Abstract
ureI encodes an integral cytoplasmic membrane protein. It is present in the urease gene cluster of Helicobacter pylori and is essential for infection and acid survival, but its role is unknown. To determine the function of UreI protein, we produced H. pylori ureI deletion mutants and measured the pH dependence of urease activity of intact and lysed bacteria and the effect of urea on the membrane potential. We also determined ureI expression, urease activity, and the effect of urea on membrane potential of several gastric and nongastric Helicobacter species. ureI was found to be present in the genome of the gastric Helicobacter species and absent in the nongastric Helicobacter species studied, as determined by PCR. Likewise, Western blot analysis confirmed that UreI was expressed only in the gastric Helicobacter species. When UreI is present, acidic medium pH activation of cytoplasmic urease is found, and urea addition increases membrane potential at acidic pH. The addition of a low concentration of detergent raised urease activity of intact bacteria at neutral pH to that of their homogenates, showing that urease activity was membrane limited. No acidic pH activation or urea induced membrane potential changes were found in the nongastric Helicobacter species. The ureI gene product is probably a pH activated urea transporter or perhaps regulates such a transporter as a function of periplasmic pH.
Helicobacter pylori infection of the human stomach results in gastritis, ulcer disease, and gastric cancer (9, 10, 17). H. pylori, a spiral shaped, gram-negative neutralophilic microorganism, is unique in its ability to colonize the normal human stomach. Being a neutralophile, it must have developed mechanisms to combat the variable acidity of the gastric environment, which ranges from very acidic to close to neutrality.
Yersinia enterocolitica uses a cytoplasmic localized urease with a sharp pH optimum of 5.0 to enable gastric survival during transit to its site of infection. By buffering its cytoplasm to pH 5.0, Y. enterocolitica is able to survive gastric acidity but is unable to colonize the human stomach since growth requires a cytoplasmic pH greater than pH 5.0 (31). Acid tolerance of H. pylori also depends on urease activity, since urease-negative species are unable to colonize animal models (2, 8, 28). H. pylori constitutively produces large amounts of a neutral pH optimum urease, a hexameric heterodimer that represents 10 to 15% of total protein synthesis (3, 26). Urease is found both in the cytoplasm and adhering to the outer membrane of the organism (4, 11, 24). Urease isolated from H. pylori has a pH optimum of 7.5, but when urease activity is assayed in the intact bacterium, maximal activity is between 6.0 and 3.0. Activity is 10- to 20-fold higher throughout this range of pH than at pH 7.0 in intact cells (25, 26). It has been suggested that surface urease is responsible for neutralizing the acidic environment of the organism by generating a cloud of NH3 (17, 24). However, surface or free urease activity is irreversibly inhibited at a pH of less than 4.0 (26). Thus, either the pH around the organism never falls to that range of pH or external urease plays a less significant role in the acid tolerance of H. pylori than generally supposed. Hence, internal (i.e., cytoplasmic) urease may play a more important role than surface urease in enabling acid tolerance. Consideration of the bioenergetics of this acid-tolerant organism suggests that this is likely to be the case.
H. pylori requires a constant proton motive force (PMF) to synthesize ATP, as do all aerobic bacteria. The PMF is the sum of the pH gradient (ΔpH) and the membrane potential (Δψ) across the inner membrane. The PMF of H. pylori is maintained at around −200 mV, as for other organisms (14, 20). Assuming a constant cytoplasmic pH, medium acidification in the absence of urea increases the ΔpH across the membrane, resulting in a decrease of membrane potential to maintain a stable PMF (20). Urea addition activates cytoplasmic urease at medium pH values of <6.2. NH3 produced by the hydrolysis of urea freely diffuses from the cytoplasm into the periplasm and elevates the pH toward 6.2 (25). This results in a constant membrane potential of about −101 mV and an inward H+ gradient of about 2 pH units (26). This process of pH regulation maintains PMF at a level consistent with normal and essential cellular functions such as ATP synthesis and H+ gradient-dependent import and export of ions and solutes. Indirectly, this maintenance of periplasmic pH also maintains the transmembrane potential at a level consistent with appropriate folding of integral inner membrane proteins that depend on an interior negative potential of sufficient magnitude, the positive inside rule (1). Hence, this mechanism not only allows acidic survival but also allows growth at acidic pH and is therefore important for gastric colonization.
Activation of cytoplasmic urease only when the external pH drops below 6.2 prevents a potentially lethal rise of cytoplasmic pH. In the presence of urea, an unregulated cytoplasmic urease would, at neutral pH, raise the pH toward 9.25, the pKa of NH3. This neutralophile cannot survive a pH of 8.5 or more (7, 22). Therefore, a neutral pH optimum urease, although advantageous for acid survival would, in the absence of regulation, be incompatible with survival when gastric acid secretion fails to acidify the site of colonization, as occurs during digestion, where gastric contents can become neutral due to the buffering action of food.
Although the phenomenon of periplasmic pH regulation by the urea-urease couple has been described (25, 26), the genes involved in acid activation of cytoplasmic urease are unknown. The urease gene cluster of H. pylori is composed of several genes: ureA and ureB, followed by a possible promoter sequence and ureI, ureE, ureF, ureG, and ureH (15, 21). The ureA and ureB genes encode for the structural subunits of the enzyme, while ureE, ureF, ureG, and ureH are accessory genes required for nickel incorporation to produce the catalytically active metalloenzyme. The function of ureI is not known, but it is not required for urease synthesis or assembly (21). Although crude extracts of H. pylori ureI mutants have levels of urease activity equal to the wild-type parental strain, ureI mutants were unable to colonize the mouse stomach (26, 27). Additionally, the ureI mutant showed much lower survival than the wild type in vitro when exposed to low pH in the presence of 10 mM urea (27).
