The BAF complex, in particular ATPase subunit SMARCA4/BRG1, is critical for maintaining pediatric H3K27M-glioma cells in an oncogenic progenitor state, and pharmacologic inhibition is a potential therapeutic avenue for these highly aggressive tumors.
Abstract
Diffuse midline gliomas are uniformly fatal pediatric central nervous system cancers that are refractory to standard-of-care therapeutic modalities. The primary genetic drivers are a set of recurrent amino acid substitutions in genes encoding histone H3 (H3K27M), which are currently undruggable. These H3K27M oncohistones perturb normal chromatin architecture, resulting in an aberrant epigenetic landscape. To interrogate for epigenetic dependencies, we performed a CRISPR screen and show that patient-derived H3K27M-glioma neurospheres are dependent on core components of the mammalian BAF (SWI/SNF) chromatin remodeling complex. The BAF complex maintains glioma stem cells in a cycling, oligodendrocyte precursor cell–like state, in which genetic perturbation of the BAF catalytic subunit SMARCA4 (BRG1), as well as pharmacologic suppression, opposes proliferation, promotes progression of differentiation along the astrocytic lineage, and improves overall survival of patient-derived xenograft models. In summary, we demonstrate that therapeutic inhibition of the BAF complex has translational potential for children with H3K27M gliomas.
Significance:
Epigenetic dysregulation is at the core of H3K27M-glioma tumorigenesis. Here, we identify the BRG1–BAF complex as a critical regulator of enhancer and transcription factor landscapes, which maintain H3K27M glioma in their progenitor state, precluding glial differentiation, and establish pharmacologic targeting of the BAF complex as a novel treatment strategy for pediatric H3K27M glioma.
See related commentary by Beytagh and Weiss, p. 2730.
See related article by Mo et al., p. 2906.
This article is highlighted in the In This Issue feature, p. 2711
INTRODUCTION
Altered epigenetic patterns leading to stalled developmental cell states is a central theme in pediatric cancer (1). Pediatric high-grade glioma, including H3K27M glioma, is a classic example of altered epigenetics underlying tumorigenesis, in which more than 70% of tumors harbor mutations in genes encoding the histone 3 variants H3.1, H3.2, and H3.3, resulting in oncohistones (2, 3). These somatic H3K27M mutations lead to inhibition of Polycomb repressive complex 2 (PRC2) activity, with a corresponding global reduction of H3K27 trimethylation (H3K27me3) and increase in H3K27 acetylation (H3K27ac) levels (4, 5). Despite a global reduction, H3K27me3 levels are maintained at gene promoters of strong Polycomb targets, in which residual PRC2 activity is required for proliferation and tumor maintenance (4, 5). Studies on isogenic H3K27M-glioma patient-derived cell lines show that the main effect of the mutation is a defect in spreading of H3K27me2/3 repressive marks, rather than affecting the recruitment of PRC2, or deposition of H3K27me3 by PRC2 catalytic subunit, EZH2 (6). Thus, the presence of H3K27M leads to the reprogramming of canonical functions of chromatin remodeling complexes, which is at the core of tumorigenesis.
The cellular context for expression of oncohistones is critical: H3K27M mutations likely arise prenatally in neural stem cells and upregulate stem-like signatures for premature activation of an oligodendrocyte precursor cell (OPC)–like state, resulting in tumor formation (7, 8). In addition to the expression of H3K27M, activation of platelet-derived growth factor receptor α (PDGFRα) and loss of p53 seem to be required for tumor formation (7). Hence, epigenetic reprogramming of early OPCs and acquisition of secondary genomic alterations cooperate to initiate H3K27M glioma. In line with this tumorigenesis model, we and others have shown that H3K27M-glioma patient tumors are mainly composed of highly proliferative stem-like cells representative of the OPC-like progenitor state, with a block in differentiation along the glial lineages (9, 10). Although a few tumor cells still differentiate toward astrocytic- like (AC-like) and oligodendrocytic-like (OC-like) cell states, only OPC-like tumor cells are able to produce tumor xenografts in mice, suggesting an inverse relationship between differentiation and tumorigenicity (10). Furthermore, depletion of H3K27M in patient-derived cell lines and xenograft models leads to differentiation of glioma stem–like cells along oligodendrocytic or astrocytic lineages (6, 11, 12).
We hypothesized that H3K27M-induced chromatin reprogramming in the predominant OPC/stem-like cells creates epigenetic dependencies that have not yet been systematically analyzed. To address this, we conducted an epigenetically focused CRISPR/Cas9 screen in glioma neurosphere models, representative of the OPC-like tumorigenic state (10). To our knowledge, this is the first study to systematically evaluate specific gene dependencies in chromatin remodeling complexes driving the oncogenesis of H3K27M glioma. Here, we identify the BAF chromatin remodeling complex, which is a crucial remodeler for self-renewal and proliferation of cancer-initiating stem cells in leukemia and adult glioblastoma (13, 14), as a novel dependency in H3K27M glioma. In this study, we show that the catalytic subunit of the mammalian SWI/SNF (BAF) chromatin remodeling complex, BRG1 (encoded by SMARCA4), maintains glioma stem cells in an OPC-like state and describe the functional impact of SMARCA4 loss in H3K27M-glioma cells. Importantly, we show that pharmacologically targeting BRG1 activity using small-molecule inhibitors and proteolysis-targeting chimera (PROTAC) degraders mimics mechanistic effects of the genetic depletion of SMARCA4 in H3K27M glioma. In summary, we show that targeting the ATPase activity of the BAF complex is a novel strategy for epigenetic targeted therapy in this lethal type of pediatric glioma.
RESULTS
Epigenetic Dependencies of H3K27M Glioma Identified through a CRISPR/Cas9 Screen in Neurosphere Models
Single-cell RNA sequencing (scRNA-seq), together with chromatin and bulk transcriptomic analyses of H3K27M-glioma patient tumors, patient-derived cell models, and patient-derived xenograft (PDX) models, has shown that the H3K27M mutation suppresses glial differentiation and maintains cancer cells in a cycling, OPC-like state (9–12). To identify chromatin remodeling complexes and histone regulators responsible for maintaining this oncogenic state, we conducted an epigenetically focused, pooled, negative selection CRISPR/Cas9 screen in H3.3K27M (n = 4, BT869, BT245, HSJD-DIPG007, HSJD-DIPG012) and H3 wild-type (H3WT; n = 1, HSJD-GBM001) patient-derived glioma neurosphere models enriched for OPC-like cells (Fig. 1A). Top-scoring hits were identified using three different algorithms to calculate gene dependency scores: Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK; ref. 15); CERES, which accounts for copy number–specific effects (16); and Chronos, which calculates the longitudinal rate of dropout for improved inference of gene dependencies over time (17). After filtering out common essential genes derived from 855 adult and pediatric cancer cell lines (18), we identified 215 genes as dependencies for the survival of H3.3K27M-glioma cells (Supplementary Fig. S1A and SB; Supplementary Tables S1–S3).
Figure 1.
Epigenetic CRISPR screen identifies BAF complex as a novel dependency in pediatric H3K27M glioma. A, Schematic of epigenetic-focused CRISPR/Cas9-negative selection screen conducted in one H3WT-glioma and four H3.3K27M-glioma neurosphere models. B, ssGSEA of top-scoring protein complexes in four H3.3K27M-glioma models using Chronos and CERES dependency scores. C, Mean difference in ssGSEA complex scores (more negative scores indicate selective dependencies) for BAF, PRC1, PRC2, and HDAC-containing complexes in four H3.3K27M-glioma models compared with 855 human adult and pediatric cancer cell lines in the DepMap (Broad Institute). D, Mean difference in ssGSEA complex scores for BAF, PRC1, PRC2, and HDAC-containing complexes in four H3.3K27M-glioma models compared with 60 adult and pediatric brain cancer cell lines in the DepMap (Broad Institute). E, Relative cell viability across three H3.3K27M-glioma neurosphere lines following single-gene CRISPR/Cas9-mediated knockout of BAF complex genes (normalized to AAVS1-negative sgRNA control, n = 3). Three bars are shown per gene to indicate three individual sgRNAs used for knockout (sgRNA sequences are provided in Supplementary Table S5). Data are shown as mean ± SEM; ****, P < 0.0001; **, P = 0.0065. F, Dependency scores (CERES) for SMARCA4 in four H3.3K27M-glioma models compared with 23 other cancer types reported in the DepMap (Broad Institute). The median is indicated by the red dashed line, and quartiles are shown in dotted lines. Types of cancer models are displayed in descending order from least sensitive to knockout to the most sensitive (most negative median CERES score).
To determine top-scoring chromatin remodeling complexes, we calculated a median single-sample gene set enrichment analysis (ssGSEA) score per chromatin complex from all top 215 gene dependencies. The CORUM protein complex database was used as a reference (19), and complexes were considered when (i) they were composed of three or more members, and (ii) at least 75% of complex members were represented in our epigenetically focused CRISPR library. Using this analysis, we identified dependencies in BAF, PRC1, PRC2, and histone deacetylase (HDAC)–containing complexes (Fig. 1B). Although dependencies in HDAC-containing, PRC1, and PRC2 complexes have previously been described in H3K27M gliomas (4, 5, 20, 21), we discovered the BAF complex as a novel dependency.
Next, we compared the chromatin complex dependencies identified in H3K27M glioma against the Cancer Dependency Map (DepMap; Broad Institute, https://depmap.org/portal/), which is a comprehensive collection of genome-scale CRISPR/Cas9 screens in adult and pediatric cancer models (16). Dependencies in BAF, PRC1, PRC2, and HDAC-containing complexes were enriched in H3K27M-glioma cell lines compared with 855 human cancer or 60 brain cancer cell lines, including 51 adult and nine pediatric brain cancer cell lines representing medulloblastoma and H3WT and H3.3G34R pediatric glioblastoma (Fig. 1C and D).
BAF complex top hits included SMARCA4, SMARCD1, ARID1A, ARID2, BRD9, and BAF complex regulators CARM1, and SMARCAL1 (Supplementary Table S4). Intriguingly, unlike SMARCA4, SMARCA2 did not score as a dependency even though our CRISPR/Cas9 screen library included six individual single-guide RNAs (sgRNA) targeting SMARCA2 (Supplementary Fig. S1C and Supplementary Tables S1–S3), suggesting that BRG1, but not its paralog BRM, is required for H3K27M tumor cell survival. Dependencies in PRC1 and PRC2 complexes included BMI1 and CBX4, and EZH2, EED, and SUZ12, respectively (Supplementary Table S4; refs. 4, 5, 10, 21). Genes within the BAF complex did not score as dependencies in our H3WT pediatric glioma model, HSJD-GBM001 (Supplementary Table S4). Dependencies that overlapped between the H3.3K27M- and H3WT-glioma models were ATRX–DAXX chromatin remodelers and regulators of RNA polymerase II–mediated transcription, such as mediator and TATA-binding protein-free TAF-containing complexes.
We next performed secondary, single-gene CRISPR/Cas9 knockout validation experiments in three patient-derived H3.3K27M-glioma neurosphere models (BT869, SU-DIPGXXV, and SU-DIPGXIIIP*) targeting 14 genes within BAF, PRC1, PRC2 complexes, as well as HDAC2 and EP300. We confirmed that selective depletion of these genes led to a significant reduction in cell viability and an increase in apoptosis and cell death (Fig. 1E; Supplementary Fig. S1D and S1E). Within the BAF complex, we decided to focus on SMARCA4 (which encodes for BRG1), because it is one of the catalytic subunits of the complex and plays a major role in BAF-mediated chromatin remodeling. Furthermore, SMARCA4 exhibited a strong dependency in H3.3K27M glioma relative to 23 other cancer types reported in the DepMap (Fig. 1F). Even when compared with 29 adult glioblastoma cell models and an immortalized normal human astrocyte cell line (NHA-hTERT), SMARCA4 was selectively essential in pediatric H3.3K27M-glioma models (Supplementary Fig. S1F and S1G). Interestingly, SMARCA4 depletion also drastically reduced cell viability of an H3.1K27M-glioma neurosphere model (Supplementary Fig. S1H).
Taken together, our epigenetically focused CRISPR/Cas9 screen and secondary validation assays identified the BAF complex and its catalytic subunit SMARCA4 as novel genetic dependencies in H3K27M glioma.
BRG1–BAF Is a Key Regulator of the OPC/Stem-like State in Pediatric H3K27M Glioma
To define the role of BRG1–BAF in H3K27M-glioma cells, we aimed at identifying SMARCA4-driven regulatory networks. Single-cell regulatory network inference and clustering (SCENIC) analysis constructs and evaluates gene modules (regulons) based on the correlated expression of transcriptional regulators and putative target genes (22). We conducted a SCENIC analysis using our H3K27M-glioma single-cell, full-length mRNA sequencing dataset to define regulatory networks that drive each distinct tumor cell subpopulation (OPC-like, AC-like, OC-like, and cycling states) previously described in H3K27M gliomas (10). Interestingly, SMARCA4 emerged as an orchestrator of gene regulatory networks of the OPC-like, OC-like, and cycling cell states (Fig. 2A). In particular, the relative activity of SMARCA4 regulons in the OPC-like cancer cell subpopulation was significantly higher than in the more mature AC-like cancer cell subpopulation (Fig. 2B).
Figure 2.
BRG1–BAF chromatin remodeling complex regulates the OPC stem cell–like state in pediatric H3K27M glioma. A, Specificity scores of all regulons as determined by SCENIC analysis of gene regulatory networks in cycling, OPC-like, AC-like, and OC-like subpopulations of pediatric H3K27M-glioma patient tumors (n = 6). B, Relative activity of SMARCA4 regulons in cycling, OPC-like, AC-like, and OC-like subpopulations of pediatric H3K27M-glioma patient tumors (n = 6). Significance was compared between the progenitor OPC-like and the mature AC-like subpopulation of H3K27M-glioma primary tumors using a Student t test. The median is marked by the middle line within the box plot, the first and third quartiles by the upper and lower limits, and the 1.5× interquartile range by the whiskers. C, Heat maps (left) depicting BRG1, H3K27ac, and H3K27me3 ChIP-seq signals in BT869 (H3.3K27M-glioma) neurospheres at BRG1 binding sites according to their distance to transcription start sites (TSS). Regions within 1 kb of the TSS were considered TSS-proximal sites (top, n = 99), whereas all others were classified as TSS distal (bottom, n = 1,236). Each row shows 6.4-kb regions, centered on BRG1 peaks and ranked by BRG1 ChIP-seq signal intensity. Color shading corresponds to ChIP-seq read counts. Distribution of BRG1 peak counts (right) according to their distance to the TSS and genome location. D, Biological processes as determined by gene ontology (GO) analysis associated with genes detected in close proximity to BRG1 peaks in the BT869 neurosphere model. The top 15 enriched GO terms are displayed. The gene ratio refers to the ratio between the number of genes associated with BRG1 peaks present in each GO term and in all GO terms. The number of genes related to BRG1 peaks and associated with each GO term is indicated by the size of the circle, and significance is depicted by the color scale. Reg., regulation. E, Single-cell expression scores of genes defined as BRG1 bound in the BT869 neurosphere model, projected onto the scRNA-seq data of H3K27M-glioma patient tumors (n = 6). OPC-like cancer cells (high stemness score) show higher expression of BRG1-bound genes than AC-like cancer cells (low stemness score and negative lineage score) or OC-like cancer cells (low stemness score and positive lineage score). F, Representation of the enrichment of H3K27M-glioma tumor transcriptional programs in BRG1-associated genes, defined by ChIP-seq in BT869 cells, overlapping with enhancer-regulated genes identified in H3.3K27M-glioma patient tumors. The largest circle indicates a higher overlap of BRG1/enhancer-regulated genes with the OPC-like transcriptional program in H3.3K27M-glioma tumors. Red color refers to significance in overlap, as determined by a hypergeometric test. G, Gene tracks display input, BRG1, and H3K27ac ChIP-seq signal at promoters and enhancers of marker genes of the OPC-like cancer cell state (EGR1, NAV1, and ETV1).
This cell-state specificity within H3K27M-glioma subpopulations shown by SCENIC suggests that the BRG1–BAF complex remodels chromatin to maintain the oncogenic OPC-like transcriptional state of H3K27M-glioma stem cells. To test this hypothesis, we performed chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) to assess BRG1 binding and its correlation to activating H3K27ac and repressive H3K27me3 marks in a patient-derived H3.3K27M-glioma neurosphere model (BT869). We observed that BRG1 binding sites partially overlapped with H3K27ac peaks, and that BRG1 was not found in regions marked by H3K27me3, suggesting that BRG1 mainly localizes to active regions (Fig. 2C; Supplementary Fig. S2A). BRG1 binding sites localized primarily to intronic and intergenic regions distal to transcription start sites (>10,000 bp), which is consistent with enhancer locations (Fig. 2C). Gene ontology analysis revealed that genes in the vicinity of BRG1 binding sites, hence likely regulated by BRG1, are involved in central nervous system (CNS) development, neurogenesis, cell development, differentiation, and morphogenesis (Fig. 2D). In addition, motif analysis demonstrated that BRG1 binding sites were enriched for ASCL1, SOX10, and TCF4 DNA binding motifs (Supplementary Fig. S2B), which are transcription factors involved in oligodendrocyte specification and differentiation during CNS development. These results suggest that BRG1–BAF regulates transcription factors essential for H3.3K27M-glioma stem-like cell proliferation and differentiation.
We next explored the overlap between genes regulated by BRG1, as identified in our ChIP-seq assays, and genes representative of the cancer cell state–specific transcriptional programs (OPC-, OC-, and AC-like) identified in H3K27M-glioma patient tumors (10). Consistent with our findings above, we observed that BRG1 binding sites were enriched in the OPC-like gene signature as compared with mature AC- or OC-like signatures (Fig. 2E).
As the BAF complex is known to regulate enhancer landscapes to orchestrate expression of developmental genes during differentiation (23, 24), we next sought to define the role of BRG1–BAF in enhancer profiles of H3.3K27M glioma. To identify putative enhancers that are regulated by BRG1 in H3K27M glioma, we determined the overlap between BRG1-associated genes (meaning genes in close proximity to BRG1 binding sites as identified by ChIP-seq) in BT869 glioma neurospheres and enhancer-regulated genes detected in H3.3K27M-glioma patient tumors (25). We then categorized these enhancer-regulated and BRG1-associated genes according to the single-cell transcriptional states in H3K27M glioma and again found that most enhancer-regulated/BRG1-associated genes were overrepresented in OPC-like versus AC-like or OC-like programs (Fig. 2F). Indeed, BRG1 peaks, together with H3K27ac, were found in the vicinity of OPC-like tumor marker genes such as BCAN, CSPG4, and TNR (Supplementary Fig. S2C), which are glycoproteins known to be highly expressed in H3K27M glioma (26, 27), or EGR1, NAV1, and ETV1 (Fig. 2G), which are involved in CNS development, migration, proliferation, and differentiation.
