Cutaneous, acral, and mucosal melanomas have alterations in components of MAPK, PI3K, p53, p16, and telomere pathways but different mechanisms of activation or inactivation, whereas uveal melanomas are genomically distinct.
Abstract
Melanoma is a cancer of melanocytes, with multiple subtypes based on body site location. Cutaneous melanoma is associated with skin exposed to ultraviolet radiation; uveal melanoma occurs in the eyes; mucosal melanoma occurs in internal mucous membranes; and acral melanoma occurs on the palms, soles, and nail beds. Here, we present the largest whole-genome sequencing study of melanoma to date, with 570 tumors profiled, as well as methylation and RNA sequencing for subsets of tumors. Uveal melanoma is genomically distinct from other melanoma subtypes, harboring the lowest tumor mutation burden and with significantly mutated genes in the G-protein signaling pathway. Most cutaneous, acral, and mucosal melanomas share alterations in components of the MAPK, PI3K, p53, p16, and telomere pathways. However, the mechanism by which these pathways are activated or inactivated varies between melanoma subtypes. Additionally, we identify potential novel germline predisposition genes for some of the less common melanoma subtypes.
Significance:
This is the largest whole-genome analysis of melanoma to date, comprehensively comparing the genomics of the four major melanoma subtypes. This study highlights both similarities and differences between the subtypes, providing insights into the etiology and biology of melanoma.
This article is highlighted in the In This Issue feature, p. 2711
INTRODUCTION
The first solid cancer analyzed by whole-genome sequencing (WGS) a decade ago was a cutaneous melanoma (CM) cell line (1). The study highlighted that among the large number of somatic mutations, those considered “driver” mutations were rare, implying that the vast majority of aberrations were passenger events. The authors also showed that, as expected, the predominant mutational signature was due to ultraviolet radiation (UVR) exposure. Since that seminal publication, there has been a wealth of studies further describing the genomic landscape of melanoma, extending beyond the most common melanoma subtype (CM; refs. 2–12) to include acral melanoma (AM; arising from the palms, soles, or nail beds; refs. 3, 4, 7, 8, 13–15), mucosal melanoma (MM; arising from mucous membranes; refs. 4, 7, 16–21), and uveal melanoma (UM; arising from the uveal tract of the eye; refs. 7, 22–27). Even so, these studies have been limited in that most relied on whole-exome sequencing (WES) rather than on WGS and often sequenced relatively small numbers of tumors. Larger studies have included The Cancer Genome Atlas (TCGA) CM study (n = 333; ref. 2) and two meta-analyses of more than 1,000 melanoma genomes (mostly WES and CM subtype; refs. 28, 29). Recent reviews of AM (30) and MM (31) genomics present comprehensive summaries of driver gene mutations and copy-number aberrations in these melanoma subtypes based on published WGS, WES, and targeted sequencing studies. Curtin and colleagues (32) performed the first genomic comparison of melanoma subtypes, looking at copy-number alterations (CNA) and BRAF and NRAS hotspot mutations in 126 tumors. A recent meta-analysis by the same group compared acral and mucosal subtypes (33), but the majority of other studies have focused on a single melanoma subtype and were thus unable to compare mutational events between subtypes.
Here, we present the largest WGS study of melanoma to date, totaling 570 tumors with matched germline DNA across the four major melanoma subtypes, and report the results of a comprehensive genomic analysis, highlighting similarities and differences that provide insights into the etiology, biology, and disease modifiers of melanoma, which impact therapeutic options for patients. In addition, we characterize how germline variants associate with the somatic mutation landscape and identify potential novel melanoma predisposition genes for some of the less common melanoma subtypes.
RESULTS
Genomic and Clinical Summary
WGS was conducted for melanomas and matched germline DNA from 570 donors (Supplementary Table S1). RNA sequencing (RNA-seq) was performed on 230 tumors and methylation arrays on 144 tumors. Tumors were of the four major melanoma subtypes: 104 UM, 87 AM, 76 MM, and 303 CM (including 22 occult primaries; Fig. 1A). Overall, the median age at diagnosis of the primary tumor was 61 years (range, 15–97), and 56% were male. WGS data for 464 tumors have been described previously (4, 14, 16, 26) and were reanalyzed here.
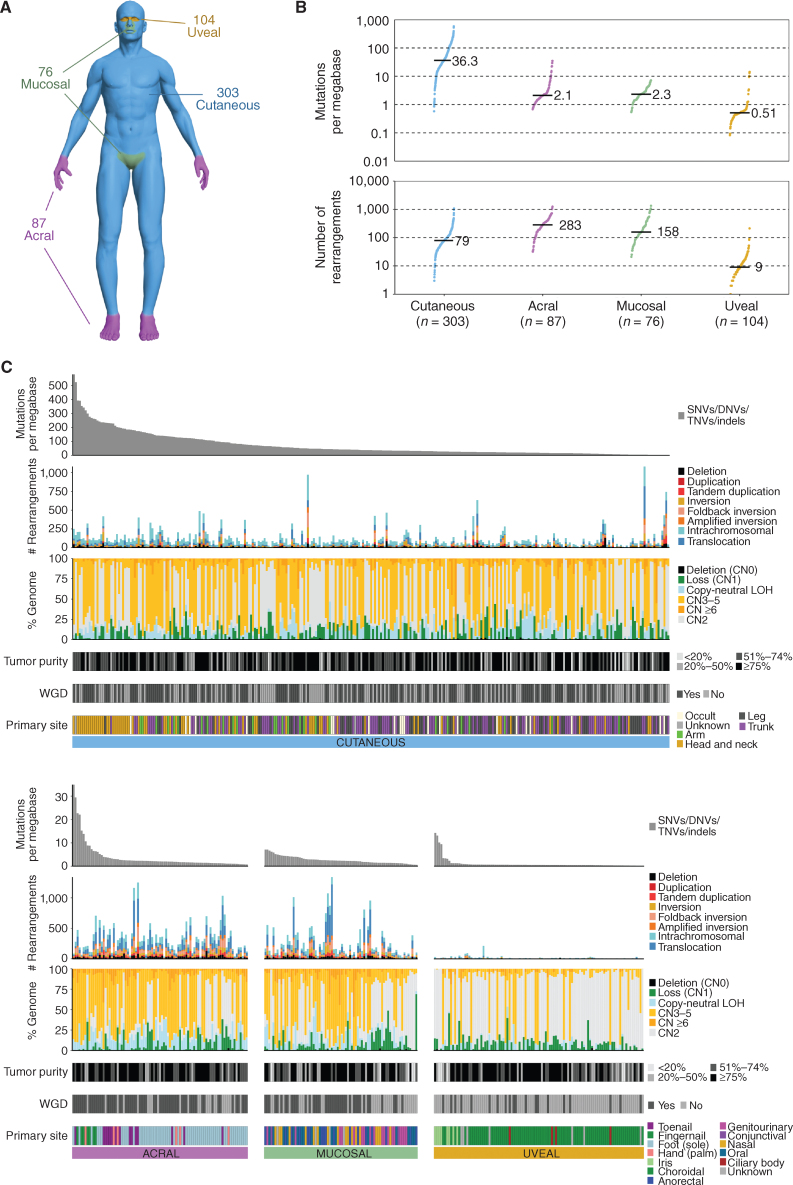
Figure 1.
Genomic overview of the cohort. A, Distribution of the melanoma subtypes in the cohort. B, Mutations per megabase of SNVs and indels (top) and number of structural rearrangements (bottom) in each subtype. Each point represents a tumor, with the black line and number representing the median for each subtype. C, Overview of the genomic alterations within each tumor sample. Patients are grouped into separate plots based on melanoma subtype (cutaneous, acral, mucosal, and uveal). The upper plot shows the mutation burden of SNVs/DNVs/TNVs and indels as mutations per megabase across the entire genome. The patients within each subtype plot are ordered from highest to lowest mutation burden. The second plot from the top shows the number of structural rearrangements per tumor, including the predicted type of rearrangements: deletion, duplication, tandem duplication, inversion, foldback inversion, amplified inversion, intrachromosomal, or translocation. The third plot shows the percentage of the genome affected by CNAs including deletion (copy number 0: CN0), loss of 1 copy (loss CN1), copy number 2 (CN2), copy-neutral loss of heterozygosity (LOH), copy-number gain with 3 to 5 copies (CN3–5), and copy-number gain (CN ≥6). Next, tumor purity in each tumor is shown, followed by the presence of WGD per tumor. Finally, the primary site of each tumor is shown.
As previously reported (4, 14, 16, 26), the number of single-nucleotide variants (SNV)/insertions and deletions (indel) and chromosome structural variants (SV)/rearrangements differed significantly between subtypes (Fig. 1B and C). CM had the highest tumor mutation burden [TMB; genome-wide SNVs/dinucleotide variants (DNV)/trinucleotide variants (TNV)/indels per megabase], followed by MM, AM, and UM (Fig. 1B). AM had the highest number of SVs, followed by MM, whereas UM had the lowest number (Fig. 1B). The proportion of tumors with whole-genome doubling (WGD) differed between subtypes (χ2, P = 2.2 × 10−12), with AM having the highest percentage (71%), followed by MM (54%), CM (54%), and UM (19%; Fig. 1B).
Within each subtype, TMB differed significantly with the primary tumor site (CM, Kruskal–Wallis, P = 4.2 × 10−11; AM, Kruskal–Wallis, P = 8.5 × 10−6; and UM, Kruskal–Wallis, P = 3.4 × 10−5) except for MM (Kruskal–Wallis, P = 0.092; Supplementary Fig. S1A–S1D). Head and neck CM had the highest TMB, as did fingernail tumors for AM and iris tumors for UM. There was no difference in the number of SVs according to primary melanoma site within each subtype (Supplementary Fig. S1E–S1H) except for a trend in MM, with oral tumors having more SVs (Kruskal–Wallis test, P = 0.05). TMB differed between CM subtypes, with desmoplastic and lentigo maligna melanomas having the highest TMB (Supplementary Fig. S1I), but there was no difference in SV burden (Supplementary Fig. S1J).
Mutational Signatures and Strand Bias
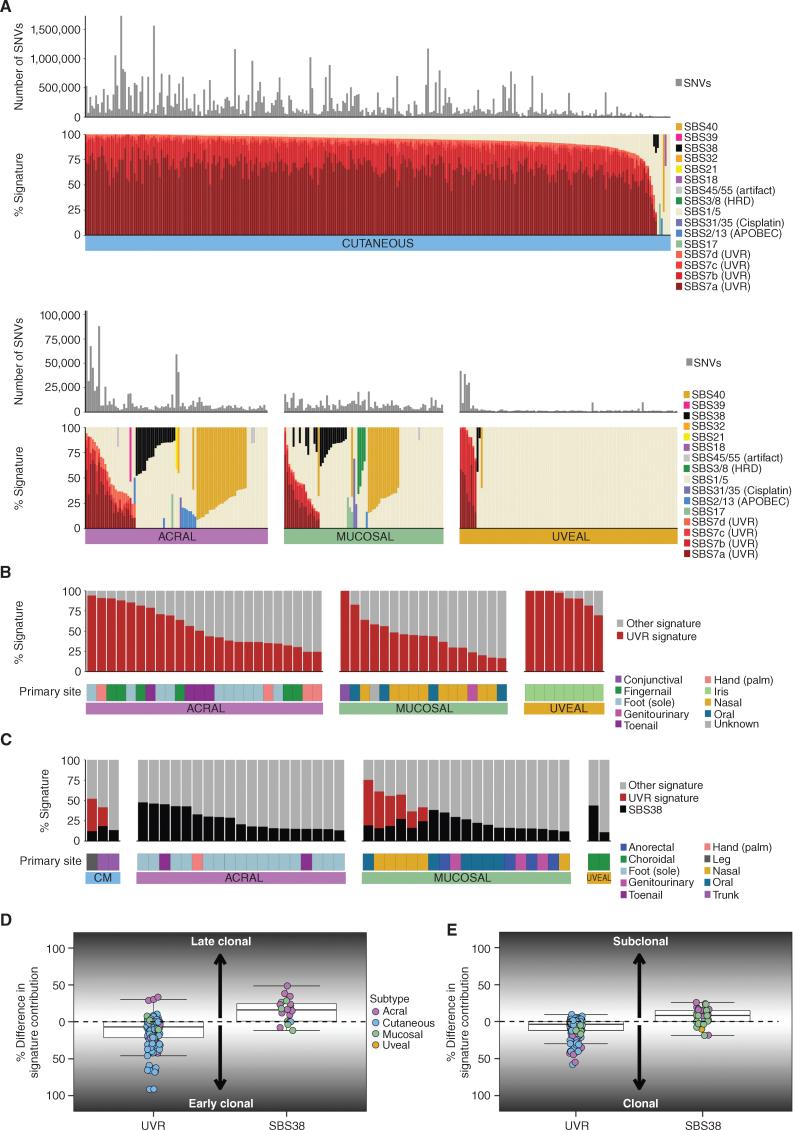
Mutational signature analysis of single base substitutions (SBS; Fig. 2A), doublet base substitutions (DBS; Supplementary Fig. S2A), and indels (ID; Supplementary Fig. S2B) was performed. De novo signature analysis did not identify any new signatures when compared with the Catalogue of Somatic Mutations in Cancer (COSMIC) database (v3). Signatures associated with UVR (SBS7a–d) were dominant in CM, with 293 of 303 having >50% UVR signature, and only 7 tumors lacking any evidence of a UVR signature. SBS7a was the dominant UVR signature in CM, with a mean contribution of 65% of total mutations per tumor, followed by SBS7b (21%), SBS7c (3%), and SBS7d (3%). Collectively, CM tumors without a UVR signature had fewer mutations (mean 1.4 per megabase, as expected in the absence of UVR) compared with tumors with a UVR signature (mean 72.1 mutations per megabase, Mann–Whitney, P = 7.3 × 10−6) and more rearrangements (Mann–Whitney, P = 0.036, mean of 105 rearrangements with UVR signature vs. 391 without UVR signature). UVR signatures were also present in subsets of UM, AM, and MM as previously reported (refs. 14, 16, 26; Fig. 2B) and were associated with more sun-exposed primary sites (AM, more common in hands than feet; MM, upper body sites; UM, iris). Non-CM tumors with a UVR signature contribution >50% had higher TMB (Supplementary Fig. S2C; AM, Kruskal–Wallis, P = 2.2 × 10−5; UM, Mann–Whitney, P = 5.5 × 10−6; MM, nonsignificant trend, Kruskal–Wallis, P = 0.089). The contribution of SBS7a was highest in CM; SBS7b was highest in UM, and SBS7d was highest in AM (Supplementary Fig. S2D). Tumors with UVR SBS signatures usually had evidence of DBS1 and ID13 (Supplementary Fig. S2A and S2B), which are similarly associated with UVR exposure (34).
