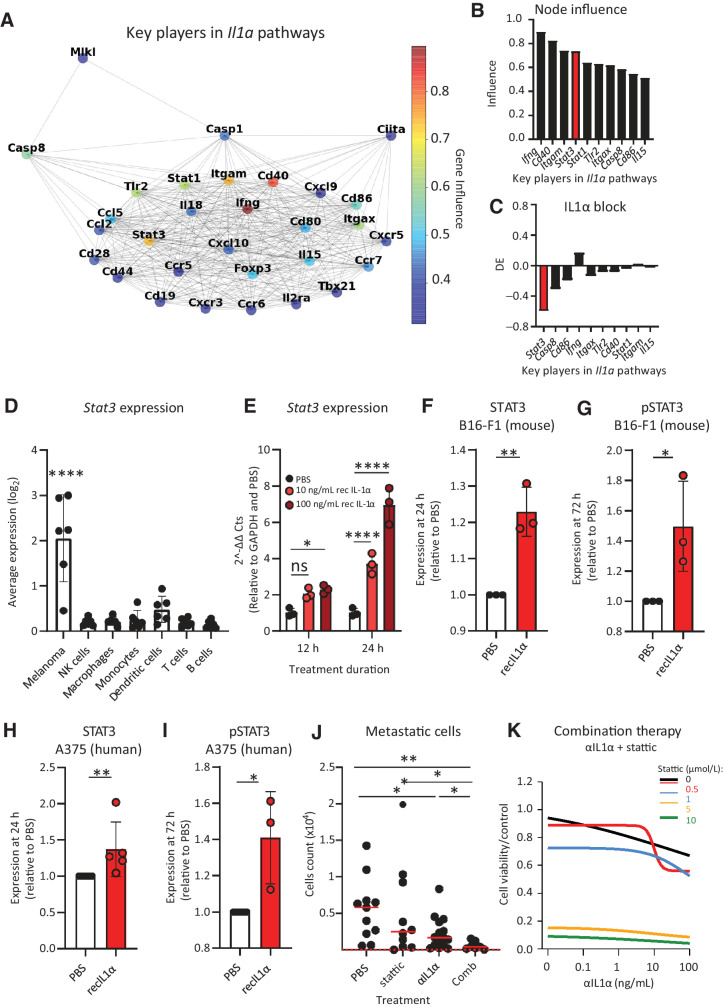
IL1α promotes aggressiveness of melanoma metastasis via STAT3
To study the pathways influenced by IL1α blocking in vivo, at three weeks p.t.i., we performed scRNA-seq of microdissected metastases from mice treated with anti-IL1α (Supplementary Fig. S5A). Next, we performed an influence analysis on the scRNA-seq data (Fig. 5A) to identify the top 10 IL1α key players, defined as the genes with the highest influence from all the Il1a-related pathways (Fig. 5B). Among them, we focused on Stat3, which was the most differentially expressed gene among the Il1a key players following IL1α blockade (Fig. 5C). This gene codifies the transcription factor STAT3, a well-characterized mediator of aggressiveness in different cancers, including melanoma (58, 59). In our scRNA-seq data set, the metastatic melanoma were the cells exhibiting the highest expression levels of this gene, excluding a major role of STAT3 in regulating the activation of immune cells (Fig. 5D). Moreover, we confirmed by qPCR the induction of the Stat3 gene in melanoma cells cultured in the presence of recombinant IL1α (Fig. 5E). To recapitulate this at a functional level, we studied STAT3 protein expression and phosphorylation in murine melanoma cells using an immunoblot assay. Treatment with recombinant IL1α induced a significant overexpression of STAT3 in comparison with untreated controls, starting at 24 hours post IL1α administration (Fig. 5F; Supplementary Fig S5B). Furthermore, exposure to recombinant IL1α induced the phosphorylation of STAT3 (Fig. 5G; Supplementary Fig. S5C). To determine whether this mechanism was also present in a human model, we quantified STAT3 and pSTAT3 in the A375 cell line post administration of human recombinant IL1α, and we observed a significant increase of both total (Fig. 5H; Supplementary Fig. S5D) and phosphorylated (Fig. 5I; Supplementary Fig. S5E) forms of the transcription factor in comparison with untreated controls. Moreover, IHC of sLNs from human patients confirmed the expression and phosphorylation of STAT3 in metastatic lesions (Supplementary Fig. S5F). In addition, analysis of human protein array data from TCPA indicated that high phosphorylation of STAT3 was correlated with reduced survival of melanoma patients, especially if the melanoma had a mutation in NF1, one of the most common genetic drivers of melanoma (Supp Supplementary Fig. S5G), further supporting the clinical relevance of this process in the progression of the disease.
Figure 5.