Overall, our BRG1 ChIP-seq data, in conjunction with enhancer profiles and extended scRNA-seq analyses of patient-derived neurosphere models and patient tumors, suggest that the BRG1–BAF complex regulates transcription factor and enhancer landscapes essential for maintaining H3K27M-glioma cells in an OPC-like progenitor state.
Genetic Depletion of SMARCA4 Abrogates Chromatin Accessibility and OPC/Stem-like State
We next investigated the biological impact of SMARCA4 loss in H3K27M-glioma cells by assessing for changes in chromatin accessibility, histone modifications (H3K27ac and H3K27me3), and gene expression at bulk and single-cell levels (Fig. 3A). As expected, 4 days after SMARCA4 knockout in a patient-derived H3.3K27M-glioma neurosphere model (BT869), we observed increased levels of apoptosis as evidenced by an increase in PARP cleavage (Supplementary Fig. S3A). As BRG1 is a chromatin remodeler, we next focused on changes in chromatin accessibility 4 days after SMARCA4 knockout by assay for transposase-accessible chromatin using sequencing (ATAC-seq) and indeed observed an overall decrease in DNA accessibility at BRG1 binding sites (Supplementary Fig. S3B). Pathway analysis revealed that genes associated with decreased accessibility upon deletion of SMARCA4 are involved in the regulation of GTPase activity, axonogenesis, neuron projection guidance, and migration (Supplementary Fig. S3C). Intriguingly, we found that SMARCA4 loss reduced chromatin accessibility specifically at OPC-like H3K27M-glioma markers and increased chromatin accessibility at mature AC-like markers (Fig. 3B), further underscoring the important role of SMARCA4 in maintaining the OPC-like state in H3K27M glioma.
Figure 3.
Genetic knockout of SMARCA4 reduces chromatin accessibility and gene expression of OPC-like markers and enriches for the AC-like subpopulation in H3K27M-glioma cells. A, Schematic showing biological assessment of SMARCA4 knockout in H3.3K27M-glioma neurospheres by ChIP-seq for histone modifications (H3K27ac and H3K27me3), assay for transposase-accessible chromatin using sequencing (ATAC-seq) for chromatin accessibility, bulk RNA sequencing (RNA-seq), and scRNA-seq for gene expression. B, Heat map depicting z-scores for chromatin accessibility of OPC-like and AC-like marker genes in AAVS1-negative sgRNA control and SMARCA4-knockout (KO) BT869 neurospheres. C, Profile plot depicting the average H3K27ac ChIP-seq signal at BRG1 binding sites (n = 1,335) in BT869 4 days after SMARCA4 knockout. The plot shows 6.4-kb regions, centered on BRG1 peaks. D, Promoter-associated H3K27ac and H3K27me3 ChIP-seq signal for genes differentially expressed [determined by bulk RNA-seq analysis, log2(fold change [FC]) ≥ |1|] in SMARCA4-knockout cells (4 days after nucleofections). E, Profile plots (left) depicting the average H3K27ac signal at H3K27ac locations divided into positive (UP) or negative (DOWN) signal changes 4 days after SMARCA4 knockout. The plots show 6.4-kb regions, centered on H3K27ac peaks. Gene ontology (GO) analyses (right) performed on H3K27ac peaks with increased (n = 104, K27acUP) or decreased (n = 2,832, K27acDOWN) acetylation levels upon SMARC4 knockout for 4 days. The top five enriched GO terms are displayed. Pos. reg. of epidermal cell diff., positive regulation of epidermal cell differentiation.F, Heat map (left) depicting fold change (as log2) of H3K27ac levels after SMARCA4 knockout for 4 days. Displayed are log2 fold changes > |1|. Genes indicated on the heat map are OPC-like markers associated with decreased H3K27ac levels after SMARCA4 depletion. Gene tracks (right) displaying BRG1 ChIP-seq signal in BT869 neurospheres, as well as H3K27ac and ATAC-seq, at two OPC-like marker genes (TNR and EPN2). G, Changes in the percentage of cycling versus noncycling (left) and OPC- and AC-like cancer cell subpopulations (right) in SMARCA4-knockout and AAVS1-negative sgRNA control cells 4 days after knockout in BT869 neurospheres as determined by scRNA-seq analysis. H, Single-cell expression scores of upregulated genes in BT869 SMARCA4-knockout or AAVS1-negative sgRNA control knockout cells projected onto scRNA-seq data of pediatric H3K27M-glioma primary tumors (n = 6).
Two major consequences of the H3K27M mutation are increased levels of H3K27ac and decreased levels of H3K27me3 in pediatric glioma cells. Therefore, we queried for changes in these histone modifications 4 days following SMARCA4 deletion. To do so, we probed for global histone modification changes after SMARCA4 depletion in BT869 neurospheres by immunoblot and for local H3K27ac and H3K27me3 changes through ChIP-seq experiments. We did not observe any changes in global H3K27me3 and H3K27ac levels following SMARCA4 depletion (Supplementary Fig. S3D). However, in accordance with BRG1–BAF being a transcriptional activator, following SMARCA4 depletion, we found reduced H3K27ac levels at BRG1 binding sites (Fig. 3C), at enhancers (Supplementary Fig. S3E), and at promoters of genes with reduced expression after SMARCA4 knockout (Fig. 3D). In contrast, only minor changes in H3K27me3 levels were observed at the same promoter regions after SMARCA4 depletion (Fig. 3D). Consistent with a decrease in acetylation following SMARCA4 loss, our bulk RNA-seq data demonstrated a clear tendency for gene downregulation 4 days after SMARCA4 knockout (Supplementary Fig. S3F).
Gene ontology analysis demonstrated that genes associated with a loss of acetylation after SMARCA4 knockout are mainly involved in cell cycle, mitosis, cell division, and microtubule formation, whereas genes with increased levels of acetylation are involved in cell adhesion, projection, and positive regulation of cell differentiation (Fig. 3E). Interestingly, similar to our ATAC-seq findings, acetylation levels in OPC-like signature genes was decreased upon loss of SMARCA4 (Fig. 3F). Among these, BRG1-bound OPC-like genes involved in CNS development and Notch signaling, such as TNR and EPN2, underwent a reduction in H3K27ac signal and chromatin accessibility after SMARCA4 knockout (Fig. 3F).
The observed reduction of chromatin accessibility and H3K27ac at OPC-like markers following SMARCA4 loss suggests that the BAF complex sustains H3K27M-glioma cells in an undifferentiated state. To test this hypothesis further, we investigated whether the loss of SMARCA4 would lead to a shift in the proportion of cancer cell subpopulations within the developmental hierarchy of H3K27M-glioma cells. To address this, we conducted scRNA-seq on BT869 glioma neurospheres 4 days following the CRISPR/Cas9-mediated knockout of SMARCA4. Indeed, we found that the loss of SMARCA4 led to a decrease in cycling and OPC-like signatures, with a corresponding increase in cells exhibiting a differentiated AC-like signature (Fig. 3G; Supplementary Fig. S3G). We then projected genes upregulated in SMARCA4-knockout neurospheres (as determined by scRNA-seq) onto single-cell gene expression data derived from patient tumor samples (10). Here again, we found that the vast majority of the genes upregulated in SMARCA4 knockout were highly expressed in the differentiated AC-like signature, whereas genes upregulated in AAVS1 control were in the cycling or progenitor OPC-like state (Fig. 3H).
In summary, our results demonstrate that loss of SMARCA4 leads to decreased levels of chromatin accessibility, H3K27 acetylation, and gene expression of the OPC/cancer stem cell–like program. Importantly, knockout of SMARCA4 led to a reduced proportion of cycling and OPC-like cancer stem cells, with a corresponding increase in AC-like mature H3K27M-glioma cells. These results strongly indicate that the BAF complex is necessary to retain H3K27M-glioma cells in a stem cell–like state, suppressing differentiation of tumor cells, and enabling tumor cell proliferation.
Genetic Loss of SMARCA4 Reduces Tumorigenicity of Orthotopic H3K27M-Glioma PDX Models
To examine whether SMARCA4 loss affects the tumorigenicity of H3K27M-glioma cells in vivo, we stereotactically implanted H3K27M-glioma cells after SMARCA4 or AAVS1 (negative control) knockout into the pons of immunocompromised mice using two H3K27M patient-derived orthotopic neurosphere models (PDX), namely, biopsy-derived BT869 and autopsy-derived SU-DIPGXIIIP*. Both models showed congruent results with significantly reduced tumor volumes based on MRI and bioluminescence signals (Fig. 4A and B; Supplementary Fig. S4A and S4B). Furthermore, overall survival significantly increased by 25% in BT869 and 44% in SU-DIPGXIIIP* PDXs after SMARCA4 knockout (Fig. 4C and D). Examining tumor cell phenotypes at end-stage disease, SMARCA4-knockout tumors displayed a marked reduction in tumor burden (Fig. 4E and F). BRG1 depletion was validated in these two models by immunofluorescence staining in tumors collected at necropsy (Supplementary Fig. S4C and S4D).
Figure 4.
Genetic knockout of SMARCA4 reduces tumorigenicity of H3K27M-glioma PDX models. A, Quantifications of MRI in PDX mice bearing tumors with either AAVS1-negative sgRNA control or SMARCA4 knockout in BT869 cells (H3.3K27M-glioma, biopsy-derived, n = 10 per group) at 110 and 117 days after injections. B, Quantifications of in vivo bioluminescence measurements in PDX mice bearing tumors with either AAVS1-negative sgRNA control or SMARCA4 knockout in SU-DIPGXIIIP* cells (H3.3K27M-glioma, autopsy-derived, n = 8 per group) until 93 days after injections. C, Kaplan–Meier survival curves of BT869 PDX mice following SMARCA4 knockout compared with AAVS1 controls. The median survival of SMARCA4-knockout mice was 144 days after injection compared with 115 days in AAVS1 controls; ****, P < 0.0001 (n = 10 mice per group). D, Kaplan–Meier survival curves of SU-DIPGXIIIP* PDX mice after the loss of SMARCA4 compared with AAVS1 controls. Median survival of SMARCA4-knockout mice was 75.5 days after injection compared with 52.5 days in AAVS1 controls; **, P = 0.0014 (n = 8 mice per group). E, Representative hematoxylin and eosin (H&E) staining in AAVS1 control and SMARCA4 knockout BT869 PDX tumors collected at end-stage disease. The number of days indicated in the figure refers to the survival endpoint for each mouse from the date of tumor cell injections. F, Representative H&E staining in AAVS1 control and SMARCA4-knockout SU-DIPGXIIIP* PDX tumors collected at end-stage disease. The number of days indicated in the figure refers to the survival endpoint for each mouse from the date of tumor cell injections.G, Representative immunostaining for nuclei (DAPI), oncohistone (H3K27M), and AC-like marker (GFAP) in AAVS1 control and SMARCA4-knockout (KO) BT869 PDX tumors collected at the end-stage disease. H, Representative immunostaining for nuclei (DAPI), oncohistone (H3K27M), and AC-like marker (SOX9) in AAVS1 control and SMARCA4-knockout BT869 PDX tumors collected at end-stage disease. I, Quantification of immunofluorescence staining for GFAP in AAVS1 control (n = 4) and SMARCA4-knockout (n = 4) BT869 PDX tumors collected at end-stage disease (**, P = 0.002, unpaired t test). J, Quantification of immunofluorescence staining for SOX9 in AAVS1 control (n = 4) and SMARCA4-knockout (n = 4) BT869 PDX tumors collected at end-stage disease (****, P < 0.0001, unpaired t test).
We next assessed the expression of tumor cell–state markers in H3K27M-glioma SMARCA4-knockout PDX models. Our previous scRNA-seq profiling of PDX tumors revealed that only OPC-like H3K27M cells can form tumors in mice, whereas orthotopic implantation of differentiated AC-like glioma cells into immunodeficient mice did not produce tumors (10). Consequently, we assessed the expression of AC-like (GFAP and SOX9) markers in the SMARCA4-knockout BT869 PDX tumors and found that SMARCA4-depleted tumors exhibited an increased proportion of mature AC-like cells compared with AAVS1 knockout–negative control tumors (Fig. 4G–J).
Collectively, we find that the loss of SMARCA4 decreases tumorigenicity of H3K27M-glioma cells and increases the proportion of the AC-like cell subpopulation in vivo. These findings suggest that SMARCA4 deletion shifts H3K27M-glioma cells away from a tumorigenic OPC/stem-like state and provide a rationale for targeting BRG1–BAF complex activity as a novel therapeutic strategy.
Therapeutically Targeting BRG1–BAF Is a Novel Strategy for Clinical Translation
To pharmacologically target dependencies identified in the BAF complex for therapeutic translation, we tested the efficacy of allosteric small-molecule ATPase inhibitors of BRG1/BRM (Compounds 11, 12, and 14; ref. 28), a bromodomain inhibitor of BRG1/BRM/PBRM1 (PFI-3; ref. 29), CARM1 inhibitors (CARM1 inhibitor and TP064), and BRD9-targeting agents (the inhibitor I-BRD9 and the degrader dBRD9-13). These chemical agents were tested on H3.3K27M (n = 5), H3.1K27M (n = 1), and H3WT (n = 3) pediatric high-grade glioma neurosphere models and two nonmalignant control models: NHA-hTERT (human astrocytes) and Oli Neu (mouse OPCs). We found that H3K27M-glioma neurospheres were more sensitive to BRG1/BRM ATPase inhibitors than H3WT-glioma and nonmalignant cell models (Fig. 5A; Supplementary Fig. S5A). Of note, among all drugs tested, BRG1/BRM ATPase inhibitors were the most efficacious in reducing viability in H3K27M-glioma models (IC50 values ranging from 50 nmol/L to 1.5 μmol/L) in comparison with the BRG1/BRM bromodomain inhibitor PFI-3 (IC50 values from 580 nmol/L to more than 10 μmol/L), CARM1 inhibitors (IC50 values from 1.3 μmol/L to more than 10 μmol/L) and BRD9-targeting agents (IC50 values from 5 μmol/L to more than 10 μmol/L).
Figure 5.
BAF complex ATPase inhibition and degradation are novel therapeutic strategies in pediatric H3K27M-glioma. A, Heat map of IC50 values comparing small-molecule inhibitors and a degrader targeting BAF complex members, and its regulators (BRG1/BRM inhibitors: Compounds 11, 12, 14, PFI-3; CARM1 inhibitors: CARM1 inhibitor, TP064; BRD9 inhibitor: I-BRD9; and BRD9 degrader: dBRD9-13) in H3.3K27M (n = 5), H3.1K27M (n = 1), and H3WT (n = 3) pediatric glioma neurosphere models and nonmalignant cell lines (n = 2, NHA-hTERT: immortalized normal human astrocytes, and Oli Neu: immortalized normal mouse OPCs). B, PRISM analysis of 694 cancer cell lines representing 23 lineages (Broad Institute), treated with a BRG1/BRM inhibitor (Compound 11) at an 8-point dose curve (3-fold dilution, with a maximum of 10 μmol/L) for 5 days. The black dashed line represents the mean AUC computed over cell lines of all lineages. Cancer lineages below this line represent those sensitive to BRG1/BRM inhibition by Compound 11. C, Chemical structures of BRG1/BRM inhibitors (Compounds 11, 12, and 14) and a novel BRG1/BRM degrader (JQ-dS-4). D, Log2 fold change (FC) of differential proteins (left) as assessed by SILAC of DMSO control (light isotope labeled) and 1 μmol/L JQ-dS-4 (heavy isotope labeled)–treated BT869 H3.3K27M-glioma neurospheres (2 days of treatment). Heat map (right) of BAF complex proteins (with encoding genes shown in parentheses) depleted upon JQ-dS-4 treatment in BT869 neurospheres. E, Immunoblot for BRG1 and BRM protein levels in BT869, HSJD-DIPG007, and SU-DIPGXIIIP* neurospheres treated with novel BRG1/BRM degraders (AU-15330 and JQ-dS-4) at indicated doses and time points. Cleaved PARP was used as a marker for apoptosis. Total H3 and GAPDH served as loading controls. F, Heat map of IC50 values comparing two BRG1/BRM degraders (JQ-dS-4 and AU-15330) in H3.3K27M (n = 5), H3.1K27M (n = 1), and H3WT (n = 3) pediatric glioma neurosphere models and nonmalignant cell lines (n = 2, NHA-hTERT: immortalized normal human astrocytes, and Oli Neu: immortalized normal mouse OPCs). G, Dose–response curves for BRG1/BRM degraders (AU-15330 and JQ-dS-4) in H3.3K27M (n = 5), H3.1K27M (n = 1), and H3WT (n = 3) pediatric glioma neurosphere models and nonmalignant cell lines (n = 2, NHA-hTERT: immortalized normal human astrocytes, and Oli Neu: immortalized normal mouse OPCs).
To test the specificity of BAF inhibition in H3K27M-glioma models, we tested isogenic models that expressed either H3K27M or H3WT and observed that human OPCs and mouse neural progenitor cells (NPC) expressing H3K27M were more sensitive to Compound 11 (BRG1/BRM ATPase inhibitor) compared with their isogenic H3WT control cells (Supplementary Fig. S5B). We confirmed that this observed change in drug sensitivity was not due to changes in cell growth because there was not a significant change in proliferation rates of H3K27M-expressing models compared with their H3WT controls (Supplementary Fig. S5C). These results suggest an increased sensitivity to BAF inhibition in the context of H3K27M oncohistones. Moreover, the reduced sensitivity observed in nonmalignant human and mouse OPCs, mouse NPCs, and human astrocytes to BRG1/BRM inhibitors suggests acceptable toxicity of BAF-targeting agents (Fig. 5A; Supplementary Fig. S5A and S5B).
Next, we comprehensively tested the ATPase inhibitors Compounds 11 and 14 in vitro against 694 and 880 cancer cell lines, respectively, representing at least 23 lineages (Broad Institute). Strikingly, H3K27M-glioma neurospheres were the most sensitive to Compound 11 compared with cancer cell lines of all other lineages and scored within the top 10 lineages most sensitive to Compound 14 (Fig. 5B; Supplementary Fig. S5D). Other than H3K27M-glioma models, cell lines of only five other lineages (prostate, salivary gland, hematopoietic and lymphoid tissue, skin, and endometrium) previously associated with BAF complex dysfunction and/or dependence were sensitive to BRG1/BRM inhibition by both Compounds 11 and 14 (30–34).