Figure 2.
Mutational signatures. A, The number of SNVs and proportion of SBS signatures in each melanoma subtype. If the etiology for a signature is known, this is listed in parentheses after the signature name. HRD, homologous repair defect. B, The contribution of UVR signature and tumor primary site in non-CM tumors that have a UVR signature present (>0% UVR contribution). C, The contribution of SBS38 in each subtype in tumors that have an SBS38 signature present (>0% SBS38 contribution). D, Difference per tumor between the percentage of early and late clonal mutations attributed to UVR or SBS38 mutational signatures. E, Difference per tumor between the percentage of clonal and subclonal mutations attributed to UVR or SBS38 mutational signatures.
SBS38, a signature reported primarily in melanomas (34), was found in tumors that generally lacked UVR signatures and were mostly non-CM, occurring in only three CM (Fig. 2C). There is no known etiology for SBS38, and we did not identify associations with other genomic or clinical features.
The Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium reported the evolutionary history of 2,778 cancer samples, including the timing of mutational signatures, by identifying clonal events present in all cells, as well as subclonal mutations present only in a subset of cells, as well as defining early and late clonal events as those preceding or succeeding copy-number (CN) gains (35). We applied similar techniques to melanomas containing UVR and SBS38 signatures in order to examine when during tumor evolution the mutational processes gave rise to these signatures. The timing of mutations with a UVR or SBS38 signature differed. UVR signatures were a very early event, with the contribution to early clonal mutations generally being higher when compared with late clonal mutations (Fig. 2D), and UVR-related mutations were predominantly clonal rather than subclonal (Fig. 2E), agreeing with previous observations (35) and confirming that UVR is the early initiating event in tumors with this signature. In contrast, the SBS38 signature often arose later, again agreeing with previous observations (35), with a higher proportion being late clonal (Fig. 2D), and in subclonal mutations rather than clonal mutations (Fig. 2E), indicating that this signature is not associated with the initiating event for tumor development in these cancers.
As previously reported (34), mutational signatures SBS7a and SBS7b showed a strong transcriptional strand bias, consistent with transcription-coupled repair (TCR) acting on UVR-associated DNA damage. To characterize this bias further, we examined C>T mutations at dipyrimidines TpC and CpC separately (34). The bias was stronger for mutations at CpC (median 34%) than for TpC sites (median 26%, Wilcoxon signed rank, P < 0.0001). This difference was mainly driven by TpC sites showing a weaker bias for intronic mutations (median 25%) than exonic mutations (median 32%, Wilcoxon signed rank, P < 0.0001), whereas for CpC mutations, the bias was similar for intronic (median 32.4%) and exonic regions (33.8%; Supplementary Fig. S2E). Consistent with strand bias being due to TCR, it was stronger in genes highly expressed in CM (ref. 2; Supplementary Fig. S2F and S2G).
The predominant mutational signatures in non-CM were SBS1, SBS5, and SBS40, which are present in most cancer types and correlated with age (ref. 34; Fig. 2A). Other signatures were present in some tumors, mostly non-CM: mismatch repair signature (SBS21, one tumor), signatures of unknown etiology (SBS17a and b, five tumors), cisplatin signatures (SBS31/SBS35, three tumors), APOBEC signatures (SBS2/13, 11 tumors), and homologous repair defect (HRD) signatures (SBS3/SBS8, four tumors). One tumor with SBS8 had methylation of the BRCA1 promoter (Supplementary Fig. S3A), BRCA1 loss of heterozygosity (LOH), low BRCA1 expression (Supplementary Fig. S3B), and a high number of nonclustered SVs (Supplementary Fig. S3C). For the other three tumors with HRD signatures, methylation data were unavailable, and no BRCA1/2 or PALB2 pathogenic germline or somatic mutations were observed.
We also ran a recently described algorithm Signature Fit Multi-Step (FitMS), which was applied to WGS from more than 12,000 tumors from the UK National Health Service to identify 40 new SBS reference signatures (36). When we applied this algorithm to our melanoma cohort (Supplementary Fig. S4), we saw roughly similar patterns for COSMIC SBS signatures that are also identified by the FitMS algorithm, as shown in Fig. 2A, with some exceptions being the UVR signatures SBS7b and d and COSMIC SBS40, which were not identified by the FitMS algorithm in Degasperi and colleagues (36), and therefore could not be assigned with this cohort. Of note, we identified an additional signature, SBS96, in two uveal tumors, both with germline MBD4 mutations, in keeping with the reported association (36, 37). There were also smaller contributions of rare signatures of unknown etiology in other tumors, including SBS99 in AM, SBS126 in AM and MM, and SBS103 in UM.
Rearrangements and CNAs
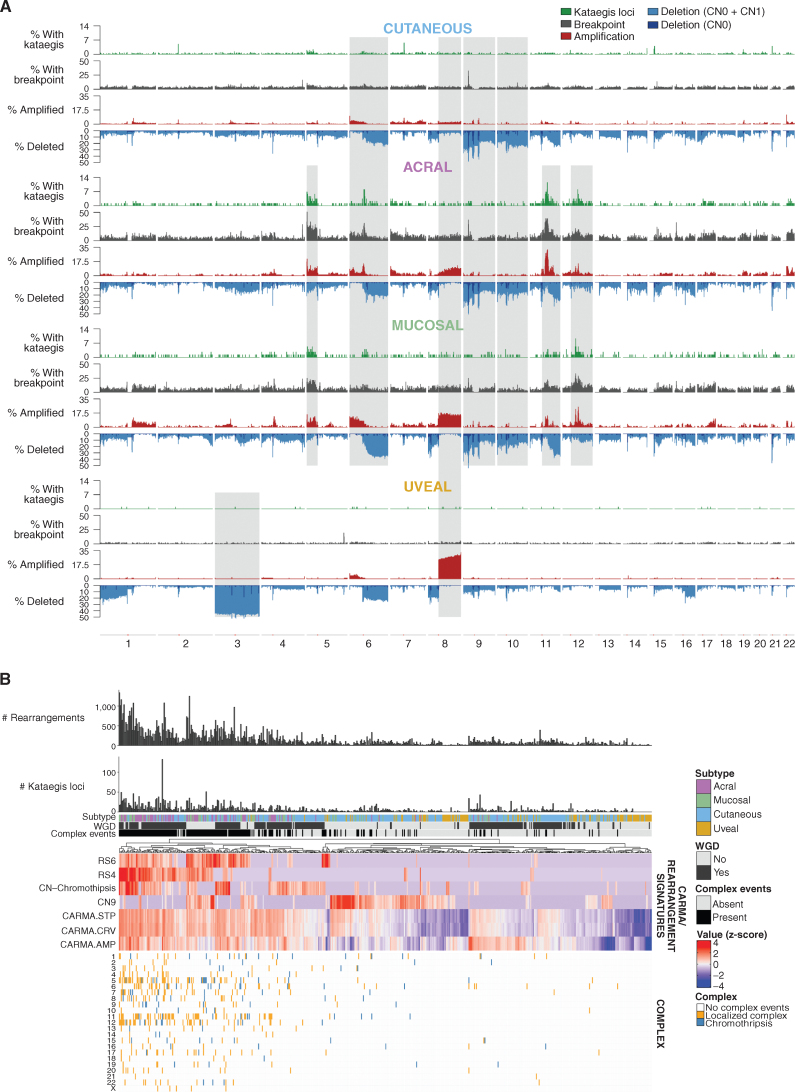
A comparison of tumors with kataegis loci, rearrangements, and CNAs across the genome in each subtype was undertaken (Fig. 3A). There were very few SVs, kataegis loci, or CNAs in UM; only chromosome 3 loss and 8q gain were common events, consistent with prior reports (25, 26). As expected (38, 39), all metastatic UM presented with chromosome 3p LOH (hemizygous or copy-neutral) or an SF3B1:p.Arg625 driver mutation compared with 73% of the primary tumors (P = 0.03). Chromosome 8q gain was also seen in AM and MM and, to a lesser extent, in CM. The CM, AM, and MM subtypes displayed a number of similar CNAs, including gain of chromosome 6p and loss of 6q, 9, and 10. AM and MM had further similarities, with frequent kataegis loci, rearrangement breakpoints, and amplifications on chromosomes 5p, pericentromeric 11q, and pericentromeric 12q.
Figure 3.
Rearrangements, CNAs, and complex rearrangement events. A, Distribution of (top to bottom) the percentage of tumors with kataegis loci (green), a rearrangement breakpoint (gray), a CNA with amplifications (red), and deletions (CN0 + CN1, blue) in 100-kb regions across the genome in each melanoma subtype. Regions of similarity and difference in the pattern of kataegis, breakpoints, and CNA between subtypes are shaded gray. B, Complex events in each tumor. From top to bottom: number of rearrangements, number of kataegis loci, melanoma subtype, presence of WGD, presence of complex rearrangement events, heat map of measures of complexity, and presence of complex events in each chromosome in each tumor. For the heat map of measures of complexity, tumors were clustered using various measures that indicate the presence of complex structural rearrangements and chromosomal instability, including chromothripsis-related CN signatures (CN chromothripsis: CN5 + CN6 + CN7 + CN8), diploid chromosomal instability CN signature CN9, clustering rearrangement signatures (RS4 and RS6), and CARMA features of amplification (AMP) and complexity (STP and CRV). For clustering, z-score–transformed values for each factor were used.
De novo analysis of CNA signatures was performed, which were then decomposed into the recently described COSMIC CNA signatures (40). Eleven COSMIC CNA signatures were identified as well as two novel signatures (Supplementary Fig. S5A–S5C). The contribution of each signature was compared with genomic features associated with the proposed etiology (Supplementary Fig. S5D). Diploid signature CN1 was most prevalent in UM and associated with the absence of WGD (Fisher exact, P < 0.001). Chromothripsis-associated signatures (CN5, CN6, CN7, and CN8) were common in AM and MM and associated with WGD (Fisher exact, P < 0.001) and complex rearrangements (Fisher exact, P < 0.001) as expected. CN9, associated with diploid chromosomal stability, was present in all subtypes and associated with a lack of WGD (Fisher exact, P < 0.001). CN20 was present in all subtypes, most frequently in AM; is of unknown etiology; and mostly occurred in tumors with WGD. Artifact signature CN23 (which occurred in tumors with a lack of CN events in which segmentation algorithms often perform poorly) was present in 12% of tumors. Novel signature CNV48C was most common in AM and MM and was characterized by heterozygous segments of 0–1 megabase, and CN gains were often present (Supplementary Fig. S5B). Novel signature CNV48D, characterized by segments of LOH <100 kb, was common in CM, with high (>50%) contribution in 18 CM, including eight CM cell lines (Supplementary Fig. S5C).
Tumors were clustered using various measures that indicate the presence of complex structural rearrangements and chromosomal instability (Fig. 3B). Highly complex tumors were common in AM and MM. These tumors had a high number of SVs, kataegis loci, and complex localized events such as chromothripsis, particularly on chromosome 5 (in the region of TERT), chromosome 11 (CCND1), and chromosome 12 (CDK4 and MDM2) as previously reported (14, 16, 21).
Recurrent breakpoints (Supplementary Fig. S6A–S6D) included regions containing tumor suppressors such as CDKN2A (in CM, AM, and MM); PTEN (CM and MM); 17q11.2 near NF1 (AM and MM); and 15q13.3 near SPRED1 (CM, AM, and MM). Some regions were close to centromeres or telomeres, including 6q12 (AM and CM) and 7p22.3 (AM). Other regions across MACROD2 (CM) and the long noncoding RNA (lncRNA) LINC00290 (CM, AM, and MM) have been suggested as fragile sites (41, 42). Regions with high numbers of SVs close to cancer genes included TERT (CM, AM, and MM), CCND1 (AM), and CDK4 (AM and CM). No recurrent SV regions were identified in UM. Focal amplifications and deletions were identified in each subtype (Supplementary Fig. S7A–S7D). Deletions in CDKN2A and NF1 (CM, MM, and AM) and PTEN (CM and MM) were identified. Amplifications were seen in regions containing TERT, CCND1, CDK4, and MDM2 (CM, MM, and AM), KIT (MM), BRAF (CM and MM), MAP2K1 (AM and MM), and EP300 (CM and AM).
