1 Summary
Bloodstream infection with Bacillus cereus/thuringiensis can be life threatening, particularly in patients who are severely immunocompromised. In this report we describe a case that progressed from asymptomatic to fatal over approximately 5 hours despite extensive resuscitation efforts. We identify the pathogen and assemble its genome, in which we find genes for toxins that may have contributed to the precipitous demise. In the context of this and other cases we discuss the possible indication for rapid appropriate antibiotic administration and potentially antitoxin treatment or toxin removal in fulminant illness in immunocompromised patients.
Keywords: Bacillus cereus/thuringiensis, sepsis, immunocompromise, induction chemotherapy, hematologic malignancy
2. History
A previously healthy 5 year old female presented with neck and facial swelling, gum bleeding and petechiae. She was found to have leukocytosis (white blood cell count 56.7 K/cu mm, reference range: 6.0 – 17.0 K/cu mm), anemia (hemoglobin 3.4 g/dL, reference range: 11.6 – 13.6 g/dL), and thrombocytopenia (platelet count 4 K/cu mm, reference range: 150 – 350 K/cu mm). Flow cytometry on peripheral blood revealed a population of blasts comprising 90% of cellularity with a phenotype consistent with precursor B-cell acute lymphoblastic leukemia (pre-B ALL). She was started on induction chemotherapy and empiric intravenous cefepime for a febrile episode during blood transfusion. Her hospital course was otherwise stable and cefepime was discontinued on hospital day 13 with plans for discharge and administration of chemotherapy as an outpatient if afebrile after 48 hours.
Approximately 18 hours after her last cefepime dose, she began to experience abdominal discomfort though she had stable vital signs (temperature 37.9 C) and physical examination. Routine labs drawn approximately 30 minutes prior showed newly elevated transaminases (aspartate amino transferase 256 U/L, reference range: 0 – 31 U/L; alanine amino transferase 194 U/L, reference range: 0 – 31 U/L). Approximately two hours from her initial abdominal discomfort, she became febrile to 38.6 C. Within thirty minutes of becoming febrile she acutely decompensated with alterations in mental status, a dusky appearance, and worsening vital signs including tachycardia (heart rate 200 beats per minute), tachypnea (respirations 54 breaths per minute), hypoxia (oxygen saturation 64% on room air), and hypotension (blood pressure 70/43 mm Hg). Blood cultures were drawn and she received a bolus of normal saline and intravenous cefepime. Pediatric advanced life support was initiated for unstable bradycardia with progression to pulseless electrical activity. Chest radiograph showed bilateral hazy opacities with new bilateral pleural effusions. Extracorporeal membrane oxygenation was initiated. Her abdomen became increasingly rigid and distended, raising concern for abdominal compartment syndrome. Bedside decompressive laparotomy was performed revealing an engorged liver without hemoperitoneum. After approximately three hours of cardiopulmonary resuscitation, the decision was made to withdraw resuscitation efforts.
Autopsy was performed at an interval of approximately 8 hours. Externally there was edema and the skin showed scattered ecchymoses and petechiae. The thoracic cavity showed bilateral pleural effusions, pneumomediastinum, and a pericardial effusion. The lungs were approximately twice normal weight and showed patchy gray-brown discoloration. The small intestine progressed from a grossly unremarkable appearance proximally to wall hemorrhage and ischemic changes distally. The large intestine had patchy hemorrhage with extensive pneumatosis intestinalis in the transverse colon. The liver was enlarged with mottling, discoloration, and firm areas. There was no thrombus in the inferior vena cava, hepatic vein, or portal vein. There was scattered soft tissue and organ hemorrhage involving the mediastinum, diaphragm, heart, mesentery, pancreas, right kidney, adrenal glands, bladder, ovaries, and brain.
On microscopic examination, bacilli were present in the trachea, bronchi, both lungs, grossly abnormal portions of small intestine, liver, pancreas, soft tissue adjacent to the thyroid, splenic red pulp, and intracranial blood vessels. The lungs also showed vascular congestion, patchy hemorrhage, and proteinaceous debris filling the alveoli. The bacilli in the liver filled sinusoids in foci, and surrounding hepatocytes showed necrosis. The scattered bacilli in the pancreas were surrounded by acinar cell necrosis. The kidneys showed diffuse acute tubular injury, consistent with prolonged hypotension. Despite the extensive involvement of multiple organs with bacilli, there was no associated acute or chronic inflammation, consistent with the patient’s neutropenic state and likely inability to mount an immune response. Figure 1 demonstrates key microscopic findings.