The ureI gene encodes for a 21.6-kDa putative integral membrane protein with six predicted membrane spanning helical sequences. The primary sequence lacks a signal peptide and the helical structure of the membrane segments predicts that this is a cytoplasmic membrane protein. The ureI gene of H. pylori has homology to ureI from Streptococcus salivarius, amiS from Pseudomonas aeruginosa, and amiS2 from Rhodococcus sp. strain R312 (5, 6, 30). The latter two genes are thought to encode amide transporters, but this has not been experimentally verified.
Here we investigate the role of the ureI gene product in the acid tolerance of H. pylori by determining the effect of ureI deletion on acid activation of internal urease and by searching for the ureI gene and its product in various gastric and nongastric Helicobacter spp. From the data presented below, we postulate that the ureI gene product is essential for the acid activation of internal urease of H. pylori as well as other gastric Helicobacter spp. Its apparent absence in the nongastric Helicobacter spp. may account, at least in part, for their absence from the stomach.
MATERIALS AND METHODS
Bacterial strains.
H. pylori strains 49503 and 43504 and H. nemestrinae were obtained from the American Type Culture Collection (ATCC). H. bilis, H. mustelae, H. troguntum, H. hepaticus, and H. muridarum were obtained from A. Lee's laboratory but were used in our laboratory for the experiments described. All bacteria were grown on Trypticase soy agar plates supplemented with 5% sheep blood (Gibco-BRL) for 24 h for use in all experiments.
ureI-negative mutants.
H. pylori ureI mutants were produced by allelic exchange as described in detail elsewhere (M. Rektorschek, A. Buhmann, D. Schwan, K. W. Bensch, G. Sachs, and K. Melchers, submitted for publication). Briefly, a plasmid was constructed for exchange of the ureI open reading frame of the H. pylori genome with a kanamycin resistance marker gene. The 3′ region of ureB and the 5′ region of ureE were amplified by PCR with plasmid pHP808 as template (10). The two PCR products were then fused via the BamHI recognition sites, which produced a ureB-ureE hybrid. A kanr open reading frame derived from the pUC4K plasmid was inserted into the ureB-ureE hybrid sequence at the BamHI restriction site. The plasmid was digested with EcoRI and HindIII for DNA fragment cloning. To select for the presence of the plasmid, cells were grown in LB medium (Luria broth base [Gibco BRL]) supplemented with 50 μg of ampicillin per ml. Plasmids were assayed for presence and orientation of the inserted DNA employing restriction analysis. Plasmid DNA of this vector was used for homologous recombination in H. pylori.
H. pylori was grown on brain heart infusion (BHI) agar plates (Difco) supplemented with 10% horse serum (Gibco-BRL) in gas pak jars under microaerophilic conditions for 24 h. Cells from one plate were harvested in 1 ml of BHI broth (Difco) supplemented with 6% fetal calf serum (Eurobio). After determination of the optical density at 578 nm (OD578), the cells were diluted to give a final OD578 of 0.1. Then, 1 ml of this suspension was incubated for 4 to 5 h at 37°C in 24-well plates in an incubator with 10% CO2. After the addition of 1 μg of DNA, the cell suspension was incubated for another 24 h. The cultures were spread on BHI agar plates containing 8 μg of kanamycin per ml and 10% horse serum for selection and growth.
PCR was used to confirm the exchange of ureI for Kanr in the H. pylori ureI mutants. The primers used were: 5′-CGG TAC CAT GGA ATA CGA TGC AAA CAT ACA-3′ (ureB4824) and 5′-CCA ACG TCG ACA ATT TCC TTC TCT TC-3′ (ureE6090). These primers correspond to the final one-sixth of the 3′ (ureB4824) end of the ureB gene and the first one-half of the 5′ region of the ureE gene (ureE6090). ExTAQ polymerase (TaKaRa) was used for the PCR for 30 cycles with each cycle consisting of 1 min at 92°C, 1 min at 56.2°C, and 1 min at 72°C. The PCR products were size separated on a 0.8% agarose gel in the presence of ethidium bromide, and φX174DNA/HaeIII markers were used as molecular weight standards.
Immunoblot analysis of ureI gene product.
Antibodies were generated against peptides in the predicted extracellular loops between membrane-spanning regions M2 and M3 and between regions M4 and M5 of the ureI gene products, CEGAEDIAQVSHHLTSFYGPATG (UP2/3) and CAILSHYSDMLDDHKVLGITEGD (UP4/5), respectively. The antibodies were generated in rabbit and affinity purified.
The bacteria were passed three times through a French press at 20,000 lb/in2, and the resulting lysate was centrifuged at 3,000 × g for 10 min to remove the unbroken cells. The samples were further centrifuged at 10,000 × g for 10 min, followed by centrifugation at 100,000 × g for 1 h to pellet the membrane fraction. The membrane pellet was suspended in 25 mM phosphate buffer (pH 7.4). The membrane proteins were size fractionated by sodium dodecyl sulfate (SDS)-tricine polyacrylamide gel electrophoresis and electrobloted onto nitrocellulose membrane followed by immunodetection by enhanced chemiluminescence (Amersham).
PCR for detection of ureI.
Genomic DNA was isolated from the different bacterial strains by the cetyltrimethylammonium bromide (CTAB) procedure (29). Briefly, the bacteria were washed with 10 mM Tris-HCl–1 mM EDTA (pH 8.0) (TE buffer) and dissolved by use of 0.5% SDS in the presence of 100 μg of proteinase K per ml at 37°C for 1 h. The solubilized bacteria were mixed with NaCl (0.5 M, final concentration), followed by 10 min of incubation at 65°C in the presence of 1% CTAB. Protein was removed by use of a chloroform-isoamyl alcohol (24:1) wash, followed by a second wash with phenol-chloroform-isoamyl alcohol (25:24:1). Genomic DNA was precipitated with ice-cold isopropanol and resuspended in TE buffer. The upper primer and lower primer used were: 5′-GCT AGG ACT TGT ATT GTT ATA ATG-3′ and 5′-CCC AGT GTT GGA TAA GAG C-3′, respectively. ExTAQ polymerase (TaKaRa) was used for the PCR for 30 cycles, with each cycle consisting of 1 min at 92°C, 1 min at 52°C, and 1 min at 72°C. The PCR products were size separated on a 0.8% agarose gel in the presence of ethidium bromide, and φX174DNA/HaeIII markers were used as molecular weight standards.