To determine the best option to therapeutically target BRG1 and evaluate whether H3K27M glioma cells are differentially sensitive to the removal of BRG1 protein versus inhibition of BRG1 ATPase activity, we designed a heterobifunctional PROTAC utilizing the BRG1/BRM inhibitors tested (Compounds 11, 12, and 14) as chemical “warheads.” We linked the core of these BRG1 ATPase inhibitors to phthalimides (thalidomide, lenalidomide, and pomalidomide, referred to as immunomodulatory imide drugs, or IMiDs), which are cereblon (CRBN) ubiquitin ligase binders, and created a degrader, JQ-dS-4, that can dimerize BRG1/BRM with CRBN, resulting in E3 ligase–induced polyubiquitylation and proteasomal degradation to eliminate BRG1/BRM (Fig. 5C). We first confirmed target engagement of our novel BRG1/BRM ATPase degrader by evaluating binding of JQ-dS-4 to BRG1. Utilizing isothermal titration calorimetry (ITC), we observed that JQ-dS-4 successfully binds BRG1 at a binding affinity of 6.3 μmol/L (Supplementary Fig. S5E). We next performed stable isotope labeling by amino acids in cell culture (SILAC) in an H3.3K27M-glioma neurosphere model (BT869) grown in the presence or absence of the degrader, as a quantitative proteomics approach to identify all proteins degraded by JQ-dS-4. BRG1 was one of the top significant proteins degraded by JQ-dS-4, and the most depleted subunit of the BAF complex, indicating on-target activity of our novel degrader (Fig. 5D).
Recently, another PROTAC targeting BRG1/BRM, AU-15330, was developed and published by Xiao and colleagues (35). In contrast to JQ-dS-4, AU-15330 combines a bait moiety binding to the bromodomain of BRG1/BRM with a ligand for the von Hippel–Lindau ubiquitin ligase (Supplementary Fig. S5F). We tested the degradation kinetics of both PROTACs in three H3.3K27M-glioma neurosphere models and observed that AU-15330 rapidly degraded BRG1/BRM from SU-DIPGXIIIP* cells within 6 hours of treatment, with concentrations as low as 500 nmol/L (Supplementary Fig. S5G). Complete degradation of BRG1/BRM by JQ-dS-4 was only achieved at 48 hours in BT869 cells and at 72 hours for both HSJD-DIPG007 and SU-DIPGXIIIP* cells (Fig. 5E). Similar to genetic knockout of SMARCA4 in H3K27M-glioma neurosphere models, degradation of BRG1/BRM induced by both PROTACs resulted in antiproliferative effects (Fig. 5F and G) and induction of apoptosis as shown by an increase of PARP cleavage (Fig. 5E; Supplementary Fig. S5G). We observed that the IC50 values obtained after treatment of H3K27M-glioma neurospheres with BRG1/BRM inhibitors were in the same range as the ones obtained with BRG1/BRM degraders (Fig. 5A and F).
To comprehensively study these degraders, we tested JQ-dS-4 and AU-15330 against 694 and 880 cancer cell lines, respectively (Supplementary Fig. S5H and S5I). Pan-cancer lineage results of AU-15330 were similar to Compounds 11 and 14, with H3K27M gliomas being among the most sensitive tumors together with cancers of the prostate, salivary gland, and hematopoietic and lymphoid tissue. On the other hand, JQ-dS-4, the CRBN-based PROTAC, seemed to behave less specifically, targeting a broader range of tumor types. These results suggest that JQ-dS-4 might have some off-target activity, which would justify the observed cleavage of PARP at early time points (Fig. 5E). Finally, to further verify on-target activity, we tested Compound 14 and JQ-dS-4 on a BRG1/BRM-deficient adrenocortical carcinoma cell line (SW-13) and did not observe sensitivity to BAF ATPase inhibition (IC50 over 10 μmol/L), but did observe reduced sensitivity to degradation (IC50 of 6 μmol/L; Supplementary Fig. S5J).
In summary, BRG1 inhibition or degradation was efficacious in reducing the viability of H3K27M-glioma neurosphere models with reduced activity in normal cells and H3WT pediatric glioma neurosphere models. In our pan-cancer lineage analysis, we observed that H3K27M gliomas are among the most sensitive tumor types to inhibition or degradation of BAF ATPases. Thus, we identify BRG1 as a druggable vulnerability in H3K27M glioma, setting the stage for future clinical translation in children diagnosed with these tumors.
Pharmacologic Targeting of the BRG1–BAF Complex Mimics Biological Effects of SMARCA4 Loss
We next investigated the biological impact of therapeutically targeting BRG1–BAF in H3K27M-glioma neurospheres by assessing global changes in chromatin accessibility, gene expression, and in vivo tumorigenicity after BRG1/BRM inhibition or degradation (Fig. 6A). we found that treatment with BRG1/BRM inhibitors (Compound 11 and Compound 14) and degraders (JQ-dS-4 and AU-15330) led to a decrease in DNA accessibility at BRG1 binding sites 24 and 48 hours after treatment in H3.3K27M-glioma neurospheres (BT869; Fig. 6B), which is in agreement with data recently reported in leukemia and mouse embryonic stem cells (36, 37). Gene ontology analysis of regions with decreased accessibility after 24 hours of BRG1/BRM antagonist treatment revealed a positive enrichment for processes involved in axonogenesis and regulation of neuron projection development (Fig. 6C). Treatment with BAF ATPase inhibitors and degraders also led to a marked reduction in chromatin accessibility at genes of the OPC- and OC-like cancer cell metaprograms (Fig. 6D). This is in line with our finding that SMARCA4 governs gene regulatory networks of the OPC-like and OC-like cancer cell states (Fig. 2A). The observed reduction in chromatin accessibility at BRG1 binding sites and at OPC-like metaprogram–related genes after drug treatment phenocopied changes detected after SMARCA4 knockout (Fig. 3). Concurrent with changes in DNA accessibility, we found that the expression of many OPC-like genes was downregulated in BRG1/BRM ATPase inhibitor (both Compounds 11 and 14)–treated H3.3K27M-glioma cells as compared with DMSO control (Fig. 6E).
Figure 6.
Pharmacologic targeting of the BAF complex in vitro and in vivo mimics the biological effects of SMARCA4 depletion in H3K27M glioma. A, Schematic showing biological assessment of BRG1/BRM ATPase inhibitor– and degrader–treated H3.3K27M-glioma neurospheres for changes in chromatin accessibility by ATAC-seq, gene expression by bulk RNA sequencing (RNA-seq), and in vivo efficacy of chemical compounds in subcutaneous xenograft mouse models. B, Profile plot (top) depicting average ATAC-seq signal at BRG1 binding sites (n = 1,335) in BT869 (H3.3K27M-glioma) neurospheres after 24 hours of treatment with 1 μmol/L of BRG1/BRM inhibitors [Compound 11 (C11), Compound 14 (C14)] and degraders [JQ-dS-4 (JQ), AU-15330 (AU)] and DMSO controls. The plot shows 6.4-kb regions, centered on BRG1 peaks. Gene tracks (bottom) exemplify changes in chromatin accessibility observed at BRG1 binding sites. Displayed are BRG1 ChIP-seq signal in control BT869 cells as well as ATAC-seq data on the same cells after 24 or 48 hours of treatment with 1 μmol/L of BRG1/BRM inhibitors or degraders and DMSO controls. C, Biological processes (gene ontology analysis) enriched in regions with decreased accessibility in BT869 neurosphere cells treated for 24 hours with 1 μmol/L of Compound (Cmpd) 14, Compound 11, JQ-dS-4, or AU-15330 as determined by ATAC-seq. The number of genes is indicated by the size of the circle, and significance is depicted by the color scale. Reg., regulation of. D, Overlap of genes with reduced chromatin accessibility in drug-treated BT869 neurospheres (as determined by bulk ATAC-seq) with single-cell transcriptional metaprograms in H3K27M glioma. BT869 cells were treated with 1 μmol/L of either Compound 11 (C11), Compound 14 (C14), JQ-dS-4 (JQ), or AU-15330 (AU) for 24 hours. The color scale indicates the number of overlapping genes. E, A heat map showing z-scores of gene expression of OPC-like metaprogram marker genes with statistically significant changes upon Compound 11 or 14 treatments in BT869 neurospheres after 24 hours. Colored bars indicate z-scores of gene expression. rep, replicate.F, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)– or BRG1/BRM degrader JQ-dS-4 (50 mg/kg i.p. daily)–treated mice bearing BT869 (H3.3K27M-glioma) subcutaneous tumors as compared with vehicle controls (n = 10 mice per group). Data are shown as mean ± SEM, P < 0.0001 (unpaired t test comparing vehicle and Compound 14 groups). Kaplan–Meier survival curves (bottom) of mice treated with either Compound 14 (20 mg/kg i.p. daily) or JQ-dS-4 (50 mg/kg i.p. daily) for 60 days in a subcutaneous BT869 xenograft model (n = 10 mice per group). The P value was calculated using the log-rank (Mantel–Cox) test. Gray bars show the duration of treatment. G, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)–treated mice bearing SU-DIPGXIIIP* (H3.3K27M) subcutaneous tumors as compared with vehicle controls (n = 8 mice per group). Data are shown as mean ± SEM, P < 0.0001 (unpaired t test). Gray bar shows the duration of the treatment. Tumor volume (bottom) measured after 30 days of Compound 14 treatment compared with vehicle control in mice bearing SU-DIPGXIIIP* (H3.3K27M) subcutaneous tumors. P = 0.0017 (unpaired t test). H, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)–treated mice bearing HSJD-GBM001 (H3WT) subcutaneous tumors as compared with vehicle controls (n = 10 mice per group). Data are shown as mean ± SEM, P = 0.578 (unpaired t test). Kaplan–Meier survival curves (bottom) of mice treated with Compound 14 (20 mg/kg i.p. daily) for 30 days in a subcutaneous xenograft model of HSJD-GBM001 (n = 10 mice per group). The P value was calculated using the log-rank (Mantel–Cox) test. Gray bars show the duration of treatment.
Based on the promising in vitro efficacy results of BRG1/BRM inhibitors and degraders in H3K27M-glioma neurosphere models, we were interested in testing these BAF-targeting compounds in PDX models. We first tested the blood–brain barrier (BBB) penetrance of BRG1/BRM inhibitors and the BRG1/BRM degrader JQ-dS-4. Unfortunately, mass spectrometry studies revealed limited penetration into brain tissue after intraperitoneal administration, precluding further investigation in orthotopic PDX models (Supplementary Fig. S6A and S6B). Therefore, we addressed the in vivo efficacy of Compound 14 and JQ-dS-4 in a subcutaneous model of H3.3K27M glioma, acknowledging that the nonorthotopic microenvironment might change transcriptional programs and response to drugs. Histologic patterns of tumor growth in our subcutaneous model (developed by injecting BT869 neurospheres into the right flank of mice) revealed that these tumors displayed hallmark features of H3K27M glioma, including presence of mitosis, necrosis, high index of proliferation, and vascularization indicative of the World Health Organization III–IV tumors (Supplementary Fig. S6C).
We treated immunocompromised mice bearing H3.3K27M-glioma subcutaneous (BT869) tumors with Compound 14 (20 mg/kg i.p. daily) or JQ-dS-4 (50 mg/kg i.p. daily) for 60 days. We monitored body weight and examined plasma, liver, and spleen collected from mice treated for 45 days, and did not observe any significant changes in body weight or toxic effects from either Compound 14 or JQ-dS-4 (Supplementary Fig. S6D). Remarkably, we observed a significant reduction in tumor volume and increased overall survival upon intraperitoneal treatment with the BRG1/BRM inhibitor Compound 14 (Fig. 6F). The efficacy of Compound 14 in preventing H3K27M-glioma tumor growth in vivo was confirmed in two additional subcutaneous tumor models using SU-DIPGXIIIP* and HSJD-DIPG007 neurospheres (Fig. 6G; Supplementary Fig. S6E; mice were treated with 20 mg/kg of Compound 14, i.p., daily for either 30 days or 45 days, respectively). In contrast, Compound 14 treatment (20 mg/kg, i.p., daily, 30 days) in an H3WT pediatric high-grade glioma subcutaneous model, HSJD-GBM001, did not result in significant changes in tumor growth or overall survival (Fig. 6H). Congruent with our in vitro findings, we found that BT869 tumors treated with Compound 14 for 10 days displayed a significant increase in the apoptosis marker cleaved caspase-3 and decrease in the proliferation marker Ki-67 (Supplementary Fig. S6F and S6G). This further confirms the in vivo efficacy of this pharmacologic compound targeting BAF complex activity in H3.3K27M glioma. A similar decrease in Ki-67 was also observed in HSJD-DIPG007 tumors collected at end-stage disease (Supplementary Fig. S6H). In agreement with the results obtained upon SMARCA4 knockout in the orthotopic BT869 model (Fig. 4G–J), BT869 and HSJD-DIPG007 subcutaneous tumors treated with Compound 14 exhibited a significant increase in the AC-like marker GFAP (Supplementary Fig. S6G and S6H).
Treatment with the BRG1/BRM degrader JQ-dS-4 only marginally reduced tumor volume and did not lead to an increase in overall survival of H3.3K27M glioma–bearing mice (Fig. 6F), likely due to poor systemic absorption and/or tissue penetrance as reflected by negligible degradation of BRG1 in treated tumors compared with vehicle controls (Supplementary Fig. S6I). Indeed, unlike the small-molecule inhibitors (Compounds 11 and 14), which showed reasonable solubility in aqueous solution (≥ 100 μmol/L), the solubility of JQ-dS-4 in PBS was lower than 1.56 μmol/L (detection limit of the method; Supplementary Fig. S6J), hence limiting its bioavailability in vivo. In addition, a single subcutaneous injection of 50 mg/kg of JQ-dS-4 resulted in plasma levels of only 240 ng/mL (∼0.3 μmol/L), suggesting an overall low bioavailability. These differences in solubility and bioavailability might be attributed to the high molecular weight of JQ-dS-4 (719.16 g/mol), as commonly described for PROTACs (38), and likely have led to poor efficacy of the degrader in reducing tumor growth in vivo. Further optimization of JQ-dS-4 is required to improve its pharmacokinetic properties allowing in vivo assessment of the therapeutic potential of this BAF degrader. Alternatively, AU-15330 can be used in future in vivo studies to assess the possible use of BAF degradation as a therapeutic strategy, as it has been shown to deplete BRG1 levels in tumors in vivo (35).
We conclude that gene programs critical for H3K27M-glioma oncogenesis were reduced in chromatin accessibility and gene expression upon BRG1/BRM inhibition or degradation, mimicking biological effects of SMARCA4 genetic depletion. Importantly, we show that pharmacologically inhibiting the ATPase activity of the BAF complex is a potent therapeutic strategy for H3K27M glioma in vivo.
DISCUSSION
Despite significant advances in pediatric oncology, diffuse midline gliomas with H3K27M mutations remain one of the few universally fatal pediatric cancers. These oncohistone-harboring tumors display dysregulated patterns of chromatin regulation, which maintain tumor cells in an undifferentiated OPC-like cycling state. However, a proportion of tumor cells can exit this stem-like state and transition toward a more mature AC- or OC-like state and lose tumorigenic potential (10). This chromatin dysregulation underlying tumorigenesis suggests a pivotal role for epigenetic therapies (reviewed in ref. 39), and indeed preclinical studies in H3K27M glioma revealed efficacy of HDAC, lysine demethylase (LSD1), BMI1, EZH2, and BET inhibitors (4, 20, 21, 40). However, these discoveries have yet to prove a benefit for patients, underscoring the need for additional efficacious and selective epigenetic-targeted therapies. To our knowledge, this study is the first to systematically evaluate epigenetic dependencies using a CRISPR/Cas9 screen in multiple representative neurosphere models of H3K27M glioma. Here, we identify the BAF complex as a major dependency in pediatric H3K27M glioma and BRG1 ATPase as a novel druggable epigenetic vulnerability.
BRG1 can function in a variety of different modes during normal development and in disease: it can promote cell viability (41) and is required for canonical cell development and differentiation (42). Here, we show that BRG1 is essential for H3K27M-glioma cell survival and proliferation. Mechanistically, BRG1 supports dysregulated patterns of chromatin accessibility, H3K27 acetylation, and expression of genes associated with self-renewal to regulate the transcription factor and enhancer landscapes in H3K27M glioma. This can be reversed, as knockout of SMARCA4, as well as chemical inhibition or degradation of BRG1, leads to a decrease in chromatin accessibility at regulatory regions associated with neuron development and expression of OPC/cancer stem–like genes. Similar mechanisms of action where treatment with BRG1/BRM inhibitors and degraders restores altered chromatin and dysregulated transcription have been described in leukemia stem cells (36, 43), prostate cancer (35), and lung cancer (44).
We show that chromatin alterations following depletion of SMARCA4 or BRG1-directed therapy in H3K27M glioma cause an enrichment of the AC-like cell subpopulation in vitro and in vivo. We rationalize that this could result from either OPC-like cancer cells differentiating into AC-like cells or from selective depletion of highly proliferative OPC-like cells. Future studies using lineage-tracing experiments are needed to comprehensively address this question. In addition, SMARCA4 knockout or treatment with BRG1 antagonists leads to an increase in tumor cell death and apoptotic markers. These antiproliferative effects might again be related to the selective killing of OPC-like cells upon BRG1 loss, which would simultaneously lead to an enrichment of AC-like cells and apoptotic markers, or result from blocking an alternate function of BRG1 unrelated to state transitions. In line with this rationale, BRG1 loss in acute myeloid leukemia (AML) has been shown to not only promote tumor cell differentiation but also increase the expression of apoptotic regulators in a differentiation-independent manner (31). Further studies investigating BRG1 regulation of H3K27M-glioma cell death mechanisms are required to fully comprehend this phenotypic change after BRG1 loss.
Recent evidence of BAF vulnerabilities in multiple cancers has supported the synthesis of novel BAF antagonists (28, 35). We tested BRG1 inhibitors and degraders against a total of over 800 cancer cell lines representing at least 23 lineages and found that H3K27M-glioma cells are among the top six cancer lineages most sensitive to BRG1/BRM inhibition. Indeed, we observed that BRG1/BRM inhibitors and degraders impair H3K27M-glioma cell proliferation in vitro (with IC50 values in the low nanomolar range) and that the BRG1/BRM inhibitor Compound 14 consistently prevented tumor growth in vivo in three H3K27M-glioma models. Furthermore, H3WT-glioma cells were less sensitive to BRG1 inhibition both in vitro and in vivo. Future studies in an expanded cohort of pediatric high-grade gliomas, as well as functional biochemical studies, will be needed to directly assess the relationship between BRG1 sensitivity and the H3K27M mutation.