Figure 1: Histology demonstrates bacilli but no inflammatory response.
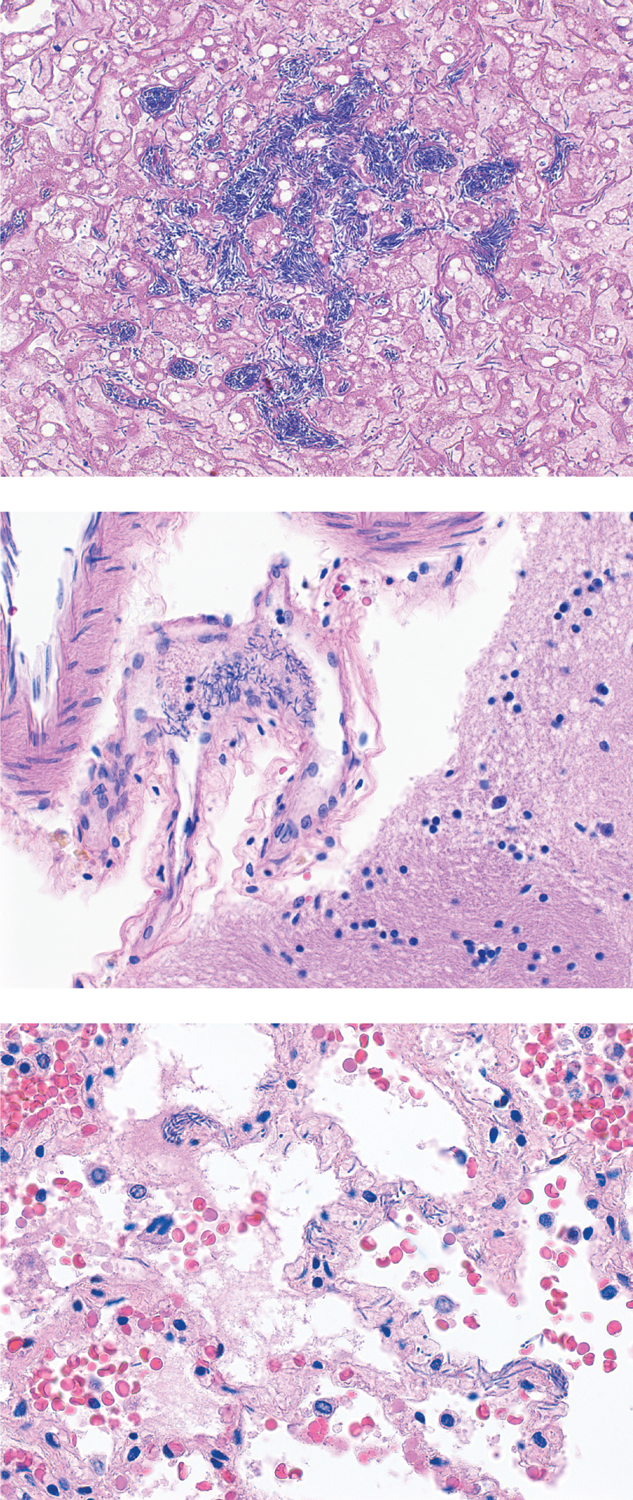
Top: Abundant organisms in hepatic sinusoids. Original magnification 160x. Middle: Bacilli in fibrin thrombi in leptomeningeal vessel, brain. Original magnification 260x. Bottom: Bacilli in alveolar membranes of lung. In these areas as well as all other areas with bacilli examined, no cellular immune response was present. Original magnification 260x. All panels hematoxylin and eosin stain.
3. Identification of the pathogen, assembly of its genome, and identification of toxin genes present
Blood cultures
Two Bacillus species were recovered from blood cultures drawn at the same time during the resuscitation effort from two separate sites. Both blood bottles (BACTEC-FX, Becton Dickinson, Sparks, MD) produced a positive growth signal at less than 24 hours of incubation at 35°C. Gram stain of the blood bottles demonstrated large gram positive rods. The blood bottles were subcultured to 5% sheep blood agar (Becton-Dickinson). Good growth was observed after approximately 24 hours of incubation at 35°C in 5% C02. Colonies demonstrated beta hemolysis and were large, dull, slightly yellow, flat, with a granular texture, and with a spready edge. Further characterization showed the isolates to be positive for catalase and motility (agar based test).