Urease activity.
Urease activity was measured radiometrically (19, 26). Bacteria or bacterial lysates were added to 100 mM sodium phosphate buffer containing 5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 1 mM glutamine, and 5 mM [14C]urea with a specific activity of 10 μCi/μmol. The range of pH of the buffer used was between pH 2.5 and 8.5. The pH of the buffer between 4.5 and 8.5 was achieved by mixing various amounts of 100 mM sodium phosphate monobasic and 100 mM sodium phosphate dibasic to the desired pH. Below a pH of 4.5 the desired pH was achieved by the addition of HCl. The pH of the buffer during the course of the experiment did not change by more than 0.1 pH units. Plastic wells containing 500 mM KOH soaked filter paper hung from rubber stoppers were used to collect the liberated 14CO2 that resulted from the hydrolysis of urea by urease. Urease activity was measured for 30 min at 37°C with constant agitation. The reaction was terminated by the addition of 5 N H2SO4 and incubated 30 min at 37°C. The wells were placed in scintillation cocktail (HiIonicFluor; Packard Instruments), and the radioactivity was measured by scintillation counting (1216 RackBeta; LKB Institute).
Bacteria scraped from plates and suspended in 1 ml of 1 mM phosphate buffer to a final concentration of 0.01 OD600 were used for the measurement of intact urease activity. Bacterial homogenates were prepared by scraping the bacteria from the plate into 3 ml of ice-cold distilled water followed by three passages through a French press (SLM Instruments, Rochester, N.Y.) at 20,000 lb/in2. Then, 10 μl of the bacterial homogenate was used to assay urease activity. In some experiments 0.01% of the nonionic detergent C12E8 was used to permeabilize the bacterial membranes without disruption of the bacteria as visualized by acridine orange fluorescence in a confocal microscope (data not shown). Urease activity is reported as micromoles of CO2 released per minute per milligram of protein. The protein concentration was determined by the method of Lowry (16).
Measurement of membrane potential.
Membrane potential (PD) was determined as previously described (20). Briefly, H. pylori were harvested from plates in 300 μl of HP buffer (1 mM phosphate buffer containing 5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 1 mM glutamine, and 10 mM glucose). The fluorescent, membrane-potential-sensitive dye, DiSC3(5), was dissolved in dimethyl sulfoxide, and 3 μl was added to 3 ml of the appropriate buffer to give a final concentration of 1 μM. The bacterial suspension was then added to 3 ml of the dye solution at different pHout in a fluorimeter cuvette to reach an OD600 of 0.160 (usually 15 to 20 μl).
Fluorescence quenching due to potential-dependent uptake of the dye was measured in a fluorimeter set at an excitation wavelength of 600 nm and an emission wavelength of 665 nm. The dye solution was added 5 min before adding the bacteria to allow temperature equilibration. With addition of the bacteria, the fluorescence quenched due to dye uptake driven by the interior negative potential. After the fluorescence reached equilibrium with the membrane potential, 5 mM urea was added, and the change in fluorescence was measured. All experiments were done at 37°C. Calibration of the membrane potential was carried out as previously described by the addition of valinomycin followed by the addition of K+ until no further change in fluorescence was observed (18, 25). This enables calculation of the K+ equilibrium potential found with the addition of the K+ selective ionophore, valinomycin, by using the Nernst equation: PD = 61 log 5/[K+]i, where [K+]i is equal to the external K+ concentration at which the potential difference becomes zero in the presence of valinomycin, i.e., where [K+]out = [K+]in, and 5 mM is the medium concentration when valinomycin is first added. The membrane potential in the absence of valinomycin can then be calculated. No change in medium pH was found in these strong buffers before or after the addition of urea over the time course of measurement.
The membrane potential is displayed based on the calibration once the dye reaches equilibrium. The initial fluorescence quench depends on the accumulation of the dye inside the bacteria, and the fluorescence present prior to equilibrium does not imply a positive interior potential.
RESULTS
Confirmation of ureI replacement by kanr.
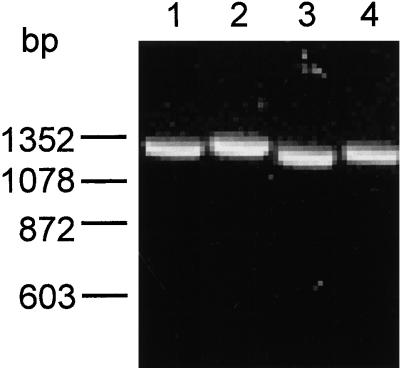
ureI deletion mutants of H. pylori strains ATCC 49503 and ATCC 43504 were constructed by double crossover between the chromosomal wild-type gene sequences and plasmid derived homologous ureB-ureE sequences with the ureI deletion replaced by the kanr sequence. The ureI deletion consisted of both the ureI gene and the ureB-ureI intergene region totaling 768 bp. The kanr sequence contains 1,325 bp. The primers used correspond to the final one-sixth of the 3′ end of the ureB gene (ureB4824) and the first one-half of the 5′ region of the ureE gene (ureE6090). The expected PCR product from the H. pylori wild-type strains would be 1,281 bp, while the PCR product of the H. pylori ureI mutant strains would be 1,325 bp, a difference of 44 bp. Figure 1 shows the results of the PCR. PCR products of about 1,281 and 1,325 bp were detected for the H. pylori wild-type strains and H. pylori ureI mutant strains, respectively. Further evidence for the deletion of the ureI gene as determined by PCR of the ureI gene itself is discussed below (see Fig. 9). Western analysis also confirmed the absence of UreI in the H. pylori ureI mutant strains (see Fig. 7).