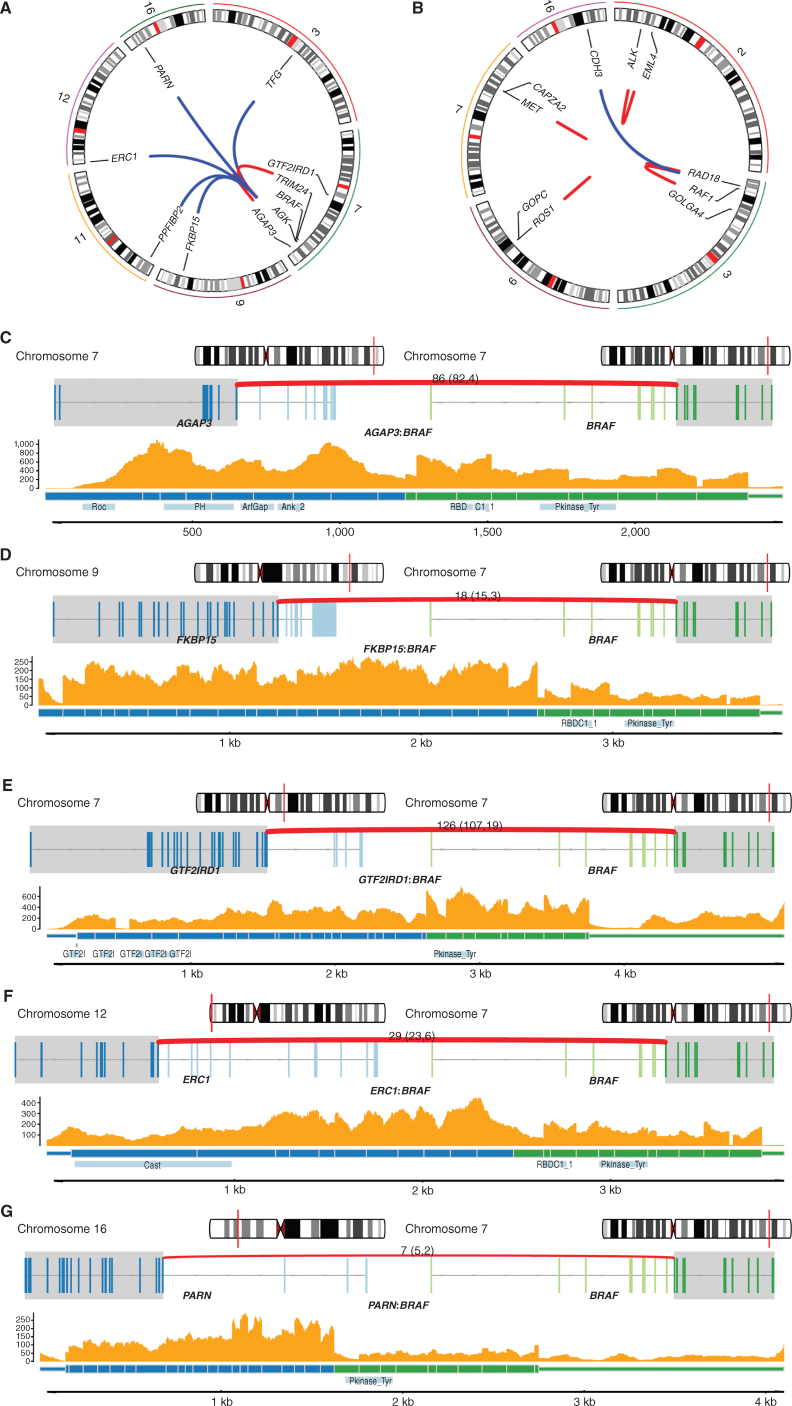
Of the 83,961 rearrangements identified using WGS, 53,506 had a breakpoint within a gene (Supplementary Table S2) and 1,279 were predicted to result in in-frame fusion genes. Fusions in cancer genes were identified (Supplementary Table S2; Fig. 4A and B) and included nine BRAF (four CM, three AM, two MM; Fig. 4A and C–G), one ALK, one MET, one ROS1 (14), and three RAF1 fusions (ref. 4; Fig. 4B; Supplementary Table S2). No NTRK1/2/3 fusions, which have been reported rarely in melanoma (43), were identified.
Figure 4.
Cancer gene fusions A, Circos plot showing the nine BRAF gene fusion events and the corresponding fusion gene partner. Only chromosomes containing the genes involved in the gene fusions are shown. B, Circos plot showing RAF1, ALK, MET, and ROS1 gene fusion events of interest and the corresponding fusion gene partner. Only chromosomes containing the genes involved in the gene fusions are shown. C–G, Examples of BRAF fusion genes in tumors with RNA-seq available. For each fusion gene, the top plot shows the location in the chromosome of each fusion gene partner, and the middle plot shows the exons involved in each gene (transcripts are collapsed). The bottom plot shows the read depth of both fusion gene partners by RNA-seq, predicted fusion protein, and the protein domains (including BRAF kinase domain) involved. C and D,AGAP3:BRAF fusion and FKBP15:BRAF fusion, respectively. E–G,GTF2IRD1:BRAF fusion, ERC1:BRAF fusion, and PARN:BRAF fusion, respectively.
Noncoding Mutations
A consensus approach using multiple tools was used to identify significantly mutated noncoding genomic elements by analyzing somatic SNVs and indels in regulatory regions [promoters, 5′ untranslated regions (UTR), 3′ UTRs, and enhancers] and lncRNAs (gene body and promoter; Supplementary Table S3). Mutations in the TERT promoter and 5′ UTR were identified in both AM and MM, and the SSBP1 promoter (mutated in four tumors) was identified in MM. In UM, the 5′ UTR of TUBB8B (RP11–683L23.1 in GRCh37) was mutated in three tumors. In CM, 491 sites were identified as significantly mutated: 333 promoter elements (including TERT), 149 5′ UTR elements, two lncRNAs, and seven lncRNA promoters. For 42 elements, mutations were associated with altered gene expression (Mann–Whitney, P < 0.05, none were significant after correction for multiple testing) and included promoters of RPS27 and RNF185, previously reported as recurrently mutated in CM (4); IQGAP1, reported to be involved in CM metastasis (44); and RTEL1, a telomere maintenance gene. Although noncoding mutations in 42 CM elements were associated with altered gene expression (Supplementary Table S3), the PCAWG study of noncoding mutations (45) showed that noncoding drivers are rare, and most CM noncoding changes are likely to be passenger mutations as a result of a high TMB due to UVR exposure. Therefore, it is likely that many of the significant noncoding mutations identified here are also nonfunctional passengers.
Telomere Maintenance Genes and Telomere Length
Mutations in telomere maintenance genes were common in all melanoma subtypes except UM (Supplementary Fig. S8A). TERT aberrations were present in 88% of CM, 46% of AM, and 34% of MM (Supplementary Fig. S8B). TERT promoter mutations were more common in CM (84%) than in AM (10%) and MM (16%). Most promoter mutations occurred at the −124 and −146 sites, and one patient with AM (MELA_0337) carried a germline TERT mutation (−57). Other types of TERT aberrations were more common in AM and MM and included TERT amplifications present in 17% of AM, 12% of MM, and 2% of CM. SVs within 20 kb upstream of TERT, postulated to result in enhancer hijacking (14, 46), were present in 29% of AM, 13% of MM, and 4% of CM, often (62%, 29/47) co-occurring with TERT amplification. ATRX and DAXX loss-of-function (LOF) mutations and mutations in TP53 and RB1 have been associated with telomere lengthening (47). ATRX LOF mutations were present in 2% of CM, 3% of AM, and 8% of MM, but only one DAXX LOF mutation was observed (in an MM tumor). Mutations in TP53 and RB1 were found in all subtypes but were more common in CM. Relative telomere length differed between melanoma subtypes (Kruskal–Wallis, P = 0.003), with shorter telomeres in CM (Supplementary Fig. S8C). Melanomas with TERT aberrations had shorter telomeres, and tumors with ATRX LOF mutations had longer telomeres (Supplementary Fig. S8D). This observation was complicated by the fact that both telomere length and prevalence of TERT aberrations correlated with subtype. In a multivariable linear regression analysis including tumor subtype and presence of ATRX, TERT, RB1, or TP53 mutations, only ATRX modification was associated with longer telomere length (P = 0.0002).
TCGA Molecular Categories and Significantly Mutated Gene Analysis
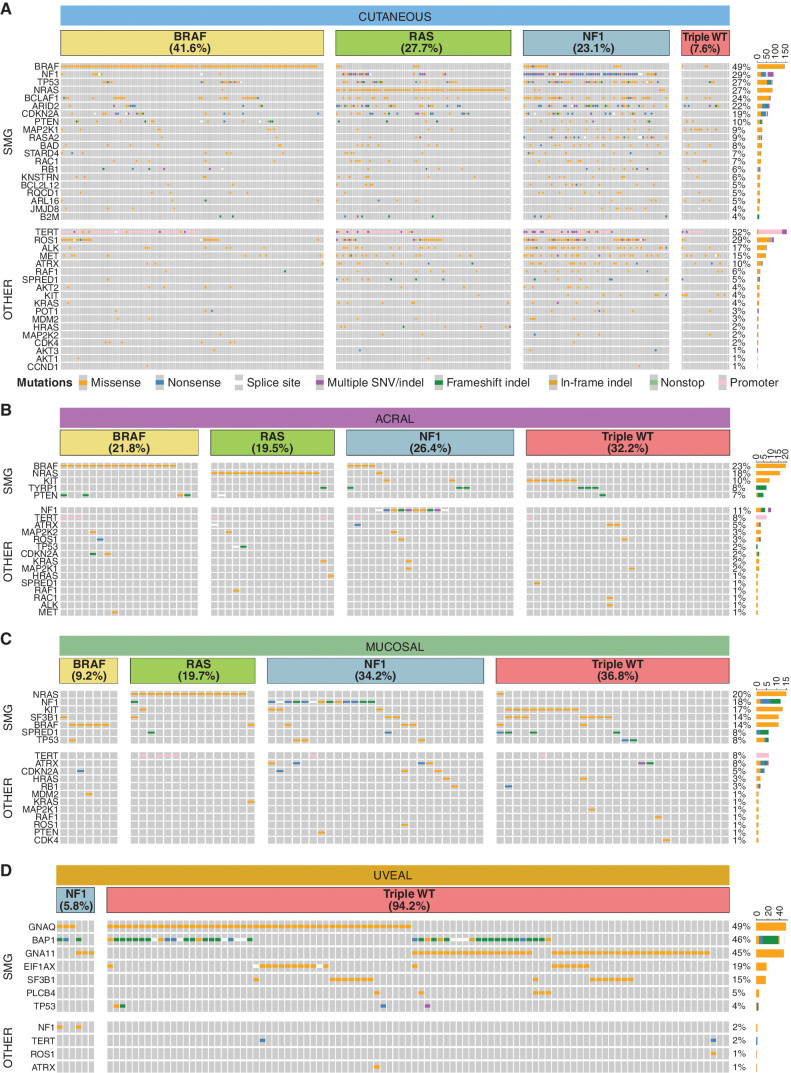
Melanoma samples were grouped by TCGA molecular categories. In CM, 42% of tumors were BRAF category, followed by RAS (28%), NF1 (23%), and triple wild-type (WT; 8%, Fig. 5A; Supplementary Table S1). The proportions differed significantly (χ2, P = 4 × 10−6) compared with the TCGA cohort (2) of 316 CMs (BRAF 47%, RAS 29%, NF1 9%, triple WT 15%), with the higher proportion of NF1 mutants in our cohort likely due to the usage of WGS, which allowed detection of SVs that lead to NF1 inactivation (n = 39). For desmoplastic melanomas (n = 28), 79% were NF1 category tumors, agreeing with previous reports (48). As expected, the proportions of TCGA molecular categories were different in the other subtypes, with 94% of UMs being triple WT. BRAF and RAS categories were less common in AM and MM, whereas NF1 and triple WT were more common than in CM (Fig. 5B–D).
Figure 5.
SMGs. Oncoplots of SMGs (top) and other melanoma genes (other, bottom) in cutaneous (A), acral (B), mucosal (C), and uveal (D) tumors. Each subtype is separated into TCGA molecular categories. Oncoplots show SNVs and indels only. Tumors without an SNV or indel in a TCGA category gene that were assigned to that TCGA category had other structural alterations of the gene (e.g., a BRAF fusion).
When comparing the TMB of the TCGA categories in CM, there was a significant difference overall (Kruskal–Wallis, P < 2 × 10−16), with the NF1 subtype having the highest TMB (Supplementary Fig. S9A). The number of SVs in CM differed across TCGA categories (Supplementary Fig. S9B, Kruskal–Wallis, P = 0.013), with more SVs in the triple-WT and NF1 tumors, but the proportion of tumors with complex rearrangements did not differ between categories (χ2, P = 0.4). In AM, there was an overall difference in TMB (Kruskal–Wallis, P = 0.042) and number of SVs (Kruskal–Wallis, P = 0.022) across TCGA categories (Supplementary Fig. S9C and S9D), but no significant difference in the proportions of tumors with complex rearrangements (χ2, P = 0.068). There was no difference between TCGA categories for TMB (Kruskal–Wallis, P = 1), SVs (Kruskal–Wallis, P = 0.1), or complex rearrangements (χ2, P = 0.09) in MM (Supplementary Fig. S9E and S9F).