Gas liquid chromatography (GLC)
Initial identification was attempted using GLC analysis of cellular fatty acid (CFA) content. Further analysis of the CFA content was performed using the MIDI system (MIDI Inc., Newark DE). This software compares CFA profiles of unknown organisms against a database of profiles associated with known organisms and produces a report of possible matches accompanied by a confidence score (i.e., similarity index) from zero to 1.0 where 1.0 is a perfect match. Both isolates were identified as Bacillus thuringiensis with similarity indices of .519 and .545 with no second choices.
MALDI TOF Mass spectrometry
Additional identification was performed using matrix assisted laser desorption ionization time of flight mass spectrometry analysis (MALDI-TOF MS, BioTyper, Bruker Daltonics, Billerica, MA). The isolate from lumen number 1 was identified as B. thuringiensis while the isolate from lumen number 2 was identified as B. cereus. Because both GLC and MALDI-TOF MS cannot reliably differentiate between B. cereus and B. thuringiensis the final identification of the 2 isolates was reported as a non-anthracis member of the Bacillus cereus/thuringiensis group. Supplementary Figure 1 shows MALDI TOF MS results.
Sequencing
Sanger sequencing was performed on the first 500bp of the 16S rRNA gene using procedures and reagents from Applied Biosystems Inc. (Thermo Fisher, Waltham MA). Sequence editing, text file generation, and database matching were performed using SmartGene software (Integrated Database Network Systems, Raleigh NC). Identification was determined following guidelines outlined in the Clinical and Laboratory Standards Institute (CLSI, Wayne, PA) document MM18. Species-level identification was considered acceptable when agreement between multiple reference strains and the clinical isolate was 99% or greater. Sanger sequencing on both isolates also indicated similarity to B. cereus, B. thuringiensis, and B. anthracis (ruled out due to phenotypic properties) as well as B. toyonensis, B. wiedmannii, B. proteolyticus, B. albus, B. tropicus, and B. pacificus, further highlighting the difficulty of distinguishing species within this group.
Assembly
A run of an Oxford Nanopore (ONT) MinION sequencer was performed using DNA isolated from the bacterium after growth in culture. This generated 6,117,476 reads with an N50 (weighted average) length of 2189 bp, for a total of 11.1 billion bases, with an average GC content of 36.0%. The ONT reads were assembled using the Canu assembler [1], after first downsampling the reads to 200X coverage, with 100x coverage for reads longer than 5 Kb. The Canu assembly contained 14 contigs ranging in size from 2,855 bp to 4,790,273 bp. The contigs were aligned to one another, overlapping contigs merged, and redundant ones removed, thereby creating a complete, circular assembly of the main chromosome and 2 plasmids. Separately, unpaired 151-bp Illumina reads were acquired from a metagenomics sample taken from tissue collected at autopsy and embedded in paraffin for microscopic examination. After removing human reads, 16.1 million reads remained. These reads were aligned to the Canu assembly, identifying 390,719 Illumina reads (59 Mbp) that matched the assembly. These high-quality Illumina reads were used together with the ONT reads to polish the assembly using Racon [2] and Pilon [3]. The final, polished assembly contains one complete chromosome and two plasmids, with the following lengths: 5,323,903 bp, 509,015 bp, and 295,504 bp. Re-alignment of reads to each of these molecules shows approximately equal coverage, suggesting that the ratio of plasmids to the chromosome is 1:1:1. The genome was aligned to all complete bacterial genomes at NCBI, and the closest match to the chromosome, at 99.75% identity, was B. cereus G9842 (accession NC_011772.1). The closest match to plasmid 1, at 98.3% identity, was B. thuringiensis serovar thuringiensis str. IS5056 plasmid pIS56–233 (accession NC_020394.1). The closest match to plasmid 2, at 93.9% identity, was B. anthracis str. CDC 684 plasmid pXO1 (accession NC_012591.1). The genome was then annotated with Prokka [4] and Prodigal [5], as well as with the NCBI automated pipeline PGAP. The PGAP annotation was chosen as the final annotation. This identified 5,376 protein coding genes, of which 4,611 had named functions and 765 were hypothetical proteins. Annotation also identified 518 pseudogenes, 39 rRNAs, 44 binding sites, 107 tRNAs, 43 regulatory regions, 4 ncRNAs, 2 CRISPR genes, and 1 transfer-messenger RNA (tmRNA). All assemblies and annotation have been deposited in NCBI under BioProject PRJNA591929.