FIG. 1.
PCR analysis confirming the replacement of the ureI gene with the kanamycin cassette. Genomic DNA was isolated from the H. pylori wild type and H. pylori ureI mutant strains and used as a template for PCR. PCR primers corresponding to regions of the ureB and ureE gene were used as described in Materials and Methods. A PCR product of 1,281 bp was detected for the H. pylori wild type strains (ATCC 43504, lane 1; ATCC 49503, lane 3). A PCR product of 1,325 bp was detected for the H. pylori ureI mutant strains (ATCC 43504-I, lane 2; ATCC 49503-I, lane 4).
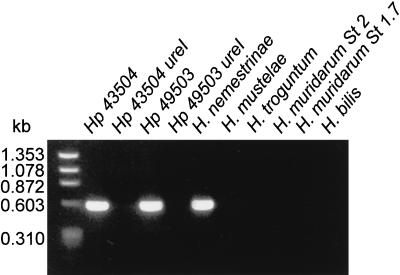
FIG. 9.
PCR analysis of ureI from genomic DNA of gastric and nongastric Helicobacter species. Genomic DNA was isolated from the different bacterial species and used as template for PCR of ureI as described in Materials and Methods. A single PCR product of about 587 bp was detected in the lanes containing genomic DNA from H. pylori ATCC 43504 and ATCC 49503 and from H. nemestrinae. No PCR product was detected in the lanes containing genomic DNA from the ureI deletion mutants, H. mustelae or the nongastric Helicobacter species.
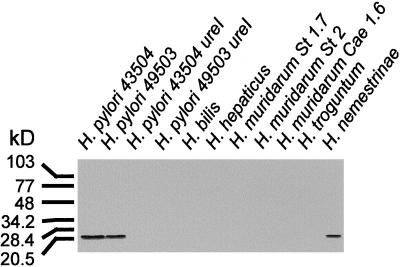
FIG. 7.
Western blot analysis of membrane protein extracts from gastric and nongastric Helicobacter species by using UreI antibodies. Membrane protein extracts were size fractionated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-UreI (UP2/3 and UP4/5 mixture) as described in Materials and Methods. A single band corresponding to a molecular size of about 21 kDa was detected in the lanes containing membrane proteins from H. pylori ATCC 43504 and ATCC 49503 and from H. nemestrinae. No immunoreactivity was detected in the lanes containing membrane proteins from the ureI deletion mutants or the nongastric Helicobacter species.
Urease activity as a function of medium pH.
The experimental system for the measurement of urease activity was calibrated to be in a linear range for the 30-min incubation time used in these experiments. The 100 mM buffer at each pH and the low bacterial numbers resulted in a stable pH (±0.1 U) throughout the time course of measurement of enzyme activity. The urease pH optimum in HP buffer of bacterial lysates from the different Helicobacter species is shown in Table 1. The pH optima of the bacterial lysates ranged from 7.0 to 7.5 for H. pylori to 8.5 for H. mustelae. The urease activity is reported as micromoles of CO2 liberated per minute per milligram of protein. The pH optima of the urease measured in the ureI mutant lysates were similar to the wild-type strain as previously reported (Fig. 2) (26). The demonstration of wild-type levels of urease activity in the ureI mutant lysates shows that the kanr for ureI substitution is nonpolar. If the ureI knockout was a polar mutation, the downstream accessory genes required for nickel insertion would not be expressed, resulting in little or no urease activity (13).
TABLE 1.
pH optima and urease activity of different Helicobacter species
| Strain | Urease activitya | pH optimum | Acid activation | Site of infection |
|---|---|---|---|---|
| H. pylori | 5.15 ± 0.27 | 7.5 | Yes | Human stomach |
| H. nemestrinae | 6.94 ± 0.92 | 8.5 | Yes | Ferret stomach |
| H. mustelae | 4.74 ± 0.09 | 7.0 | Yes | Monkey stomach |
| H. troguntum | 0.23 ± 0.01 | 7.0 | No | Rat intestine |
| H. bilis | 0.57 ± 0.04 | 7.0 | No | Mouse liver |
| H. hepaticus | 3.01 ± 0.19 | 7.0 | No | Mouse liver |
| H. muridarum | 2.85 ± 0.11 | 7.0 | No | Mouse intestine |
Micromoles of CO2 liberated per minute per milligram of protein ± SEM.
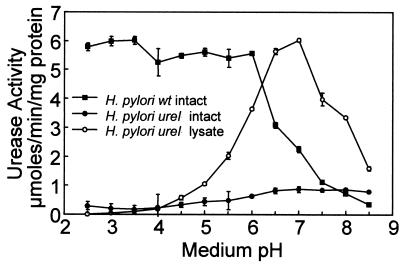
FIG. 2.
pH dependence of urease activity of H. pylori and H. pylori ureI mutant. The urease activity of the intact H. pylori wild type (■) increased rapidly as the medium pH decreased. The urease activity remained high down to a medium pH of 2.5. In contrast, there was no increase of urease activity in the H. pylori ureI mutant (●). The urease activity of the H. pylori ureI mutant lysates displayed a single pH optimum (○). Urease activity was measured radiometrically under strong buffering conditions in the presence of 5 mM urea as described in Materials and Methods (micromoles of CO2 liberated per minute per milligram of protein; n = 3, ± the standard error of the mean [SEM]).
The urease activity as a function of pH in intact H. pylori and the H. pylori ureI mutant bacteria is shown in Fig. 2. H. pylori showed a rapid 10-fold increase in urease activity when the pH dropped below 6.5 with a half-maximal activation at pH 6.2. This activity remained relatively constant down to pH 2.5 (Fig. 2). In contrast, there was no increase in activity of the H. pylori ureI mutant as the pH decreased (Fig. 2).