IL 1α induces STAT3 expression and phosphorylation in tumor. We described that IL1α in the sLN promotes STAT3 expression and phosphorylation in metastatic cells and that IL1α blockade synergizes with STAT3 inhibition in controlling metastatic cell proliferation. A, STRING graph representing the most influential genes obtained by node influence analysis of Il1a-enriched pathways using the scRNA-seq data of the metastatic area of a representative sLN. The influence of each node is expressed in a colorimetric scale. B, Bar plot showing node influence of the 10 most influential genes in Il1a pathways 3 weeks p.t.i. STAT3 is highlighted (red; n = 6). C, Bar plot indicating differential expression (DE) of the 10 most influential nodes in tumor following IL1α block in comparison with untreated mice (n = 4). Stat3 is highlighted (red). D, Average Stat3 expression in each cell population of the metastatic sLN, as analyzed using the scRNA-seq data (n = 6). E, qPCR quantification of Stat3 expression in B16-F1 following recombinant IL1α administration (n = 3). Quantification of (F) STAT3 and (G) pSTAT3, measured by immunoblot, in B16-F1 after IL1α treatment (n = 3). Immunoblot quantifications of (H) STAT3 (n = 5) and (I) pSTAT3 (n = 3) in human melanoma A375 following exposure to IL1α. J, Flow-cytometric quantification of metastatic cells in mouse sLNs of mice treated with anti–IL1α antibody, the STAT3 inhibitor stattic or their combination, in comparison with untreated (n > 10 for all groups). K, Proliferation of mouse B16-F1 melanoma cells upon combination therapy with anti–IL1α and stattic at different concentrations, measured by MTT assay (n = 3). Data in all the graphs indicate the mean. Lines and dots, where present, represent standard deviation and individual values, respectively. The red dashed line in J indicates the background, calculated on the number of events in nontumor-bearing mice. All results have been confirmed by at least three independent experiments. Data have been analyzed with one-way ANOVA or unpaired t test in case of normal distribution, and Kruskal–Wallis or Mann–Whitney test for groups that did not present a normal distribution. In all statistical tests, P value is indicated as *, <0.05; **, <0.01; ***, <0.001; ****, < 0.0001.
Once we confirmed a connection between IL1α and STAT3, we evaluated whether therapy with a STAT3 inhibitor (STAT3i) could improve the efficiency of the previously described IL1α blocking therapy in the model of metastatic melanoma. First, we observed that the administration of both therapies in vivo was able to contain the growth of the metastases more effectively than either of the two individual treatments (Fig. 5J). The STAT3i therapy did not alter the activation status of macrophages (Supplementary Fig. S5H) or NK cells (Supplementary Fig. S5I), supporting the hypothesis of a more critical role of STAT3 in tumor aggressiveness rather than in immune suppression. Additionally, we evaluated the combinatorial effect of these two therapies using an in vitro system, which highlighted a synergistic effect of the anti-IL1α blocking therapy combined with the STAT3i stattic (Fig. 5K; Supplementary Fig. S5J and S5K). In more detail, the addition of stattic improved both the efficacy (Supplementary Fig. S5L) and potency (Supplementary Fig. S5M and S5N) of the IL1α blocking therapy.
Discussion
In the present work, we characterized the protumoral role of SSMs, the first immune cells to encounter melanoma metastasis in the sLNs. Upon phagocytosis of malignant cells, SSMs released IL1α, which increased the aggressiveness of the tumor by promoting STAT3 signaling and metastasic cell proliferation.
Melanoma metastasis is the leading cause of mortality associated with skin cancer (60). However, the initial phases of metastasis formation in the LNs remain poorly understood. In patients with cancer, large numbers of tumor cells are released from the primary site daily. However, melanoma studies using murine models have observed that less than 0.1% of the tumor cells that leave the primary tumor metastasize (61). This is mainly due to the role played by immune cells around the tumor, which influence the metastatic potential of the disseminating cells (62). It is, therefore, clear that the development of metastases in LNs requires that cancer cells escape immune surveillance. However, it is also evident that both innate and adaptive immunity can promote tumor progression. In this regard, it is known that chronic inflammation, which has long been associated with increased tumor risk, is involved in the polarization of the immunity that facilitates tumor growth (63).
The absence of an evident implication of LN macrophages in the immunosuppressive tumor microenvironment is in contrast with the classic functions attributed to tumor-associated macrophages (21). However, immune suppression is only one of the heterogeneous mechanisms by which macrophages can support tumor (64). These protumoral mechanisms have been recently demonstrated to depend on different factors, including the anatomic location of macrophages and the tumor (22), and therefore they might vary between lesions in LNs and other organs. Moreover, some genes, including SIGLEC1 and CX3CR1, which are highly expressed in SSM (10), seem to be major modulators of the function of macrophages in cancer, activating protumoral feedback loops independently from immune suppression (22), similar to what we observed here. Our results imply that different cell types might be regulators of immune activation. For example, tumor cells can directly induce immune suppression by different mechanisms, including through their expression of checkpoint ligands (65). Additionally, DCs (66) and Tregs (67) are widely studied mediators of immune suppression in cancer and might play major roles. The elimination of LN macrophages prevented tumor growth in the sLNs, suggesting a complex role for these cells, probably not only restricted to IL1α release. For instance, macrophages can promote tumors by many different mechanisms, including release of growth factors, matrix remodeling, and induction of neoangiogenesis (21). In particular, angiogenesis might be especially relevant in the sLNs, where cancer cells gain access to blood circulation (8, 9).
