Abstract
The surface glycoprotein gp82, expressed in the insect-stage metacyclic trypomastigotes of Trypanosoma cruzi, has been implicated in mammalian cell invasion. Here we have characterized the cell adhesion site of gp82 by using recombinant proteins and synthetic peptides based on gp82. The recombinant protein Del-4/8, lacking 65 amino acids of gp82 central domain (at positions 257 to 321), was virtually devoid of cell-binding activity and lacked the ability to inhibit parasite invasion, in contrast to J18, the construct containing the full-length gp82 sequence (amino acids 1 to 516). Constructs with shorter deletions, i.e., Del-4 (deleted from 257 to 271) and Del-8 (deleted from 293 to 321), bound to target cells to a significantly lesser degree than did J18. The sites deleted in recombinant proteins Del-4 and Del-8 contained acidic amino acids critical for cell adhesion. Thus, the cell-binding capacity of protein Del-E/D, lacking the glutamic acid (259/260) and aspartic acid (303/304) pairs, was negligible, as was its capacity to inhibit parasite internalization. Of a set of synthetic peptides spanning the gp82 central domain, a 22-mer hybrid peptide, p4/8, formed by two noncontiguous sequences (at positions 257 to 273 and 302 to 306) and containing the four acidic residues, competed with the binding of J18 protein to target cells and significantly inhibited (∼60%) the penetration of parasites. This peptide, generated by the juxtaposition of sequences that are separated by a hydrophobic stretch in the linear molecule, appears to be mimicking a conformation-dependent cell-binding site of gp82. Experiments of antibody competition with a set of 20-mer overlapping peptides mapped the epitope for 3F6, a monoclonal antibody directed to gp82 that inhibits parasite invasion, to the sequence represented by peptide p3 (244 to 263), which has a partial overlap with the cell adhesion site.
Infection by Trypanosoma cruzi, a protozoan parasite that causes Chagas disease in humans, is initiated by metacyclic trypomastigotes which develop in and are transmitted by insect vectors. Successful establishment of T. cruzi infection in mammalian hosts is critically dependent on parasite entry into target cells. Metacyclic parasite forms express a set of surface glycoproteins that interact with mammalian cells (21, 22). One of these molecules, a metacyclic-stage-specific 82-kDa glycoprotein (gp82), has been implicated in host cell invasion on the basis that this process can be inhibited by native or recombinant gp82 (21, 23) and by a monoclonal antibody (MAb 3F6) that recognizes metacyclic forms but not the noninfective epimastigote or other developmental stages of T. cruzi (28).
The metacyclic stage gp82 is an adhesion molecule that binds to host cells in a receptor-mediated manner and triggers Ca2+ mobilization (7, 22), an event that is essential for parasite penetration (3, 7, 15, 27, 32). Recent studies have indicated that gp82 also induces the activation of metacyclic trypomastigote protein tyrosine kinase (8) and an increase in the parasite intracellular Ca2+ concentration (22). The requirement of these signal transduction events for T. cruzi infectivity has been reported by several authors (15, 29, 33). Among other surface glycoproteins in the size range of gp82, such as gp83 (14), gp85 (19), and Tc-85 (1), which are expressed in bloodstream or tissue-culture derived trypomastigotes and which are implicated in mammalian cell adhesion/invasion, gp83 has been reported to signal through the mitogen-activated protein kinase pathway to up regulate T. cruzi entry in macrophages (30). Although gp82 is a member of a multigene family (2) that shares a considerable degree of homology with members of the gp85-trans-sialidase superfamily (5, 6, 24), it differs from these molecules with regard to its adhesive properties. In tissue-culture-derived trypomastigotes, a gp85 with affinity for fibronectin (19), a laminin-binding Tc-85 (10, 11), a collagen- and heparan sulfate-binding molecule of unknown sequence designated penetrin (18), and an undefined heparin-binding Ca2+ signaling factor (4) have been reported. In contrast to all of these other T. cruzi proteins that have been implicated in the cell adhesion and invasion process, the metacyclic stage gp82 apparently does not bind to extracellular matrix components (21).
The interaction of metacyclic trypomastigote gp82 with target cells is mediated by the peptide portion of the molecule, the carbohydrate moiety playing no role in parasite invasion (21, 23). The gp82 cell-binding site and the epitope for MAb 3F6 appear to be located in the central domain of the molecule, but they have not been precisely defined. In the present study, we sought to define these functional sites of gp82.
MATERIALS AND METHODS
Expression of gp82 recombinant proteins in pGEX-3.