As both catalytic subunits of the ATPase (BRG1 and BRM) are broadly expressed and participate in diverse cell homeostasis processes, potential consequences of BRG1/BRM-directed therapy need to be carefully evaluated. Here, we show that BRG1 inhibitors were selectively effective in H3K27M-glioma in vitro models compared with nonmalignant mouse OPCs and human astrocytes. Moreover, we and others have observed that BRG1/BRM antagonists are well tolerated in vivo over several months (35, 43, 44).
Another concern related to BAF inhibition is that, intriguingly, SMARCA4 was first described as a tumor suppressor gene (45, 46), being altered in 5% to 7% of all human malignancies (47). Indeed, inactivating mutations of SMARCA4 are a hallmark of certain cancers such as small cell carcinoma of the ovary hypercalcemic type, thoracic sarcomatoid tumors, and malignant rhabdoid cancer of the uterus (47, 48). Based on these observations, it might be argued that treatment with BRM/BRG1 antagonists could lead to an increased long-term risk of developing a secondary malignancy. However, overexpression of nonmutant SMARCA4 has also been described across diverse tumor types, such as in neuroblastoma and colorectal cancer (49), and its elevated expression has been correlated with a worse prognosis in some cases (50). Recent studies have demonstrated that missense mutations of SMARCA4 can lead to enzymes with not only loss but also gain of function (51). SMARCA4 also promotes tumor development in a diverse set of tumor types such as Ewing sarcoma (52), glioblastoma (13), and AML (31). Consequently, the role of BRG1 as a tumor suppressor or oncogene seems to be highly context dependent. In our studies and other recently published data, no oncogenic sequelae have been observed in mice, even after treating immunodeficient and tumor-prone NOD/SCID gamma (NSG) mice with BAF antagonists over several months (35, 43). Taken together, these in vitro and in vivo studies highlight the potential for a therapeutic window in which tumor growth inhibition is achieved with limited toxicity. Nevertheless, long-term in vivo toxicity studies are required to fully appreciate the consequences of systemic dual BRM/BRG1 inhibition.
Although we show the potent efficacy of BAF inhibitors and degraders against H3K27M glioma in vitro and in subcutaneous in vivo models, these chemical agents exhibit poor BBB penetration and are thus not yet clinically applicable for the treatment of brain tumors. However, PROTACs have recently been advanced to clinical-stage drugs (reviewed in ref. 53) and developed to target proteins within the CNS, indicating their potential application as future brain tumor therapeutics (reviewed in ref. 54). Furthermore, a chemical analogue of one of the inhibitors used in this study was very recently tested in a first-in-human, adult clinical trial for metastatic uveal melanoma (FHD-286, Foghorn Therapeutics) underscoring the potential for BRG1-targeting agents for cancer therapy.
In summary, we identified the BAF chromatin remodeling complex as a novel vulnerability in pediatric H3K27M glioma. We show that genetic perturbation of SMARCA4, as well as pharmacologic catalytic inhibition or degradation of BRG1, leads to chromatin rewiring, resulting in a shift from oncogenic stem-like to differentiating glioma cell subpopulations, and increased cell death. Moreover, treatment with BRG1-targeting inhibitors leads to a significant survival benefit in patient-derived H3K27M in vivo models, highlighting BRG1 as a potential novel target for epigenetic therapy in pediatric H3K27M gliomas.
METHODS
Cell Culture
Patient-derived H3K27M- and H3WT-glioma neurosphere lines were established at Boston Children's Hospital [BT869 (RRID:CVCL_C1MH), BT245 (RRID:CVCL_IP13), BT1160], Stanford University [SU-DIPGXIII (RRID:CVCL_IT41), SU-DIPGXIIP*, SU-DIPGXXV, SU-DIPGIV (RRID:CVCL_IT39), SU-pcGBM2 (RRID:CVCL_IT42)], and Hospital Sant Joan de Deu Barcelona [HSJD-DIPG007 (RRID:CVCL_VU70), HSJD-DIPG012 (RRID:CVCL_VU71), HSJD-GBM001] as previously described (10, 55). H3K27M- and H3WT-glioma cells were acquired from 2015 to 2020 and grown as neurospheres in tumor stem media (TSM) base (55) supplemented with B27 minus vitamin A (1:50, cat. #12587010, Thermo Fisher Scientific), human growth factors (20 ng/mL EGF, cat. #100-26-100 μg, 20 ng/mL FGF, cat. #100-146-100 μg, 10 ng/mL PDGF-AA, cat. #100-16-100 μg, 10 ng/mL PDGF-BB, cat #100-18-100 μg, Shenandoah Biotechnology) and 2 μg/mL heparin (cat. # 07980, STEMCELL Technologies) in ultralow attachment flasks. BT1160 neurospheres were acquired in 2022 from Boston Children's Hospital and grown in an NS-A Human Proliferation Kit (cat. #05750, STEMCELL Technologies) supplemented with human growth factors (EGF, cat. #100-26-100 μg, FGF, cat. #100-146-100 μg, Shenandoah Biotechnology) and heparin (cat. #07980, STEMCELL Technologies) in ultralow attachment flasks. Neurosphere cultures were dissociated for passaging using Accutase cell detachment solution (cat. #07920, STEMCELL Technologies) for 3 to 5 minutes at 37°C. Cells were passaged 1 to 6 times before being used in the described experiments. All neurosphere models were authenticated by high-resolution short tandem repeat profiling (Molecular Diagnostics Core, Dana-Farber Cancer Institute), with the latest testing dating to June 2021. Whole-exome or whole-genome sequencing was conducted on neurosphere models to obtain copy-number alterations. Mycoplasma testing was regularly performed every 3 months using the MycoAlert Mycoplasma Detection Assays kit (cat. #LT07-218, Lonza).
Immortalized normal human astrocytes (NHA-hTERT) were acquired in 2019 from the Bernstein lab at Massachusetts General Hospital and grown as adherent cultures in high-glucose DMEM (cat. #11965092, Thermo Fisher Scientific) containing 10% fetal bovine serum (cat. #A3160402, Thermo Fisher Scientific) and 1% antibiotic–antimycotic (cat. #15240096, Thermo Fisher Scientific) on tissue culture–treated flasks. Cells were detached using TrypLE Express Enzyme without phenol red (cat. #12604013, Thermo Fisher Scientific). SW-13 cells were purchased from ATCC in 2019 (cat. #CCL-105, RRID:CVCL_0542) and grown according to the manufacturer's instructions.
E13.5 NPCs were extracted from timed pregnancies of C57BL/6J mice (cat. #000664, The Jackson Laboratory, RRID:IMSR_JAX:000664) using the Papain Dissociation System by Worthington Biochemical Corp. NPCs were cultured in a NeuroCult Proliferation Kit (cat. #005702, STEMCELL Technologies) supplemented with heparin (cat. #07980, STEMCELL Technologies), basic FGF (cat. #78003, STEMCELL Technologies) and EGF (cat. #78006, STEMCELL Technologies). NPCs were grown as adherent lines on Laminin-coated plates and transduced with empty vector pLV, wild-type H3F3A, or H3F3A K27M, with both wild-type H3F3A and H3F3A K27M tagged with C-terminal HA. Both WTH3 and H3K27M plasmids had a T2A EGFP tag. Seventy-two hours later, cells were sorted for EGFP. EC50 curve analysis was performed using raw fluorescent Alamar blue values that were input into PRISM 7.0. Drug concentrations were log10 transformed and values normalized to percent viability with respect to vehicle-treated cells. EC50 values were interpolated from a log inhibitor versus normalized response with variable slope using a least square fit model.
Previously characterized immortalized murine oligodendroglial precursor cells (Oli neu RRID:CVCL_IZ82, mouse OPCs used in Fig. 5; Supplementary Fig. S5) were acquired from Stanford University in 2021 and cultured in suspension in DMEM/F12 with 4.5 g/L glucose (cat. #11330057, Thermo Fisher), 1× penicillin–streptomycin (cat. #15140122, Thermo Fisher Scientific), B27 supplement without vitamin A (cat. #12587010, Thermo Fisher Scientific), and 1 μmol/L NT3 (cat. #450-03, PeproTech). The media were supplemented with 1× SATO, consisting of Neurobasal A (cat. #10888-022, Thermo Fisher Scientific), 100 μg/mL transferrin (cat. #T-1147, Sigma), 100 μg/mL BSA (cat. #A9418, Sigma), 16 μg/mL putrescine (cat. #P5780, Sigma), 60 ng/mL progesterone (cat. #P8783, Sigma), and 40 ng/mL sodium selenite (cat. #S5261, Sigma).
All cells were grown in a humidified atmosphere with 5% CO2 at 37°C.
Induced Pluripotent Stem Cell Models
OPCs with inducible H3.3K27M expression were derived from the healthy control induced human pluripotent stem cell (hPSC; ref. 56). Two previously described hPSC subclones with stably integrated H3.3K27M cDNA under the control of a Tet regulatory element were utilized (25). OPC differentiation was performed as previously described (25, 57). Starting on day 30 of differentiation, cells remained in PDGF media with or without 1.5 μg/mL doxycycline for at least 14 days to induce H3.3K27M expression and its downstream effects. Prior to conducting cell viability assays, hPSC-derived OPC spheres with/without H3.3K27M expression were dissociated into single cells for FACS isolation. Spheres were first treated with Accutase at 37°C for 20 minutes under constant rotation. Aggregates were then triturated with a p1000 followed by a p200 pipette 15 to 20 times each in the presence of DNAse. Undissociated aggregates were allowed to settle, and the single-cell suspension was removed. The leftover aggregates were then triturated in 1% BSA in HBSS, and the above steps were repeated until all cells were dissociated. The single-cell suspension was sorted on a BD FACSAria II for DAPI-, RFP+ (H3.3K27M) live cells. The viability of hPSC-derived OPCs in the presence of varying concentrations of the BRG1/BRM inhibitor Compound 11 was assessed by CellTiter-Glo. Five thousand freshly sorted cells were plated in each well of a 96-well White Polystyrene Microplate precoated with poly-L-ornithine/laminin. The appropriate drugs were added in duplicate and redosed after 72 hours. At 144 hours, cells were lysed, and viability was assayed by luminescence imaging.
Epigenetically Focused CRISPR/Cas9 Screen
H3K27M-glioma neurosphere models stably expressing Cas9 (BT869, BT245, HSJD-DIPG007, and HSJD-DIPG012) and the H3WT neurosphere model (HSJD-GBM001) were transduced with an epigenetically focused sgRNA lentiviral library at a target multiplicity of infection of 0.3 to 0.5 and scaled to a minimum posttransduction coverage of 1,500 cells per sgRNA. The epigenetically focused CRISPR library targeted 1,350 genes, and six different sgRNAs (Avana library, Broad Institute) were used to target each gene (total of 9,100 sgRNAs). One thousand nontargeting sgRNAs were used as negative controls (Supplementary Fig. S1A and S1B). Forty-eight hours following transduction, infected cells were selected by culturing in the presence of 2 μg/μL puromycin (Gold Biotechnology) for 72 hours. Selected cells were then pooled and split into triplicate flasks, resulting in a minimum of 500 cells per sgRNA per replicate. Cells were grown under standard neurosphere conditions as described above for the duration of the screen. For each screen time point, a minimum of 5.0 × 106 cells were harvested for genomic DNA isolation. sgRNA sequences were amplified by PCR and sequenced on Illumina HiSeq 2500. Three different scoring algorithms, MAGeCK (15), Chronos (17), CERES (16), were used to identify gene dependencies. Dependency scores (using MAGeCK, CERES, Chronos algorithms) were calculated from time points 2, 3, and 4, indicative of days 23, 30, and 37 after infection (Supplementary Tables S1–S3 display dependency scores obtained for top 215 hits).
ssGSEA Analysis
ssGSEA is an extension of GSEA, which calculates separate enrichment scores for each sample and gene set. To identify top-scoring protein complex dependencies in H3.3K27M glioma, we compared gene dependencies from our primary CRISPR/Cas9 screen to core protein complexes in the CORUM database (ref. 19; RRID:SCR_002254, http://mips.helmholtz-muenchen.de/corum/#download). We included complexes with at least three members, and minimally 75% of the members were present in the epigenetically focused library. Common essential genes (as defined by the DepMap, Broad Institute, RRID:SCR_017655, https://depmap.org/portal/) were excluded. H3K27M-glioma complex dependencies were ranked by either Chronos or CERES metrics, and an enrichment score was calculated for each derived protein complex using ssGSEA (58). Complexes with strong ssGSEA scores calculated for both Chronos and CERES ranks or specifically enriched in H3K27M-glioma cells as compared with other DepMap cell lines were defined as strong or enriched dependencies, respectively.
Secondary CRISPR Validations
Single-gene CRISPR/Cas9-mediated knockouts were conducted as previously described (10) using the 4D-Nucleofector system (Lonza Group) with the program DS-150. Six sgRNAs [two each from validated genome-scale CRISPR/Cas9 sgRNA Avana and Brunello libraries (ref. 59; RRID:SCR_017655, https://depmap.org/portal/), and two from custom sgRNAs designed using ChopChop (RRID:SCR_015723, https://chopchop.cbu.uib.no/)] were used to knock out individual target genes of the BAF, PRC1, PRC2 complexes as well as EP300 and HDAC2. All experiments were conducted in triplicate in a minimum of two H3K27M-glioma neurosphere models. Cell viability was assessed at days 7, 14, and 21 after nucleofection using the standard CellTiter-Glo assay system (cat. #G7572, Promega). The top three best-performing sgRNAs were chosen for subsequent gene-knockout experiments using homology-directed repair (HDR) templates (200 pmol/L) designed for each sgRNA (Supplementary Table S5). All custom sgRNAs (20 bp) and single-stranded ultramer HDR templates (∼200 bp) were purchased from Integrated DNA Technologies. Cas9 only and an sgRNA targeting the AAVS1 safe harbor locus (AGGGAGACATCCGTCGGAGA) were used as negative controls; an sgRNA against PDGFRA was used as a positive control (10). Viability detected 7 days after nucleofection was compared with AAVS1 control by ordinary one-way ANOVA with multiple comparisons to obtain adjusted P values. Experiments were conducted in technical triplicates, and a minimum of two biological replicates per cell line were done. Induction of apoptosis and necrosis was also confirmed 7 days after nucleofection by flow cytometry for Annexin V (cat. #422201 and #640924, BioLegend) and 7-AAD (cat. #420403, BioLegend) staining according to the manufacturer's instructions.
Gene Regulatory and Transcription Factor Network Reconstruction
To characterize underlying transcription factor activities in H3K27M glioma, the SCENIC package (RRID:SCR_017247) was used to identify gene regulatory modules (termed regulons) in our lab's previously published scRNA-seq dataset (10, 22). For each cell subpopulation, a regulon specificity score (RSS) was computed to determine the specificity of the regulons across different cell subpopulations (60). For a given cell subpopulation, the RSS for all predicted regulons was ranked from high to low, and the highest-ranked ones were highly cell type–specific regulons.
BRG1, H3K27ac, and H3K27me3 ChIP-seq in H3.3K27M-Glioma Cells
BT869-untreated neurospheres were analyzed for BRG1, H3K27ac, and H3K27me3 ChIP-seq; whereas BT869 SMARCA4-knockout neurospheres were analyzed for H3K27ac and H3K27me3 ChIP-seq. BT869 SMARCA4-knockout cells were collected 4 days after nucleofection (described above). Cell pellets were frozen at −80°C and sent to Active Motif Services to be processed for ChIP-seq. In brief, cells were fixed with 1% formaldehyde for 15 minutes and quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated, and the DNA sheared to an average length of 300 to 500 bp. Genomic DNA (input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for de-cross-linking, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield. An aliquot of chromatin (25–30 μg) was precleared with protein A agarose beads (Invitrogen). The aliquots of chromatin used for H3K27ac and H3K27me3 reactions were spiked in with 200 ng of Drosophila chromatin in both control and knockout experiments. Genomic DNA regions of interest (ROI) were isolated using 4 μg of antibody against BRG1 (Abcam, cat. #ab110641, lot #GR3208604-19, RRID:AB_10861578), H3K27ac (Active Motif, cat. #39133, lot #16119013, RRID:AB_2561016), and H3K27me3 (Active Motif, cat. #39155, lot #26720022, RRID:AB_2561020). Antibody against H2Av (0.4 μg) was also present in the H3K27ac and H3K27me3 reactions to ensure efficient pulldown of the spike-in chromatin (61). Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Cross-links were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol–chloroform extraction and ethanol precipitation. Quantitative PCR reactions were carried out in triplicate on specific genomic regions using SYBR Green Supermix (Bio-Rad). The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using input DNA.
Illumina sequencing libraries were prepared from the ChIP and input DNAs by the standard consecutive enzymatic steps of end polishing, dA addition, and adapter ligation. Steps were performed on an automated system (Apollo 342, Wafergen Biosystems/Takara). After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on Illumina's NextSeq 500 (75 nt reads, single end). Reads were aligned to the human genome (hg38) and to the Drosophila genome (dm3) using the BWA algorithm (ref. 62; default settings, RRID:SCR_010910). Duplicate reads were removed, and only uniquely mapped reads (mapping quality ≥25) were used for further analysis. The number of human alignments used in the analysis was adjusted according to the number of Drosophila alignments that were counted in the samples that were compared (BT869 control and SMARCA4 knockout). Human alignments were extended in silico at their 3′-ends to a length of 200 bp, which is the average genomic fragment length in the size-selected library and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in bigWig files, and genome tracks were visualized by loading such files into Integrative Genomics Viewer (IGV; https://software.broadinstitute.org/software/igv/; RRID:SCR_011793; ref. 63).
For H3K27ac and BRG1, peak locations were determined using the MACS algorithm (v2.1.0, RRID:SCR_013291; ref. 64) with a cutoff of P = 1e−7. H3K27me3-enriched regions were identified using the SICER algorithm (RRID:SCR_010843) at a cutoff of FDR 1e−10 and a max gap parameter of 600 bp. Peaks that were on the ENCODE (RRID:SCR_006793) blacklist of known false ChIP-seq peaks were removed. Signal maps and peak locations were used as input data to Active Motifs proprietary analysis program, which creates Excel tables containing detailed information on sample comparison, peak metrics, peak locations, and gene annotations.