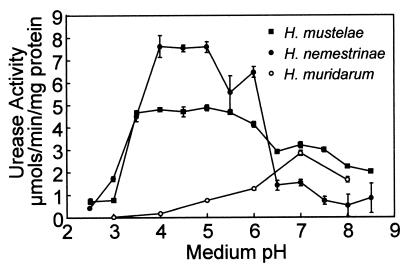
The urease activity of H. mustelae and H. nemestrinae also rapidly increased as the pH dropped below pH 7.0 and 6.5, respectively (Fig. 3). The urease activity of intact H. bilis, H. troguntum, and H. hepaticus was not detectable with the radiometric urease activity assay. There was no increase in the activity of urease in intact H. muridarum as pH declined; however, as can be seen in Fig. 3, there were high levels of urease activity near neutral pH similar to the pH dependence of urease activity of the lysed organism (data not shown).
FIG. 3.
The pH dependence of urease activity in intact Helicobacter species. Urease activity was measured radiometrically under strong buffering conditions in the presence of 5 mM urea as described in Materials and Methods. Urease activity of intact H. mustelae (■), H. nemestrinae (●), and H. muridarum (○) (micromoles of CO2 liberated per minute per milligram of protein; n = 3, ±SEM).
Effect of detergent permeabilization on urease activity.
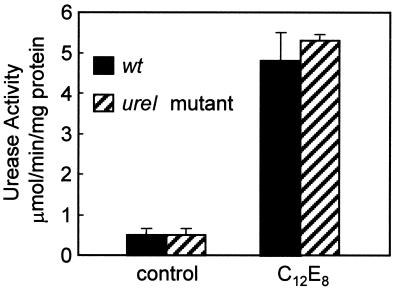
Since the intact ureI mutant was deficient in activating its cytoplasmic urease under acidic conditions, we reasoned that UreI was either directly linked to urease activity or was needed for urea entry into the cytoplasm. The urease activities of the whole-cell lysate of the ureI mutant and wild-type H. pylori strains were nearly identical. This finding would make it unlikely that UreI played a role in inhibiting urease activity at neutral and alkaline pH levels or activated urease at acidic pH. To test the hypothesis that UreI was required for urea to access the cytoplasm, we permeabilized both H. pylori and the H. pylori ureI mutant with the nonionic detergent C12E8.
Addition of 0.01% C12E8 did not affect urease activity of the bacterial lysates (data not shown), but it raised urease activity at neutral pH in both the wild-type and ureI mutant intact bacteria to the level seen in lysates (Fig. 4 and Table 1). Visualization of the bacteria by acridine orange fluorescence showed that this concentration of detergent did not affect their morphology (data not shown). Hence, membrane permeabilization was sufficient to fully activate cytoplasmic urease in intact H. pylori, showing that the cytoplasmic membrane impedes the diffusion of urea into the cytoplasm.
FIG. 4.
Effect of detergent permeabilization on the urease activity of intact H. pylori and H. pylori ureI mutant. Addition of 0.01% C12E8 to intact H. pylori and H. pylori ureI mutant increases urease activity at neutral pH. Urease activity was measured radiometrically under strong buffering conditions in the presence of 5 mM urea as described in Materials and Methods (micromoles of CO2 liberated per minute per milligram of protein; n = 3, ±SEM).
Effect of urea on membrane potential at acidic pH.
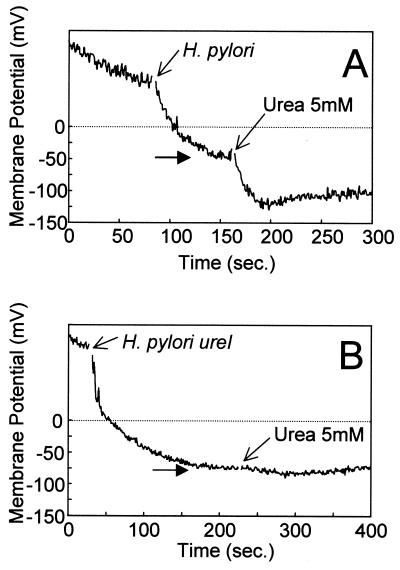
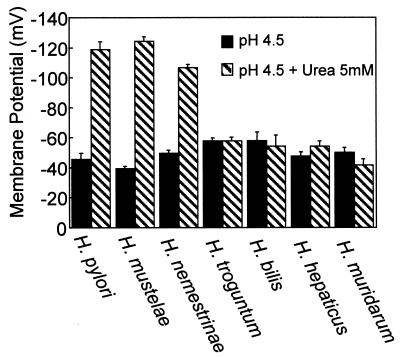
We have shown that the addition of 5 mM urea to H. pylori in 100 mM phosphate buffer at pH 6.0 and below results in an increase in the transmembrane potential to approximately −100 mV (26). Figure 5 shows the effect of 5 mM urea addition on the membrane potential of the H. pylori ureI-negative mutant and the parent strain at pH 4.5 under pH clamp conditions. The membrane potential of the wild-type strain increased following urea addition as previously reported (26). However, the ureI deletion mutant showed no increase in membrane potential after the addition of 5 mM urea. The membrane potential of H. mustelae and H. nemestrinae increased after 5 mM urea was added to HP buffer at pH 4.5, whereas urea addition to the nongastric Helicobacter species H. bilis, H. troguntum, H. hepaticus, and H. muridarum had no effect on membrane potential (Fig. 6).
FIG. 5.
Effect of urea on the membrane potential of H. pylori wild type and ureI mutants. Membrane potential was measured by using DiSC3(5) at pH 4.5 under pH clamp conditions as described in Materials and Methods. Urea was added as indicated by the arrow and resulted in an increase of membrane potential in H. pylori (A) but not in the H. pylori ureI mutant (B). The horizontal arrow at the point of equilibration of the dye indicates bacterial membrane potential prior to urea addition.