Significantly mutated gene (SMG) analysis of SNVs and indels using a consensus approach from multiple tools identified 20 CM, five AM, seven MM, and seven UM SMGs (Fig. 5; Supplementary Tables S4 and S5). Additionally, mutations occurred in other genes in important melanoma development pathways (MAPK, PI3K, p53, cell cycle, and telomere maintenance; Fig. 5, bottom). CM SMGs (Fig. 5A) included well-known melanoma drivers: BRAF, NF1, NRAS, MAP2K1, PTEN, RASA2, RAC1, CDKN2A, TP53, RB1, B2M, and ARID2. Other genes previously reported as SMGs (2, 28, 29) included RQCD1 (CNOT9) a transcription (co)factor that is a potential oncogene (49) and KNSTRN, a kinetochore gene that is recurrently mutated in cutaneous squamous cell carcinoma (50). Other SMGs included three BCL-2 associated genes—BCLAF1, BCL2L12, and BAD; however, most mutations in these genes were missense (Supplementary Fig. S10A–S10C). BCLAF1 is a transcription factor that activates apoptosis and can form a DNA damage-induced BRCA1–mRNA splicing complex that contains another melanoma driver gene, SF3B1 (51, 52). BCLAF1 mutations occurred across the protein and included four nonsense mutations, one in-frame insertion, and 89 missense mutations, with 65% of missense predicted as possibly or probably damaging by PolyPhen and 60% predicted as potentially damaging by SIFT. BCLAF1 variants were present in 24.6% (nonsense 1.5%) of TCGA skin cutaneous melanoma (TCGA-SKCM) tumors and 24.6% (nonsense 0.9%) of tumors used in the Conway and colleagues (29) CM meta-analysis. BCL2L12 mutations in CM were concentrated in the first 18 amino acids, with eight at R18W; in addition, there were 15 of the F17F synonymous mutation (53). Interestingly, these recurrent mutations lie within the promoter and 5′ UTR, respectively, of the IRF3 gene on the opposite strand. It is, therefore, possible that the functional consequence of this mutation is on IRF3 rather than BCL2L12, a scenario that warrants further scrutiny. Mutations in BAD affected a noncanonical transcript (ENST00000544785), and there was low evidence for expression of this transcript in CM tumors; therefore, BAD could potentially be a false positive. The functions of the remaining genes—STARD4, ARL16, and JMJD8—do not currently have a confirmed role in cancer; therefore, these genes could potentially be false positives resulting from the high TMB in CM. It has been reported that ETS binding sites can exhibit high neutral mutation rates in melanoma, leading to recurrent mutations that do not confer a selective advantage and therefore resulting in potential false-positive SMGs (28). The genes BAD, BCL2L12, KNSTRN, ARL16, and JMJD8 all had a high proportion of mutations overlapping ETS sites, and this is further evidence for these genes being false positives.
Missense mutations were common in ROS1, ALK, and MET; these mutations were not recurrent and are of unknown functional consequence. Mutations (SNVs/indels) in TP53 were significantly enriched (68%) in desmoplastic melanomas (χ2, P = 2.5 × 10−5). In CM, TP53 gene aberrations were associated with TCGA categories (χ2, P = 4.1 × 10−10), with 70% of NF1 category tumors also having a TP53 aberration [includes SNVs, indels, homozygous deletion, LOH (CN1), and rearrangements]. In comparison, 39% of RAS category, 23% of BRAF category, and 17% of triple-WT category tumors had TP53 aberrations. The prevalence of SNVs, indels, CNAs, and rearrangements in a more comprehensive list of previously identified SMGs, as well as CM SMGs identified in this study, are listed in Supplementary Table S4.
The SMGs identified in non-CM were NRAS, BRAF, TYRP1, PTEN, and KIT in AM (Fig. 5B); NRAS, SPRED1, BRAF, NF1, KIT, TP53, and SF3B1 in MM (Fig. 5C); and BAP1, GNAQ, GNA11, SF3B1, EIF1AX, PLCB4, and TP53 in UM (Fig. 5D)—all previously identified as melanoma drivers (14, 16, 26, 30, 31). In UM, driver mutations in the G-protein signaling pathway were mutually exclusive except for two PLCB4 D630 mutations that co-occurred with GNA11/GNAQ R183H mutations, which have been described previously (25, 26). For the 144 tumors with methylation array data, promoter methylation of tumor suppressor genes TP53, CDKN2A, RB1, ARID2, and PTEN was examined. Methylation of RB1 was observed in one CM tumor (Supplementary Fig. S11A) with low expression (Supplementary Fig. S11B), and CDKN2A (p16 isoform) promoter methylation was observed in 12 tumors (six CM, three AM, and three MM; Supplementary Fig. S11C), with expression of the p16 transcript reduced in almost all (11/12) tumors with methylation (Supplementary Fig. S11D).
Germline Predisposition Genes
Germline WGS data were assessed for variants in cancer predisposition genes to identify potential associations with susceptibility for different melanoma subtypes (Supplementary Table S6). Four patients with CM carried known pathogenic germline CDKN2A variants (54), three of which had a second CDKN2A hit in the tumor. Two POT1 pathogenic variant carriers were seen (one CM/one AM). Four patients (three CM/one AM) carried the pathogenic (55) MITF p.E318K variant. One patient with AM carried a PTEN LOF variant and had clinical features consistent with Cowden syndrome. Another AM case carried a TERT promoter variant (position –57 upstream of the translation start site) that was frequently somatically mutated in CM, previously identified as a germline variant in a large CM family (56), and is here for the first time reported in AM. LOF variants in WRN were identified in two AM cases (both tumors showed LOH) and one patient with UM (without LOH). Four CHEK2 frameshift variants associated with breast and pancreatic cancer (57) were carried by three AM cases (all c.1100del) and one patient with CM (c.1263del). A BRCA1 frameshift deletion (c.70_80del) was seen in an individual with CM, but the tumor had lost the variant allele. In UM, two cases had BAP1 LOF variants and two had MBD4 LOF variants, all with second hits (LOH) in the tumors.
To identify potential novel predisposition genes, we searched for other germline LOF variants associated with somatic second hits. Two AM cases carried nonsense mutations in AIM1 (absent in melanoma 1), a gene associated with the control of tumorigenicity in CM (58), and both tumors had lost the WT allele, an association that has not previously been reported.
A study of 5,954 tumors across 22 cancer types correlated germline polymorphisms and somatic events (59). Of the 36 loci associated with somatic mutations, none were confirmed in our melanoma cohort.
Factors Associated with TMB
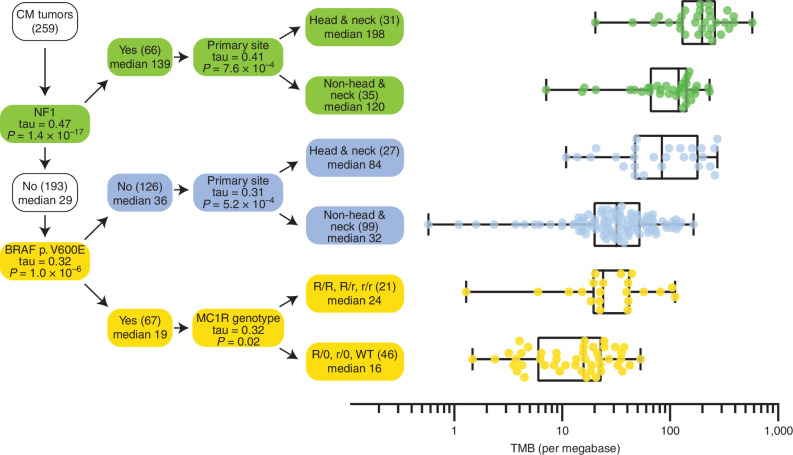
High TMB is associated with favorable response in patients treated with immune-checkpoint inhibitors; therefore, we explored factors correlated with TMB in CM. TMB was strongly associated with the type of MAPK pathway driver mutation as previously reported (60). NF1 melanomas had the highest TMB, whereas BRAF melanomas [odds ratio (OR), 0.33; 95% confidence interval (CI), 0.24–0.45], particularly p.V600E mutations (OR, 0.25; 95% CI, 0.16–0.39), and RAS p.Q61 melanomas (OR, 0.72; 95% CI, 0.42–0.88) had lower TMB. High TMB was also associated with head and neck tumors (OR, 2.9; 95% CI, 2.4–3.6), old age (OR, 1.7; 95% CI, 1.3–2.3), and being male (OR, 1.7, 95% CI, 1.3–2.4; Supplementary Fig. S12A). We built a decision tree to determine the strongest predictor of TMB in each subgroup (Fig. 6). Primarily, TMB is associated with TCGA categories, splitting melanomas into three groups: NF1 melanomas with high TMB, BRAF p.V600E melanomas with low TMB, and melanomas with other drivers (including RAS and BRAF p.V600K), with intermediate TMB. Among the CM with a BRAF p.V600E mutation, TMB was associated with the MC1R genotype—that is, patients with R or r variants in both copies of MC1R had significantly higher TMB than patients with at least one WT allele (Kendall's τ, Benjamini–Hochberg (BH)-adjusted P = 0.02). Although TMB is affected by the MC1R genotype, it is more strongly affected by the type of MAPK driver somatic mutation, which was not assessed in previous studies of TMB and MC1R association (61, 62).
Figure 6.
Decision tree predicting TMB in CM. Starting with CM tumors in the root node (top left), the variable that had the highest correlation with TMB was identified (i.e., NF1 mutation status). In the branching node, the correlation (Kendall's tau) with TMB and associated Bonferroni corrected P value is displayed. The branching node splits the cohort into two groups with size (in brackets), and median TMB is displayed in the daughter nodes. The splitting is repeated recursively until no more significant variables can be found. For each leaf node in the decision tree, the distribution of TMB is displayed on the right-hand side in the form of box-and-whisker plots with whiskers from minimum to maximum. Medians are displayed as a line within the box defined by the quartiles.
Associations with Age of Onset
The age at diagnosis for CM varied between different TCGA categories, with NF1 (mean, 66.0 years) and RAS (mean, 64.6 years) patients being significantly older than BRAF patients (mean, 51.5 years, Mann–Whitney test, P = 2 × 10−9; Supplementary Fig. S12B). The early onset for BRAF melanomas was more strongly associated with p.V600E mutation (mean, 48.6 years) than p.V600K (mean, 59.2 years, Mann–Whitney test, P = 0.01). As reported in a subset of this cohort (61), patients with variants in MC1R developed CM earlier. An additive model counting both R and r alleles gave a slightly better fit than a model counting only R alleles, implying that r alleles also affect age at onset. Multivariable regression analysis showed that somatic BRAF mutation and germline MC1R genotype are independently associated with age at onset, which is in line with MC1R genotype not being significantly associated with BRAF mutations (χ2, P = 0.74), confirming a meta-analysis (63).
Regression analysis of the age of onset in AM identified three significant factors (Supplementary Fig. S12C). As observed in other cancers (64), patients with greater numbers of pathogenic germline variants in “cancer” genes had earlier onset (2 years per variant, 95% CI, 0.7–3.2, BH-adjusted P = 0.002). Men were younger (mean, 65.9 years) compared with women (mean, 72.3 years, BH-adjusted P = 0.036) and AM with a BRAF hotspot mutation were on average 10.8 years younger (BH-adjusted P = 0.002). Multivariable analysis confirmed that BRAF mutation, gender, and germline variant burden are independently associated with early onset (P < 0.01).
As high nevus count is a major risk factor for CM (65), we calculated nevus polygenic risk scores (PRS) from the germline WGS data. PRS did not correlate with TCGA category (Kruskal–Wallis, P = 0.93); however, within the BRAF category, patients with a p.V600E mutation had higher nevus PRS than those with a p.V600K mutation (Mann–Whitney, P < 0.0001).
Survival
Overall survival (OS) was analyzed separately for patients with CM (n = 284) and AM (n = 83) with available survival data (Supplementary Table S7). In CM, the presence of ulceration, high mitotic rate, and lymphovascular invasion were clinical features associated with poor OS. In AM, ulceration was associated with poor OS. The only genomic features associated with poor OS were long telomeres in CM and TCGA RAS category in AM.