Known transcription factor motifs were identified using the findMotifsGenome program of the HOMER package version 4.11.1 (RRID:SCR_010881; ref. 65) using default parameters (-size 200). Annotation of genome locations as well as genome ontology analysis of such locations was done using the HOMER program annotatePeaks.pl with the -go parameter. Genome ontology results were plotted either in R version 4.1.1 using ggplot2 geom_point (ref. 66; https://ggplot2.tidyverse.org; RRID:SCR_014601) or in GraphPad Prism (8.4.2, RRID:SCR_002798). Overlap between identified ChIP-seq peaks of different samples was obtained using the program intersect from the BEDTools package version 2.26.0 (RRID:SCR_006646; ref. 67) with the -u parameter or subtract using the -A parameter. Heat maps and profile plots of ChIP-seq data were obtained using the deepTools package version 3.5.1 (RRID:SCR_016366; ref. 68), specifically the computeMatrix program (–referencePoint center, –beforeRegionStartLength 3200, –afterRegionStartLength 3200, –binSize 32) combined with either the plotHeatmap program (–colorMap, –missingDataColor 1, –plotFileFormat pdf) or the plotProfile profile program (–perGroup, –plotFileFormat pdf). Enhancer locations were determined using the Rank Order of Super Enhancers (ROSE) package (version 0.1, snapshot 11/27/18, http://younglab.wi.mit.edu/super_enhancer_code.html, RRID:SCR_017390), specifically ROSE_main.py and ROSE_geneMapper.py with default parameters. Gene tracks were plotted using the IGV genome browser version 2.11.0 (https://software.broadinstitute.org/software/igv/, RRID:SCR_011793).
To perform differential H3K27ac/me3 ChIP-seq signal analysis between BT869 cells after SMARCA4 knockout and untreated control cells, ChIP-seq peaks of both knockout and control samples were merged. Then, reads were counted for each merged peak in each sample, and fold changes between knockout and control were calculated. Regions with low signals were discarded, and regions with differential acetylation levels were defined as log2(fold change) ≥ |1|.
Enhancer Analysis
Active enhancer-associated genes previously derived by H3K27ac ChIP-seq in H3.3K27M-glioma primary tumors (25) were compared with genes adjacent to BRG1 genomic binding sites. Overlapping genes were integrated with scRNA-seq–derived H3K27M glioma cell subpopulation-specific marker genes by overrepresentation analysis using the Fisher exact t test. Bonferroni corrected P values are shown by color scale, and Jaccard similarity coefficients computed between overlapping H3.3K27M enhancer/BRG1-associated genes and cell type–specific markers are depicted by circle size.
Immunoblotting in BT869 SMARCA4-Knockout Cells
Western blot was used to quantify BRG1 protein levels in SMARCA4- knockout cells compared with untreated and AAVS1-negative controls. Cells were collected 4 days after SMARCA4 knockout by nucleofection (described above). Proteins were extracted using 1× RIPA buffer (supplemented with protease inhibitor, DTT, and benzonase) and quantified by the BCA Protein Assay Kit (cat. #23225, Thermo Fisher Scientific). Protein lysate was separated by NuPAGE Tris-Acetate protein gels (Invitrogen). The following antibodies were used: BRG1 (#49360S, Cell Signaling Technology, RRID:AB_2728743), PARP (#9542S, Cell Signaling Technology, RRID:AB_2160739), and β-Actin (#3700S, Cell Signaling Technology, RRID:AB_2242334).
For the immunoblots of histone modifications, cells were also collected 4 days following nucleofection. Histone enrichment was performed using the Abcam Histone Extraction Kit (ab113476). The following primary antibodies were utilized for western blots: Histone H3 (clone 1b1b2, Cell Signaling Technology, #14269s, RRID:AB_2756816), Acetyl K27 Histone H3 (clone D5E4, Cell Signaling Technology, #8173T, RRID:AB_10949503), and Trimethyl K27 Histone H3 (clone C36B11, Cell Signaling Technology, #9733T, RRID:AB_2616029). LI-COR secondary antibodies were used for detection (IRDye 680RD goat anti-mouse #926-68070, RRID:AB_10956588 and IRDye 800CW goat anti-rabbit #926-32211, RRID:AB_621843) on the Odyssey CLx Infrared Imaging System. Images were quantified using ImageStudio software, and values were plotted in GraphPad Prism (RRID:SCR_002798).
ATAC-seq Analysis of SMARCA4-Knockout and BAF Inhibitor/Degrader–Treated Cells
CRISPR/Cas9-mediated knockout of SMARCA4 or AAVS1 was performed on BT869 neurosphere cells as described above. Four days after nucleofection, neurospheres were dissociated with Accutase and FACS sorted into PBS/0.04% BSA by staining for calcein AM (cat. #C3100MP, Life Technologies) and TO-PRO-3 iodide (cat. #T3605, Thermo Fisher Scientific) to identify live and dead cell populations, respectively. For the drug treatment assays, BT869 cells were treated with either DMSO (control samples) or 1 μmol/L of BRG1/BRM inhibitors (Compounds 11 and 14) and BRG1/BRM degraders (JQ-dS-4 and AU-15330). Cells were collected 24 or 48 hours after treatment. FACS sorting of viable cells for drug-treated samples was not necessary, as roughly 90% of the cells were viable. Two technical replicates were processed for each biological replicate in all experiments. Data from two biological replicates for AAVS1 control, SMARCA4 knockout, and drug treatment assays (Compounds 11, 14, JQ-dS-4, AU-15330) in BT869 neurospheres are depicted in Figs. 3 and 6, respectively.
For ATAC-seq library preparation, the Omni-ATAC protocol was used (69). A minimum of 40,000 to 50,000 live cells per treatment condition and technical replicate were pelleted by centrifugation at 500 × g, 4°C for 5 minutes, and lysed in 0.1% NP40 (cat. #11332473001, Sigma/Roche), 0.1% Tween-20 (cat. #11332465001, Sigma/Roche), and 0.01% digitonin (cat. #G9441, Promega). Lysed nuclei were washed and pelleted at 500 × g in 4°C for 10 minutes, which was followed by tagmentation in 1× TD buffer, 100 nmol/L Tn5 transposase (cat. #15027865, Illumina), and 0.3× PBS in a 50 μL reaction volume for 30 minutes at 37°C, with 1,000 rpm mixing. Tagmented DNA was purified using the MinElute PCR Cleanup kit (cat. #28004, Qiagen) and PCR preamplified in 5 cycles using NEBNext-Fidelity 2× PCR Master Mix (cat. #M0541S, NEB) and primers containing Illumina adapter sequences before the number of additional cycles was assessed by quantitative PCR using SYBR Green (cat. #S9430, Sigma). On average, 5 to 9 additional PCR cycles were conducted, and the final library was purified by a MinElute PCR Cleanup kit (cat. #28004, Qiagen), quantified by a Qubit dsDNA HS Assay kit (cat. #Q32854, Thermo Fisher Scientific), and evaluated on a Bioanalyzer 2100 system (Agilent). Libraries were prepared and sequenced on a NextSeq 500 sequencer (Illumina) using NextSeq High Output Cartridge kits by paired-end reads.
We processed bulk ATAC-seq data as previously validated (70). In brief, raw fastq reads were trimmed of adapters and aligned using Bowtie2 version 2.4.2 (ATAC-seq, RRID:SCR_016368; ref. 71) to the hg19 reference genome. PCR duplicates were removed such that only one representative read was selected per unique pair of transposition events. Chromatin accessibility peaks were called using MACS2 (RRID:SCR_013291) with custom parameters for ATAC-seq (–nomodel –nolambda –call-summits; ref. 64), and a signal track in .bdg form was emitted from the peak calling function that was subsequently converted into a bigwig file for visualization. To establish a uniform feature set for the ATAC-seq data, we used all 1-bp summits from the MACS2 peak calls per population and expanded these to a uniform 500 bp, similar to a strategy implemented previously (69). Summits overlapping the same window were iteratively centered at the summit with the strongest signal (measured by −log10 FDR) until all summits were accounted. A peak × sample fragment counts matrix was completed using the getCounts function in chromVAR (72). Profile plots of ATAC-seq data were obtained using the deepTools package version 3.5.1 (RRID:SCR_016366; ref. 68), specifically the computeMatrix program (–referencePoint center, –beforeRegionStartLength 3200, –afterRegionStartLength 3200, –binSize 32) combined with the plotProfile profile program (–perGroup, –plotFileFormat pdf). Gene tracks were plotted using the IGV genome browser version 2.11.0 (RRID:SCR_011793). Differential peaks between SMARCA4- and AAVS1-knockout samples, as well as between inhibitors/degraders and DMSO-treated cells, were determined genome-wide using the counts matrix via DESeq2 version 1.30.1 (RRID:SCR_015687) with default parameters (73). Genes linked to differentially accessible peaks were tested for enrichment in gene ontology (GO) terms by the hypergeometric test using the R package clusterProfiler's enrichGO function (RRID:SCR_016884) and for overlap with published OPC/OC/AC metaprograms. Selected GO terms with an adjusted P value less than 0.05 are shown in Supplementary Figs. S3C and S6C, whereas the overlap between genes linked to differentially accessible peaks and single-cell metaprograms is shown in Fig. 6D.
Bulk RNA-seq Analysis of SMARCA4-Knockout and BRG1/BRM Inhibitor–Treated Cells
CRISPR/Cas9-mediated knockout of SMARCA4 or AAVS1 was performed on BT869 neurosphere cells as described above. Four days after nucleofection, neurospheres were collected and frozen at −80°C for future RNA isolation. BT869 cells were treated with either DMSO (control samples) or 1 μmol/L of BRG1/BRM inhibitors (Compounds 11 and 14), and cells were collected 24 hours after treatment and frozen at −80°C for future RNA isolation. Total RNA was extracted from frozen cell pellets using the Qiagen RNeasy Plus Mini Kit (cat. #74134), and RNA concentrations were determined using a NanoDrop spectrophotometer (Thermo Scientific). Bulk RNA-seq data from one biological replicate were analyzed for SMARCA4 knockout, and two biological replicates were analyzed for drug treatments, as shown in Fig. 3 and Supplementary Figs. S3 and S6. Library preparation and sequencing were performed by Novogene. In brief, messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first-strand cDNA was synthesized using random hexamer primers, followed by the second-strand cDNA synthesis. The library was ready after end repair, A-tailing, adapter ligation, size selection, amplification, and purification. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina platforms according to effective library concentration and data amount. The original raw data from Illumina platform were transformed to Sequenced Reads, known as Raw Data or RAW Reads, by base calling. Raw data were recorded in a FASTQ file, which contained sequencing reads and corresponding sequencing quality.
We processed bulk RNA-seq data similarly to scRNA-seq data described below. In brief, raw fastq reads were aligned using bowtie (v.0.12.7, RRID:SCR_005476) to the hg19 reference genome, and gene counts were quantified using RSEM (v1.2.19, RRID:SCR_013027) as expected counts. Differentially expressed genes between SMARCA4-knockout and untreated cells, as well as between inhibitor and DMSO-treated cells, were determined using DESeq2 and default parameters (v1.30.1, RRID:SCR_015687). Gene counts of each sample were normalized by DESeq2's median of ratios method and fitted with a negative binomial regression model. Differentially expressed genes were assessed using the Wald test. Relative expressions of OPC metaprogram genes (10) in DMSO- and inhibitor-treated samples are displayed in a heat map in Fig. 6E. Each row represents a gene and each column a sample. Color bars correspond to relative expression as z-scores. For Fig. 3D, BRG1-regulated genes were considered as genes that expression changed [log2(fold change) ≥ |1|] after 4 days of SMARCA4 knockout. Rows are ordered based on average expressions of these genes in the DMSO samples.
scRNA-seq Analysis of SMARCA4-Knockout Cells
Four days after nucleofection, AAVS1 control and SMARCA4-knockout neurospheres generated by CRISPR/Cas9 nucleofection as described above were dissociated with Accutase and FACS sorted for live cells using calcein AM (cat. #C3100MP, Life Technologies) and TO-PRO-3 iodide (cat. #T3605, Thermo Fisher). Both live and dead single cells were sorted individually into 96-well plates containing TCL lysis buffer (Qiagen). Plates were frozen at −80°C prior to processing for Smart-seq2 as previously described (74). SMARCA4-knockout BT869 cells were combined computationally for analysis of knockout cells compared with AAVS1 and Cas9 controls.
scRNA-seq data were preprocessed as previously described with some modifications (75). Raw sequencing reads were aligned to the hg19 genome by bowtie (RRID:SCR_005476), and gene counts were quantified using RSEM (RRID:SCR_013027) as transcript per million (TPM; ref. 76). Expression levels were calculated as Ei,j = log2(TPMi,j/10 + 1) for gene i in sample j. TPM values were divided by 10 for better approximation of the estimated complexity of single-cell libraries. We filtered out low-quality cells with fewer than 2,500 detected genes or an average housekeeping expression level below 2.5 and removed low expressed genes with TPM greater than 16 in fewer than five cells. We defined relative expression as centered expression levels, Eri,j = Ei,j − average [Ei,1…n] for the remaining cells and genes.
We adapted graph-based clustering with data integration to identify cellular clusters and gene signatures. Seurat (RRID:SCR_016341) was used to identify highly variable genes, and relative expression values of these highly variable genes were used for principal component analysis (77). We applied Harmony (RRID:SCR_022206) to the first 100 principal components with default parameters for data integration from multiple samples (78). The first 20 Harmony corrected dimensions were selected for computing t-distributed stochastic neighbor embeddings, and graph-based clustering of cells was used to group cells into subpopulations. Differentially expressed genes of each cluster were derived using Seurat's FindAllMarkers function. Cell type identities (e.g., OPC-like, AC-like, and cycling) were assigned to each cell cluster based on top differentially expressed genes.
Generation of Single-Cell Expression Scores and Plotting
Single-cell expression scores were computed in a similar way as described previously (75). Given a set of genes (Gj) for a gene set (e.g., a DAM signature), a score, SCj(i), which quantifies the scaled expression (z-score) of Gj for each cell i, was computed as the average scaled expression (Er) of the genes in Gj compared with the average scaled expression of a control gene set Gcont: SC (i) = average[Er(G, i)]− average[Er(Gcont, i)]. For each gene of the gene set, its control gene set contains 100 genes with the most similar aggregated expression level to that gene. Therefore, the control gene set represents a 100-fold larger but comparable distribution of expression levels to that of the considered gene set.
Then, we examined the enrichment of the scores calculated (BRG1 score − expression of BRG1-bound genes; AAVS1/SMARCA4-knockout scores − genes upregulated after each knockout) in the H3K27M-glioma scRNA-seq data (GSE102130) and OPC/OC/AC metaprograms previously identified (10). Cells were ordered by a stemness (as expression of the OPC program minus the maximal expression of the two differentiation AC/OC programs) and differentiation score (as expression of the OC program minus expression of the AC program) and labeled by scores of BRG1-bound genes (Fig. 2E) or genes upregulated after AAVS1/SMARCA4 knockout (Fig. 3H).
H3.3K27M-Glioma SMARCA4-Knockout Orthotopic Xenograft Models
SU-DIPGXIIIP* and BT869 neurosphere cells stably expressing luciferase (40) were dissociated and nucleofected to knockout SMARCA4 or AAVS1 control guide RNA using CRISPR/Cas9 as described above. Prior to injections, luciferase expression was confirmed using the ONE-Glo EX Luciferase Assay System (cat. #E8110, Promega). Twelve hours after nucleofection, cells were injected stereotactically into the right pons of 6-week-old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory, RRID:IMSR_JAX:005557) treated with 0.05 mg/kg buprenorphine and anesthetized with 2% to 3% isoflurane. The skull of the mouse was exposed through a small skin incision, and a small burr hole was made using a 25-gauge needle at the selected stereotactic coordinates zeroed on lambda: −1.0 mm X, −0.8 mm Y and 5.0 mm Z. SU-DIPGXIIIP* (100,000 cells in 3 μL PBS per mouse, n = 8 per group) and BT869 (200,000 cells in 3 μL PBS per mouse, n = 10 per group) cells were loaded into a 33-gauge Hamilton syringe and injected at a rate of 0.5 μL/minute with use of an infusion pump. Upon completing injection, the needle was left in place for another minute and then withdrawn slowly to help reduce cell reflux. Mice were returned to their cages after closing the scalp with suture and staple, placed on a warming pad, and visually monitored until full recovery. Mice were then checked daily for signs of distress, including seizures, weight loss, and tremors, and euthanized as they developed neurologic symptoms, including head tilt, seizures, sudden weight loss, loss of balance, and/or ataxia. All animal studies were performed according to Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (IACUC)–approved protocols (13-053).
Tumor growth was monitored weekly for SU-DIPGXIIIP* mice and monthly for BT869 mice using the IVIS Spectrum In Vivo Imaging System (PerkinElmer), starting either 7 days (for SU-DIPGXIIIP*) or 1 month (for BT869) after cell injections. Briefly, mice were injected subcutaneously with 75 mg/kg D-luciferin potassium salt (cat. #E1605, Promega) in sterile PBS and anesthetized with 2% isoflurane in medical air. Serial bioluminescence images were acquired using the automated exposure setup. The peak bioluminescence signal intensity within selected ROIs was quantified using Living Image Software (PerkinElmer) and expressed as photon flux (p/sec/cm2/sr). Representative planar bioluminescence images were displayed with indicated adjusted minimal and maximal thresholds. In vivo bioluminescence signal in mice injected with SU-DIPGXIIIP* SMARCA4-knockout cells was compared with AAVS1 controls using a Mann–Whitney test to determine a significant reduction. Unfortunately, mice injected with BT869 neurosphere cells lost expression of luciferase in vivo about 3 months after injections, and tumor growth was monitored by MRI. For both SU-DIPGXIIIP* and BT869 in vivo models, MRI was performed on SMARCA4-knockout and AAVS1 control mice as previously described (10).
Mouse brains collected at the survival endpoint were fixed in 4% paraformaldehyde for 24 hours and subsequently stored in 70% ethanol. Fixed mouse brains were cut in half mid-sagittally and embedded in paraffin to generate formalin-fixed, paraffin-embedded (FFPE) blocks. FFPE blocks were sectioned into 4-μm-thick sections and stained for hematoxylin and eosin (H&E) by iHisto histopathology support.