FIG. 6.
Effect of urea on membrane potential of other Helicobacter spp. Membrane potential was measured by using DiSC3(5) at pH 4.5 under pH clamp conditions as described in Materials and Methods. Urea addition resulted in an increase in membrane potential in the gastric Helicobacter species H. pylori, H. mustelae, and H. nemestrinae. Urea addition had no effect on the membrane potential of H. bilis, H. troguntum, H. hepaticus, or H. muridarum (n = 3, ±SEM).
Western blot analysis of UreI.
Western blot analysis with a mixture of antibodies against synthetic peptides of the putative external loops between transmembrane-spanning regions 2/3 and 4/5 was performed to determine whether UreI protein was expressed in both the gastric and nongastric Helicobacter species. A single band corresponding to a molecular size of about 21 kDa was detected in the lanes containing membrane proteins from H. pylori ATCC 43504 and ATCC 49503 and from the gastric pathogen of the Macaque monkey, H. nemestrinae (Fig. 7). No immunoreactivity was detected in the lanes containing purified membranes from the ureI mutants of H. pylori ATCC 43504 and ATCC 49503. Similarly, no bands were detected from any of the nongastric Helicobacter species (Fig. 7).
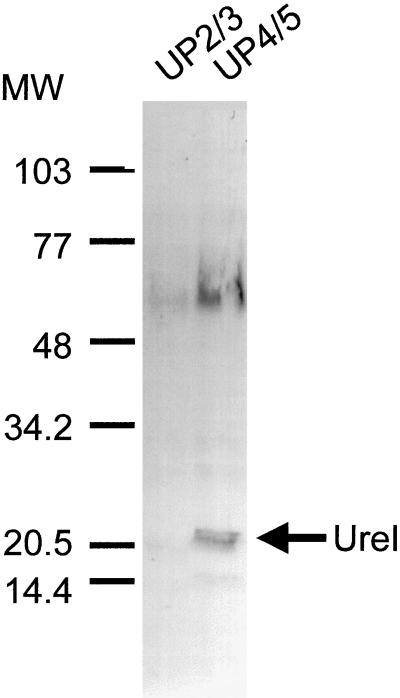
Western blot analysis of H. mustelae purified membrane proteins with the mixture of the two antibodies failed to detect the presence of UreI (data not shown). Additional Western blot analysis following affinity separation of the two antibodies was performed. Immunoblots with antibody UP4/5 resulted in the detection of a band of about 21 kDa corresponding to the UreI protein and a diffuse immunoreactivity with a molecular size of about 60 to 65 kDa (Fig. 8, lane 2). No immunoreactivity was detected when the blots were probed with antibody UP2/3 (lane 1).
FIG. 8.
Western blot analysis of membrane protein extracts from H. mustelae with anti-UreI UP2/3 or UP4/5. Membrane protein extracts of H. mustelae were size fractionated on an SDS-polyacrylamide gel, transferred to nitrocellulose, and probed with anti-UreI UP2/3 and UP4/5 as described in Materials and Methods. A band corresponding to a molecular size of about 21 kDa was detected in the lane probed with anti-UreI UP4/5 (lane 2). It was not possible to detect UreI with anti-UreI UP2/3 alone (lane 1).
PCR analysis of ureI.
PCR amplification of the ureI gene from genomic DNA was performed to determine if the gene was deleted from the deletion mutants of H. pylori ATCC 43504 and ATCC 49503 strains and to determine if the gene was present but not translated in the nongastric Helicobacter spp. A single PCR product of about 587 bp was detected in the gastric H. pylori strains ATCC 43504 and ATCC 49503 and the H. nemestrinae strain (Fig. 9, lanes 1, 3, and 5, respectively). It was not possible to generate a PCR product from the H. pylori ureI mutants, showing that the gene had been deleted (Fig. 9, lanes 2 and 4). No PCR product was detected with H. mustelae or the nongastric Helicobacter species.
DISCUSSION
H. pylori is a neutralophilic bacterium that lives in the acidic environment of the stomach. H. pylori must be able to withstand the high acid of the stomach and survive those time periods when its environment is close to neutrality. Further, it has to grow in the gastric milieu. The organism expresses high levels of a neutral pH optimum urease, and previous data have shown that the cytoplasmic component of this urease is inactive at neutral pH but activated at acidic pH (3, 25, 26). The H. pylori urease gene cluster contains a unique gene, ureI, which appears to be required for gastric infection (27). Here we have confirmed that ureI gene expression is not required for urease activity in vitro but is required for the acidic pH induced increase in cytoplasmic urease activity in the intact organism. Expression of ureI is also required for the urea-dependent increase in cytoplasmic membrane potential at acidic medium pH levels.
Urea has a diffusion permeability coefficient through lipid bilayers of 4 × 10−6 cm/s (23). This low permeability is modified in animals when high urea permeability is required by the expression of a urea transporter (12). The rate of entry of urea at neutral pH in intact H. pylori is unable to saturate urease activity with urea concentrations as high as 200 mM (26). Since the urea concentration of the gastric juice is 1 to 3 mM, a urea transporter would be essential for acid survival. The increase in cytoplasmic urease activity with medium acidification appears to be the result of increased urea permeability across the inner membrane of the bacterium, since the addition of low concentrations of a nonionic detergent was able to duplicate acid activation of cytoplasmic urease. The low levels of urease activity observed in the intact H. pylori ureI mutant and at neutral pH and above in intact wild-type H. pylori are likely due to the limited diffusion of urea across the membrane. A contribution of surface urease cannot be excluded at neutral pH, although this component is inactive at pH 4.0 and below. It is possible that the hydrolysis of urea produced internally through the urea cycle may contribute to the acid survival of H. pylori (18). However, the radiometric urease assay used in the experiments reported here would not detect hydrolysis of urea produced through the urea cycle since it would not be radiolabeled.