We confirmed the protumoral function of a specific subtype of macrophages, the SSMs, during melanoma metastasis, although we did not detect any indication of an immunoregulatory role for these cells. However, previous studies reported controversial functions of this cell type in tumor biology, highlighting their capacity to be either pro- or antitumoral (6, 68, 69). CD169+ macrophages, for example, have been described to trigger activation of CD8+ T cells against melanoma through cross-talk with DCs in the spleen (70) and to control antitumoral B-cell responses in the LN (19). However, these functions, which are apparently in contrast with our results, might be explained by different conditions of exposure to tumor antigens. For instance, macrophage activation in a different target organ like the spleen might follow different patterns in comparison with the LN, and tumor vesicles drained from peripheral tissues might trigger different reactions in comparison with the active migration and proliferation of metastatic cells in the LN (71). Indeed, it has been proposed that, as a consequence of their cellular plasticity, macrophages are capable of activating both pro- and antitumoral responses (64). In this regard, it has been previously observed that MARCO+ macrophages, which are located in a different compartment of the LN from SSMs and MMs, promote tumor killing (72). Consequently, targeting a specific protumoral pathway, such as IL1α, might reveal a better strategy than depleting the entire macrophage population, avoiding the hampering of possible antimetastatic functions carried out by these cells or by other macrophage subsets (18–20). Moreover, the elimination of the whole macrophage population could predispose patients toward infections (13). Additionally, the development of drugs capable of specifically targeting SSMs might be useful for modifying protumoral pathways only in these cells and to boost their antitumoral properties in a process of macrophage repolarization (73). For example, specific TLR stimulation has shown promising results in directing CD169+ macrophages toward antigen presentation, DC activation, and T-cell responses against cancer (74). Unfortunately, despite studies describing compounds capable of localizing differentially in the SS and the medullary area of the LNs (75), a therapy able to target specific macrophage subsets in the LNs is still missing. Moreover, according to our results, we cannot discard the fact that neither the other subsets of LN macrophages nor other molecules produced by SSMs might also be relevant in this process.
SSMs initiate the inflammatory response in sLNs by different mechanisms, including cell death associated with the release of prestored IL1α (16), which typically functions as an alarmin molecule (76). In a previous study, we associated the release of this cytokine in LNs with the necrotic death of LN macrophages following influenza vaccination (16). The disappearance of the SSM layer in the metastatic region observed in this work might suggest a similar mechanism that needs to be further evaluated. In accordance with previous studies (26), the release of IL1α was associated with a prominent secretion of different chemokines. Among them, a significant upregulation of CCL2 was observed. The increased levels of this chemokine might explain the elevated numbers of infiltrated monocytes, which could differentiate to SSMs. This process would possibly explain the constant numbers of macrophages observed, despite the specific disappearance of the CD169+ layer associated with the tumor. This hypothesis is supported by a recent study in which the authors described a mechanism by which SSMs originate from bone marrow–derived monocytes in the case of SS layer disrupture (77).
We observed prominent recruitment of NK cells, expressing high levels of IL1R1, in the metastatic region. These cells were not able to prevent melanoma metastasis growth and exhibited impaired maturation and activation, similar to what was previously shown (78). The absence of IL1R1 in these cells did not significantly affect the metastasis growth in the sLNs. Conversely, it was previously reported that cytosolic IL1α can induce tumor regression by the activation of NK cells in different tumor models, including fibrosarcoma (79), lymphoma (80), and hepatocellular carcinoma (81). This divergence could be associated with differences related to the type of tumor, metastatic environment as well as potential exhaustion of NK cells. In this perspective, other works have described the capacity of the tumor and macrophages to exhaust NK cells (82).
Despite previous works having highlighted the protumoral activity of IL1α in primary tumors (83–85), the relevance of this cytokine in the context of metastasis remained uncharacterized. Similar to the inflammatory process, IL1α can play a dual role that can be either pro- or antitumoral (25, 83). This dual function is shaped according to the tissue microenvironment (86) or its mechanism of action (87). For instance, IL1α can initiate signal transduction by binding to IL1R1, or it can also act intracellularly, serving as a transcription factor (88). Different authors have suggested that this second process is associated with the protumoral role of IL1α, activating autocrine loops that might lead to protumoral cytokine secretion (89, 90). Instead, we observed that IL1α increases the aggressiveness and proliferation of tumor cells via phosphorylation of the transcriptional factor STAT3, which has been previously associated with melanoma progression (91). A connection between IL1α and STAT3 has been previously observed in immune cells (92) and other tumor models (86, 93). However, here, we have described this relation for the first time in association with the metastatic melanoma context. Additionally, we observed that in vitro phosphorylation of STAT3 in tumor cells occurred 12 hours after the administration of recombinant IL1α. This time frame suggests that other cytokines, possibly secreted by the tumor cells, might participate in this pathway. This idea has been previously proposed in other studies (90, 94). Interestingly, it has already been observed that the activation of the STAT3 pathway is mediated by IL6 (95). However, we could not measure an upregulation of this cytokine in the tumor microenvironment.