Immunofluorescent Staining and Image Analysis
Sections of FFPE blocks containing tumor samples were deparaffinized and rehydrated. Antigen retrieval was performed using a pressure cooker and 1× citrate buffer, pH 6.0 (cat. #C9999, Sigma). Sections were then quenched for autofluorescence using a protocol adapted from ref. 79. Subsequently, sections were stained with the following primary antibodies overnight at 4°C: anti-BRG1 (Abcam, EPNCIR111A, ab110641, RRID:AB_10861578, 1:50), anti-GFAP (Invitrogen, MA5-12023, RRID:AB_10984338, 1:200), anti–Histone H3 (mutated K27M; Abcam, ab240310, 1:100), anti–Ki-67 (BD, 550609, RRID:AB_393778, 1:100), and anti-SOX9 (Abnova, H00006662-M02, RRID:AB_875842, 1:100). After repeated washes in PBS, sections were incubated with the following secondary antibodies for 2 hours at room temperature: goat-anti-rabbit Alexa 555 (Invitrogen, A27039, RRID:AB_2536100, 1:200) and goat-anti-mouse Cy5 (Invitrogen, A10524, RRID:AB_2534033, 1:200). Imaging was performed using a Keyence BZ-X800E fluorescent microscope equipped with a BZ Nikon Objective Lens (20×). Following image acquisition, digital images were analyzed using Visiopharm.
Drug Screens in H3K27M and H3WT Pediatric Glioma, Normal, and Control Cell Models
Compounds targeting BRG1/BRM and its associated proteins CARM1 and BRD9 were tested on H3.3K27M (BT869, SU-DIPGXIII, SU-DIPGXIIIP*, SU-DIPGXXV, and HSJD-DIPG007), H3.1K27M (SU-DIPGIV), and H3WT (HSJD-GBM001, SUpcGBM2, and BT1160) pediatric glioma neurosphere cells, as well as in immortalized nonmalignant human astrocytes (NHA-hTERT) and mouse OPCs (Oli Neu). BRG1/BRM inhibitor (Compound 11) was tested on nonmalignant mouse NPCs and human induced pluripotent stem cell–derived OPCs expressing H3K27M and H3WT as described above. BRG1/BRM inhibitor (Compound 11) and degrader (JQ-dS-4) were tested on the BRG1/BRM-deficient adrenocortical carcinoma control cell line SW-13.
Dual ATPase BRG1/BRM inhibitors, Compounds 11, 12, and 14, were synthesized at Dana-Farber Cancer Institute as previously described (28). The on-target effects and mechanism of action of these BRG1/BRM inhibitors have previously been described (28, 36, 37). The novel ATPase and BRD9 degraders (80) were designed to ablate BRG1/BRM or BRD9 using an E3 ligase system by linking the BRG1/BRM ATPase or BRD9 with CRBN binders (J. Qi's laboratory at Dana-Farber Cancer Institute). The structure and purity of these compounds were confirmed by nuclear magnetic resonance and LC-MS. Further details on the chemical synthesis and on-target effect testing of JQ-dS-4 are described below. AU-15330 was synthesized by J. Qi's laboratory according to a recently published report by Xiao and colleagues (35). The BRG1 and PBRM1 inhibitor PFI-3 was purchased from SelleckChem. The BRD9 inhibitor I-BRD9 was purchased from Sigma. CARM1 inhibitors (CARM1 inhibitor and TP064) were purchased from Sigma and Tocris Biosciences, respectively.
Cells were plated in 384-well, white-wall, and clear-bottom untreated plates (800 cells per well for pediatric high-grade glioma cells, and 500 cells per well for NHA-hTERT cells and Oli Neu) and dosed at 24 and 96 hours after plating with compounds (concentrations ranging from 10 nmol/L to 10 μmol/L) using the D300e digital dispenser (HP). Cell viability was determined 7 days after drug exposure using the CellTiter-Glo system (cat. #G7572, Promega). Drug concentrations were log10 transformed, and values were normalized to percent viability with respect to DMSO-treated control cells. Log IC50 values were interpolated from a log inhibitor concentration versus normalized response curves with variable slope using a least square fit model in GraphPad Prism (RRID:SCR_002798).
Profiling Relative Inhibition Simultaneously in Mixtures Testing
Compound 11 and JQ-dS-4 were tested at eight concentrations following a 5-day treatment period against 694 cancer cell lines and Compound 14 and AU-15330 in 880 cancer cell lines as previously described (81). Two cell line collections, PR500 (including only adherent cell lines) and PR300 (including adherent and suspension cell lines), were analyzed. Three benchmark compounds—included in the profiling relative inhibition simultaneously in mixtures (PRISM) validation set—were also tested to ensure high data quality. All compounds were run in triplicate, and each plate contained positive (bortezomib, 10 μmol/L) and negative (DMSO) controls. Each cell line is represented by its AUC and plotted by lineage (primary disease and subtype) to visualize lineage-based sensitivity patterns. In particular, we calculated effect size as the difference between that lineage and all others and P values based on a t test. We used the R package “dr4pl” (version 2.0.0) to fit dose–response curves for four H3K27M-glioma cell lines treated with Compound 11, Compound 14, JQ-dS-4, and AU-15330. To calculate AUC for each cell line, we input these dose–response parameters into the same custom R function used to construct the PRISM validation set. Finally, we calculated the mean AUC value across H3K27M-glioma cell lines to facilitate comparison with other cancer types.
Synthesis of JQ-dS-4
The synthesis of JQ-dS-4 [3-((4-(3-(2-chloropyridin-4-yl)ureido)pyridin-2-yl)ethynyl)-N-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl) amino)butyl)benzamide] was perfomed in a 10-mL flask fitted with a stir bar. Compound 15 (ref. 28; 6 mg, 0.015 mmol/L, 1.0 eq.) and Int-2 (5.8mg, 0.017 mmol/L, 1.1 eq.) were dissolved in N,N-dimethylformamide (DMF, 0.6 mL, 0.025 M), followed by the addition of di-isopropyl ethyl amine (DIPEA; 11.2 μL, 0.067 mmol/L, 4 eq.) and hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU; 12.7 mg, 0.033 mmol/L, 2.2 eq.). The reaction mixture was stirred at 25 °C for 3 hours. After the reaction was complete, the mixture was purified directly by silica gel chromatography (Ethyl Acetate/Hexane as mix solvent with 20%–90% gradient) to produce 7 mg of JQ-dS-4 as a yellow powder (yield: 65%).
ITC
For cloning, residues 537 to 1,393 of human BRG1 were inserted into the pET28PP (N-terminal HIS-3C tag) vector (82) with a final plasmid sequence described in GenBank. For protein expression and purification, human BRG1 (537–1393, HIS-tagged) was overexpressed in Escherichia coli BL21 (DE3) and purified using affinity chromatography and size-exclusion chromatography. Briefly, cells were grown at 37°C in TB medium in the presence of 50 μg/mL of kanamycin to an optical density of 0.8, cooled to 17°C, induced with 500 μmol/L isopropyl-1-thio-D-galactopyranoside, incubated overnight at 17°C, collected by centrifugation, and stored at −80°C. Cell pellets were lysed in buffer A (20 mmol/L HEPES, pH 7.5, 500 mmol/L NaCl, 10% glycerol, 7 mmol/L mercaptoethanol, and 20 mmol/L imidazole) using Microfluidizer (Microfluidics), and the resulting lysate was centrifuged at 30,000 × g for 40 minutes. Ni-NTA beads (Qiagen) were mixed with cleared lysate for 30 minutes and washed with buffer A. Beads were transferred to an FPLC-compatible column, and the bound protein was washed further with buffer A for 10 column volumes and eluted with buffer B (20 mmol/L HEPES, pH 7.5, 500 mmol/L NaCl, 10% glycerol, 7 mmol/L mercaptoethanol, and 400 mmol/L imidazole). The eluted sample was concentrated and purified further using a Superdex 200 16/600 column (Cytiva) in buffer SEC (20 mmol/L HEPES, pH 7.5, 500 mmol/L NaCl, 10% glycerol, and 10 mmol/L DTT). Protein peak fractions were pooled and concentrated to ∼200 μmol/L (∼20 g/L) and stored at −80°C. ITC experiments were carried out using an Affinity ITC from TA Instruments equipped with autosampler in a buffer containing 20 mmol/L HEPES, pH 7.5, 500 mmol/L NaCl, and 10 mmol/L DTT, with 2% DMSO at 25°C. A concentration of 20 μmol/L protein solution or buffer (control) in the calorimetric cell was titrated by injecting 3 μL of 200 μmol/L ligand solution in 200-second intervals with stirring speed at 75 rpm. Resulting isotherms were subtracted against buffer runs and fitted with a single site model to yield thermodynamic parameters of ΔH, ΔS, stoichiometry, and Kd using NanoAnalyze software (TA Instruments).
SILAC in BT869, an H3.3K27M-Glioma Model
BT869 cells were grown in custom TSM media adjusted for heavy (13C615N2 L-lysine and 13C615N4 L-arginine) or light (L-lysine and L-arginine) isotope labeling for 5 passages, and baseline incorporation was confirmed >95%. For the JQ-dS-4 proteomics experiment, a total of 1.5 × 106 BT869 cells (light and heavy labeled, respectively) were seeded in a T25 flask 1 day before treatment. Cells were treated with DMSO (light) or JQ-dS-4 1 μmol/L (heavy) for 48 hours. Cells were then washed twice with ice-cold PBS and lysed in RIPA buffer supplemented with protease inhibitors, benzonase, and 1 mmol/L DTT. Protein concentration was measured in triplicate using a BCA assay (cat. #23225, Thermo Fisher Scientific), and each heavy/light replicate was combined 1:1 in equal protein concentration. Samples were diluted in 2× reducing sample buffer (Thermo Fisher), followed by boiling at 95°C for 5 minutes. Samples were then loaded and run on a NuPAGE Novex 4% to 12% Bis-Tris gel (Life Technologies) with MOPS running buffer and separated by SDS-PAGE. Gels were stained with InstantBlue Coomassie Protein Stain (Abcam) and washed with distilled water. Each lane was then cut into two bands based on proteins of interest (∼90–240, 40–90 kDa) and stored in cold PBS for short-term storage.
Excised gel bands were cut into approximately 1-mm3 pieces. The samples were reduced with 1 mmol/L DTT for 30 minutes at 60°C and then alkylated with 5 mmol/L iodoacetamide for 15 minutes in the dark at room temperature. Gel pieces were washed and dehydrated with acetonitrile for 10 minutes, followed by the removal of acetonitrile. Pieces were then completely dried in a speed-vac. Rehydration of the gel pieces was done with 50 mmol/L ammonium bicarbonate solution containing 12.5 ng/μL modified sequencing-grade trypsin (Promega) at 4°C. Samples were then placed in a 37°C room overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried in a speed-vac (∼1 hour), and samples were stored at 4°C until analysis.
On the day of analysis, the samples were reconstituted in 5 to 10 μL of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 2.6-μm C18 spherical silica beads into a fused silica capillary (100-μm inner diameter × ∼30 cm length) with a flame-drawn tip (2). After equilibrating the column, each sample was loaded via a Famos autosampler (LC Packings) onto the column. A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As each peptide was eluted, they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program Sequest (ThermoFinnigan). The differential modification of 8.0142 and 10.0083 mass units for lysine and arginine, respectively, were included in the database searches to find SILAC-labeled peptides. All databases include a reversed version of all the sequences, and the data were filtered to a 1% or lower peptide FDR.
For downstream analysis, only proteins with a minimum of four unique peptides per protein were included per replicate, and only proteins meeting this threshold in at least two replicates were included. Significance was determined using a heteroscedastic two-tailed t test.
Immunoblotting for BRG1/BRM in Degrader-Treated Samples
BT869, HSJD-DIPG007, and SU-DIPGXIIIP* cells were collected after 24, 48, and 72 hours of treatment with increasing concentrations of the BRG1/BRM degraders JQ-dS-4 and AU-15330. For SU-DIPGXIIIP* cells, nuclear extracts were collected using the NE-PER nuclear and cytoplasmic extraction reagents (cat. #78833, Thermo Fisher Scientific) according to the manufacturer's instructions, and samples were also collected 6 hours after treatment. For BT869 and HSJD-DIPG007 cells, whole-cell lysates were collected using a standard lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mmol/L EGTA, 50 mmol/L NaF, 5 mmol/L Na4P2O7, 1 mmol/L Na3VO4, and 10 mmol/L b-glycerol phosphate in the presence of protease inhibitors, cOmplete Mini, Roche). Protein concentration was measured using the Pierce BCA Protein Assay Kit (cat. #23225, Thermo Fisher Scientific) according to the manufacturer's instructions. Then, proteins were separated using NuPAGE 4% to 12% Bis-Tris precast gels (cat. #NP0323BOX, Thermo Fisher Scientific) and transferred into PVDF western blotting membranes (cat. #3010040001, Sigma). Membranes were blocked with 5% milk in TBST, incubated overnight at 4°C with primary antibodies diluted 1:1,000, and then incubated for 1 hour with HRP-linked secondary antibodies at room temperature. Finally, proteins were detected by chemiluminescence. The following primary and secondary antibodies were used: BRG1 (D1Q7F, #49360, Cell Signaling Technology, RRID:AB_2728743), BRM (#6889, Cell Signaling Technology, RRID:AB_10831818), PARP (#9542S, Cell Signaling Technology, RRID:AB_2160739), cleaved PARP (D64E10, #5625, Cell Signaling Technology, RRID:AB_10699459), GAPDH (D4C6R, #97166, Cell Signaling Technology, RRID:AB_2756824), and H3 (96C10, #3638, Cell Signaling Technology, RRID:AB_1642229).
BBB Penetration Analyses
For BBB testing shown in Supplementary Fig. S6A, NSG mice (RRID:BCBC_1262) were treated with Compound 11 via intraperitoneal injection and euthanized after 1 hour; whole brains were collected and flash frozen for further processing. For MALDI tissue preparation, mouse brain tissue homogenates were spiked with concentrations of Compound 11 ranging from 1.9 to 27 μmol/L, dispensed into a 40% gelatin tissue microarray mold containing 1.5-mm core diameter channels and kept frozen at −80°C. Mouse brains were sectioned coronally at 10-μm thickness and thaw mounted onto indium tin oxide slides adjacent to a 10-μm thick section of the tissue drug mimetic, and placed in a vacuum desiccator to dry prior to matrix deposition. The matrix solution consisted of 2,5-dihydroxybenzoic acid (160 mg/mL) in 70:30 methanol:0.1% TFA with 1% DMSO and was applied using a TM sprayer (HTX Technologies). The matrix solution was applied using a two-pass cycle at a flow rate of 0.18 mL/minute, spray nozzle velocity of 1,200 mm/minute, nitrogen gas pressure of 10 psi, spray nozzle temperature of 75°C, and track spacing of 2 mm. A recrystallization step was performed by fixing the matrix-coated slide to the inside of a Petri dish lid, with the dish containing a 5% acetic acid solution–soaked filter. The chamber was then heated at 85°C for 6 minutes prior to matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI MSI) data acquisition. H&E staining was performed on serial sections, and digital microscopy images were acquired using an X objective (Zeiss Observer Z.1).
A multiple reaction monitoring (MRM) method for the detection of Compound 11 was used for quantitative imaging by monitoring the transition of precursor to product ion m/z 320.018/170.011. A timsTOF fleX mass spectrometer (Bruker Daltonics) equipped with a dual MALDI and electrospray ionization (ESI) source was operated in positive ion mode over an m/z range of 100 to 650. The drug detection method was optimized using the ESI source with a direct infusion of Compound 11 and adjusting the ion transfer funnels, quadrupole, collision cell, and focus pre-TOF parameters. Initiating the MRM feature, optimal collision energy of 23 eV with an isolation width of 3 m/z was determined to be optimal to monitor the mass transition. The method was mass calibrated using an Agilent tune mix solution (Agilent Technologies). Using the MALDI source, the imaging run parameters were defined for a laser repetition rate of 10,000 Hz and a spatial resolution of 50 μm, in which each pixel consisted of 1,000 laser shots. For data visualization and quantification, the SCiLS Lab software (version 2020a premium, Bruker Daltonics) was used without data normalization. A linear regression for ion intensity and concentration was established for a concentration range of 1.9 to 27 μmol/L with a correlation coefficient of 0.998, resulting in a limit of detection of 1.8 μmol/L (S/N ratio of >3) and limit of quantification of 6.3 μmol/L (S/N ratio of >10).
For the pharmacokinetic studies shown in Supplementary Fig. S6B, Crl:CD1(ICR) mice (Charles River Laboratories, cat. #022, RRID:IMSR_CRL:022) were used. All animal studies were performed according to Dana-Farber Cancer Institute IACUC-approved protocols (13-053). Animals were maintained according to institutional guidelines. A total of nine female CD1 mice (three mice per group) were injected intraperitoneally with single doses of Compounds 11, 12, and 14 and JQ-dS-4 solubilized in 10% hydroxypropyl β-cyclodextrin (Sigma-Aldrich) in sterile water. After injections, mice were euthanized, and drug concentrations in animal plasma and brain were measured by LC/MS-MS analysis at 30 minutes (for the first group), 2 hours (for the second group), and 4 hours (for the third group) using an LC/MS-MS method and pharmacokinetic parameters calculated with WinNonlin V 6.2 statistics software (Pharsight Corporation) using a noncompartmental model.
Subcutaneous Xenografts in NSG Mice
Studies were conducted in female NOD.Cg-PrkdcscidIl2rgtm1Wjl/Sz (NSG) mice (The Jackson Laboratory, RRID:IMSR_JAX:005557) of ∼6 to 8 weeks of age on the day for tumor implantation. Animals were transplanted with BT869 (H3.3K27M), SU-DIPGXIIIP* (H3.3K27M), HSJD-DIPG007 (H3.3K27M), and HSJD-GBM001 (H3WT) cells subcutaneously into the right flank (3 × 106 cells in 50 μL PBS and 50 μL Matrigel). When tumors reached ∼100 to 200 mm3, mice were randomized into Compound 14 [20 mg/kg once a day (q.d.) i.p.], JQ-dS-4 (50 mg/kg q.d. i.p., only for the BT869 tumor model), and vehicle groups. The time between tumor cell injection and the start of treatment differed for each tumor model because the tumor engraftment period is different for each model. Tumor engraftment took ∼3 months for BT869, ∼30 days for SU-DIPGXIIIP*, ∼3 weeks for HSJD-DIPG007, and ∼10 days for HSJD-GBM001. Compound 14 and JQ-dS-4 were dissolved in DMSO first and then diluted at the final ratio: 10% DMSO + 10% Kolliphor HS 15+80% (10% HP-β-CD) sterile water. For the BT869 model, 13 mice were enrolled per group (pharmacodynamic studies n = 3 per group, efficacy studies n = 10 per group). For HSJD-DIPG007, nine mice were enrolled per group, for SU-DIPGXIIIP*, eight mice were enrolled per group, and for HSJD-GBM001, 10 mice were enrolled per group—all being used for efficacy studies. For efficacy studies, mice were treated for 60 (BT869), 45 (HSJD-DIPG007), or 30 (SU-DIPGXIIIP* and HSJD-GBM001) days and then followed for survival. Tumors were measured by calipers, and mice were weighed every 3 days. Mice were humanely euthanized when the tumor volume reached 2,000 mm3, weight loss was more than 15% of body weight, or animals became moribund. All animal studies were performed according to Dana-Farber Cancer Institute IACUC-approved protocols (13-053).