The construct J18, comprising the full-length gp82 sequence in frame with the glutathione S-transferase (GST) gene of plasmid pGEX-3 (25), as well as the constructs J18a, encoding 156 amino acids of the N-terminal domain, and J18b, which encodes 293 amino acids of the C-terminal domain, were prepared as previously described (23). To obtain gp82 recombinants with various deletions in the central domain of the molecule, novel restriction sites were first added to the gp82 gene by using the Quikchange Mutagenesis kit (Stratagene) as follows. Restriction sites for XhoI (in boldface) and SalI (underlined) were inserted by using complementary pairs of oligonucleotides corresponding to the gp82 coding strand (5′-ATTTCACTTGCCCGCTCGAGCGAGGAGCTGAAGACG-3′, 5′-GTCCTCAGCACTTGGTCGACGCTGGATGCCTCCTTC-3′, 5′-CTGGTTGGACTCCTCTCGAGTTCGCCAGCGGTGAC-3′, 5′-AAGGTCCATGACGGGTCGACATTCACGGGGTTTGGT-3′) to prime amplification of plasmid J18 with Pfu DNA polymerase (Stratagene) for 18 cycles of elongation at 68°C for 14 min 30 s per cycle. Reaction products were treated with DpnI to digest the input plasmid templates then were purified and used to transform Escherichia coli. Recovered plasmids were digested with BamHI (restriction site present in the original preparation) plus either XhoI or SalI (restriction sites introduced by PCR) to confirm the presence of the new sites. DNA samples were then subjected to a new PCR cycle for the introduction of a second restriction site, according to the protocol described above. The sequence flanked by the new sites was deleted by digestion of plasmids with XhoI plus SalI and religation with T4 DNA ligase. To generate the gp82 mutants lacking the contiguous pairs of acidic amino acids (see Fig. 2A), PCR amplification was carried out as described above by using the primers 5′-GGTGACGCGTGGATCTACCGTTCCGTGAAT-3′ and 5′-CTTGCCCGCCTGACCCTGAAGACGATCAAG-3′. All constructs were sequenced in order to ascertain that the desired deletion mutants were obtained.
FIG. 2.
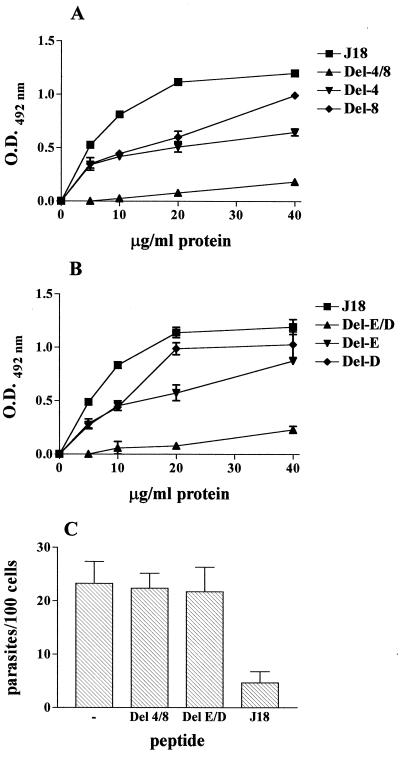
Binding of T. cruzi gp82 recombinant proteins to host cells and effect on parasite invasion. In panels A and B, increasing concentrations of protein were added to wells in enzyme-linked immunosorbent assay (ELISA) plates containing adherent HeLa cells. After washes, cells were sequentially incubated with anti-J18a antibodies and anti-mouse immunoglobulin conjugated to peroxidase. The bound enzyme was revealed by using o-phenylenediamine as substrate. In panel A, the assays were performed with the deletion mutants Del-4, Del-8, and Del-4/8, lacking 15, 29, and 65 amino acid residues, respectively. Panel B shows the results of experiments with the constructs Del-E, Del-D, and Del-E/D lacking either or both of the glutamic acid and aspartic acid doublets. The recombinant J18 containing the full-length gp82 sequence was used as control in both assays. Representative results of one of four comparable experiments are shown. The values are the means ± the standard deviations (SD) of triplicates. Panel C shows the result of an invasion assay in which the metacyclic forms were incubated with HeLa cells in the presence of the indicated recombinant protein at 80 μg/ml. After 3 h at 37°C, the cells were stained with Giemsa, and the number of intracellular parasites were counted in a total of 500 cells. Values are the means ± SD of three assays performed in duplicate. O.D., optical density.
Purification of gp82 recombinant proteins.
The recombinant GST fusion proteins were purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli cells essentially as described (23), followed by electroelution (31). To assess the purity of the proteins, the purified protein samples were analyzed in sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gel by staining with Coomassie blue and by immunoblotting as detailed (35). The purified recombinant proteins appeared as bands of expected size. The bands of smaller size and lower intensity seen in some preparations were probably products of protein degradation and, as such, were recognized by MAb 3F6 (data not shown).
Synthetic peptides.
Peptides were synthesized with the 9-fluorenylmethoxy carbonyl methodology (12), by using an automated benchtop simultaneous multiple solid-phase peptide synthesizer (PSSM 8 system; Shimadzu, Tokyo, Japan). The final peptides were deprotected in trifluoroacetic acid (TFA) and were purified by semipreparative high-pressure liquid chromatography (HPLC) by using an Econosil C18 column (10 μ, 22.5 by 250 mm) and a two-solvent system (TFA/H2O [1:1,000] and TFA/acetonitrile [ACN]/H2O [1:900:100]). The column was eluted at a flow rate of 5 ml/min with a 10 (or 30) to 50 (or 60)% gradient of solvent B over 30 or 45 min. Analytical HPLC was performed by using a binary HPLC system from Shimadzu with a SPD-10AV Shimadzu UV-visual light detector, coupled to an Ultrasphere C18 column (5 μ, 4.6 by 150 mm) which was eluted with solvent systems A1 (H3PO4/H2O, 1:1,000) and B1 (ACN/H2O/H3PO4, 900:100:1) at a flow rate of 1.0 ml/min and a 10 to 80% gradient of B1 over 20 min. The HPLC column eluates were monitored by their absorbance at 220 nm. The molecular weight and purity of synthesized peptides were verified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (TofSpec-E; Micromass).
Parasites, mammalian cells, and cell invasion assay.