Pathways Important in Melanoma Development
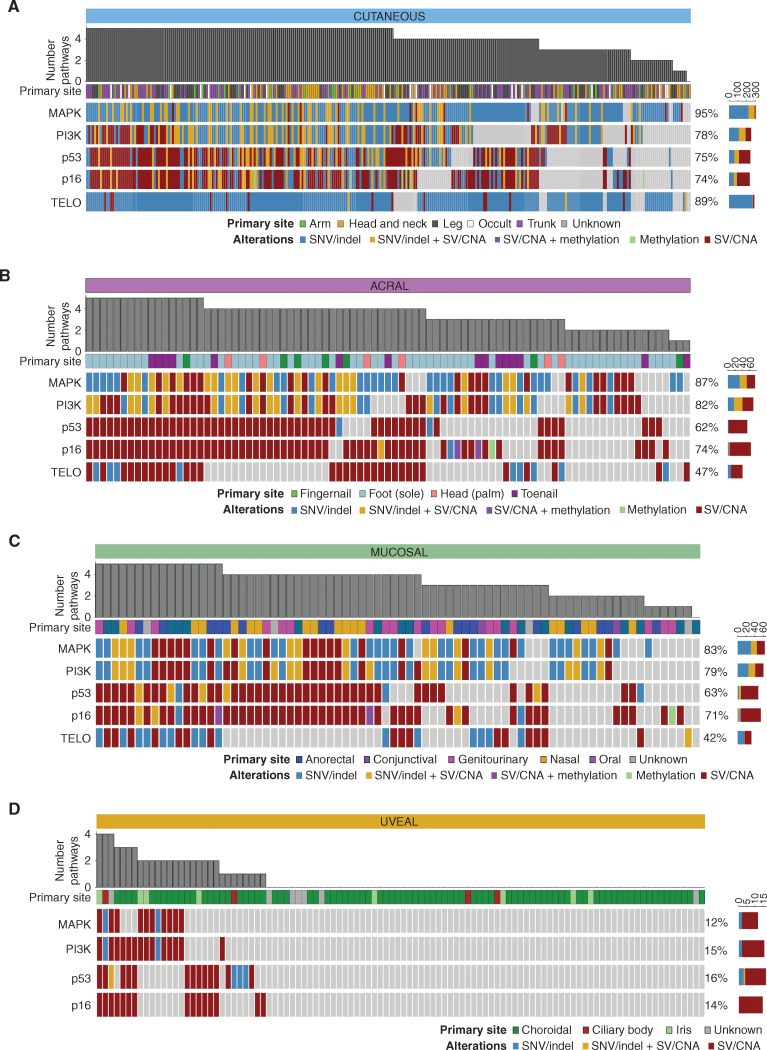
Five major pathways are important in the development of the CM, AM, and MM subtypes: MAPK, PI3K, p16, p53, and telomere maintenance (Fig. 7A–D; Supplementary Fig. S13A–S13D). UM is distinct, as few tumors have aberrations in these pathways, and barring LOH of the TP53 region of chromosome 17, it is unlikely that other LOH events in components of these pathways are functionally relevant. Instead, the vast majority (98%) of UM tumors harbor driver mutations in the G-protein signaling pathway (GNAQ, GNA11, PLCB4, and CYSLTR2), which leads to the activation of Protein Kinase C and its downstream effectors. The canonical MAPK pathway (RTK>RAS>RAF>MEK>ERK) was highly mutated (83%–95%) in all subtypes except UM. There were similar levels of PI3K and p16/cell-cycle pathway aberrations, whereas the telomere maintenance pathway was more highly mutated in CM (89%) than AM (47%) and MM (42%), mostly due to the high prevalence of TERT promoter mutations in CM. Overall, CM had more SNV/indel mutations in these pathways, likely driven by a higher TMB, whereas in MM and AM, a larger proportion of pathway mutations were SV/CNA events.
Figure 7.
Pathways important in melanoma development. Genomic aberrations are shown in five signaling pathways that are important in melanoma development: MAPK, PI3K, p53, p16–CDK–RB (p16), and telomere maintenance (TELO). Included genes and aberrations for each pathway are listed in Supplementary Table S8. Oncoplots show the type of aberration(s) in a gene within each pathway, and plots are grouped by subtype. Cutaneous tumors (A) and acral tumors (B), respectively. Mucosal tumors (C) and uveal tumors (D), respectively. The number of pathways mutated and the primary site of the tumor are shown. The color sidebars show the percentage of melanomas with the aberrant pathway.
DISCUSSION
Here we present the largest WGS analysis of melanoma and comprehensively compare genomic characteristics of the four major subtypes. We show that UM is genomically distinct from the other subtypes, as it harbors the lowest TMB and contains SMGs in different pathways (principally G-protein signaling). In contrast, the majority of CM, AM, and MM share similar requirements for tumorigenesis, with largely requisite alterations in components of the MAPK, PI3K, TP53, p16, and telomere pathways. However, the mechanism by which these pathways are altered differs between subtypes.
Of all cancers (66), CM has one of the highest TMB, whereas AM and MM have intermediate TMB, and UM has one of the lowest. AM and MM are among cancers with the highest number of SVs, and AM has high rates of WGD. CM and MM have similar lower proportions of WGD, whereas WGD is almost absent from UM. In CM, high TMB is associated with several parameters that may shape tumor development, including NF1 mutation, head and neck primary site, age of onset, and dominant UVR signature. In contrast, low TMB is associated with BRAF V600E mutation. The subsets of non-CM tumors with high TMB are those with primary sites exposed to the sun and that have UVR signatures. There is also a different distribution of UVR subsignatures (SBS7a–d) in each melanoma subtype, with SBS7a highest in CM, SBS7b highest in UM, and SBS7d highest in AM, which may be a reflection of the different intensity of UVR exposure at different body sites or that the same mutational process has diverse effects at different anatomic sites. UVR mutational signatures account for a high proportion of early clonal mutations, confirming that UVR is a very early event (1). In contrast to the previous speculation that SBS38 is associated with UVR in CM (34), we find no evidence for this being due to UVR because it occurs predominantly in non-CM (AM and MM) and is a late clonal mutational process. We also identified the presence of other rare signatures—of note, SBS96 in two UM cases with MBD4 mutations.
We confirm previous reports that germline MC1R genotype is associated with high TMB and early age of onset (61, 62), highlighting that the patient germline can influence the tumor mutation profile. Although a small number of CM and UM cases carried germline pathogenic variants in known high-penetrance melanoma susceptibility genes (54, 67–69), we found several novel cancer-predisposing germline variants in patients with AM. The most notable of these is the CHEK2 c.1100del variant carried by three of 87 AM cases, thus extending the spectrum of cancers associated with CHEK2 and pointing to CHEK2 potentially being the most common known genetic cause of AM susceptibility. Similarly, we extend the cancer spectrum associated with germline variants in POT1, PTEN, and TERT to include AM. We also found germline LOF variants in WRN in two patients with AM, adding further support for an association between this gene and predisposition to AM (70, 71). Analysis of germline LOF variants with second somatic hits in the tumors identified two AM cases carrying germline nonsense mutations in AIM1, with their tumors showing LOH. Functional assessment of AIM1 in CM demonstrated that it influences tumorigenicity (58); hence, we propose it as a strong candidate for a new susceptibility gene for AM that requires validation in independent series of cases.
CN signature analysis identified a number of recently reported CN signatures, including chromothripsis-associated signatures, which are highest in AM and MM. AM and MM share several regions of complex rearrangements accompanied by the focal amplification of oncogenes such as CCND1, CDK4, MDM2, and TERT. There are also similarities between CM, AM, and MM, which share a number of chromosome arm CNAs, highlighting a common requirement of specific gene dysregulation in their tumorigenesis. This includes the main driver genes shared by these subtypes, including oncogenes (BRAF and RAS) and tumor suppressor genes (CDKN2A, NF1, and PTEN). Through WGS analysis, we were able to better characterize the prevalence of these key driver genes. We found that BRAF fusions account for 6% of activating BRAF mutations and that these occur across multiple melanoma subtypes. As these fusions and NF1 inactivation through SVs are underreported by WES, the TCGA triple-WT group is overestimated in such studies (2, 29). Inactivation of other genes including PTEN and SPRED1 through LOF rearrangements is also underreported in WES studies, as are TERT promoter mutations and upstream SVs. Using WGS, we highlight different processes but similar end results between CM and AM/MM, with TERT promoter mutations being common in CM and SVs common in AM and MM.
Current first-line drug treatment for metastasized CM is checkpoint inhibitor immunotherapy, which has shown considerably less success in AM and MM (72, 73) for which targeted therapies may be the preferred option, particularly in tumors with low TMB and hence fewer neoantigens. The remarkable similarity in pathway alterations between CM, AM, and MM points to a number of common potential targeted treatments—for example, MAPK, PI3K, TERT, MDM2, and CDK4 inhibitors.
METHODS
Human Melanoma Specimens Including Cell Lines
Fresh-frozen tissue and matched normal germline samples for 570 donors were obtained from the biospecimen banks of Melanoma Institute Australia, QIMR Berghofer Medical Research Institute, Lions Eye Institute, Royal Perth Hospital, St John of God Hospital, Ludwig Institute for Cancer Research, Rigshospitalet, University of Colorado, Peking University Cancer Hospital, Skane University Hospital, and University of Zurich. All samples were collected with the written informed consent of donors. The study was approved by institutional ethics committees of the Melanoma Institute Australia, the Sydney Local Health District Royal Prince Alfred Hospital, QIMR Berghofer Medical Research Institute, University of Colorado, University of Zurich, University of Western Australia, the Capitol Region of Denmark, Peking University Cancer Hospital, Skane University Hospital, and the Ludwig Institute for Cancer Research. Tumor/normal pairs for 464 of the 570 donors have been described (4, 14, 16, 26) and were reanalyzed for this study (details in Supplementary Table S1).
The cohort included 18 cell lines. Cell line MELA_0577 was obtained from Mitch Levesque, University of Zurich (M140325, RRID:CVCL_A1UE). Cell lines MELA_0102, MELA_0103, MELA_0104, MELA_0105, MELA_0107, MELA_0108, MELA_0110, MELA_0114, MELA_0687, MELA_0116, MELA_0118, MELA_0119, MELA_0121, MELA_0122, MELA_0220, MELA_0349, and MELA_0135 were obtained from Chris Schmidt, QIMR Berghofer Medical Research Institute. There was matched germline DNA for all cell lines and all were subject to short tandem repeat profiling at QIMR Berghofer Medical Research Institute, and tumor–normal pairs were confirmed as matching via the subsequence sequencing using qSignature (https://github.com/AdamaJava/adamajava/tree/master/qsignature). For all cell lines except MELA_0577, these were additionally matched to STR profiling performed when the cell lines were established. Mycoplasma testing was performed using an in-house service at QIMR Berghofer Medical Research Institute prior to sequencing. The DNA was extracted from cell lines immediately after they reached confluence for use in sequencing.
DNA and RNA Extraction
Fresh-frozen tumor DNA and RNA were extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen #80224). Blood DNA was extracted from whole blood using the QIAamp DNA Blood Kit (#51126). DNA samples were quantified using NanoDrop (ND-1000, Thermo Scientific) and the Qubit dsDNA HS Assay (#Q32851, Life Technologies), with DNA size and quality tested using gel electrophoresis. RNA samples were quantified using the Qubit RNA HS Assay (#Q32852, Life Technologies). Most DNA and RNA samples were from the same vial of tissue. Six RNA (MELA_0267, MELA_0015, MELA_0004, MELA_0268, MELA_0068, and MELA_0010) samples were from the same lesion but from a different vial of tissue than the DNA sample.
WGS
Sequencing library construction was performed according to the manufacturer's instructions using TruSeq DNA Sample Preparation kits (Illumina). Whole-genome paired-end sequencing was carried out on HiSeq2000, HiSeq X-Ten, or NovaSeq instruments (Illumina) at Macrogen (Geumcheon-gu, Seoul, South Korea) or sequencing facilities in Australia (Australian Genomic Research Facility, Ramaciotti Centre for Genomics, John Curtin School of Medical Research, Kinghorn Cancer Centre, Garvan Institute of Medical Research). Tumor samples underwent WGS to an average coverage of 63× (range, 8–140×) and normal germline samples to an average coverage of 36× (range, 21–140×). Sequence reads were adapter trimmed using Cutadapt (RRID:SCR_011841, version 1.9; ref. 74) and aligned using BWA-MEM (version 0.7.12; ref. 75) to the GRCh37 assembly. Duplicate reads were marked with Picard MarkDuplicates (RRID:SCR_006525; https://broadinstitute.github.io/picard; version 1.129). Tumor purity was assessed using ascatNgs (76), and if the purity assessment from ascatNgs was found to be unreliable after manual review, mean variant allele frequency was used (Supplementary Table S1). All tumors had a purity of >10%, with 534 of the 570 (94%) tumors having a purity of >30%.
RNA-seq Analysis
Libraries were prepared from RNA using the TruSeq Stranded mRNA kit (n = 182) or the TruSeq RNA Library Prep Kit (n = 48) and sequenced with 100-bp, paired-end reads. RNA-seq reads were aligned using STAR (version 2.5.2a, RRID:SCR_004463) to the GRCh37 assembly with the gene, transcript, and exon features of the Ensembl (release 70) gene model after trimming for adapter sequences using Cutadapt (RRID:SCR_011841, version 1.11). Gene expression was estimated using RSEM (version 1.2.30; ref. 77), and quality control was carried out using RNA-SeQC (version 1.1.8; ref. 78). Trimmed mean of M-values (TMM) normalization was performed using the R package “edgeR” (RRID:SCR_012802) for the 182 samples that were sequenced using the Stranded TruSeq mRNA kit. TMM-normalized values of these 182 samples were used for the association of expression with methylation and with significant noncoding gene elements for CM (n = 88). Fragments per kilobase of transcript per million mapped reads (FPKM) values for all 230 samples were used in SMG analysis to determine if a gene was expressed for SMG analysis, and RNA-seq BAM files for all 230 samples were used for fusion analysis.