To identify toxins that could have contributed to the rapid course, we compared the genes identified in our assembly with recognized B. cereus toxins [6]. Present were genes for hemolysin B subunit A (hblA), hemolysin B subunit C (hblC), nonhemolytic enterotoxin B (nheB), nonhemolytic enterotoxin C (nheC), cytotoxin K (cytK), and phospholipase C (plcA). While the pXO1 plasmid has been associated with toxicity edema factor (cya), lethal factor (lef), and protective antigen (pagA) [7], we did not find genes for those toxins.
4. Discussion
The Bacillus cereus group consists of eight species of spore-forming, gram-positive bacilli with highly similar genomes [8], which can preclude definitive identification to the species level. While species in this group are found endemic to soil, the most notable member, B. anthracis, is a zoonotic infectious agent most often spread by human contact with infected animals or animal products [9]. In contrast to B. anthracis infection, which may result in cutaneous, gastrointestinal, or inhalational disease, Bacillus species other than anthracis were previously regarded as nonpathogenic. However, these species can act as human pathogens [10], and B. cereus and B. thuringiensis are increasingly recognized as etiologic agents of non-gastrointestinal human disease, including local cutaneous and ocular infections, respiratory infections, endocarditis, [10], and sepsis [11].
Non-gastrointestinal B. cereus infections are most often seen in patients who have undergone organ transplantation [12] or with other immunocompromised states such as solid organ malignancy, end-stage renal disease, polytrauma, intravenous drug use, or hematologic malignancy [13]. Patients in this last group, particularly those undergoing induction chemotherapy, are at increased risk for rapidly-progressive infection [14–17]. Several of the reported cases with fulminant B. cereus septicemia had clinical presentations similar to our patient’s, with a rapidly progressive course, sudden onset of fever, abdominal pain, and vomiting. B. cereus sepsis was associated with hemolysis in two leukemic patients, both of whom had abdominal pain prior to rapid clinical deterioration [16]. The phenomenon may not be widely appreciated, and it has been suggested that referring to this clinical picture in neutropenic patients as ‘fulminant septicemia syndrome of Bacillus’ could increase awareness [18]. ‘Bacillus sepsis’ might be a simpler alternative.
Identifying the source of B. cereus infection can be difficult. Our patient had an indwelling line, which is associated with increased risk for disseminated infection due to colonization [19]. However, often no definitive source is identified, and many cases are attributed to the patient’s endogenous flora or gastrointestinal colonization [13, 14]. Hospital outbreaks of Bacillus sepsis have been traced to contaminated vials of calcium gluconate [19, 20], bananas (for leukemic patients) [21], tea [22], linens [23, 24], and colonization of mechanical ventilation equipment [25–27].
While the pathogenicity of Bacillus species may be attributed to any of multiple toxins and virulence factors, B. cereus produces no unique pathogenic factors allowing for distinction from other members of the group [28]. Among its potential toxins are hemolysins, phospholipases, cereulide, and enterotoxins (HBL, nhe, and cytK) [10]. Both HBL and nhe are tripartite toxins, comprised of three proteins, each encoded by a separate gene. B. cereus hemolysin BL has hemolytic, dermonecrotic, vascular permeability, and enterotoxic activity [29]. Large doses of the emetic cereulide [30] can be fatal [31]. Phylogenetic analysis has suggested a degree of relationship among B. cereus strains that cause disease [32]. Hemolysin BL shows a high degree of heterogeneity among B. cereus strains [33,34] and the presence or absence of HBL genes correlates closely with groupings of B. cereus isolates [35]. However, in studies comparing genome and clinical behavior of a number of pathologic B. cereus isolates, no relation was found between the presence of toxins and the clinical manifestations [36], highlighting the difficulty of attributing a particular clinical course to individual toxin genes. In addition to chromosomally encoded proteins and virulence factors, strains of B. cereus containing the B. anthracis plasmid pXO1 have been recovered from patients with pulmonary infections resembling inhalational anthrax [37].