T. cruzi G (34) was used in this study. Parasites were maintained by cyclic passage in mice and in axenic cultures in liver infusion tryptose medium. Metacyclic trypomastigotes were purified by passage through DEAE-cellulose columns, as described (35). HeLa cells, the human-carcinoma-derived epithelial cells, were grown at 37°C in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum (FCS), streptomycin (100 μg/ml), and penicillin (100 U/ml) in a humidified 5% CO2 atmosphere. Mammalian cell invasion assays were carried out as previously described (35) by seeding 106 metacyclic forms onto each well of 24-well plates containing 13-mm-diameter, round, glass coverslips coated with 2 × 105 HeLa cells. After 3 h of incubation at 37°C, the duplicate coverslips were washed in phosphate buffered saline (PBS), were pulsed for 1 min with water to lyse externally associated parasites, and were washed three times with PBS. The coverslips were fixed with methanol and were stained with Giemsa, and the number of intracellular parasites was counted in a total of 500 cells.
Antibodies.
MAb 3F6, directed to metacyclic trypomastigote gp82 (28), was purified on a protein A Sepharose column. Immune sera to recombinant protein J18a, containing the N-terminal gp82 sequence fused to GST, were prepared by immunizing BALB/c mice with 4 doses of purified antigen (10 μg per mouse) plus Al(OH)3 as adjuvant, at 2-week intervals. Ten days after the last immunizing dose, the animals were bled and the sera were stored at −20°C. This antiserum recognized all recombinant proteins analyzed in this study.
HeLa-cell-binding assay.
HeLa cells (3 × 104), placed in 96-well microtiter plates (Costar), were grown overnight at 37°C. After fixation with 4% paraformaldehyde in PBS, the cells were washed with PBS, were blocked with PBS containing 10% FCS (FCS-PBS) for 1 h at room temperature, and were incubated with varying concentrations of recombinant proteins in FCS-PBS. After 1 h at 37°C, the cells were washed in PBS and were sequentially incubated for 1 h at 37°C with anti-J18a antibody diluted in FCS-PBS, and anti-mouse immunoglobulin G conjugated to peroxidase. Following washes with PBS, the bound enzyme was revealed by using o-phenylenediamine, as described (23).
Inhibition of binding of recombinant gp82 to HeLa cells with synthetic peptides.
The assay was performed as described above in 96-well microtiter plates, with each well containing 3 × 104 HeLa cells. Cells were incubated for 1 h at 37°C with the recombinant protein J18 (10 μg/ml) in FCS-PBS, in the absence or presence of individual synthetic peptides (200 μg/ml) representing the gp82 central domain. Bound antibody was detected as described above.
Competitive antibody binding between gp82 recombinant protein and synthetic peptides.
Wells of microtiter plates were coated with recombinant protein J18b (1 μg/well), which contains the epitope for MAb 3F6. The plates were blocked with FCS-PBS for 1 h at room temperature and were then incubated with MAb 3F6 (1.6 μg/ml) in FCS-PBS, in the absence or presence of individual synthetic peptides based on the gp82 central domain. After 1 h at 37°C, the plates were washed in PBS and were incubated for 1 h at 37°C with anti-mouse immunoglobulin G conjugated to peroxidase. Following washes in PBS, the bound enzyme was revealed by using o-phenylenediamine as substrate.
RESULTS
Gp82 sequences containing acidic amino acid doublets are involved in host-cell adhesion.
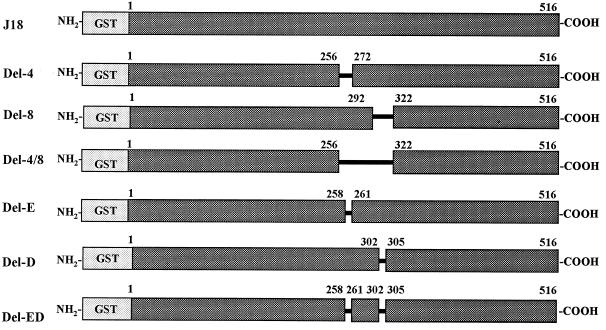
To define the cell-binding site of gp82, known to be located in the central domain of the molecule (23), binding assays were performed with recombinant constructs lacking different sequences of that domain (Fig. 1). Binding was assessed by incubating HeLa cells immobilized on the bottom of microtiter plates with increasing concentrations of purified recombinant proteins. As shown in Fig. 2A, the construct J18, containing the full-length gp82 sequence, bound to HeLa cells in a dose-dependent and saturable manner, whereas the construct Del-4/8, deleted of 65 residues (amino acids 257 to 321), had a negligible binding activity. Two other constructs with shorter deletions, Del-4 (257 to 271) and Del-8 (293 to 321), bound to HeLa cells, but to a significantly lesser degree than did J18 (Fig. 2A), suggesting that the portions deleted in these constructs participate in cell adhesion. Since the presence of a pair of contiguous acidic amino acids was a common feature of the deleted sequences in constructs Del-4 and Del-8, we investigated whether these residues played a role in cell adhesion by analyzing the binding capacity of recombinant proteins lacking either one or both pairs of acidic residues (Fig. 1). Removal of four residues in the construct Del-E/D, namely the glutamic acid (259/260) and aspartic acid (303/304) doublets, virtually abolished the cell adhesion capacity (Fig. 2B). A significant decrease in binding was also observed in Del-E, which lacks the glutamic acid pair, whereas Del-D, devoid of the aspartic acid pair (303/304), retained most of its binding capacity (Fig. 2B).