It has been previously demonstrated that STAT3 favors the spread of melanoma cells to distant organs, and STAT3 is particularly expressed in melanoma metastasis (58, 59). For this reason, the IL1α–STAT3 axis is a promising target to treat metastases at their first stage, gaining particular clinical relevance (34). Indeed, this concept is sustained by the reduced survival of NF1-mutated melanoma patients expressing high pSTAT3. In addition, the involvement of STAT3 associated with the secretion of IL1α has further relevance in the context of combined therapies, which represent a promising approach to target cancer cells at different levels, including the tumor microenvironment (96). In this context, blocking multiple immune pathways, such as IL1α and IL6, might improve the efficacy of STAT3i in comparison with single or dual therapy, as suggested by other studies indicating the synergistic effect of these two cytokines (97). In addition, considering the variability of cytokine levels and responses to cytokine-based therapies in patients (98), IL1α blockade could be envisaged as an alternative to IL6 inhibition for boosting STAT3i (99) in those patients with low levels of IL6 and low sensitivity to IL6 blockade (100). Thus, specific cytokine expression profiling in patients might be a useful tool to predict response to the treatment and to design the best therapeutic strategy, according to the concept of personalized medicine, as previously proposed (101). IL1α-blocking agents have already been tested in clinical trials on patients with various tumors and with different grading, showing promising results (29, 30, 32, 102). Moreover, previous evidence has described a possible connection between PD-1 and IL1 (27) or STAT3 (103) in various tumors, indicating that IL1α blockade in combination with checkpoint inhibitors might be a promising therapeutic strategy. The idea of combining IL1α blockade and checkpoint inhibitors might be based on the high prevalence in the metastatic sLN of exhausted T cells, which are not reduced during IL1α blockade and represent a relevant target for checkpoint inhibitors. Moreover, the high prevalence of exhausted and regulatory T cells explains why high concentrations of T-cell chemokines, including CXCL9 and CXCL10, are not associated with tumor clearance.
In conclusion, our results provide evidence of a novel role for SSMs in melanoma metastasis progression by inducing tumor aggressiveness. Importantly, IL1α blockade decreased metastasis growth and acted synergistically with a STAT3i in controlling tumor growth. Taken together, these findings provide new opportunities to improve the currently available immunotherapies against metastatic melanoma.
Supplementary Material
Acknowledgments
The present research was funded by the KREBSLIGA Foundation (KFS-4274-08-2017), the Swiss National Science Foundation (SNSF 176124), the IBSA Foundation Fellowship 2019 award to T. Virgilio, the Israel Ministry of Science (MOS) jointly with the Deutsches Krebsforschungscentrum (DKFZ), Heidelberg, Germany, the Israel Science Foundation funded by the Israel Academy of Sciences and Humanities (ISF), the Israel Cancer Association, the Israel Ministry of Health Chief Scientist's Office, the Binational (Israel-USA) Science Foundation (BSF), the German-Israeli Foundation (GIF), the Italian Association for Cancer Research (AIRC, MFAG2019-ID-22818), the Cariplo Foundation (ID-2019-1609), and the Italian Ministry of Health Ricerca Corrente—IRCCS MultiMedica. We thank Federica Sallusto (IRB, Bellinzona, Switzerland), Carlo Catapano (IOR, Bellinzona, Switzerland), and Greta Guarda (IRB, Bellinzona, Switzerland) for providing, respectively, Il1r1 KO mice, A375, B16-F1, and B16-F10 cell lines. We also thank David Jarossay (IRB, Bellinzona, Switzerland) for assistance in flow cytometry and live-cell sorting experiments, Diego Ulisse Pizzagalli (IRB, Bellinzona, Switzerland) for the support in Imaris Software usage, Kevin Ceni (IRB, Bellinzona, Switzerland) for Supplementary Movie S1 editing, Rocco D'Antuono (IRB, Bellinzona, Switzerland, now Francis Crick Institute, London, United Kingdom) for microscopy support, and Luca Lepore for assistance in IHC. The present research was funded by the KREBSLIGA Foundation (KFS-4274-08-2017), the Swiss National Science Foundation (SNSF 176124), the IBSA Foundation Fellowship 2019 award to T.V., the Israel Ministry of Science (MOS) jointly with the Deutsches Krebsforschungscentrum (DKFZ), Heidelberg, Germany, the Israel Science Foundation funded by the Israel Academy of Sciences and Humanities (ISF), the Israel Cancer Association, the Israel Ministry of Health Chief Scientist's Office, the Binational (Israel–USA) Science Foundation (BSF), the German–Israeli Foundation (GIF), the Italian Association for Cancer Research (AIRC, MFAG2019-ID-22818), the Cariplo Foundation (ID-2019-1609), and the Italian Ministry of Health Ricerca Corrente—IRCCS MultiMedica.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