BT869 tumors for pharmacodynamic studies were collected after 10-day treatment, dissected out, cut into small pieces, and fixed in a 10% formaldehyde solution followed by transfer to 70% ethanol. Subcutaneous HSJD-DIPG007 (n = 2) mouse tumors collected at the survival endpoint were fixed in 4% paraformaldehyde for 24 hours and subsequently stored in 70% ethanol. Fixed mouse tumors were embedded in paraffin to generate FFPE blocks by iHisto histopathology support. FFPE blocks were used for immunofluorescent staining (described above). FFPE blocks of BT869 tumors were also sectioned into 4-μm-thick sections and stained for H&E, cleaved caspase-3 (cat. #MA5-32015, Invitrogen, RRID:AB_2809309), and BRG1 (cat. #GTX35204, GeneTex) by iHisto histopathology support. Following IHC staining and imaging, IHC quantification was performed using QuPath v0.3.0 (ref. 83; RRID:SCR_018257). Four ROIs were selected from the center of the PDX for each slide. The number of positive cells was calculated using “Hematoxylin OD” as the Detection Image with a single intensity threshold of 0.2. For cleaved caspase-3, the Score Compartment was defined as “Cytoplasm: DAB OD Mean,” and for BRG1, the Score Compartment was defined as “Nucleus: DAB OD Mean.”
Solubility Analysis
Solubility studies of Compound 11, Compound 14, and JQ-dS-4 were performed by Creative Bioarray. In brief, compounds (10 mmol/L in DMSO) were diluted in 50 mmol/L PBS (pH 7.4) to a final concentration of 200 μmol/L in lower chambers of Whatman Mini-UniPrep vials. Amiodarone hydrochloride, carbamazepine, and chloramphenicol were used as controls. Samples and controls were vortexed for 2 minutes, shaken for 24 hours (800 rpm, room temperature), centrifuged (20 minutes, 4,000 rpm, room temperature), and filtered before being analyzed by HPLC.
Data Availability
The data generated in this study are available within the article and its supplementary files. Sequencing data generated for this study (including ChIP-seq, bulk RNA-seq, scRNA-seq, and bulk ATAC-seq) are publicly available in the Gene Expression Omnibus with the accession number GSE212786. Additional data or resources related to this article are available upon reasonable request from the corresponding authors.
Supplementary Material
Acknowledgments
We thank Dr. Ross Tomaino of the Taplin Mass Spectrometry facility at Harvard Medical School for assistance with mass spectrometry, and Drs. Jennifer Roth, Ginevra Botta, and Andrew Boghossian of the PRISM screening group at the Broad Institute for assistance with collecting PRISM data. This work was supported by generous funding from the V Foundation for Cancer Research (T2020-001, to M.G. Filbin and J. Qi), the Mark Foundation (J. Qi), the Cuming Family Fund for Pediatric Brain Tumor Research (M.G. Filbin), the Sajni Fund (M.G. Filbin), and the DIPG All-In initiative from the Pediatric Brain Tumor Foundation/Musella Foundation for Brain Tumor Research and Information Inc. (E. Panditharatna and M.G. Filbin).
M.G. Filbin holds an NIH Director New Innovator Award (1DP2 NS127705-01), a Career Award for Medical Scientists from the Burroughs Wellcome Fund, the Distinguished Scientist Award from the Sontag Foundation, the “A” Award from Alex's Lemonade Stand Foundation, a Hyundai Hope on Wheels Hope Scholar Award, and a Developmental Research Program award of the Dana-Farber/Harvard Cancer Center Brain Cancer SPORE (5P50 CA165962–08). E. Panditharatna holds a fellowship funded by the Michael Mosier Defeat DIPG Foundation, the ChadTough Foundation, and the SoSo Strong Pediatric Brain Tumor Foundation. E. Panditharatna was also funded by Pedals for Pediatrics and the David Abraham Foundation. J.G. Marques is funded by an Early Postdoc Mobility grant awarded by the Swiss National Science Foundation (P2ZHP3_199691). M.C. Trissal is funded by NIH T32 training grant 5T32CA136432–12. I. Liu was supported by the German Research Foundation (DFG, #LI-3486/1–1). The BBB testing work was funded in part by NIH U54 CA210180/MIT/Mayo Physical Science Oncology Center for Drug Distribution and Drug Efficacy in Brain Tumors (N.Y.R. Agar), NIH P41-EB-015898/the Ferenc Jolesz National Center for Image Guided Therapy (N.Y.R. Agar), and the Dana-Farber Cancer Institute PLGA Fund (N.Y.R. Agar). During this study, S. Stopka was in receipt of an NIH T32 (award number:1T32EB025823-01A1) Fellowship. Cancer research in the Shi lab was/is supported by the NCI (R35 CA210104) and the Ludwig Institute for Cancer Research, respectively. J.N. Anastas received a fellowship funded by the Michael Mosier Defeat DIPG Foundation and the ChadTough Foundation. Y. Shi is an American Cancer Society Research Professor. S. Dhe-Paganon acknowledges funding from the Linde Family Foundation, the Doris Duke Charitable Foundation, Deerfield 3DC, and Taiho Pharmaceuticals. K. Stegmaier is funded by NIH R35 CA210030.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
J.G. Marques reports grants from the Swiss National Science Foundation during the conduct of this study. N.V. Dharia reports grants from St. Baldrick's Foundation Julia's Legacy of Hope during the conduct of the study, as well as other support from Genentech, Inc., a member of the Roche Group, outside the submitted work. F. Vazquez reports grants from Novo Ventures and the Dependency Map Consortium during the conduct of the study. F. Piccioni reports personal fees from Merck Research Laboratories outside the submitted work. J.G. Doench reports personal fees from Pfizer, Tango Therapeutics, Microsoft Research, Sangamo, Maze Therapeutics, BioNTech, Servier, and Abata Therapeutics outside the submitted work. D.E. Root reports grants from AbbVie, Bristol Myers Squibb, Janssen, Merck, and Vir outside the submitted work. N.D. Mathewson reports grants from the American Cancer Society and the NIH education loan repayment program funded by the NCI during the conduct of the study; personal fees from Immunitas Therapeutics and other support from Asher Biotherapeutics outside the submitted work; and a patent for methods and compositions for treating cancer by targeting the CLEC2D–KLRB1 pathway issued, a patent for monoclonal antibodies that bind human CD161 and uses thereof issued, and a patent for a targeted cytokine construct for engineered cell therapy issued. M.L. Suva reports personal fees from Immunitas Therapeutics outside the submitted work. Y. Shi reports personal fees from K36 Therapeutics, CBio-X, and Active Motif outside the submitted work. S. Agnihotri reports grants from the NIH during the conduct of the study. N.Y.R. Agar reports other support from EMD Serono outside the submitted work, as well as support from the DNA Damage Response Consortium of the National Brain Tumor Society. K. Stegmaier reports grants from the NCI during the conduct of the study, as well as other support from Auron Therapeutics (scientific advisory board member and stock options), personal fees from Auron Therapeutics, Bristol Myers Squibb, and AstraZeneca, grants from Novartis, and grants and personal fees from Kronos Bio outside the submitted work. T.R. Golub reports grants from the Howard Hughes Medical Institute and the NIH during the conduct of the study, as well as personal fees from Forma Therapeutics, Sherlock Biosciences, GSK, Anji Pharmaceuticals, and Dewpoint Therapeutics and grants from Novo Holdings, Calico Life Sciences, and Bayer HealthCare outside the submitted work. J. Qi reports grants from the V Foundation during the conduct of the study; other support from Epiphanes and Talus outside the submitted work; and a patent for small-molecule degraders of SMARCA2/BRM proteins and uses thereof pending, a patent for chemical targeting SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 (SMARCA4) and use in diffuse intrinsic pontine glioma (DIPG) pending, and a patent for compounds, compositions and methods for protein degradation pending. M.G. Filbin reports a patent for small-molecule degraders of SMARCA2/BRM proteins and uses thereof pending and a patent for chemical targeting SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 (SMARCA4) and use in DIPG pending. No disclosures were reported by the other authors.
Authors’ Contributions
E. Panditharatna: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.G. Marques: Formal analysis, validation, investigation, visualization, writing–review and editing. T. Wang: Formal analysis, validation, investigation, visualization, methodology. M.C. Trissal: Formal analysis, validation, investigation, methodology, writing–original draft. I. Liu: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. L. Jiang: Data curation, software, formal analysis, validation, investigation, visualization, methodology. A. Beck: Formal analysis, validation, investigation, methodology. A. Groves: Formal analysis, validation, investigation, visualization, methodology. N.V. Dharia: Data curation, formal analysis, visualization. D. Li: Resources, investigation. S.E. Hoffman: Data curation, software, formal analysis, visualization. G. Kugener: Data curation, formal analysis. M.L. Shaw: Investigation. H.M. Mire: Formal analysis, validation. O.A. Hack: Investigation, visualization. J.M. Dempster: Resources, data curation, formal analysis. C. Lareau: Formal analysis. L. Dai: Validation, visualization. L.H. Sigua: Resources, methodology. M.A. Quezada: Investigation. A.-C.J. Stanton: Investigation. M. Wyatt: Investigation. Z. Kalani: Investigation. A. Goodale: Supervision. F. Vazquez: Supervision. F. Piccioni: Supervision. J.G. Doench: Supervision. D.E. Root: Supervision. J.N. Anastas: Resources, validation. K.L. Jones: Investigation, methodology. A. Saur Conway: Investigation, methodology. S. Stopka: Investigation, methodology. M.S. Regan: Investigation, methodology. Y. Liang: Resources, methodology. H.-S. Seo: Investigation, methodology. K. Song: Investigation, methodology. P. Bashyal: Investigation, methodology. W.P. Jerome: Investigation. N.D. Mathewson: Resources, investigation. S. Dhe-Paganon: Supervision. M.L. Suva: contributed ideas. A.M. Carcaboso: Resources. C. Lavarino: Resources. J. Mora: Resources. Q.-D. Nguyen: Supervision. K.L. Ligon: Resources. Y. Shi: Resources, supervision. S. Agnihotri: Supervision, methodology. N.Y.R. Agar: Supervision, methodology. K. Stegmaier: contributed ideas. C.D. Stiles: Writing–review and editing, contributed ideas. M. Monje: Resources, supervision, contributed ideas. T.R. Golub: Resources, supervision, project administration. J. Qi: Resources, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. M.G. Filbin: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Filbin M, Monje M. Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat Med 2019;25:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31. [DOI] [PubMed] [Google Scholar]
- 3. Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 2017;23:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 2017;23:483–92. [DOI] [PubMed] [Google Scholar]
- 6. Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun 2019;10:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haag D, Mack N, Benites Goncalves da Silva P, Statz B, Clark J, Tanabe K, et al. H3.3-K27M drives neural stem cell-specific gliomagenesis in a human iPSC-derived model. Cancer Cell 2021;39:407–22. [DOI] [PubMed] [Google Scholar]
- 8. Pathania M, De Jay N, Maestro N, Harutyunyan AS, Nitarska J, Pahlavan P, et al. H3.3(K27M) cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell 2017;32:684–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 2017;31:635–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018;360:331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol 2019;137:637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, et al. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 2019;51:1702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganguly D, Sims M, Cai C, Fan M, Pfeffer LM. Chromatin remodeling factor BRG1 regulates stemness and chemosensitivity of glioma initiating cells. Stem Cells 2018;36:1804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood 2014;123:1720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol 2021;22:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015;163:1515–26. [DOI] [PubMed] [Google Scholar]
- 19. Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res 2019;47:D559–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015;21:827. [DOI] [PubMed] [Google Scholar]
- 21. Kumar SS, Sengupta S, Lee K, Hura N, Fuller C, DeWire M, et al. BMI-1 is a potential therapeutic target in diffuse intrinsic pontine glioma. Oncotarget 2017;8:62962–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 2017;8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narayanan R, Pirouz M, Kerimoglu C, Pham L, Wagener RJ, Kiszka KA, et al. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep 2015;13:1842–54. [DOI] [PubMed] [Google Scholar]
- 25. Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, et al. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol Cell 2019;76:965–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yadavilli S, Scafidi J, Becher OJ, Saratsis AM, Hiner RL, Kambhampati M, et al. The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget 2015;6:12141–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qi J, Esfahani DR, Huang T, Ozark P, Bartom E, Hashizume R, et al. Tenascin-C expression contributes to pediatric brainstem glioma tumor phenotype and represents a novel biomarker of disease. Acta Neuropathol Commun 2019;7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papillon JPN, Nakajima K, Adair CD, Hempel J, Jouk AO, Karki RG, et al. Discovery of orally active inhibitors of brahma homolog (BRM)/SMARCA2 ATPase activity for the treatment of brahma related gene 1 (BRG1)/SMARCA4-mutant cancers. J Med Chem 2018;61:10155–72. [DOI] [PubMed] [Google Scholar]
- 29. Vangamudi B, Paul TA, Shah PK, Kost-Alimova M, Nottebaum L, Shi X, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res 2015;75:3865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jancewicz I, Siedlecki JA, Sarnowski TJ, Sarnowska E. BRM: the core ATPase subunit of SWI/SNF chromatin-remodelling complex-a tumour suppressor or tumour-promoting factor? Epigenetics Chromatin 2019;12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013;27:2648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med 2016;6:a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mehrotra A, Mehta G, Aras S, Trivedi A, de la Serna IL. SWI/SNF chromatin remodeling enzymes in melanocyte differentiation and melanoma. Crit Rev Eukaryot Gene Expr 2014;24:151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y, Hoang L, Ji JX, Huntsman DG. SWI/SNF complex mutations in gynecologic cancers: molecular mechanisms and models. Annu Rev Pathol 2020;15:467–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiao L, Parolia A, Qiao Y, Bawa P, Eyunni S, Mannan R, et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 2022;601:434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schick S, Grosche S, Kohl KE, Drpic D, Jaeger MG, Marella NC, et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat Genet 2021;53:269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iurlaro M, Stadler MB, Masoni F, Jagani Z, Galli GG, Schubeler D. Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet 2021;53:279–87. [DOI] [PubMed] [Google Scholar]
- 38. Durbin AD, Wang T, Wimalasena VK, Zimmerman MW, Li D, Dharia NV, et al. EP300 selectively controls the enhancer landscape of MYCN-amplified neuroblastoma. Cancer Discov 2022;12:730–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Panditharatna E, Filbin MG. The growing role of epigenetics in childhood cancers. Curr Opin Pediatr 2020;32:67–75. [DOI] [PubMed] [Google Scholar]
- 40. Anastas JN, Zee BM, Kalin JH, Kim M, Guo R, Alexandrescu S, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell 2019;36:528–44. [DOI] [PubMed] [Google Scholar]
- 41. Padilla-Benavides T, Nasipak BT, Imbalzano AN. Brg1 controls the expression of Pax7 to promote viability and proliferation of mouse primary myoblasts. J Cell Physiol 2015;230:2990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenetics Chromatin 2019;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rago F, Rodrigues LU, Bonney M, Sprouffske K, Kurth E, Elliott G, et al. Exquisite sensitivity to dual BRG1/BRM ATPase inhibitors reveals broad SWI/SNF dependencies in acute myeloid leukemia. Mol Cancer Res 2022;20:361–72. [DOI] [PubMed] [Google Scholar]
- 44. Jagani Z, Chenail G, Xiang K, Bushold G, Bhang HEC, Li A, et al. In-depth characterization and validation in BRG1-mutant lung cancers define novel catalytic inhibitors of SWI/SNF chromatin remodeling. BioRxiv 812628 [Preprint]. 2019. Available from: https://doi.org/10.1101/812628. [Google Scholar]
- 45. Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994;79:119–30. [DOI] [PubMed] [Google Scholar]
- 46. Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mardinian K, Adashek JJ, Botta GP, Kato S, Kurzrock R. SMARCA4: implications of an altered chromatin-remodeling gene for cancer development and therapy. Mol Cancer Ther 2021;20:2341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mittal P, Roberts CWM. The SWI/SNF complex in cancer: biology, biomarkers and therapy. Nat Rev Clin Oncol 2020;17:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Q, Lian JB, Stein JL, Stein GS, Nickerson JA, Imbalzano AN. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017;9:919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng L, Li J, Wu J, Xu B, Wang Z, Giamas G, et al. A pan-cancer analysis of SMARCA4 alterations in human cancers. Front Immunol 2021;12:762598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clapier CR, Verma N, Parnell TJ, Cairns BR. Cancer-associated gain-of-function mutations activate a SWI/SNF-family regulatory hub. Mol Cell 2020;80:712–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 2017;171:163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wimalasena VK, Wang T, Sigua LH, Durbin AD, Qi J. Using chemical epigenetics to target cancer. Mol Cell 2020;78:1086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Farrell K, Jarome TJ. Is PROTAC technology really a game changer for central nervous system drug discovery? Expert Opin Drug Discov 2021:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin GL, Monje M. A protocol for rapid post-mortem cell culture of diffuse intrinsic pontine glioma (DIPG). J Vis Exp 2017:55360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 2017;95:779–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Douvaras P, Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat Protoc 2015;10:1143–54. [DOI] [PubMed] [Google Scholar]
- 58. Krug K, Mertins P, Zhang B, Hornbeck P, Raju R, Ahmad R, et al. A curated resource for phosphosite-specific signature analysis. Mol Cell Proteomics 2019;18:576–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suo S, Zhu Q, Saadatpour A, Fei L, Guo G, Yuan GC. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep 2018;25:1436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Egan B, Yuan CC, Craske ML, Labhart P, Guler GD, Arnott D, et al. An alternative approach to ChIP-Seq normalization enables detection of genome-wide changes in histone H3 lysine 27 trimethylation upon EZH2 inhibition. PLoS One 2016;11:e0166438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wickham H. ggplot2: Elegant graphics for data analysis. Use R!. 2nd ed.Cham (Switzerland): Springer International Publishing; 2016. [Google Scholar]
- 67. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ludwig LS, Lareau CA, Bao EL, Nandakumar SK, Muus C, Ulirsch JC, et al. Transcriptional states and chromatin accessibility underlying human erythropoiesis. Cell Rep 2019;27:3228–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9:171–81. [DOI] [PubMed] [Google Scholar]
- 75. Gojo J, Englinger B, Jiang L, Hubner JM, Shaw ML, Hack OA, et al. Single-cell RNA-Seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell 2020;38:44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Du Z, Lin JR, Rashid R, Maliga Z, Wang S, Aster JC, et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat Protoc 2019;14:2900–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Remillard D, Buckley DL, Paulk J, Brien GL, Sonnett M, Seo HS, et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew Chem Int Ed Engl 2017;56:5738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yu C, Mannan AM, Yvone GM, Ross KN, Zhang YL, Marton MA, et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol 2016;34:419–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Varca AC, Casalena D, Auld D, Buhrlage SJ. Identification of deubiquitinase inhibitors via high-throughput screening using a fluorogenic ubiquitin-rhodamine assay. STAR Protoc 2021;2:100896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PP, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary files. Sequencing data generated for this study (including ChIP-seq, bulk RNA-seq, scRNA-seq, and bulk ATAC-seq) are publicly available in the Gene Expression Omnibus with the accession number GSE212786. Additional data or resources related to this article are available upon reasonable request from the corresponding authors.