FIG. 1.
Schematic representation of the structure of T. cruzi gp82 recombinant proteins. Shown are the recombinant proteins fused to GST containing the full-length gp82 sequence (J18) or lacking varying numbers of amino acid residues at different positions in the central domain. The constructs Del-4, Del-8, and Del 4/8 lack, respectively, 15, 29, and 65 amino acid residues. Del-E and Del-D constructs are devoid of two glutamic and aspartic acid residues, respectively. Four acidic residues are deleted in the construct Del-E/D.
Consistent with the loss of cell-binding sites, the recombinant proteins Del-4/8 and Del-E/D failed to inhibit the parasite internalization, when added to HeLa cells in metacyclic trypomastigote invasion assays, in contrast to what is seen with construct J18 (Fig. 2C). The inhibitory effect of the recombinant protein J18 was dose dependent: at concentrations of 80, 40, 20, and 10 μg/ml, the inhibition of HeLa cell invasion was on the order of 80, 55, 40, and 15%, respectively.
A synthetic peptide formed by two noncontiguous gp82 sequences inhibits cell binding of gp82 and the parasite entry into host cells.
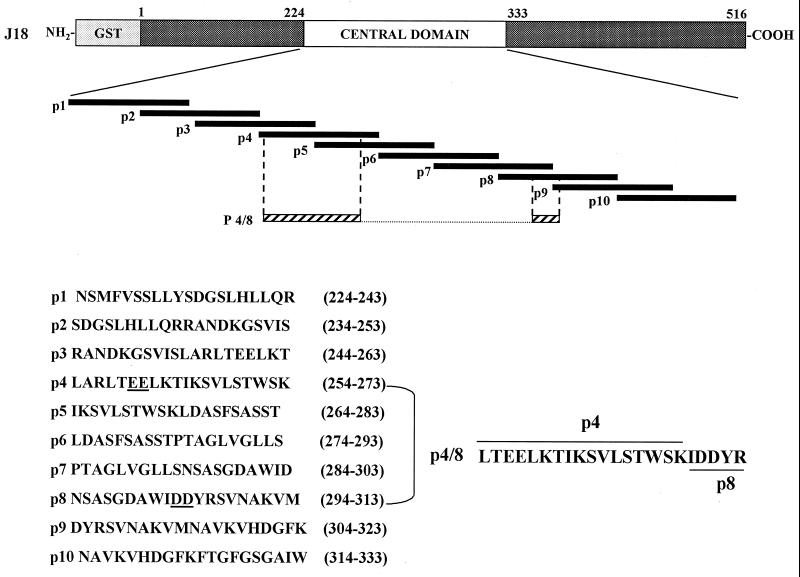
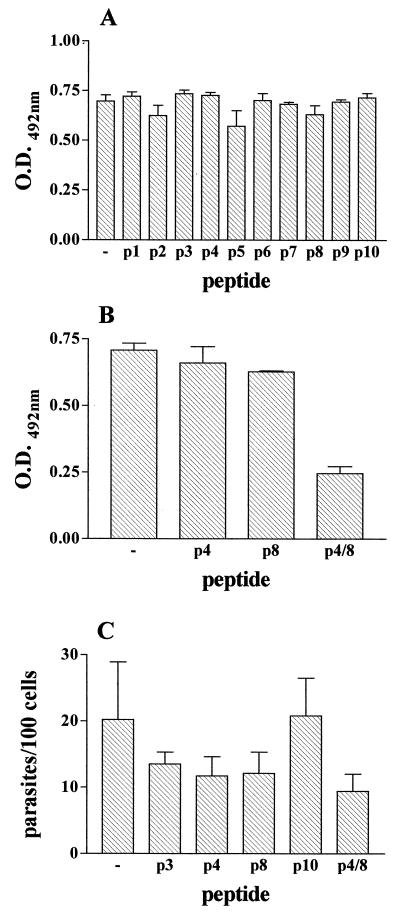
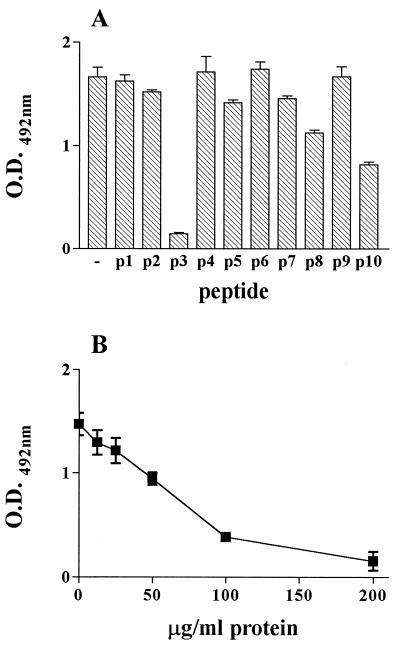
To further characterize the cell-binding site of gp82, we performed inhibition binding experiments by incubating HeLa cells with gp82 recombinant protein in the presence of individual peptides spanning the gp82 central domain (Fig. 3). None of the peptides significantly inhibited the binding of recombinant J18 to HeLa cells (Fig. 4A), including the peptides p4 and p8 which overlap with the sequences deleted in the recombinant proteins Del-4 and Del-8, respectively. As our data have shown that each of these sequences individually contributes to gp82 adhesion to target cells, but more efficient binding requires both sites (Fig. 2), we investigated the binding efficacy of a composite of p4 and p8 sequences. A 22-mer hybrid peptide p4/8 formed by a portion of p8 fused to p4, and containing the four acidic amino acids, was synthesized (Fig. 3) and was tested in experiments of inhibition of binding. We observed a significantly diminished binding of recombinant J18 to HeLa cells in the presence of peptide p4/8 (Fig. 4B). When added to HeLa cells in parasite invasion assays, in addition to p4/8, the peptides p4 and p8 displayed inhibitory activity (Fig. 4C). To determine whether peptides with the same amino acid composition of p4, p8, or p4/8, but with scrambled sequences, had any effect on host cell invasion, the following peptides: p4* (EWATIKVLTKSTERSLKSLL), p8* (GNMKWDVDNSRAVDASAIYS), p4/8* (SKTRLLDTELDVTKSIEYWKIS), and p4/8** (VSDLRITKDSYETKSKELWITL) were tested. Excepting p8*, which displayed some inhibitory activity, the tested peptides did not significantly affect parasite entry into HeLa cells, suggesting that binding is sequence dependent.