T. Virgilio reports a grant from the IBSA Foundation Fellowship 2019 during the conduct of the study. F. Bertoni reports other support from Helsinn, Menarini Ricerche, Oncternal Therapeutics, and AstraZeneca outside the submitted work. S.F. Gonzalez reports grants from the KREBSLIGA Foundation and the Swiss National Science Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
T. Virgilio: Conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. J. Bordini: Data curation, software, formal analysis, methodology. L. Cascione: Data curation, formal analysis, visualization, methodology. G. Sartori: Investigation. I. Latino: Investigation. D. Molina Romero: Investigation. C. Leoni: Investigation, methodology. M. Akhmedov: Resources, data curation, software. A. Rinaldi: Data curation, investigation. A.J. Arribas: Investigation. D. Morone: Investigation, methodology. S.M. Seyed Jafari: Investigation, methodology. M. Bersudsky: Investigation. A. Ottolenghi: Investigation. I. Kwee: Data curation, software, investigation. A.M. Chiaravalli: Investigation, methodology. F. Sessa: Investigation. R.E. Hunger: Conceptualization, resources, validation, investigation, methodology, project administration. A. Bruno: Resources, investigation, methodology. L. Mortara: Resources, investigation, methodology. E. Voronov: Resources, investigation, methodology. S. Monticelli: Resources, investigation, methodology. R.N. Apte: Conceptualization, resources, investigation. F. Bertoni: Resources, investigation, methodology. S.F. Gonzalez: Conceptualization, resources, data curation, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg Clin North Am 2020;100:1–12. [DOI] [PubMed] [Google Scholar]
- 2. Karlsson MC, Gonzalez SF, Welin J, Fuxe J. Epithelial-mesenchymal transition in cancer metastasis through the lymphatic system. Mol. Oncol. 2017;11:781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Damsky WE, Rosenbaum LE, Bosenberg M. Decoding melanoma metastasis. Cancers 2011;3:126–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: evidence-based changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:472–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das S, Sarrou E, Podgrabinska S, Cassella M, Mungamuri SK, Feirt N, et al. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. J Exp Med 2013;210:1509–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh R, Choi BK. Siglec1-expressing subcapsular sinus macrophages provide soil for melanoma lymph node metastasis. Elife 2019;8:e48916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kretschmer L, Mitteldorf C, Hellriegel S, Leha A, Fichtner A, Ströbel P, et al. The sentinel node invasion level (SNIL) as a prognostic parameter in melanoma. Mod. Pathol 2021;34:1839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown M, Assen FP, Leithner A, Abe J, Schachner H, Asfour G, et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 2018359:1408–11. [DOI] [PubMed] [Google Scholar]
- 9. Pereira ER, Kedrin D, Seano G, Gautier O, Meijer EFJ, Jones D, et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 2018;359:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gray EE, Cyster JG. Lymph node macrophages. J Innate Immun 2012;4:424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuka M, Iannacone M. The role of lymph node sinus macrophages in host defense. Ann N Y Acad Sci 2014;1319:38–46. [DOI] [PubMed] [Google Scholar]
- 12. Carrasco Y, Batista F. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity 2007;27:160–71. [DOI] [PubMed] [Google Scholar]
- 13. Iannacone M, Moseman EA, Tonti E, Bosurgi L, Junt T, Henrickson SE, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature 2010;465:1079–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonzalez SF, Lukacs-Kornek V, Kuligowski MP, Pitcher LA, Degn S¸RE, Kim Y-A, et al. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat Immunol 2010;11:427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaya M, Castello A, Montaner B, Rogers N, Reis e Sousa C, Bruckbauer A, et al. Host response. Inflammation-induced disruption of SCS macrophages impairs B cell responses to secondary infection. Science 2015;347:667–72. [DOI] [PubMed] [Google Scholar]
- 16. Chatziandreou N, Farsakoglu Y, Palomino-Segura M, D'Antuono R, Pizzagalli DU, Sallusto F, et al. Macrophage death following influenza vaccination initiates the inflammatory response that promotes dendritic cell function in the draining lymph node. Cell Rep 2017;18:2427–40. [DOI] [PubMed] [Google Scholar]
- 17. Asano K, Nabeyama A, Miyake Y, Qiu C-H, Kurita A, Tomura M, et al. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 2011;34:85–95. [DOI] [PubMed] [Google Scholar]
- 18. Moalli F, Proulx ST, Schwendener R, Detmar M, Schlapbach C, Stein JV. Intravital and whole-organ imaging reveals capture of melanoma-derived antigen by lymph node subcapsular macrophages leading to widespread deposition on follicular dendritic cells. Front. Immunol 2015;6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C, et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 2016;352:242–6,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tacconi C, Commerford CD, Dieterich LC, Schwager S, He Y, Ikenberg K, et al. CD169+ lymph node macrophages have protective functions in mouse breast cancer metastasis. Cell Rep 2021;35:108993. [DOI] [PubMed] [Google Scholar]