DNA Methylation Analysis
Bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research) and was hybridized to the Infinium MethylationEPIC Array (Illumina) following the manufacturer's protocol at the Queensland University of Technology. The arrays were scanned on the iScan system (Illumina). Raw idat files for 144 tumors were imported, filtered, and normalized using the ChAMP R package (RRID:SCR_012891; ref. 79; version 3.5.1). Probes were filtered out where there were fewer than three beads in 5% or more of the samples or where there was a detection P > 0.01. A β-mixture quantile normalization (80) was performed to account for probe type 1 and type 2 biases, followed by quantile normalization. Additional filtering was performed to remove probes located in non-CpG sites, on chromosome X or Y, single-nucleotide polymorphism (SNP)–related polymorphisms as per Zhou and colleagues (81), and probes that map to multiple locations as per Nordlund and colleagues (82). Promoter methylation of selected tumor suppressor genes—TP53, CDKN2A, RB1, BRCA1, PTEN, ARID2, NF1, and SPRED1—was examined. For each gene, promoter CpG sites were examined (TSS200, TSS1500, 5′ UTR, first exon, with annotation according to the RefSeq canonical transcript). Tumors with homozygous deletions of the gene of interest were not included. Tumors with multiple CpG sites that had a beta value >0.2 above the median beta value of the site for tumors of the same melanoma subtype were considered to be methylated after confirmation by a manual review of methylation beta value plots. For tumors that had matching RNA-seq, expression of a gene in the presence or absence of methylation was examined using TMM-normalized RNA-seq data.
Somatic Substitution and Indel Calling
Somatic SNV and indels were detected with a previously described pipeline (4), which uses a dual calling strategy to detect single-nucleotide, dinucleotide, and trinucleotide variants. The consensus of two tools was used for downstream analysis: qSNP (version 2.0; ref. 83) and GATK HaplotypeCaller (version 3.3–0; ref. 84). GATK was used to detect indels (1–50 bp). SnpEff (85) was used for variant annotation. To avoid false negatives in known mutation hotspots, the following regions were called with higher sensitivity as previously described (26): TERT−57, −124, −137, and −146; BRAF p.597, p.600–601; (N/K/H) RAS p.12–13 and p.59–62; SF3B1 p.625, p.666, and p.700; GNAQ/11 p.183, p.209; CYSLTR2:p.129; PLCB4:p.630; and EIF1AX p.1–20. For each genomic position and sample, the Fisher exact test was used to compare the read counts in the tumor sample with the pool of 570 normal samples. A BH-adjusted P value (86) below 0.01 was considered significant.
Germline
The consensus of two different tools was used to call germline variants—qSNP (version 2.0; ref. 83) and GATK HaplotypeCaller (RRID:SCR_001876, version 3.3–0; ref. 84)—and detection of indels (1–50 bp) was carried out using GATK. Germline variants were annotated using Ensembl Variant Effect Predictor (VEP) v99.2 (87), and the relevant transcript was selected by cross-referencing with the NCBI RefSeq Select database. To estimate the predisposing germline burden, we counted the number of predicted pathogenic germline variants in cancer genes (88). If the variant existed in ClinVar (RRID:SCR_006169; ref. 89) without “conflicting interpretation” or “no assertion criteria provided,” we used the prediction in ClinVar—that is, LOF variants (categorized as HIGH impact by VEP) were counted unless classified as “Benign” or “Likely Benign.” Missense variants and in-frame indels (categorized as MODERATE impact by VEP) were counted as pathogenic if categorized as “Pathogenic” or “Likely Pathogenic” in ClinVar; categorized as “Uncertain Significance” or “conflicting interpretation” if absent; and counted as pathogenic if at least one of SIFT (90) and PolyPhen2 (91) predicted them as deleterious/damaging and none as tolerated/benign.
Mutational Signatures
SigProfiler (34) was used for the de novo discovery of SBS, DBS, and indel signatures. De novo signatures were compared against COSMIC (RRID:SCR_002260; ref. 92) version 3 signatures using cosine similarity. For SBS signatures, the contribution of mutational signatures to individual samples was determined by SigProfilerSingleSample (34) using the signatures identified by de novo analysis as input. As there were insufficient mutations for DNVs and indels to extract de novo signatures with confidence, signatures were assigned using deconstructSigs. There was a limit of four signatures per sample and a minimum of 15% contribution of mutations was required for the signature to be assigned. Only samples with more than >50 mutations underwent assignment; the remaining samples were defined as unassigned/other.
For mutation signature timing, pyclone-VI (93), with CN and purity input from ascatNgs, was run to obtain information about the number of subclones (clusters) and their cellular prevalence. MutationTimeR (https://github.com/gerstung-lab/MutationTimeR) is an R package that times somatic mutations relative to clonal and subclonal CN states. MutationTimeR was run using SNVs, CN segments from ascatNgs and cluster information from pyclone-VI as input to assign point mutations to these clonal states: early clonal, late clonal, unspecified clonal, or subclonal. The contribution of COSMIC v3 UVR (SBS7a,b,c,d) or SBS38 signature to the mutations assigned to these clonal states: early clonal, late clonal, clonal (combining early, late and unspecified clonal mutations), and subclonal were determined for each clonal state using SigProfilerSingleSample (https://github.com/AlexandrovLab/SigProfilerSingleSample). The percent contribution of the signature was then compared between early and late clonal events in tumors where there was a UVR or SBS38 signature present (>0%), and there were at least 100 early and 100 late clonal mutations. The percent contribution of the UVR and SBS38 signatures was also compared between clonal events (combining early, late, and not otherwise assigned clonal mutations) and subclonal events in tumors where there was a UVR or SBS38 signature present (>0%), there was at least one subclone, and there were at least 100 clonal and 100 subclonal mutations.
The presence of common and rare SBS reference mutational signatures was identified using Signature FitMS (36) in the R package signature.tools.lib available at https://github.com/Nik-Zainal-Group/signature.tools.lib and the RefSig SBS v2.03 reference signatures. The contribution of signatures was estimated using the FitMS method. This was run with default parameters with the useBootstrap option set to TRUE and exposureFilterType option set to “giniScaledThreshold.” The common and rare signatures were selected by choosing the organ option “Skin.”
SMG and Pathway Analysis
To identify SMGs in each subtype (CM, AM, MM, and UM) with respect to substitution and indel mutations, a consensus approach was employed using MutPanning (94), ActiveDriverWGS (95), DriverPower (96), OncodriveCLUSTL (97), OncodriveFML (98), dNdScv (99), and MutSigCV (100). All tools were run with default parameters unless otherwise stated. OncodriveFML was run using CADD v1.0 through the Web interface at https://bbglab.irbbarcelona.org/oncodrivefml/home. MutPanning was run through the GenePattern module at https://www.genepattern.org/modules/docs/MutPanning. The ActiveDriverWGS R package was run using CDS as defined by the PCAWG consortium (45) and was downloaded from https://dcc.icgc.org/releases/PCAWG/drivers/metadata/genomic_intervals_lists (June 12, 2020). ActiveDriverWGS was run with a hypermutation filter (filter_hyper_MB) of 600. DriverPower was run in python using the provided feature and elements files, and a background mutation rate model was generated using the gradient boosting machines (GBM) algorithm. Driver candidates were inferred using functional information in the form of CADD scores with a functional score cutoff of 0.01. A gene was considered significant in each tool if it had a q-value or FDR of <0.1, and a gene was considered to be significantly mutated if it was significant in three or more tools. Genes were further filtered by examining the expression of the gene within the subtype using matching RNA-seq (except for UM, as only six tumors had RNA-seq). A gene was considered to be expressed if ≥40% of tumors with RNA-seq had an FPKM value (from RSEM) of >3, and genes that were not expressed were removed from the final SMG list.
For CM, the prevalence of mutations in BCLAF1 was determined in the TCGA-SKCM cohort and the Conway and colleagues (29) meta-analysis cohort. The TCGA mutation annotation format (MAF; n = 468) was obtained using the tcga_load() function in the “TCGAmutations” R package (https://github.com/PoisonAlien/TCGAmutations), and the Conway meta-analysis MAF was obtained from Supplementary Data S1 of that article. For the Conway meta-analysis, tumors overlapping with TCGA or with our CM cohort or tumors with a histology type of “acral,” “mucosal,” or “no info” were removed, leaving a total of 463 tumors. For CM, the prevalence of all types of gene aberrations in the SMGs identified in the current analysis, as well as a list of previously published SMGs (2–4, 7, 28, 29), was determined. This includes the prevalence of SNV and indel mutations, homozygous deletions, LOH (CN1), amplifications, and for rearrangements, predicted intergene fusions and predicted LOF events. Expression of these genes was also determined in the CM cohort and in the TCGA-SKCM cohort (2). A gene was considered to be expressed if ≥40% of tumors with RNA-seq had an FPKM value (from RSEM) of >3.
For pathway analysis (Fig. 7; Supplementary Fig. S13), gene-specific criteria were used to determine whether a tumor contained an aberration in a gene assigned to five different pathways: MAPK, PI3K, p53 (p53), p16–CDK–RB (p16), and telomere maintenance. The criteria applied to each gene are listed in Supplementary Table S8.
Significant Noncoding Regions
To identify significantly mutated noncoding genomic elements in each subtype (CM, AM, MM, and UM) with respect to substitution and indel mutations, a consensus approach was employed using ActiveDriverWGS (95), DriverPower (96), and OncodriveFML (98), and LARVA (101). Genomic elements were downloaded from https://dcc.icgc.org/releases/PCAWG/drivers/metadata/genomic_intervals_lists (June 12, 2020) and were defined by the PCAWG consortium (45). Functional genomic elements were defined based on GENCODE v.19 (102) and other genomic resources and included 5′ UTR, 3′ UTR, and promoters of protein-coding genes, gene body and promoters of lncRNAs, and enhancers. ActiveDriverWGS was run with a hypermutation filter (filter_hyper_MB) of 600. DriverPower was run in python using the provided feature and elements files, and a background mutation rate model was generated using the GBM algorithm. Driver candidates were inferred using functional information in the form of CADD scores with a functional score cutoff of 0.01. A noncoding genomic element was considered significant in each tool if it had a q-value of <0.05 for AM, MM, and UM. Due to the high tumor burden in CM, a more stringent q-value of <0.001 was used. Overall, a noncoding element was considered significantly mutated if it was significant in two or more tools. For CM tumors with matching RNA-seq (n = 88, stranded library RNA-seq only), a difference in expression using TMM-normalized gene expression values and Mann–Whitney U tests was determined for genomic elements when there were more than three tumors with mutations in the region.
CNAs and Structural Rearrangement Variants
CNAs were identified using ascatNgs (76) and gene-specific CN was determined by annotation with Ensembl known genes (version 75). Amplifications were defined using the following criteria previously defined by COSMIC (RRID:SCR_002260; ref. 92): (i) total DNA segment CN ≥5 for tumors with average genome ploidy ≤2.7 as inferred by ascatNgs or (ii) total DNA segment CN ≥9 for tumors with average genome ploidy >2.7. WGD was defined as >50% of the autosomal tumor genome had a major CN ≥2 (103). Detection of significant focal regions of amplification and deletion was assessed using GISTIC2.0 using a confidence level of 0.95 and a q-value of <0.05.
Structural rearrangements/variants were identified using qSV (4), and the likely consequence of rearrangements, including predicted in-frame gene fusions and LOF variants, was determined using in-house scripts and annotation against Ensembl known genes (version 75). RETREAD (https://github.com/UCL-Research-Department-of-Pathology/RETREAD) was used to identify recurrent breakpoint regions in 100-kb bins across the genome, and a q-value of <0.2 was considered significant. For tumors with matching RNA-seq, STAR-Fusion (104) and Arriba (105) were used to detect fusions in RNA-seq.
TCGA Molecular Category Assignment
We classified melanomas into TCGA subtypes based on their mutations in BRAF, (H/K/N)RAS, and NF1 (2). Mutations (including in-frame indels) in hotspot codons 597, 600, and 601 of BRAF were counted as well as in-frame fusions in which BRAF was the 3′-partner. Mutations in codons 12, 13, and 61 of (N/K/H)RAS were counted. For NF1, SNVs, small indels, breakpoints due to SVs, LOH, and larger deletions were counted. If a melanoma had mutations in multiple genes and one mutation had low variant allele frequency (<1:7), suggesting mutation was only present in a subclone, the melanoma was classified as the mutation present in the major clone. Otherwise, BRAF had precedence and RAS had precedence over NF1.
Inference of Kataegis
Regions of local hypermutation (kataegis) were determined using piecewise constant fitting of intermutation distances using the method defined by the PCAWG consortium (106). Sets of at least six adjacent SNVs were flagged as candidate kataegis events if the segmented intermutation distance dropped below a threshold d. The threshold was determined based on the TMB (mb) of a sample using the method described by PCAWG (106), with d calculated as
 where K = 6, and the intermutation distance (X) is modeled assuming an exponential distribution with
where K = 6, and the intermutation distance (X) is modeled assuming an exponential distribution with  . If d was greater than 1 kb, then d of 1 kb was used in the calculation.
. If d was greater than 1 kb, then d of 1 kb was used in the calculation.
Rearrangement and CN Signatures
Rearrangement signatures were identified using nonnegative matrix factorization. Rearrangement signatures were determined based on cosine similarity to the signatures used for breast cancer as described by Nik-Zainal and colleagues (107). Rearrangements were classified into 32 categories based on deletion, duplication, inversion, and interchromosomal translocation event types; the size of the event; and if the breakpoints were clustered or nonclustered. To determine if a breakpoint was clustered, the BEDTools (RRID:SCR_006646) cluster function was used (108). To reduce overfitting, the R package decontructSigs was used to estimate the exposure of each identified signature in each tumor, and a minimum of 15% contribution of mutations was required for the signature to be assigned. Clustered signatures RS4 and RS6 were used in further analysis (Fig. 3B).