FIG. 3.
Amino acid composition and position in T. cruzi gp82 central domain of synthetic peptides. Shown are the 20-mer peptides with an overlap of 10 residues and 22-mer hybrid peptide p4/8 formed by two noncontiguous sequences containing parts of peptides p4 and p8.
FIG. 4.
Inhibitory effect of hybrid peptide p4/8 on binding of T. cruzi gp82 recombinant proteins to HeLa cells and on parasite invasion. In panels A and B, HeLa cells were incubated with the recombinant proteins J18 containing the full-length gp82 sequence, in the absence or presence of the indicated peptide, at 200 μg/ml. Binding was measured by ELISA. Representative results of one of three comparable experiments are shown. The values are the means ± SD of triplicates. (C) HeLa cells were incubated with metacyclic trypomastigotes in the presence of the peptide. After 3 h at 37°C, the cells were stained with Giemsa, and the number of intracellular parasites were counted in a total of 500 cells. Values are the means ± SD of three assays performed in duplicate. O.D., optical density.
We have noted that peptides p4 and p8 were unable to affect binding of gp82 to HeLa cells (Fig. 4B) but partially inhibited parasite invasion (Fig. 4C). This apparent discrepancy could be due to the fact that, in the binding assay, the peptides p4 and p8 are competing with soluble recombinant gp82, whereas in the invasion assay, the petides are competing with the gp82 molecule inserted in the parasite membrane, a circumstance that poses a considerable constraint and which may restrict the gp82 competitive capacity.
3F6 epitope has partial overlap with gp82 cell-binding sequence.
In order to map the 3F6 epitope of gp82, we performed assays of inhibition of antibody binding with the same set of synthetic peptides used for inhibition of gp82 binding to HeLa cells (Fig. 3). Microtiter plates coated with recombinant protein J18b (amino acids 224 to 516) were incubated with MAb 3F6 in the absence or presence of individual peptides at 200 μg/ml. Peptide p3 (244 to 263) inhibited binding of MAb 3F6 by ∼90% (Fig. 5A). Lesser peptide p3 concentrations had proportionally lower inhibitory effects (Fig. 5B). It should be noted that there is an overlap of 10 amino acids between peptide p3, representing the MAb 3F6 epitope, and peptide p4 containing the cell adhesion site (Fig. 3). This explains how MAb inhibits the penetration of metacyclic trypomastigotes into host cells (21). Although p3 contains the glutamic acid pair also found in p4, these amino acids do not appear to be part of 3F6 epitope. The reactivity of MAb 3F6 with the recombinant protein Del-4 or Del-E, which lacks the glutamic acid doublet, was similar to that with J18 or with other controls, such as recombinant proteins Del-8 and Del-D.
FIG. 5.
Mapping of 3F6 epitope. Assays of inhibition of MAb 3F6 binding were performed in microtiter plates coated with recombinant protein J18b, which were incubated in the absence or presence of the indicated synthetic peptide at 200 μg/ml (A) or with varying concentrations of peptide p3 (B). Binding of MAb 3F6 was measured by ELISA. Representative results of one out of three experiments are shown, and values are the means ± SD of triplicate samples. O.D., optical density.
Gp82 does not bind to serum components or to extracellular matrix proteins.
Since the cell-binding assay was performed in the presence of 10% FCS, in parallel experiments we determined whether the gp82 recombinant protein eventually bound to serum components. J18, at the same concentrations used in the HeLa cell-binding assays, was added to FCS-coated plates, and the reaction proceeded according to the standard protocol. No significant binding of J18 to FCS-coated plates was observed. On the other hand, as cell adhesion molecules of bloodstream-tissue culture trypomastigotes, such as Tc-85 and gp85 (1, 19), reportedly bind to laminin or fibronectin (10, 19), we investigated whether the metacyclic stage gp82 also bound to these extracellular matrix proteins. Laminin or fibronectin, at various concentrations, were added to microtiter plates coated with recombinant J18, and the binding was assessed with antibodies to laminin or fibronectin. Neither of these proteins bound to recombinant gp82 (data not shown). These results, added to the previous observation that gp82 does not bind heparan sulfate or collagen (21), confirm the lack of affinity of metacyclic trypomastigote gp82 for extracellular matrix proteins.