- 21. Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 2005;7:211–7. [DOI] [PubMed] [Google Scholar]
- 22. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 2019;35:588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birmingham KG, O'Melia MJ, Bordy S, Reyes Aguilar D, El-Reyas B, Lesinski G, et al. Lymph node subcapsular sinus microenvironment-on-a-chip modeling shear flow relevant to lymphatic metastasis and immune cell homing. iScience 2020;23:101751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trujillo JA, Sweis RF, Bao R, Luke JJ. T cell–inflamed versus Non-T cell–inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol Res 2018;6:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Apte RN, Dotan S, Elkabets M, White MR, Reich E, Carmi Y, et al. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev 2006;25:387–408. [DOI] [PubMed] [Google Scholar]
- 26. Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 2019;50:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaplanov I, Carmi Y, Kornetsky R, Shemesh A, Shurin GV, Shurin MR, et al. Blocking IL-1β reverses the immunosuppression in mouse breast cancer and synergizes with anti–PD-1 for tumor abrogation. Proc Natl Acad Sci U S A 2019;116:1361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lust JA, Lacy MQ, Zeldenrust SR, Dispenzieri A, Gertz MA, Witzig TE, et al. Induction of a chronic disease state in patients with smoldering or indolent multiple myeloma by targeting interleukin 1β-induced interleukin 6 production and the myeloma proliferative component. Mayo Clin Proc 2009;84:114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong DS, Hui D, Bruera E, Janku F, Naing A, Falchook GS, et al. MABp1, a first-in-class true human antibody targeting interleukin-1α in refractory cancers: An open-label, phase 1 dose-escalation and expansion study. Lancet Oncol 2014;15:656–66. [DOI] [PubMed] [Google Scholar]
- 30. Hong DS, Janku F, Naing A, Falchook GS, Piha-Paul S, Wheler JJ, et al. Xilonix, a novel true human antibody targeting the inflammatory cytokine interleukin-1 alpha, in non-small cell lung cancer. Invest New Drugs Jun. 2015;33:621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lust JA, Lacy MQ, Zeldenrust SR, Witzig TE, Moon-Tasson LL, Dinarello CA, et al. Reduction in C-reactive protein indicates successful targeting of the IL-1/IL-6 axis resulting in improved survival in early stage multiple myeloma. Am J Hematol 2016;91:571–4. [DOI] [PubMed] [Google Scholar]
- 32. Hickish T, Andre T, Wyrwicz L, Saunders M, Sarosiek T, Kocsis J, et al. MABp1 as a novel antibody treatment for advanced colorectal cancer: a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 2017;18:192–201. [DOI] [PubMed] [Google Scholar]
- 33. Garner H, de Visser KE. Immune crosstalk in cancer progression and metastatic spread: a complex conversation. Nat Rev Immunol 2020;20:483–97. [DOI] [PubMed] [Google Scholar]
- 34. Edwards SC, Hoevenaar WHM, Coffelt SB. Emerging immunotherapies for metastasis. Br J Cancer 2021;124:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michielin O, Atkins MB, Koon HB, Dummer R, Ascierto PA. Evolving impact of long-Term survival results on metastatic melanoma treatment. J Immunother Cancer 2020;8:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horai R, Asano M, Sudo K, Kanuka H, Suzuki M, Nishihara M, et al. Production of mice deficient in genes for interleukin (IL)-1α, IL-1β, IL-1α/β, and IL-1 receptor antagonist shows that IL-1β is crucial in turpentine-induced fever development and glucocorticoid secretion. J Exp Med 1998;187:1463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glaccum MB, Stocking KL, Charrier K, Smith JL, Willis CR, Maliszewski C, et al. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J Immunol 1997;159:3364–71, [PubMed] [Google Scholar]
- 38. Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 2000;20:4106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Catlin PA, Chartrand G. Introductory graph theory. Am Math Mon 1987;94:483. [Google Scholar]
- 42. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mao C, Xiao W. A comprehensive algorithm for evaluating node influences in social networks based on preference analysis and random walk. complex 2018;2018. [Google Scholar]
- 45. Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using R and Bioconductor. Statistics for biology and health. New York: Springer; 2005 10.1007/0-387-29362-0_23. [DOI] [Google Scholar]
- 46. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 47. Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics 2005;21:2789–90. [DOI] [PubMed] [Google Scholar]
- 48. Blighe K, Rana S, Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling [cited 2022 Sep 27]. Available from: https://bioconductor.org/packages/release/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html ().