CN signatures (40) were identified using AlexandrovLab R packages (https://github.com/AlexandrovLab): SigProfilerExtractor, SigProfilerMatrixGenerator, and SigProfilerAssignment. SigProfilerMatrixGenerator, with ascatNgs CN profiles as input, was used to summarize the counts of segments into 48 categories based on total CN, whether a segment was heterozygous or not, and the size of the segment. De novo signature analysis was carried out using SigProfilerExtractor, and eight signatures were identified as the optimal solution. These signatures were then decomposed into the 24 CN signatures identified by Steele and colleagues (40), using SigProfilerAssignment, and these signatures were assigned for each tumor. This identified 11 COSMIC CN signatures and two novel signatures that did not have high similarity to any known signatures.
To further profile the regional characteristics of CN profiles, the tool CARMA was used (109). CN segments for each chromosome arm were used to generate a genome-wide CARMA score for each tumor for six categories that reflect the degree of CN amplification (AMP), deletion (DEL), complexity such as chromothripsis (STP and CRV), LOH, and allelic imbalance or asymmetry (ASM). Three scores, AMP, STP, and CRV, were used for further analysis (Fig. 3B).
Inference of Chromothripsis and Other Localized Structural Rearrangements
Localized structural rearrangements were determined for each chromosome using previously described methods (14, 110) to identify the presence of chromothripsis, clustered rearrangements, or a high number of rearrangements. Several characteristics were used: (i) Chromosomes with high numbers of rearrangement events were identified as outliers if they had a breakpoint per megabase rate exceeding 1.5 times the length of the interquartile range from the 75th percentile for each sample, with a minimum threshold of 35 breakpoints per chromosome. (ii) Chromosomes with at least 10 translocations were considered to have a high number of translocations. (iii) Chromosomes with a significant nonrandom distribution of breakpoints (Kolmogorov–Smirnov test, P < 10−5) were considered to be clustered. (iv) Chromothripsis was identified using the R package “Shatterseek” (ref. 111; version 0.4) with default parameters, and structural rearrangements called by qSV and CNA called by ascatNgs. Using Shatterseek, we identified high-confidence and low-confidence calls as described by Cortés-Ciriano and colleagues (111). P values for the fragment joins test, chromosomal enrichment, and the exponential distribution of breakpoints test were corrected using the FDR method. A high-confidence chromothripsis event was defined as at least six interleaved intrachromosomal SVs or at least three interleaved intrachromosomal SVs and four or more interchromosomal SVs; seven contiguous segments oscillating between two CN states; and passed the fragment joins test (q-value < 0.2) and either the chromosomal enrichment (q-value < 0.2) or the exponential distribution of breakpoints test (q-value < 0.2). A low-confidence chromothripsis event was defined as at least six interleaved intrachromosomal SVs; four to six contiguous segments oscillating between two CN states; and passed the fragment joins (q-value < 0.2) and either the chromosomal enrichment (q-value < 0.2) or the exponential distribution of breakpoints test (q-value < 0.2). Chromosomes that had one or more of these four characteristics were manually reviewed. Chromosomes that passed manual review were assigned as localized complex, and if they were also identified by Shatterseek as having chromothripsis, they were considered to have chromothripsis.
Telomere Length
Estimation of telomere length from WGS was performed using qMotif (112), which counts reads containing telomeric repeats in the tumor and matched normal sample and normalizes this to the mean genomic coverage of the sample. A relative telomere length was expressed as the log2 ratio of the number of read counts in the tumor to the matched normal read counts.
TMB Associations
We explored which variables were associated with TMB by building negative binomial regression models. To explore how variables are associated with TMB, we built a decision tree. In each branching point, the variable most strongly associated with TMB was identified using Kendall's tau. A branch was created by dichotomizing the cohort based on the identified variable. The procedure was repeated for both branches, recursively, until no Bonferroni corrected P value was smaller than 0.05.
Regression Analysis
Linear regression was used to determine which variables correlated with age of onset. Univariate regression was used to identify predictive variables. Multivariate regression with forward feature selection was used to test whether identified variables were independent predictors.
Strand Bias
Strand bias of C>T substitutions at TpC and CpC dinucleotides (mutated base underlined) was explored separately. The strand bias in each region of interest (exonic, intronic, etc.) was estimated as follows: We counted the occurrence of the dinucleotide of interest on the coding strand and what fraction of these sites harbored a C>T substitution. This was repeated for the antisense strand, and the strand bias was quantified as one minus the ratio of the fraction on the transcribed strand to fraction on the untranscribed strand. Samples with >50% of the mutations explained by UVR-associated SBS7a–d were included in the analysis. Genes were categorized into four groups based on the expression in the TCGA-SKCM cohort (2). For each gene, the median expression (FPKM-upper quartile) was calculated across samples and genes were categorized based on the quartiles.
MC1R Genotype Categories
As previously described (61), nonsynonymous variants in MC1R were categorized into those strongly (R) or weakly (r) associated with red hair, and patients were categorized into six MC1R compound genotypes based on their germline sequence: WT, r/0, r/r, R/0, R/r, and R/R.
Polygenic Risk Score
The polygenic risk score (PRS) for CM and nevus count were calculated using independent [linkage disequilibrium (LD) r2 < 0.05 in the 1000 Genomes European samples as determined by FUMA; ref. 113] lead SNPs with a P < 5 × 10−8 from previously published loci (114). The PRS were calculated as the sum of effect alleles weighted by the per-allele natural log of the OR for CM, or beta for nevus count; each individual's PRS is reported in Supplementary Table S1.
Inference of Ancestry
SNP data from the 1000 Genomes phase III (115) were prepared using plink 1.90 (RRID:SCR_001757; ref. 116) by first applying filters –snps-only and –geno 0.9 and pruned using the indep-pairwise option with window size 1,000, window step 10, and correlation threshold 0.02. The genotypes for the same 25,285 SNP positions were inferred in our cohort of germline samples using bcftools (117). Each sample was then compared with the 20 populations represented in 1000 Genomes using the t-distributed stochastic neighbor embedding score with perplexity set to 10 (118). In short, the conditional probability that sample i would pick sample j as its neighbor was calculated as pj|i = exp(−(xi − xj)2/2σi2)/Z, where Z is a normalization constant and we calculated the similarity score between sample i and each population k as sk|i = Σpj|i, where the sum runs over samples from population k.
Association between Germline Variants and Somatic Events
The associations between germline polymorphisms and somatic events previously proposed (59) were assessed. The genotypes of the listed polymorphic loci were inferred with bcftools (117). For each listed gene, somatic nonsynonymous mutations, high copy gain (CN ≥6), or both were collated, depending on whether the alteration had been proposed to be mutation, CNA, or either, respectively. Individuals with a European ancestry score below 80% were excluded to avoid confounding effects from ethnicity. Associations between germline genotype and somatic events were assessed with the Fisher exact test under a dominant model. P values were Bonferroni corrected based on the number of gene and LD-block pairs that were tested.
Survival
Univariable Cox regressions were performed to assess the association between variables (genomic and clinical features) and OS. CM and AM were analyzed separately. MM and UM were not analyzed, as detailed survival data were unavailable. OS was recorded during the period patients remained naïve from immunotherapy treatment. Survival time was calculated from the date of primary diagnosis to the date of death, start of immunotherapy, or last contact. Factors with a statistically significant association with subtypes were provided.
Statistical Analysis
Statistical analyses were performed using R (RRID:SCR_001905, version 4.0.2) and were two-sided, with a P value or an FDR-adjusted P value less than 0.05 considered significant. Differences in continuous variables between two conditions were evaluated using Mann–Whitney U tests. Continuous variable differences between three or more conditions were calculated using Kruskal–Wallis tests with pairwise Mann–Whitney U tests with adjustment for FDR to compare between each condition pair. Fisher exact tests or χ2 tests were used to compare categorical variables. The box boundaries of box plots show the first to third quartiles, the median is the center line, and the whiskers represent 1.5 times the interquartile range.
Data Availability
Sequence data (WGS and RNA-seq) that support the findings of this study have been deposited in the European Genome-phenome Archive (EGA; RRID:SCR_004944) and are available under study accession EGAS00001001552 (https://www.ebi.ac.uk/ega/studies/EGAS00001001552), with data set accessions EGAD00001008798 (WGS) and EGAD00001008837 (RNA-seq). Illumina EPIC DNA methylation array data analyzed in this study are publicly available in the Gene Expression Omnibus (RRID:SCR_005012) under accession number GSE202097. All other data are available in the article, in the Supplementary Data, or from the authors upon reasonable request.
Code Availability
In-house tools that were used in this publication are available from the GitHub public code repository under the AdamaJava project (https://github.com/AdamaJava).
Supplementary Material
Acknowledgments
R.A. Scolyer is supported by a National Health and Medical Research Council (NHMRC) Practitioner Fellowship. G.V. Long is supported by an NHMRC Investigator Grant. G.V. Long is also supported by The University of Sydney Medical Foundation. J.S. Wilmott is supported by an NHMRC investigator fellowship, Melanoma Research Alliance Team Science Young Investigator Fellowship, and The University of Sydney. N. Waddell and N.K. Hayward are supported by an NHMRC Research Fellowship. R.P.M. Saw and S.N. Lo are supported by Melanoma Institute Australia. P.M. Ferguson is supported by the McMurtrie Fellowship in Melanoma Pathology and Melanoma Institute Australia. We thank Hazel Burke, Valerie Jakrot, Ping Shang, and colleagues at Melanoma Institute Australia, Royal Prince Alfred Hospital, and NSW Health Pathology for their assistance. This project was funded in part by the NHMRC, the Ainsworth Foundation, the CLEARbridge Foundation, the Cameron Family, Jan Brown, the Buck Off Melanoma community, and the memorial donors honoring Donald McHenry and Nicola Laws. We are indebted to the patients for their participation and support of this study. We gratefully acknowledge the contributors of the Australian Melanoma Genome Project consortium for the provision of samples and data in previous related studies.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
I.A. Vergara reports other support from Melanoma Institute Australia during the conduct of the study, as well as a patent for US11145412B2 issued. G.V. Long reports personal fees from Agenus, Amgen, Array Biopharma, Boehringer Ingelheim International GmbH, Bristol Myers Squibb, Evaxion Biotech A/S, Hexal AG, Highlight Therapeutics S.L., Innovent Biologics USA Inc., Merck Sharp & Dohme, Novartis Pharma AG, OncoSec Medical Australia, PHMR Limited, Pierre Fabre, Provectus Australia, QBiotics, and Regeneron Pharmaceuticals outside the submitted work. R.P.M. Saw reports personal fees from Novartis, MSD, QBiotics, and Bristol Myers Squibb outside the submitted work. J.F. Thompson reports grants from the National Health and Medical Research Council during the conduct of the study, as well as personal fees from Bristol Myers Squibb Australia and MSD Australia, personal fees and nonfinancial support from GSK Australia and Provectus, and nonfinancial support from Novartis outside the submitted work. J.V. Pearson is a cofounder of, an equity holder in, and a director for genomiQa, a genome analytics company. R.A. Scolyer reports grants from the National Health and Medical Research Council during the conduct of the study, as well as personal fees from F. Hoffmann-La Roche Ltd., Evaxion, Provectus Biopharmaceuticals Australia, QBiotics, Novartis, Merck Sharp & Dohme, NeraCare, Amgen, Bristol Myers Squibb, Myriad Genetics, and GSK outside the submitted work. N. Waddell reports grants from the National Health and Medical Research Council during the conduct of the study, as well as other support from genomiQa outside the submitted work. N.K. Hayward reports grants from the National Health and Medical Research Council during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
F. Newell: Data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. P.A. Johansson: Data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.S. Wilmott: Resources, data curation, formal analysis, methodology, project administration, writing–review and editing. K. Nones: Formal analysis, investigation, methodology, writing–review and editing. V. Lakis: Formal analysis, investigation, methodology, writing–review and editing. A.L. Pritchard: Resources, data curation, methodology, writing–review and editing. S.N. Lo: Formal analysis, investigation, methodology, writing–review and editing. R.V. Rawson: Resources, data curation, writing–review and editing. S.H. Kazakoff: Data curation, software, methodology, writing–review and editing. A.J. Colebatch: Resources, writing–review and editing. L.T. Koufariotis: Data curation, software, methodology, writing–review and editing. P.M. Ferguson: Resources, data curation, writing–review and editing. S. Wood: Resources, data curation, software, methodology, writing–review and editing. C. Leonard: Data curation, software, methodology, writing–review and editing. M.H. Law: Resources, data curation, formal analysis, investigation, writing–review and editing. K.M. Brooks: Resources, data curation, formal analysis, writing–review and editing. N. Broit: Resources, data curation, writing–review and editing. J.M. Palmer: Data curation, project administration, writing–review and editing. K.L. Couts: Resources, writing–review and editing. I.A. Vergara: Investigation, writing–review and editing. G.V. Long: Resources, writing–review and editing. A.P. Barbour: Resources, writing–review and editing. O.E. Nieweg: Resources, writing–review and editing. B. Shivalingam: Resources, writing–review and editing. W.A. Robinson: Resources, writing–review and editing. J.R. Stretch: Resources, writing–review and editing. A.J. Spillane: Resources, writing–review and editing. R.P.M. Saw: Resources, writing–review and editing. K.F. Shannon: Resources, writing–review and editing. J.F. Thompson: Conceptualization, resources, funding acquisition, writing–review and editing. G.J. Mann: Conceptualization, supervision, funding acquisition, writing–review and editing. J.V. Pearson: Resources, supervision, writing–review and editing. R.A. Scolyer: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing. N. Waddell: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing. N.K. Hayward: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2010;463:191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012;44:1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175–80. [DOI] [PubMed] [Google Scholar]