DISCUSSION
We scrutinized the central domain of the metacyclic trypomastigote surface glycoprotein gp82 in order to map the cell adhesion site(s) of this stage-specific molecule, which has been implicated in the process of parasite entry into mammalian cells (21, 23). The results of experiments of cell adhesion with deletion mutants of gp82 revealed two noncontiguous sequences as cell-binding sites. Deletion of either one of these regions affected the binding capacity of recombinant gp82, and deletion of the segment containing both regions virtually abolished the cell adhesion property (Fig. 2A). The first region encompasses amino acids 257 to 271 and the second region encompasses amino acids 293 to 321. These results with the deletion mutants are in agreement with a recent study with insertion mutants of LamB, an integral outer membrane protein of E. coli (17). In that study, gp82-derived 20-mer peptides, namely peptides p4 (254 to 273) and p8 (294 to 313), which display a considerable overlap with the cell-binding regions identified by the deletion mutants, were expressed as fusions to the LamB protein in a region known to be exposed on the protein surface (20). LamB carrying either peptide p4 or p8 bound to HeLa cells in a dose-dependent fashion, whereas the wild-type LamB, or LamB with an unrelated T. cruzi peptide inserted in the same position, lacked binding activity (20).
The gp82 segments LTEELKTIKSVLSTW and NSASGDAWIDDYRSVNAKVM, shared by peptides p4 and p8 and the sequences removed from the Del-4 and Del-8 mutants, contain several charged amino acids. Positively charged residues, interspersed in the two segments, were also present elsewhere in the gp82 central domain. However, the presence of doublets of acidic residues was a characteristic of these segments (Fig. 3). The relevance of these negatively charged residues in cell binding became evident in experiments with gp82 recombinant proteins with these amino acids deleted. Deletion of both glutamic acid and aspartic acid doublets effectively precluded binding of gp82 to HeLa cells (Fig. 2B). The effect of removing these acidic residues was comparable to that resulting from deletion of all 65 amino acids comprising segments p4 and p8 (Figs. 2A and 2B).
Of the two cell adhesion sites, p4 appears to contribute more to gp82 binding capacity than does p8. LamB carrying peptide p4 binds more efficiently to HeLa cells than LamB carrying peptide p8, and more bacteria expressing Lamb/p4 adhere to HeLa cells than do bacteria expressing LamB/p8 (20). This is consistent with our results showing that deletion of either the p4 segment or the glutamic acid doublet had a greater effect on the binding capacity of recombinant gp82 than deletion of the p8 segment or the aspartic acid pair (Fig. 2). Of further significance was the pattern of interaction of bacteria expressing LamB/p4 or LamB/p8 with HeLa cells, which was found to mimic the attachment of trypomastigotes to the edges of HeLa cells (16). Another pertinent observation regarding the p4 cell-binding site is its considerable overlap with peptide p3 (Fig. 3), which contains the epitope for MAb 3F6 (Fig. 5), an antibody that inhibits metacyclic trypomastigote entry into host cells (21). Based on evidences that the 3F6-epitope is contiguous to the cell-binding site of gp82, we presume that MAb 3F6 interferes with parasite interaction with host cells by steric hindrance of gp82 cell-binding site.
One question raised during our study was whether the native gp82 cell-binding site is conformation dependent. The cell adhesion sites represented by peptides p4 and p8 are separated in the primary sequence by a segment of 20 amino acids, of which 12 are hydrophobic residues and only one is charged. As each of these two sequences appeared to contribute to target cell adhesion, but more efficient binding required both sequences, we envisaged a scenario in which the intervening hydrophobic segment forms a loop in the protein interior in order to allow the juxtaposition of p4 and p8 sequences on the surface. Our finding that the hybrid peptide p4/8, formed by parts of p4 and p8 sequences, inhibited binding of the full-length gp82 recombinant protein, and parasite entry into target cells (Fig. 4) gives support to our hypothesis that the actual cell-binding site of gp82 depends on the conformation of this region.
To our knowledge, this is the first characterization of a cell-binding site of a T. cruzi surface glycoprotein. Other surface molecules with adhesive properties, including those in the size range of metacyclic trypomastigote gp82, have been described in tissue-culture-derived trypomastigotes (10, 11, 14, 19), but their cell recognition sites have not been defined. As these trypomastigote proteins, unlike the metacyclic gp82, bind to one or more components of the extracellular matrix such as laminin, fibronectin, heparan sulfate, and collagen, it is possible that their interaction with host cells is established through matrix proteins.
The gp82 sequence shares a considerable degree of homology with members of the gp85 family expressed in tissue culture or blood trypomastigotes (9, 11, 13, 26). Alignments by BLAST algorithm have revealed 60 to 65% identities between the central domain of metacyclic trypomastigote gp82 and the C-terminal region glycoproteins of the gp85 family and 69% identity with the C-terminal region of an amastigote surface protein. However, the differences are also significant, including substitutions of uncharged residues for lysine or arginine, substitutions of acidic residues for uncharged or positively charged amino acids, and substitutions of amino acids with polar side chains for those with nonpolar side chains and vice-versa. Of note is that such substitutions can be found within sequences identified as relevant for cell binding. Members of the trans-sialidase family, which share ∼40% identity with the metacyclic gp82 central region, showed differences which were more extensive.