- 49. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021;060012, bioRxiv. [Google Scholar]
- 50. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun 2021;12:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tarantelli C, Gaudio E, Arribas AJ, Kwee I, Hillmann P, Rinaldi A, et al. PQR309 is a novel dual PI3K/mTOR inhibitor with preclinical antitumor activity in lymphomas as a single agent and in combination therapy. Clin Cancer Res 2018;24:120–9. [DOI] [PubMed] [Google Scholar]
- 53. Chou T. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 54. Meyer CT, Wooten DJ, Paudel BB, Bauer J, Hardeman KN, Westover D, et al. Quantifying drug combination synergy along potency and efficacy axes. Cell Syst 2019;8:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee C-K, Jeong S-H, Jang C, Bae H, Kim YH, Park I, et al. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 2019;363:644–9. [DOI] [PubMed] [Google Scholar]
- 57. Pizzagalli DU, Latino I, Pulfer A, Palomino-Segura M, Virgilio T, Farsakoglu Y, et al. Characterization of the dynamic behavior of neutrophils following influenza vaccination. Front Immunol 2019;10:2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fu X-Q, Liu B, Wang Y-P, Li J-K, Zhu P-L, Li T, et al. Activation of STAT3 is a key event in TLR4 signaling-mediated melanoma progression. Cell Death Dis 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Swoboda A, Soukup R, Eckel O, Kinslechner K, Wingelhofer B, Schörghofer D, et al. STAT3 promotes melanoma metastasis by CEBP-induced repression of the MITF pathway. Oncogene 2021;40:1091–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ma MW, Medicherla RC, Qian M, Vega-Saenz de Miera E, Friedman EB, Berman RS, et al. Immune response in melanoma: an in-depth analysis of the primary tumor and corresponding sentinel lymph node. Mod Pathol 2012;25:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 1998;153:865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther 2020;5:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ostrand-Rosenberg S. Immune surveillance: a balance between pro- and anti-tumor immunity. Curr Opin Genet Dev 2008;18:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mantovani A, Marchesi F, Jaillon S, Garlanda C, Allavena P. Tumor-associated myeloid cells: diversity and therapeutic targeting. Cellular and Molecular Immunology 2021;18:566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Munn DH, Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr Opin Immunol 2016;39:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 2014;26:638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression—implications for anticancer therapy. Nat Rev Clin Oncol 2019;16:356–71. [DOI] [PubMed] [Google Scholar]
- 68. Louie DAP, Liao S. Lymph node subcapsular sinus macrophages as the frontline of lymphatic immune defense. Front Immunol 2019;10:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hu J, Xu J, Li M, Zhang Y, Yi H, Chen J, et al. Targeting lymph node sinus macrophages to inhibit lymph node metastasis. Mol Ther Nucleic Acid 2019;16:650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van Dinther D, Lopez Venegas M, Veninga H, Olesek K, Hoogterp L, Revet M, et al. Activation of CD8 + T cell responses after melanoma antigen targeting to CD169 + antigen presenting cells in mice and humans. Cancers (Basel) 2019;11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ma F, Vayalil J, Lee G, Wang Y, Peng G. Emerging role of tumor-derived extracellular vesicles in T cell suppression and dysfunction in the tumor microenvironment. J Immunother Cancer 2021;9:3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Eisinger S, Sarhan D, Boura VF, Ibarlucea-Benitez I, Tyystjärvi S, Oliynyk G, et al. Targeting a scavenger receptor on tumor-associated macrophages activates tumor cell killing by natural killer cells. Proc Natl Acad Sci U S A 2020;117:32005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cassetta L, Pollard JW. Repolarizing macrophages improves breast cancer therapy. Cell Res 2017;27:963–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nijen Twilhaar MK, Czentner L, Bouma RG, Olesek K, Grabowska J, Wang AZ, et al. Incorporation of toll-like receptor ligands and inflammasome stimuli in GM3 liposomes to induce dendritic cell maturation and T cell responses. Front Immunol 2022;13:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crecente-Campo J, Virgilio T, Morone D, Calviño-Sampedro C, Fernández-Mariño I, Olivera A, et al. Design of polymeric nanocapsules to improve their lympho-targeting capacity. Nanomedicine 2019;14:3013–33. [DOI] [PubMed] [Google Scholar]
- 76. Rider P, Kaplanov I, Romzova M, Bernardis L, Braiman A, Voronov E, et al. The transcription of the alarmin cytokine interleukin-1 alpha is controlled by hypoxia inducible factors 1 and 2 alpha in hypoxic cells. Front Immunol 2012;3:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mondor I, Baratin M, Lagueyrie M, Saro L, Henri S, Gentek R, et al. Lymphatic endothelial cells are essential components of the subcapsular sinus macrophage niche. Immunity 2019;50:1453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bruno A, Mortara L, Baci D, Noonan D, Albini A. Myeloid derived suppressor cells interactions with natural killer cells and pro-angiogenic activities: roles in tumor progression. Front Immunol 2019;10:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Douvdevani A, Huleihel M, Zöller M, Segal S, Apte RN. Reduced tumorigenicity of fibrosarcomas which constitutively generate il-1α either spontaneously or following IL-1α gene transfer. Int J Cancer 1992;51:822–30. [DOI] [PubMed] [Google Scholar]