- 5. Wilmott JS, Johansson PA, Newell F, Waddell N, Ferguson P, Quek C, et al. Whole genome sequencing of melanomas in adolescent and young adults reveals distinct mutation landscapes and the potential role of germline variants in disease susceptibility. Int J Cancer 2019;144:1049–60. [DOI] [PubMed] [Google Scholar]
- 6. Vergara IA, Mintoff CP, Sandhu S, McIntosh L, Young RJ, Wong SQ, et al. Evolution of late-stage metastatic melanoma is dominated by aneuploidy and whole genome doubling. Nat Commun 2021;12:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 2012;485:502–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014;4:94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2011;44:133–9. [DOI] [PubMed] [Google Scholar]
- 11. Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet 2011;44:165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 2011;43:442–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furney SJ, Turajlic S, Fenwick K, Lambros MB, MacKay A, Ricken G, et al. Genomic characterisation of acral melanoma cell lines. Pigment Cell Melanoma Res 2012;25:488–92. [DOI] [PubMed] [Google Scholar]
- 14. Newell F, Wilmott JS, Johansson PA, Nones K, Addala V, Mukhopadhyay P, et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat Commun 2020;11:5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furney SJ, Turajlic S, Stamp G, Thomas JM, Hayes A, Strauss D, et al. The mutational burden of acral melanoma revealed by whole-genome sequencing and comparative analysis. Pigment Cell Melanoma Res 2014;27:835–8. [DOI] [PubMed] [Google Scholar]
- 16. Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 2019;10:3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hintzsche JD, Gorden NT, Amato CM, Kim J, Wuensch KE, Robinson SE, et al. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res 2017;27:189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mundra PA, Dhomen N, Rodrigues M, Mikkelsen LH, Cassoux N, Brooks K, et al. Ultraviolet radiation drives mutations in a subset of mucosal melanomas. Nat Commun 2021;12:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, et al. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol 2013;230:261–9. [DOI] [PubMed] [Google Scholar]
- 20. Lyu J, Song Z, Chen J, Shepard MJ, Song H, Ren G, et al. Whole-exome sequencing of oral mucosal melanoma reveals mutational profile and therapeutic targets. J Pathol 2018;244:358–66. [DOI] [PubMed] [Google Scholar]
- 21. Zhou R, Shi C, Tao W, Li J, Wu J, Han Y, et al. Analysis of mucosal melanoma whole-genome landscapes reveals clinically relevant genomic aberrations. Clin Cancer Res 2019;25:3548–60. [DOI] [PubMed] [Google Scholar]
- 22. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010;330:1410–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 2013;45:933–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016;7:4624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017;32:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johansson PA, Brooks K, Newell F, Palmer JM, Wilmott JS, Pritchard AL, et al. Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat Commun 2020;11:2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 2013;3:1122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alkallas R, Lajoie M, Moldoveanu D, Hoang KV, Lefrancois P, Lingrand M, et al. Multi-omic analysis reveals significantly mutated genes and DDX3X as a sex-specific tumor suppressor in cutaneous melanoma. Nat Cancer 2020;1:635–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conway JR, Dietlein F, Taylor-Weiner A, AlDubayan S, Vokes N, Keenan T, et al. Integrated molecular drivers coordinate biological and clinical states in melanoma. Nat Genet 2020;52:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Broit N, Johansson PA, Rodgers CB, Walpole ST, Hayward NK, Pritchard AL. Systematic review and meta-analysis of genomic alterations in acral melanoma. Pigment Cell Melanoma Res 2022;35:369–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Broit N, Johansson PA, Rodgers CB, Walpole ST, Newell F, Hayward NK, et al. Meta-analysis and systematic review of the genomics of mucosal melanoma. Mol Cancer Res 2021;19:991–1004. [DOI] [PubMed] [Google Scholar]
- 32. Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135–47. [DOI] [PubMed] [Google Scholar]
- 33. Wang M, Banik I, Shain AH, Yeh I, Bastian BC. Integrated genomic analyses of acral and mucosal melanomas nominate novel driver genes. Genome Med 2022;14:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Degasperi A, Zou X, Dias Amarante T, Martinez-Martinez A, Koh GCC, Dias JM, et al. Substitution mutational signatures in whole-genome–sequenced cancers in the UK population. Science 2022;376:abl9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanders MA, Chew E, Flensburg C, Zeilemaker A, Miller SE, Al Hinai AS, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood 2018;132:1526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Invest Ophthalmol Vis Sci 2009;50:3048–55. [DOI] [PubMed] [Google Scholar]
- 39. Yavuzyigitoglu S, Koopmans AE, Verdijk RM, Vaarwater J, Eussen B, van Bodegom A, et al. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology 2016;123:1118–28. [DOI] [PubMed] [Google Scholar]
- 40. Steele CD, Abbasi A, Islam SMA, Bowes AL, Khandekar A, Haase K, et al. Signatures of copy number alterations in human cancer. Nature 2022;606:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019;575:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rajaram M, Zhang J, Wang T, Li J, Kuscu C, Qi H, et al. Two distinct categories of focal deletions in cancer genomes. PLoS One 2013;8:e66264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lezcano C, Shoushtari AN, Ariyan C, Hollmann TJ, Busam KJ. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol 2018;42:1052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hebert JD, Tian C, Lamar JM, Rickelt S, Abbruzzese G, Liu X, et al. The scaffold protein IQGAP1 is crucial for extravasation and metastasis. Sci Rep 2020;10:2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sieverling L, Hong C, Koser SD, Ginsbach P, Kleinheinz K, Hutter B, et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat Commun 2020;11:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet 2017;49:349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wiesner T, Kiuru M, Scott SN, Arcila M, Halpern AC, Hollmann T, et al. NF1 mutations are common in desmoplastic melanoma. Am J Surg Pathol 2015;39:1357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wong SQ, Behren A, Mar VJ, Woods K, Li J, Martin C, et al. Whole exome sequencing identifies a recurrent RQCD1 P131L mutation in cutaneous melanoma. Oncotarget 2015;6:1115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee CS, Bhaduri A, Mah A, Johnson WL, Ungewickell A, Aros CJ, et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat Genet 2014;46:1060–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015;29:1092–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell 2014;54:445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A 2013;110:13481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006;66:9818–28. [DOI] [PubMed] [Google Scholar]
- 55. Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011;480:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959–61. [DOI] [PubMed] [Google Scholar]
- 57. Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002;31:55–9. [DOI] [PubMed] [Google Scholar]
- 58. Ray ME, Su YA, Meltzer PS, Trent JM. Isolation and characterization of genes associated with chromosome-6 mediated tumor suppression in human malignant melanoma. Oncogene 1996;12:2527–33. [PubMed] [Google Scholar]
- 59. Carter H, Marty R, Hofree M, Gross AM, Jensen J, Fisch KM, et al. Interaction landscape of inherited polymorphisms with somatic events in cancer. Cancer Discov 2017;7:410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shoushtari AN, Chatila WK, Arora A, Sanchez-Vega F, Kantheti HS, Zamalloa JAR, et al. Therapeutic implications of detecting MAPK-activating alterations in cutaneous and unknown primary melanomas. Clin Cancer Res 2021;27:2226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Johansson PA, Pritchard AL, Patch AM, Wilmott JS, Pearson JV, Waddell N, et al. Mutation load in melanoma is affected by MC1R genotype. Pigment Cell Melanoma Res 2017;30:255–8. [DOI] [PubMed] [Google Scholar]
- 62. Robles-Espinoza CD, Roberts ND, Chen S, Leacy FP, Alexandrov LB, Pornputtapong N, et al. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun 2016;7:12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zanna I, Caini S, Raimondi S, Saieva C, Masala G, Massi D, et al. Germline MC1R variants and frequency of somatic BRAF, NRAS, and TERT mutations in melanoma: literature review and meta-analysis. Mol Carcinog 2021;60:167–71. [DOI] [PubMed] [Google Scholar]
- 64. Qing T, Mohsen H, Marczyk M, Ye Y, O'Meara T, Zhao H, et al. Germline variant burden in cancer genes correlates with age at diagnosis and somatic mutation burden. Nat Commun 2020;11:2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Olsen CM, Carroll HJ, Whiteman DC. Estimating the attributable fraction for cancer: a meta-analysis of nevi and melanoma. Cancer Prev Res (Phila) 2010;3:233–45. [DOI] [PubMed] [Google Scholar]
- 66. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Derrien AC, Rodrigues M, Eeckhoutte A, Dayot S, Houy A, Mobuchon L, et al. Germline MBD4 mutations and predisposition to uveal melanoma. J Natl Cancer Inst 2021;113:80–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 2014;46:478–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 2011;43:1018–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol Biomarkers Prev 1996;5:239–46. [PubMed] [Google Scholar]
- 71. Shibuya H, Kato A, Kai N, Fujiwara S, Goto M. A case of Werner syndrome with three primary lesions of malignant melanoma. J Dermatol 2005;32:737–44. [DOI] [PubMed] [Google Scholar]
- 72. Altieri L, Eguchi M, Peng DH, Cockburn M. Predictors of mucosal melanoma survival in a population-based setting. J Am Acad Dermatol 2019;81:136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. SP D'Angelo, Larkin J, Sosman JA, Lebbe C, Brady B, Neyns B, et al. Efficacy and safety of nivolumab alone or in combination with ipilimumab in patients with mucosal melanoma: a pooled analysis. J Clin Oncol 2017;35:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:3. [Google Scholar]
- 75. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [Preprint]. 2013. Available from: https://doi.org/10.48550/arXiv.1303.3997.
- 76. Raine KM, Van Loo P, Wedge DC, Jones D, Menzies A, Butler AP, et al. ascatNgs: identifying somatically acquired copy-number alterations from whole-genome sequencing data. Curr Protoc Bioinformatics 2016;56:15 9 1– 9 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012;28:1530–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics 2014;30:428–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 2017;45:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nordlund J, Backlin CL, Wahlberg P, Busche S, Berglund EC, Eloranta ML, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol 2013;14:r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kassahn KS, Holmes O, Nones K, Patch AM, Miller DK, Christ AN, et al. Somatic point mutation calling in low cellularity tumors. PLoS One 2013;8:e74380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990;9:811–8. [DOI] [PubMed] [Google Scholar]
- 87. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pritchard AL, Johansson PA, Nathan V, Howlie M, Symmons J, Palmer JM, et al. Germline mutations in candidate predisposition genes in individuals with cutaneous melanoma and at least two independent additional primary cancers. PLoS One 2018;13:e0194098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–81. [DOI] [PubMed] [Google Scholar]
- 91. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013; Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gillis S, Roth A. PyClone-VI: scalable inference of clonal population structures using whole genome data. BMC Bioinf 2020;21:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dietlein F, Weghorn D, Taylor-Weiner A, Richters A, Reardon B, Liu D, et al. Identification of cancer driver genes based on nucleotide context. Nat Genet 2020;52:208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhu H, Uuskula-Reimand L, Isaev K, Wadi L, Alizada A, Shuai S, et al. Candidate cancer driver mutations in distal regulatory elements and long-range chromatin interaction networks. Mol Cell 2020;77:1307–21. [DOI] [PubMed] [Google Scholar]
- 96. Shuai S, Drivers P, PCAWG Drivers and Functional Interpretation Working Group, Gallinger S, Stein L, Consortium PCAWG. Combined burden and functional impact tests for cancer driver discovery using DriverPower. Nat Commun 2020;11:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Arnedo-Pac C, Mularoni L, Muinos F, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers. Bioinformatics 2019;35:4788–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol 2016;17:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell 2017;171:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lochovsky L, Zhang J, Fu Y, Khurana E, Gerstein M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res 2015;43:8123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol 2019;20:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res 2021;31:448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pladsen AV, Nilsen G, Rueda OM, Aure MR, Borgan O, Liestol K, et al. DNA copy number motifs are strong and independent predictors of survival in breast cancer. Commun Biol 2020;3:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489–94. [DOI] [PubMed] [Google Scholar]
- 111. Cortés-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang L, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet 2020;52:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Nones K, Waddell N, Wayte N, Patch AM, Bailey P, Newell F, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 2014;5:5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Landi MT, Bishop DT, MacGregor S, Machiela MJ, Stratigos AJ, Ghiorzo P, et al. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat Genet 2020;52:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data (WGS and RNA-seq) that support the findings of this study have been deposited in the European Genome-phenome Archive (EGA; RRID:SCR_004944) and are available under study accession EGAS00001001552 (https://www.ebi.ac.uk/ega/studies/EGAS00001001552), with data set accessions EGAD00001008798 (WGS) and EGAD00001008837 (RNA-seq). Illumina EPIC DNA methylation array data analyzed in this study are publicly available in the Gene Expression Omnibus (RRID:SCR_005012) under accession number GSE202097. All other data are available in the article, in the Supplementary Data, or from the authors upon reasonable request.