Invasion of mammalian cells by T. cruzi requires the activation of signal transduction pathways in the parasite and the host cell (3, 7, 15, 27, 33). This requirement is fulfilled in metacyclic trypomastigotes by the binding of gp82 to its host cell receptor. In the target cells, gp82 triggers the signaling cascade leading to Ca2+ mobilization (7, 22), and it also signals to the parasite interior, inducing the activation of protein tryrosine kinase (8) and an increase in cytosolic free Ca2+ concentration (22). By engaging this key molecule to interact with host cells, the metacyclic trypomastigotes, which are deposited by the insect on the skin of the host and then reach the subjacent layer through the biting site, may have found the most efficient way to enter cells.
ACKNOWLEDGMENTS
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
We thank Victor Nussenzweig for critical reading of the manuscript.
REFERENCES
- 1.Alves M J M, Abuin G, Kuwajima V Y, Colli W. Partial inhibition of trypomastigote entry into cultured mammalian cells by monoclonal antibodies against a surface glycoprotein of Trypanosoma cruzi. Mol Biochem Parasitol. 1986;21:75–82. doi: 10.1016/0166-6851(86)90081-2. [DOI] [PubMed] [Google Scholar]
- 2.Araya J E, Cano M I, Yoshida N, Franco da Silveira J. Cloning and characterization of a gene for the stage-specific 82 kDa surface antigen of metacyclic trypomastigotes of Trypanosoma cruzi. Mol Biochem Parasitol. 1994;65:161–169. doi: 10.1016/0166-6851(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 3.Barr S C, Han W, Andrews N W, Lopez J W, Ball B, Pannabecker T L, Gilmour R F., Jr A factor from Trypanosoma cruzi induces repetitive cytosolic free Ca2+ transients in isolated primary canine cardiac myocytes. Infect Immun. 1996;64:1770–1777. doi: 10.1128/iai.64.5.1770-1777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burleigh B A, Andrews N W. A 120-kDa alkaline oligopeptidase from Trypanosoma cruzi is involved in the generation of a novel Ca2+-signaling factor for mammalian cells. J Biol Chem. 1995;270:5172–5180. doi: 10.1074/jbc.270.10.5172. [DOI] [PubMed] [Google Scholar]
- 5.Colli W. Trans-sialidase: a unique enzyme activity discovered in the protozoan Trypanosoma cruzi. FASEB J. 1993;7:1257–1264. doi: 10.1096/fasebj.7.13.8405811. [DOI] [PubMed] [Google Scholar]
- 6.Cross G A M, Takle G B. The surface trans-sialidase family of Trypanosoma cruzi. Annu Rev Microbiol. 1993;46:385–411. doi: 10.1146/annurev.mi.47.100193.002125. [DOI] [PubMed] [Google Scholar]
- 7.Dorta M L, Ferreira A T, Oshiro M E M, Yoshida N. Ca2+ signal induced by Trypanosoma cruzi metacyclic trypomastigote surface molecules implicated in mammalian cell invasion. Mol Biochem Parasitol. 1995;73:285–289. doi: 10.1016/0166-6851(94)00123-5. [DOI] [PubMed] [Google Scholar]
- 8.Favoreto S, Dorta M L, Yoshida N. Trypanosoma cruzi 175-kDa protein tryrosine phosphorylation is associated with host cell invasion. Exp Parasitol. 1998;89:188–194. doi: 10.1006/expr.1998.4285. [DOI] [PubMed] [Google Scholar]
- 9.Fouts D L, Ruef B J, Ridley P T, Wrightsman R A, Peterson D S, Manning J E. Nucleotide sequence and transcription of a trypomastigote surface antigen gene of Trypanosoma cruzi. Mol Biochem Parasitol. 1991;46:189–200. doi: 10.1016/0166-6851(91)90043-6. [DOI] [PubMed] [Google Scholar]
- 10.Giordano R, Chammas R, Veiga S S, Colli W, Alves M J M. An acidic component of the heterogeneous Tc-85 protein family from the surface of Trypanosoma cruzi is a laminin binding glycoprotein. Mol Biochem Parasitol. 1994;65:85–94. doi: 10.1016/0166-6851(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 11.Giordano R, Fouts D L, Tewari D, Colli W, Manning J E, Alves M J M. Cloning of a surface membrane glycoprotein specific for the infective form of Trypanosoma cruzi having adhesive properties to laminin. J Biol Chem. 1999;274:3461–3468. doi: 10.1074/jbc.274.6.3461. [DOI] [PubMed] [Google Scholar]
- 12.Hirata I Y, Cezari M H S, Nakaie C R, Boschcov P, Ito A S, Juliano M A, Juliano L. Internally quenched fluorogenic protease substrates: solid-phase synthesis and fluorescence spectroscopy of peptides containing ortho-amino benzoyl/dinitrophenyl groups as donor-acceptor pairs. Lett Pep Sci. 1994;1:299–308. [Google Scholar]
- 13.Khan S, Van Voorhis W C, Eisen H. The major 85-kDa surface antigen of the mammalian form of Trypanosoma cruzi is encoded by a large heterogeneous family of simultaneusly expressed genes. J Exp Med. 1990;172:589–597. doi: 10.1084/jem.172.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima M F, Villalta F. Host-cell attachment by Trypanosoma cruzi identification of an adhesion molecule. Biochem Biophys Res Commun. 1988;155:256–262. doi: 10.1016/s0006-291x(88)81077-5. [DOI] [PubMed] [Google Scholar]