- 80. Voronov E, Weinstein Y, Benharroch D, Cagnano E, Ofir R, Dobkin M, et al. Antitumor and immunotherapeutic effects of activated invasive T lymphoma cells that display short-term interleukin 1α expression. Cancer Res 1999;59:1029–35. [PubMed] [Google Scholar]
- 81. Lin D, Lei L, Liu Y, Zhang Y, Hu B, Bao G, et al. Microenvironment and immunology membrane IL1a inhibits the development of hepatocellular carcinoma via promoting T- and NK-cell activation. Cancer Res 2016;76:3179–88. [DOI] [PubMed] [Google Scholar]
- 82. Cao Y, Wang X, Jin T, Tian Y, Dai C, Widarma C, et al. Immune checkpoint molecules in natural killer cells as potential targets for cancer immunotherapy. Signal Transduct. Target. Ther. 2020;5:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Voronov E, Dinarello C, Apte R. Interleukin-1α as an intracellular alarmin in cancer biology. Semin Immunol 2018;38:3–14. [DOI] [PubMed] [Google Scholar]
- 84. Malik A, Kanneganti T-D. Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol Rev 2018;281:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev 2018;281:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Voronov E, Dotan S, Krelin Y, Song X, Elkabets M, Carmi Y, et al. Unique versus redundant functions of IL-1α and IL1β in the tumor microenvironment. Front Immunol 2013;4:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Werman A, Werman-Venkert R, White R, Lee J-K, Werman B, Krelin Y, et al. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc Natl Acad Sci U S A 2004;101:2434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dinarello CA. An interleukin-1 signature in breast cancer treated with interleukin-1 receptor blockade: Implications for treating cytokine release syndrome of checkpoint inhibitors. Cancer Res 2018;78:5200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chiu JW, Hanafi ZB, Chew LCY, Mei Y, Liu H. IL-1α processing, signaling and its role in cancer progression. Cells 2021;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Georgouli M, Herraiz C, Crosas-Molist E, Fanshawe B, Maiques O, Perdrix A, et al. Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 2019;176:757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Malvehy J, Puig S. Expression of Stat3 and Src proteins in mucosal, cutaneous, and metastatic melanoma. J Am Acad Dermatol 2005;52:P144. [Google Scholar]
- 92. Whitley SK, Balasubramani A, Zindl CL, Sen R, Shibata Y, Crawford GE, et al. IL-1R signaling promotes STAT3 and NF-B factor recruitment to distal cis-regulatory elements that regulate Il17a/f transcription. J Biol Chem 2018;293:15790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu S, Lee JS, Jie C, Park MH, Iwakura Y, Patel Y, et al. HER2 overexpression triggers an IL1α proinflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res 2018;78:2040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kuan EL, Ziegler SF. A tumor-myeloid cell axis, mediated via the cytokines IL-1α and TSLP, promotes the progression of breast cancer. Nat Immunol 2018;19:366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer 2020;19:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hoeller C. The future of combination therapies in advanced melanoma. memo - Mag. Eur Med Oncol 2020;13:309–13. [Google Scholar]
- 97. Tengesdal IW, Menon DR, Osborne DG, Neff CP, Powers NE, Gamboni F, et al. Targeting tumor-derived NLRP3 reduces melanoma progression by limiting MDSCs expansion. Proc Natl Acad Sci U S A 2021;118:e2000915118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Berraondo P, Sanmamed MF, Ochoa MAC, Etxeberria IA, Aznar MA, Pérez-Gracia JL, et al. Cytokines in clinical cancer immunotherapy. Br J Cancer 2018;120:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15:234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lippitz BE, Harris RA. Cytokine patterns in cancer patients: A review of the correlation between interleukin 6 and prognosis. Oncoimmunology 2016;5:e1093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kauffmann-Guerrero D, Kahnert K, Kiefl R, Sellmer L, Walter J, Behr J, et al. Systemic inflammation and pro-inflammatory cytokine profile predict response to checkpoint inhibitor treatment in NSCLC: a prospective study. Sci. Reports 2021;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kurzrock R, Hickish T, Wyrwicz L, Saunders M, Wu Q, Stecher M, et al. Interleukin-1 receptor antagonist levels predict favorable outcome after bermekimab, a first-in-class true human interleukin-1α antibody, in a phase III randomized study of advanced colorectal cancer. Oncoimmunology 2019;8:1551651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zerdes I, Wallerius M, Sifakis E, Wallmann T, Betts S, Bartish M, et al. STAT3 activity promotes programmed-death ligand 1 expression and suppresses immune responses in breast cancer. Cancers (Basel) 2019;11:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files or from the corresponding author upon reasonable request. scRNA-seq and NanoString data generated in this study are publicly available in Gene-Expression Omnibus (GEO) at GSE212227 and GSE208247, respectively. Data used for the survival analysis were obtained from the TCPA Portal at TCGA-SKCM-L3-S64.