Bacillus thuringiensis is a widely used biopesticide [38] and can cause food poisoning, which is often misattributed to B. cereus [39]. Early epidemiologic study in areas where B. thuringiensis was sprayed for crop protection raised concerns that immunocompromised individuals may have become infected [40], and it is now recognized as critical to monitor the strains of B. thuringiensis used in pest control to prevent human exposure to toxins [41]. Study of insecticidal properties of proteins produced by B. cereus/thuringiensis group bacteria have identified many toxins, and suggest that additional proteins not yet discovered also act as such to insects [42]. These or other proteins could contribute to toxicity during overwhelming infection in the setting of immunocompromise. Further work is needed to better identify toxins contributing to the fulminant course of B. cereus/thuringiensis sepsis in immunocompromised patients, so that appropriate therapies [43] can be developed.
Supplementary Material
Footnotes
The authors affirm that they have no conflicts of interest to report.
The parents of the child presented in this report affirmed in writing their support for its publication.
References
- [1].Koren Sergey, Walenz Brian P, Berlin Konstantin, Miller Jason R, Bergman Nicholas H, and Phillippy Adam M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res, 27(5):722–736, 05 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vaser Robert, Sović Ivan, Nagarajan Niranjan, and Šikić Mile. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res, 27(5):737–746, 05 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Walker Bruce J, Abeel Thomas, Shea Terrance, Priest Margaret, Abouelliel Amr, Sakthikumar Sharadha, Cuomo Christina A, Zeng Qiandong, Wortman Jennifer, Young Sarah K, and Earl Ashlee M. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One, 9(11):e112963, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seemann Torsten. Prokka: rapid prokaryotic genome annotation. Bioinformatics, 30(14):2068–9, Jul 2014. [DOI] [PubMed] [Google Scholar]
- [5].Hyatt Doug, Chen Gwo-Liang, Locascio Philip F, Land Miriam L, Larimer Frank W, and Hauser Loren J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11:119, Mar 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wehrle Esther, Moravek Maximilian, Dietrich Richard, Bürk Christine, Didier Andrea, and Märtlbauer Erwin. Comparison of multiplex PCR, enzyme immunoassay and cell culture methods for the detection of enterotoxinogenic Bacillus cereus. J Microbiol Methods, 78(3):265–70, Sep 2009. [DOI] [PubMed] [Google Scholar]
- [7].Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, Manter D, Martinez Y, Ricke D, Svensson R, and Jackson PJ. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol, 181(20):6509–15, Oct 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ehling-Schulz Monika, Lereclus Didier, and Koehler Theresa M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol Spectr, 7(3), May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Spencer RC. Bacillus anthracis. J Clin Pathol, 56(3):182–7, Mar 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bottone Edward J. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev, 23(2):382–98, Apr 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Drobniewski FA. Bacillus cereus and related species. Clin Microbiol Rev, 6(4):324–38, Oct 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Flavelle Shauna, Tyrrell Gregory J, and Forgie Sarah E. A ‘serious’ bloodstream infection in an infant. Can J Infect Dis Med Microbiol, 18(5):311–2, Sep 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tusgul S, Prod’hom G, Senn L, Meuli R, Bochud P-Y, and Giulieri SG. Bacillus cereus bacteraemia: comparison between haematologic and nonhaematologic patients. New Microbes New Infect, 15:65–71, Jan 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christenson JC, Byington C, Korgenski EK, Adderson EE, Bruggers C, Adams RH, Jenkins E, Hohmann S, Carroll K, Daly JA, and Pavia AT. Bacillus cereus infections among oncology patients at a children’s hospital. Am J Infect Control, 27(6):543–6, Dec 1999. [DOI] [PubMed] [Google Scholar]