It has been suggested that ureI is involved in ammonia or ammonium transport (27). If ureI was involved in the regulation of transport of these molecules then one or both should inhibit urease and they do not. If NH3 efflux was prevented at neutral pH, the cell would alkalinize and die. Moreover, NH3 is a gas that is highly permeable across lipid bilayers, and regulation of its permeability has never been observed. Export of NH4+ would not alkalize the periplasm. Since NH4+ is a cation it would have to be exported against an outside positive membrane potential by an active process such as an ATPase, a proton-ammonium antiport, or a urea-ammonium antiport. UreI has no signature sequence reminiscent of an ATPase and a proton antiporter would not be able to regulate pH in either the cytoplasm or periplasm. A urea-ammonium antiporter at normal gastric levels would not facilitate urea entry given an interior negative potential at pH 6.2 of −101 mV. Based on these arguments, the ureI gene product is either directly responsible for urea permeability and is active at acidic pH or it regulates the urea permeability of another cytoplasmic membrane protein. It is homologous to the amiS genes described as amide transporters (30). Since urea is an amide, the ureI gene product is possibly a urea transporter.
The large surface-area-to-volume ratio of prokaryotes has prevented measurement of passive permeability of a variety of solutes, and we were unable to show a time or pH dependence of urea uptake into H. pylori. Recently, we expressed UreI in Xenopus oocytes, which, in contrast, have a large volume-to-surface ratio (D. L. Weeks, S. Eskandari, D. R. Scott, and G. Sachs, submitted for publication). This resulted in the appearance of urea transport at acidic pH. This transport of urea had the same pH activation profile as cytoplasmic urease in the intact organism. Urea transport in these oocytes is selective, nonsaturable, and temperature independent and did not require ATP or counter ions, characteristics of a high-capacity, channel-like transporter.
All of the gastric Helicobacter species examined in this study expressed the UreI protein, although the pH required for the activation of the cytoplasmic urease was not identical. Both H. pylori and H. nemestrinae cytoplasmic urease activated at pH 6.2, and their membrane potential increased to a value consistent with a pHout of approximately 6.2 after the addition of urea at acidic pH. Immunodetection of UreI and PCR of the ureI gene detected bands of equal intensity.
H. mustelae cytoplasmic urease was activated at pH 7.0, and urea addition at acidic pH increased membrane potential to that found at pH 7.0. Although PCR analysis failed to detect ureI, UreI was detected by Western blot analysis. The results of the Western analysis of H. mustelae membrane proteins are of interest. UreI was detected with the UP4/5 antibody but not with the UP2/3 antibody. The former antibody is raised against the loop connecting TM4 and TM5, which contains two histidines and several carboxylic acids as well as a lysine residue. Many of these could determine the antigenic specificity of this region and be common to both bacterial species. The UP2/3 antibody was raised against the extracellular loop between the putative membrane-spanning helices TM2 and TM3 of H. pylori. Here, this extracytoplasmic loop region in H. pylori contains two adjacent histidine residues which have a presumed pKa of ∼6.0, about the same pH that activates the cytoplasmic urease. In addition, there is a pair of carboxylic acids in this loop. These four amino acids are predicted to be the major immunogenic determinants in this region. Protonation of these histidines could result in hydrogen bonding with the carboxylic acids enabling urea transport across the membrane domain. Substitution of one or the other of these particular amino acids might result in the loss of immunoreactivity. If this has occurred in H. mustelae, it may explain the difference in the urease activation pH. Cloning and sequencing of the ureI gene of H. mustelae would be able to support this hypothesis. Failure to generate a ureI PCR product from H. mustelae is likely due to the lack of sequence similarity between the H. pylori ureI primers used and H. mustelae DNA. The activation of cytoplasmic urease, the increase in membrane potential with the addition of urea at acidic pH and the positive Western blot analysis strongly suggest that ureI is present in H. mustelae.
UreI was not detected by Western analysis nor was there a PCR product for the ureI gene in any of the nongastric Helicobacter spp. Although urease activity was present in their lysates, no increase of urease activity at acidic pH was detected in the intact nongastric Helicobacter species. Additionally, there was no increase in transmembrane potential following urea addition at acidic pH in any of the nongastric species. Taken together, these findings suggest that UreI is absent or is nonfunctional in the nongastric Helicobacter species.
The pH optimum of urease activity in intact H. muridarum was similar to the pH optimum of its total cell lysate. This finding may imply relatively rapid diffusion of urea into the cytoplasm of H. muridarum in contrast to the other Helicobacter spp. This organism is able to colonize the stomach only in the transitional zone between antrum and fundus, and its high apparent urea permeability may enable the organism to occupy this unusual and specific niche in the absence of activation of cytoplasmic urease by UreI. Since H. muridarum urease has a pH optimum of 7.0, it could grow when the rate of cytoplasmic alkalization by urease activity is balanced by proton diffusion into the cytoplasm. This hypothesis would predict that the external pH must be held in a very narrow range and may explain its infection of only the transitional zone of the gerbil gastric mucosa.
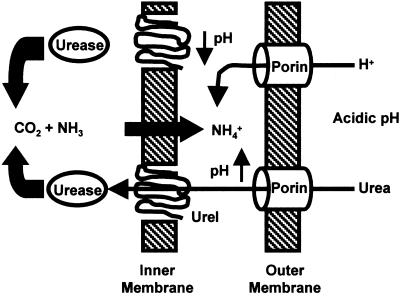
A model of the proposed mechanism of activation of cytoplasmic urease is shown in Fig. 10. Urea and H+ diffuse into the periplasm probably through porins in the outer membrane. Periplasmic acidification results in a conformational change in UreI, permitting the entry of urea into the cytoplasm, where it is rapidly hydrolyzed, producing CO2 and NH3. The ammonia rapidly diffuses into the periplasmic space, where it becomes protonated, raising the pH to about 6.2, a level consistent with survival and growth.