- 15.Moreno S N J, Silva J, Vercesi A E, Docampo R. Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J Exp Med. 1994;180:1535–1540. doi: 10.1084/jem.180.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortara R A. Trypanosoma cruzi: amastigotes and trypomastigotes interact with different structures on the surface of HeLa cells. Exp Parasitol. 1991;73:1–14. doi: 10.1016/0014-4894(91)90002-e. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega-Barria E, Pereira M E A. A novel Trypanosoma cruzi heparin binding protein promotes fibroblast adhesion and penetration on engineered bacteria and trypanosomes into mammalian cells. Cell. 1991;67:411–421. doi: 10.1016/0092-8674(91)90192-2. [DOI] [PubMed] [Google Scholar]
- 19.Ouaissi M A, Cornette J, Capron A. Identification and isolation of Trypanosoma cruzi trypomastigote cell surface protein with properties expected of a fibronectin receptor. Mol Biochem Parasitol. 1986;19:201–211. doi: 10.1016/0166-6851(86)90002-2. [DOI] [PubMed] [Google Scholar]
- 20.Pereira C M, Favoreto S, Jr, Silveira J F, Yoshida N, Castillho B A. Adhesion of Escherichia coli to HeLa cells mediated by Trypanosoma cruzi surface glycoprotein-derived peptides inserted in the outer membrane protein LamB. Infect Immun. 1999;67:4908–4911. doi: 10.1128/iai.67.9.4908-4911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez M I, Ruiz R C, Araya J E, Franco da Silveira J, Yoshida N. Involvement of the stage-specific 82-kilodalton adhesion molecule of Trypanosoma cruzi metacyclic trypomastigotes in host cell invasion. Infect Immun. 1993;61:3636–3641. doi: 10.1128/iai.61.9.3636-3641.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz R C, Favoreto S, Jr, Dorta M L, Oshiro M E M, Ferreira A T, Manque P M, Yoshida N. Infectivity of Trypanosoma cruzi strains is associated with differential expression of surface glycoproteins with differential Ca2+ signaling activity. Biochem J. 1998;330:505–511. doi: 10.1042/bj3300505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santori F R, Dorta M L, Juliano L, Juliano M A, Franco da Silveira J, Ruiz R C, Yoshida N. Identification of a domain of Trypanosoma cruzi metacyclic trypomastigote surface molecule gp62 required for attachment and invasion of mammalian cells. Mol Biochem Parasitol. 1996;78:209–216. doi: 10.1016/s0166-6851(96)02626-6. [DOI] [PubMed] [Google Scholar]
- 24.Schenkman S, Eichinger D, Pereira M E A, Nussenzweig V. Structural and functional properties of Trypanosoma trans-sialidase. Annu Rev Microbiol. 1994;48:499–523. doi: 10.1146/annurev.mi.48.100194.002435. [DOI] [PubMed] [Google Scholar]
- 25.Smith D B, Johnson K S. Single step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;6:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 26.Takle G B, Cross G A M. An 85-kilodalton surface antigen gene family of Trypanosoma cruzi encodes polypeptides homologous to bacterial neuraminidases. Mol Biochem Parasitol. 1991;48:185–198. doi: 10.1016/0166-6851(91)90114-l. [DOI] [PubMed] [Google Scholar]
- 27.Tardieux I, Nathanson M H, Andrews N W. Role in host cell invasion of Trypanosoma cruzi-induced cytosolic free Ca2+ transients. J Exp Med. 1994;179:1017–1022. doi: 10.1084/jem.179.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teixeira M M G, Yoshida N. Stage-specific surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi identified by monoclonal antibodies. Mol Biochem Parasitol. 1986;18:271–282. doi: 10.1016/0166-6851(86)90085-x. [DOI] [PubMed] [Google Scholar]
- 29.Vieira M C F, Carvalho T U, Souza W. Effect of protein kinase inhibitors on the invasion process of macrophages by Trypanosoma cruzi. Biochem Biophys Res Commun. 1994;203:967–971. doi: 10.1006/bbrc.1994.2276. [DOI] [PubMed] [Google Scholar]
- 30.Villalta F, Zhang Y, Bibb K E, Burns J M, Lima M F. Signal transduction in human macrophages by gp83 ligand of Trypanosoma cruz: trypomastigote gp83 ligand up-regulates trypanosome entry through the MAP kinase pathway. Biochem Biophys Res Commun. 1998;249:247–252. doi: 10.1006/bbrc.1998.9127. [DOI] [PubMed] [Google Scholar]
- 31.Wheatley M. Peptide mapping and the generation and isolation of sequentiable peptides from receptors. In: Hulme E C, editor. Receptor biochemistry: a practical approach. Oxford, England: IRL Press; 1993. pp. 213–261. [Google Scholar]
- 32.Wilkowsky S E, Wainszelbaum M J, Isola E L D. Trypanosoma cruzi: participation of intracellular Ca2+ during metacyclic trypomastigote-macrophage interaction. Biochem Biophys Res Commun. 1996;222:386–389. doi: 10.1006/bbrc.1996.0753. [DOI] [PubMed] [Google Scholar]
- 33.Yakubu M A, Majumder S, Kierszenbaum F. Changes in Trypanosoma cruzi infectivity by treatments that affect calcium ion levels. Mol Biochem Parasitol. 1994;66:119–125. doi: 10.1016/0166-6851(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida N. Surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi. Infect Immun. 1983;40:836–839. doi: 10.1128/iai.40.2.836-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida N, Mortara R A, Araguth M F, Gonzalez J C, Russo M. Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect Immun. 1989;57:1663–1667. doi: 10.1128/iai.57.6.1663-1667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]