- [15].Chou Ya-Ling, Cheng Shin-Nan, Hsieh Kao-Hsian, Wang Chih-Chien, Chen Shyi-Jou, and Lo Wen-Tsung. Bacillus cereus septicemia in a patient with acute lymphoblastic leukemia: A case report and review of the literature . Journal of Microbiology, Immunology and Infection, 49(3):448–451, 2016. [DOI] [PubMed] [Google Scholar]
- [16].Arnaout MK, Tamburro RF, Bodner SM, Sandlund JT, Rivera GK, Pui CH, and Ribeiro RC. Bacillus cereus causing fulminant sepsis and hemolysis in two patients with acute leukemia. J Pediatr Hematol Oncol, 21(5):431–5, 1999. [DOI] [PubMed] [Google Scholar]
- [17].Katsuya Hiroo, Takata Tohru, Ishikawa Takahiko, Sasaki Hidenori, Ishitsuka Kenji, Takamatsu Yasushi, and Tamura Kazuo. A patient with acute myeloid leukemia who developed fatal pneumonia caused by carbapenem-resistant Bacillus cereus. J Infect Chemother, 15(1):39–41, Feb 2009. [DOI] [PubMed] [Google Scholar]
- [18].Akiyama N, Mitani K, Tanaka Y, Hanazono Y, Motoi N, Zarkovic M, Tange T, Hirai H, and Yazaki Y. Fulminant septicemic syndrome of Bacillus cereus in a leukemic patient. Intern Med, 36(3):221–6, Mar 1997. [DOI] [PubMed] [Google Scholar]
- [19].Kutsuna Satoshi, Hayakawa Kayoko, Kita Kazuya, Katanami Yuichi, Imakita Natsuko, Kasahara Kei, Seto Masami, Akazawa Kenichiro, Shimizu Minoru, Kano Toshikazu, Nei Takahito, Hayashi Tetsuro, Mori Nobuaki, Yabuki Taku, and Ohmagari Norio. Risk factors of catheter-related bloodstream infection caused by Bacillus cereus: Case-control study in 8 teaching hospitals in Japan. Am J Infect Control, 45(11):1281–1283, Nov 2017. [DOI] [PubMed] [Google Scholar]
- [20].Thuler LC, Velasco E, de Souza Martins CA, de Faria LM, da Fonseca NP, Dias LM, and Gonçalves VM. An outbreak of Bacillus species in a cancer hospital. Infect Control Hosp Epidemiol, 19(11):856–8, Nov 1998. [DOI] [PubMed] [Google Scholar]
- [21].Rhee Chanu, Klompas Michael, Tamburini Fiona B, Fremin Brayon J, Chea Nora, Epstein Lauren, Halpin Alison Laufer, Guh Alice, Gallen Rachel, Coulliette Angela, Gee Jay, Hsieh Candace, Desjardins Christopher A, Pedamullu Chandra Sekhar, DeAngelo Daniel J, Manzo Veronica E, Folkerth Rebecca Dunn, Milner Danny A Jr, Pecora Nicole, Osborne Matthew, Chalifoux-Judge Diane, Bhatt Ami S, and Yokoe Deborah S. Epidemiologic Investigation of a Cluster of Neuroinvasive Bacillus cereus Infections in 5 Patients With Acute Myelogenous Leukemia. Open Forum Infect Dis, 2(3):ofv096, Sep 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].El Saleeby CM, Howard SC, Hayden RT, and McCullers JA. Association between tea ingestion and invasive Bacillus cereus infection among children with cancer. Clin Infect Dis, 39(10):1536–9, Nov 2004. [DOI] [PubMed] [Google Scholar]
- [23].Sasahara T, Hayashi S, Morisawa Y, Sakihama T, Yoshimura A, and Hirai Y. Bacillus cereus bacteremia outbreak due to contaminated hospital linens. Eur J Clin Microbiol Infect Dis, 30(2):219–26, Feb 2011. [DOI] [PubMed] [Google Scholar]
- [24].Barrie D, Wilson JA, Hoffman PN, and Kramer JM. Bacillus cereus meningitis in two neurosurgical patients: an investigation into the source of the organism. J Infect, 25(3):291–7, Nov 1992. [DOI] [PubMed] [Google Scholar]
- [25].Van Der Zwet WC, Parlevliet GA, Savelkoul PH, Stoof J, Kaiser AM, Van Furth AM, and Vandenbroucke-Grauls CM. Outbreak of Bacillus cereus infections in a neonatal intensive care unit traced to balloons used in manual ventilation. J Clin Microbiol, 38(11):4131–6, Nov 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bryce EA, Smith JA, Tweeddale M, Andruschak BJ, and Maxwell MR. Dissemination of Bacillus cereus in an intensive care unit. Infect Control Hosp Epidemiol, 14(8):459–62, Aug 1993. [DOI] [PubMed] [Google Scholar]
- [27].Kalpoe JS, Hogenbirk K, van Maarseveen NM, Gesink-Van der Veer BJ, Kraakman MEM, Maarleveld JJ, van der Reyden TJK, Dijkshoorn L, and Bernards AT. Dissemination of Bacillus cereus in a paediatric intensive care unit traced to insufficient disinfection of reusable ventilator air-flow sensors. J Hosp Infect, 68(4):341–7, Apr 2008. [DOI] [PubMed] [Google Scholar]