FIG. 10.
Model of the activation of cytoplasmic urease under acidic conditions. Urea and H+ diffuse into the periplasmic space probably through porins in the outer membrane. Periplasmic acidification results in a conformational change in UreI, permitting the entry of urea into the cytoplasm, where it is rapidly hydrolyzed, producing CO2 and NH3. The ammonia diffuses into the periplasmic space, where it becomes protonated, raising the pH to about 6.5, a level consistent with survival and growth.
Given that ureI gene expression is essential for activation of H. pylori cytoplasmic urease and for gastric colonization, it is tempting to speculate that inhibition of this putative transporter would prevent gastric habitation of this pathogen. Hence, studies of inhibitors of the function of this protein might result in the development of specific monotherapy for eradication of H. pylori.
ACKNOWLEDGMENTS
This work was supported in part by USVA SMI and NIH grants DK4697, 53462, 41301, and 17294.
REFERENCES
- 1.Anderson H, von Heijne G. Membrane protein topology: effects of delta mu H+ on the translocation of charged residues explain the “positive inside” rule. EMBO J. 1994;13:2267–2272. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrutis K A, Fox J G, Schauer D B, Marini R P, Murphy J C, Yan L, Solnick J V. Inability of an isogenic urease-negative mutant strain of Helicobacter mustelae to colonize the ferret stomach. Infect Immun. 1995;63:3722–3725. doi: 10.1128/iai.63.9.3722-3725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauerfeind P, Garner R, Dunn B E, Mobley H L T. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode G, Malfertheimer P, Lenhardt G, Nilius M, Dischuneit H. Ultrastructural localization of urease of Helicobacter pylori. Med Microbiol Immun. 1993;182:223–242. doi: 10.1007/BF00579622. [DOI] [PubMed] [Google Scholar]
- 5.Chebrou H, Bigey F, Arnaud A, Galzy P. Amide metabolism: a putative ABC transporter in Rhodococcus sp. R312. Gene. 1996;182:215–218. doi: 10.1016/s0378-1119(96)00478-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y-Y M, Clancy K A, Burne R A. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect Immun. 1996;64:585–592. doi: 10.1128/iai.64.2.585-592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669–1673. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin C S, Carrick J. Peptic ulcer disease and Helicobacter pylori infection. Curr Opin Gastroenterol. 1991;7:108–115. [Google Scholar]
- 10.Graham D Y. Campylobacter pylori and peptic ulcer disease. Gastroenterology. 1989;96:615–625. doi: 10.1016/s0016-5085(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 11.Hawtin P R, Stacey A R, Newell D G. Investigation of the structure and localization of the urease of Helicobacter pylori using monoclonal antibodies. J Gen Microbiol. 1990;136:995–2000. doi: 10.1099/00221287-136-10-1995. [DOI] [PubMed] [Google Scholar]
- 12.Hediger M A, Smith C P, You G, Lee W S, Kanai Y, Shayakul C. Structure, regulation and physiological roles of urea transporters. Kidney Int. 1996;49:1615–1623. doi: 10.1038/ki.1996.235. [DOI] [PubMed] [Google Scholar]
- 13.Hu L-T, Mobley H L T. Expression of catalytically active recombinant Helicobacter pylori urease at wild-type levels in Escherichia coli. Infect Immun. 1993;61:2563–2569. doi: 10.1128/iai.61.6.2563-2569.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashket E R. The proton motive force in bacteria: a critical assessment of methods. Annu Rev Microbiol. 1985;39:219–242. doi: 10.1146/annurev.mi.39.100185.001251. [DOI] [PubMed] [Google Scholar]
- 15.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 17.Marshall B J. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 18.Mendz G L, Hazell S L. The urea cycle of Helicobacter pylori. Microbiology. 1996;142:2959–2967. doi: 10.1099/13500872-142-10-2959. [DOI] [PubMed] [Google Scholar]
- 19.McDonald J A, Speeg K V, Jr, Campbell J V. Urease, a specific and sensitive radiometric assay. Enzmologia. 1972;42:1–9. [PubMed] [Google Scholar]
- 20.Meyer-Rosberg K, Scott D R, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111:886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 21.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neithercut W D, Greig M A, Hossack M, McColl K E L. Suicidal destruction of Helicobacter pylori: metabolic consequence of intracellular accumulation of ammonia. J Clin Pathol. 1991;44:380–384. doi: 10.1136/jcp.44.5.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orbach E, Finkelstein A. The nonelectrolyte permeability of planar lipid bilayer membranes. J Gen Physiol. 1980;75:427–436. doi: 10.1085/jgp.75.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phadnis S H, Parlow M H, Levy M, Llver D, Caulkins C M, Conners J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial lysis. Infect Immun. 1995;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rektorschek M, Weeks D, Sachs G, Melchers K. Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology. 1998;115:628–641. doi: 10.1016/s0016-5085(98)70142-8. [DOI] [PubMed] [Google Scholar]
- 26.Scott D R, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 27.Skouloubris S, Thiberge J, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuda M, Karita M, Morshed M G, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. pp. 2.4.1–2.4.2. [DOI] [PubMed] [Google Scholar]
- 30.Wilson S A, Williams R J, Pearl L H, Drew R E. Identification of two new genes in the Pseudomonas aeruginosa amidase operon, encoding an ATPase (AmiB) and a putative integral membrane protein (AmiS) J Biol Chem. 1995;270:18818–18824. doi: 10.1074/jbc.270.32.18818. [DOI] [PubMed] [Google Scholar]
- 31.Young G M, Amid D, Miller V L. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J Bacteriol. 1996;178:6487–6495. doi: 10.1128/jb.178.22.6487-6495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]