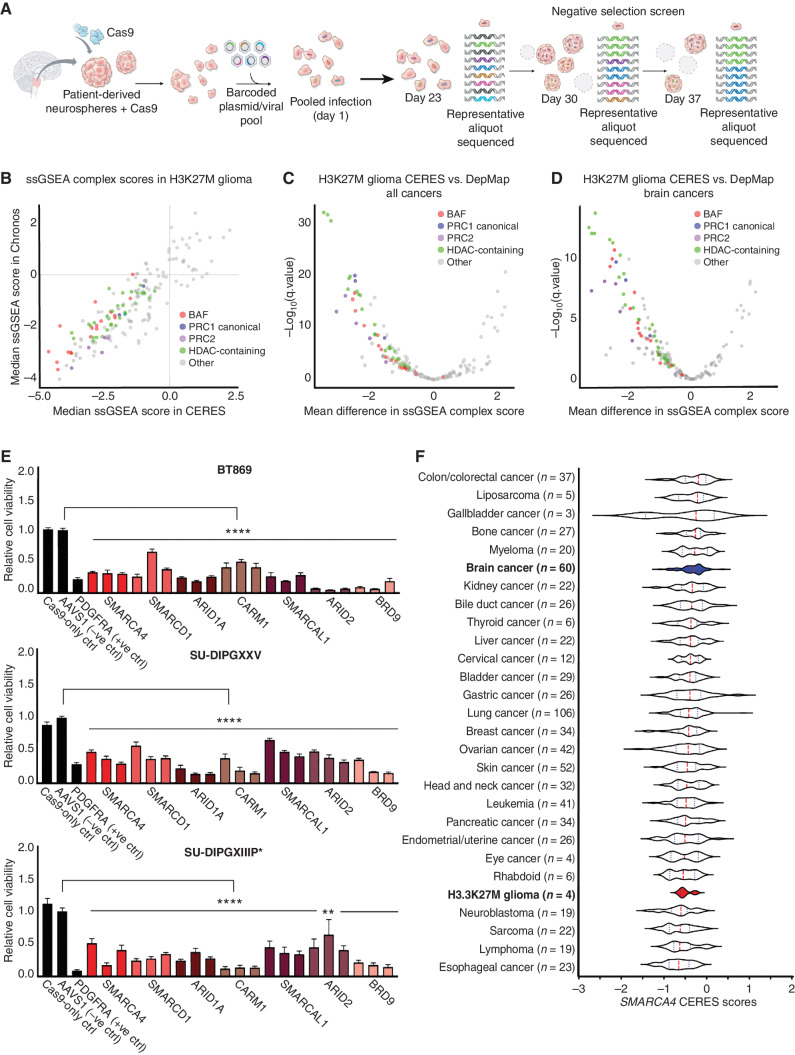
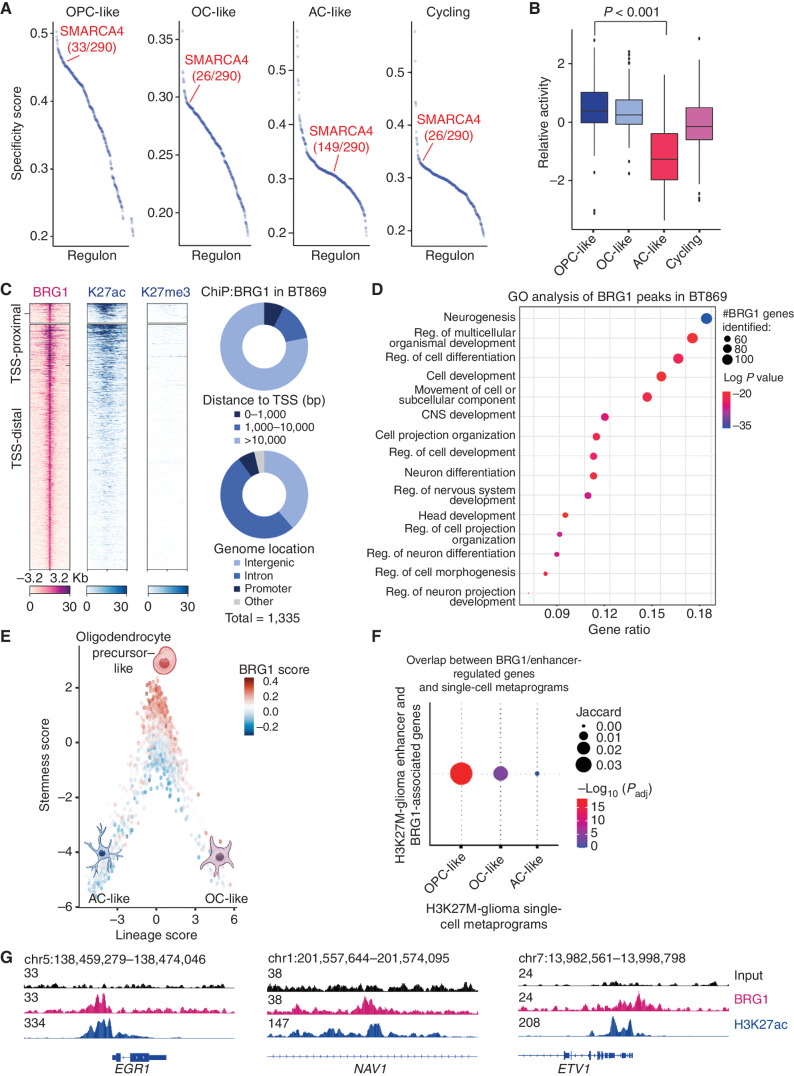
![Figure 3. Genetic knockout of SMARCA4 reduces chromatin accessibility and gene expression of OPC-like markers and enriches for the AC-like subpopulation in H3K27M-glioma cells. A, Schematic showing biological assessment of SMARCA4 knockout in H3.3K27M-glioma neurospheres by ChIP-seq for histone modifications (H3K27ac and H3K27me3), assay for transposase-accessible chromatin using sequencing (ATAC-seq) for chromatin accessibility, bulk RNA sequencing (RNA-seq), and scRNA-seq for gene expression. B, Heat map depicting z-scores for chromatin accessibility of OPC-like and AC-like marker genes in AAVS1-negative sgRNA control and SMARCA4-knockout (KO) BT869 neurospheres. C, Profile plot depicting the average H3K27ac ChIP-seq signal at BRG1 binding sites (n = 1,335) in BT869 4 days after SMARCA4 knockout. The plot shows 6.4-kb regions, centered on BRG1 peaks. D, Promoter-associated H3K27ac and H3K27me3 ChIP-seq signal for genes differentially expressed [determined by bulk RNA-seq analysis, log2(fold change [FC]) ≥ |1|] in SMARCA4-knockout cells (4 days after nucleofections). E, Profile plots (left) depicting the average H3K27ac signal at H3K27ac locations divided into positive (UP) or negative (DOWN) signal changes 4 days after SMARCA4 knockout. The plots show 6.4-kb regions, centered on H3K27ac peaks. Gene ontology (GO) analyses (right) performed on H3K27ac peaks with increased (n = 104, K27acUP) or decreased (n = 2,832, K27acDOWN) acetylation levels upon SMARC4 knockout for 4 days. The top five enriched GO terms are displayed. Pos. reg. of epidermal cell diff., positive regulation of epidermal cell differentiation.F, Heat map (left) depicting fold change (as log2) of H3K27ac levels after SMARCA4 knockout for 4 days. Displayed are log2 fold changes > |1|. Genes indicated on the heat map are OPC-like markers associated with decreased H3K27ac levels after SMARCA4 depletion. Gene tracks (right) displaying BRG1 ChIP-seq signal in BT869 neurospheres, as well as H3K27ac and ATAC-seq, at two OPC-like marker genes (TNR and EPN2). G, Changes in the percentage of cycling versus noncycling (left) and OPC- and AC-like cancer cell subpopulations (right) in SMARCA4-knockout and AAVS1-negative sgRNA control cells 4 days after knockout in BT869 neurospheres as determined by scRNA-seq analysis. H, Single-cell expression scores of upregulated genes in BT869 SMARCA4-knockout or AAVS1-negative sgRNA control knockout cells projected onto scRNA-seq data of pediatric H3K27M-glioma primary tumors (n = 6).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3854/9716260/4ec3fa4b5a56/2880fig3.jpg)
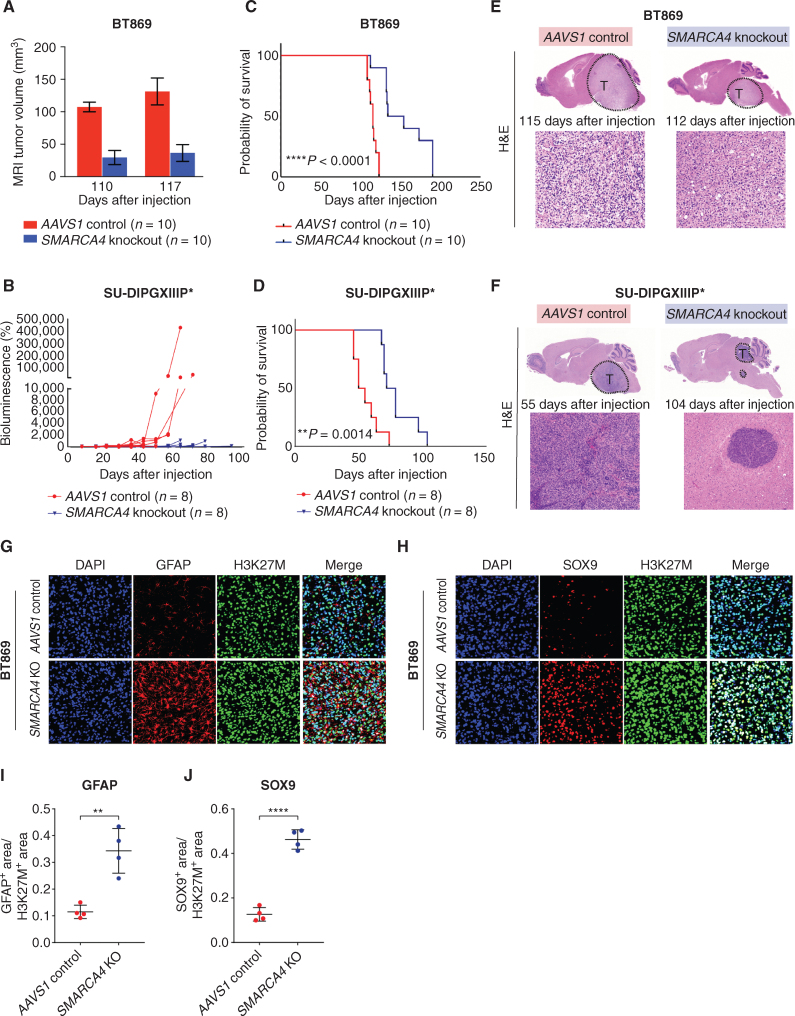
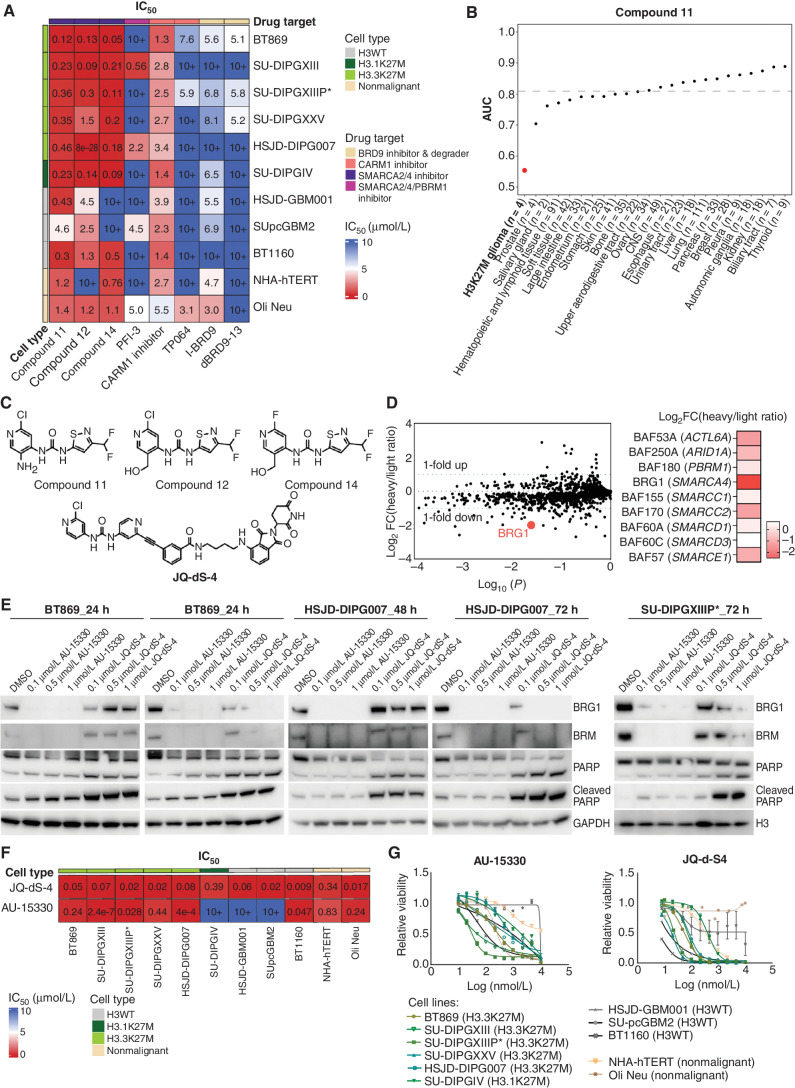
![Figure 6. Pharmacologic targeting of the BAF complex in vitro and in vivo mimics the biological effects of SMARCA4 depletion in H3K27M glioma. A, Schematic showing biological assessment of BRG1/BRM ATPase inhibitor– and degrader–treated H3.3K27M-glioma neurospheres for changes in chromatin accessibility by ATAC-seq, gene expression by bulk RNA sequencing (RNA-seq), and in vivo efficacy of chemical compounds in subcutaneous xenograft mouse models. B, Profile plot (top) depicting average ATAC-seq signal at BRG1 binding sites (n = 1,335) in BT869 (H3.3K27M-glioma) neurospheres after 24 hours of treatment with 1 μmol/L of BRG1/BRM inhibitors [Compound 11 (C11), Compound 14 (C14)] and degraders [JQ-dS-4 (JQ), AU-15330 (AU)] and DMSO controls. The plot shows 6.4-kb regions, centered on BRG1 peaks. Gene tracks (bottom) exemplify changes in chromatin accessibility observed at BRG1 binding sites. Displayed are BRG1 ChIP-seq signal in control BT869 cells as well as ATAC-seq data on the same cells after 24 or 48 hours of treatment with 1 μmol/L of BRG1/BRM inhibitors or degraders and DMSO controls. C, Biological processes (gene ontology analysis) enriched in regions with decreased accessibility in BT869 neurosphere cells treated for 24 hours with 1 μmol/L of Compound (Cmpd) 14, Compound 11, JQ-dS-4, or AU-15330 as determined by ATAC-seq. The number of genes is indicated by the size of the circle, and significance is depicted by the color scale. Reg., regulation of. D, Overlap of genes with reduced chromatin accessibility in drug-treated BT869 neurospheres (as determined by bulk ATAC-seq) with single-cell transcriptional metaprograms in H3K27M glioma. BT869 cells were treated with 1 μmol/L of either Compound 11 (C11), Compound 14 (C14), JQ-dS-4 (JQ), or AU-15330 (AU) for 24 hours. The color scale indicates the number of overlapping genes. E, A heat map showing z-scores of gene expression of OPC-like metaprogram marker genes with statistically significant changes upon Compound 11 or 14 treatments in BT869 neurospheres after 24 hours. Colored bars indicate z-scores of gene expression. rep, replicate.F, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)– or BRG1/BRM degrader JQ-dS-4 (50 mg/kg i.p. daily)–treated mice bearing BT869 (H3.3K27M-glioma) subcutaneous tumors as compared with vehicle controls (n = 10 mice per group). Data are shown as mean ± SEM, P < 0.0001 (unpaired t test comparing vehicle and Compound 14 groups). Kaplan–Meier survival curves (bottom) of mice treated with either Compound 14 (20 mg/kg i.p. daily) or JQ-dS-4 (50 mg/kg i.p. daily) for 60 days in a subcutaneous BT869 xenograft model (n = 10 mice per group). The P value was calculated using the log-rank (Mantel–Cox) test. Gray bars show the duration of treatment. G, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)–treated mice bearing SU-DIPGXIIIP* (H3.3K27M) subcutaneous tumors as compared with vehicle controls (n = 8 mice per group). Data are shown as mean ± SEM, P < 0.0001 (unpaired t test). Gray bar shows the duration of the treatment. Tumor volume (bottom) measured after 30 days of Compound 14 treatment compared with vehicle control in mice bearing SU-DIPGXIIIP* (H3.3K27M) subcutaneous tumors. P = 0.0017 (unpaired t test). H, Tumor volume measurements (top) in BRG1/BRM inhibitor Compound 14 (20 mg/kg i.p. daily)–treated mice bearing HSJD-GBM001 (H3WT) subcutaneous tumors as compared with vehicle controls (n = 10 mice per group). Data are shown as mean ± SEM, P = 0.578 (unpaired t test). Kaplan–Meier survival curves (bottom) of mice treated with Compound 14 (20 mg/kg i.p. daily) for 30 days in a subcutaneous xenograft model of HSJD-GBM001 (n = 10 mice per group). The P value was calculated using the log-rank (Mantel–Cox) test. Gray bars show the duration of treatment.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3854/9716260/9c554723f22a/2880fig6.jpg)