- [28].Senesi Sonia and Ghelardi Emilia. Production, secretion and biological activity of bacillus cereus enterotoxins. Toxins (Basel), 2(7):1690–703, July 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Beecher DJ, Schoeni JL, and Wong AC. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect Immun, 63(11):4423–8, Nov 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Agata N, Ohta M, Mori M, and Isobe M. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol Lett, 129(1):17–20, Jun 1995. [DOI] [PubMed] [Google Scholar]
- [31].Naranjo María, Denayer Sarah, Botteldoorn Nadine, Delbrassinne Laurence, Veys Jean, Waegenaere Jacques, Sirtaine Nicolas, Driesen Ronald B, Sipido Karin R, Mahillon Jacques, and Dierick Katelijne. Sudden death of a young adult associated with Bacillus cereus food poisoning. J Clin Microbiol, 49(12):4379–81, Dec 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vassileva Maria, Torii Keizo, Oshimoto Megumi, Okamoto Akira, Agata Norio, Yamada Keiko, Hasegawa Tadao, and Ohta Michio. Phylogenetic analysis of Bacillus cereus isolates from severe systemic infections using multilocus sequence typing scheme. Microbiol Immunol, 50(9):743–9, 2006. [DOI] [PubMed] [Google Scholar]
- [33].Schoeni JL and Wong AC. Heterogeneity observed in the components of hemolysin BL, an enterotoxin produced by Bacillus cereus. Int J Food Microbiol, 53(2–3):159–67, Dec 1999. [DOI] [PubMed] [Google Scholar]
- [34].Thaenthanee Suwicha, Lee Wong Amy C, and Panbangred Watanalai. Phenotypic and genotypic comparisons reveal a broad distribution and heterogeneity of hemolysin BL genes among Bacillus cereus isolates. Int J Food Microbiol, 105(2):203–12, Nov 2005. [DOI] [PubMed] [Google Scholar]
- [35].Okutani Akiko, Inoue Satoshi, Noguchi Akira, Kaku Yoshihiro, and Morikawa Shigeru. Whole-genome sequence-based comparison and profiling of virulence-associated genes of Bacillus cereus group isolates from diverse sources in Japan. BMC Microbiol, 19(1):296, 12 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chang T, Rosch JW, Gu Z, Hakim H, Hewitt C, Gaur A, Wu G, and Hayden RT. Whole-Genome Characterization of Bacillus cereus Associated with Specific Disease Manifestations. Infect Immun, 86(2), February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hoffmaster Alex R, Ravel Jacques, Rasko David A, Chapman Gail D, Chute Michael D, Marston Chung K, De Barun K, Sacchi Claudio T, Fitzgerald Collette, Mayer Leonard W, Maiden Martin C J, Priest Fergus G, Barker Margaret, Jiang Lingxia, Cer Regina Z, Rilstone Jennifer, Peterson Scott N, Weyant Robbin S, Galloway Darrell R, Read Timothy D, Popovic Tanja, and Fraser Claire M. Identification of anthrax toxin genes in a bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A, 101(22):8449–54, Jun 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ibrahim Mohamed A, Griko Natalya, Junker Matthew, and Bulla Lee A. Bacillus thuringiensis: a genomics and proteomics perspective. Bioeng Bugs, 1(1):31–50, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].te Giffel MC, Beumer RR, Klijn N, Wagendorp A, and Rombouts FM. Discrimination between Bacillus cereus and Bacillus thuringiensis using specific DNA probes based on variable regions of 16S rRNA. FEMS Microbiol Lett, 146(1):47–51, Jan 1997. [DOI] [PubMed] [Google Scholar]
- [40].Green M, Heumann M, Sokolow R, Foster LR, Bryant R, and Skeels M. Public health implications of the microbial pesticide Bacillus thuringiensis: an epidemiological study, Oregon, 1985–86. Am J Public Health, 80(7):848–52, Jul 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Granum PE and Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol Lett, 157(2):223–8, Dec 1997. [DOI] [PubMed] [Google Scholar]
- [42].Palma Leopoldo, Muñoz Delia, Berry Colin, Murillo Jesús, and Caballero Primitivo. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel), 6(12):3296–325, Dec 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gaur AH, Patrick CC, McCullers JA, Flynn PM, Pearson TA, Razzouk BI, Thompson SJ, and Shenep JL. Bacillus cereus bacteremia and meningitis in immunocompromised children. Clin Infect Dis, 32(10):1456–62, May 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


