Abstract
Humans infected with the dimorphic fungus Blastomyces dermatitidis develop strong T-lymphocyte responses to WI-1, an immunodominant antigen that has been shown to elicit protective immunity in mice. In the present study, the T-cell epitopes of WI-1 and human leukocyte antigen (HLA) restricting elements that display them were investigated. Peripheral blood mononuclear cells (PBMC) from 37 patients with a confirmed history of blastomycosis were tested for a response to WI-1 in primary proliferation assays; PBMC from 35 (95%) responded. Six patients whose PBMC proliferated strongly in response to WI-1 (defined as a stimulation index greater than 50) were tested further for responses to subcloned, recombinant fragments of the antigen. These patients responded chiefly to sequences within the N terminus and the 25-amino-acid tandem repeat. Cloned CD4+ T cells from an infected individual were used to delineate more precisely the peptide epitopes in the fragments and HLA restricting elements that present them. A majority of the T-cell clones recognized an epitope spanning amino acids 149 to 172 within the N terminus, displayed by HLA-DR 15. A minority of the clones, which have been shown to perform a cytolytic function in vitro, recognized an epitope in the tandem repeat displayed by HLA-DPw4, an uncommon restricting element. Tandem repeat epitopes required display by the β chain of DPw4 heterodimers. Thus, human T cells with different functions in vitro also recognize distinct regions of WI-1, raising the possibility that HLA restricting elements that present them could modulate immunity during blastomycosis by selection and display of WI-1 peptides.
Blastomyces dermatitidis is a dimorphic fungus that causes disease in both healthy and immunodeficient hosts. The fungus is endemic to the Mississippi and Ohio River valleys and northern Wisconsin. The spectrum of infection includes asymptomatic disease, acute or chronic pneumonia, and disseminated disease, especially in immunodeficient patients, who are at higher risk for developing widely disseminated blastomycosis (19, 20).
The growing frequency of invasive fungal diseases and the challenge of treating them have stimulated interest in developing ways to prevent fungal infections. The immunodominant and protective antigens for many fungal pathogens have not been elucidated and are actively being investigated (7). For B. dermatitidis, we previously described a 120-kDa protein antigen, WI-1, expressed abundantly on yeast phase cells. WI-1 is an adhesin that binds the fungus to receptors on human macrophages (17) and confers virulence on the yeast (2) and is also an immunodominant antigen that stimulates humoral and cell-mediated immune responses during natural infection (11, 12). Immunization of mice with WI-1 significantly enhances their resistance against a lethal pulmonary challenge with B. dermatitidis (25), an indication that anti-WI-1 immune responses benefit the host. Thus, WI-1 may serve as a candidate for developing a vaccine against blastomycosis.
Studies of mice and humans have established the central importance of delayed-type hypersensitivity in acquired resistance to B. dermatitidis (5). Since CD4+ T cells are a critical constituent of this response, a deeper understanding of T-cell recognition of WI-1 in people infected with B. dermatitidis will help elucidate how humans defend against the pathogen and how protective immune responses might be harnessed to prevent infection. In a prior study of a small number of blastomycosis patients, mononuclear cells obtained from their peripheral blood were shown to proliferate in vitro in response to WI-1 (12). These responding T cells were cloned and analyzed functionally: all had a CD4+ phenotype, and a majority of them responded by proliferating in the presence of WI-1, but a small proportion lysed antigen-presenting cells that displayed WI-1 on their surfaces. In the present study, our goals were to (i) investigate peripheral blood mononuclear cell (PBMC) responses to WI-1 in a larger number of patients with blastomycosis, (ii) determine the segments of WI-1 antigen chiefly recognized by T cells, (iii) delineate in these segments the peptide epitopes recognized by cloned T cells, and (iv) determine human leukocyte antigen (HLA) molecules that display these epitopes to T cells.
MATERIALS AND METHODS
Antigen preparations. (i) Native WI-1.
WI-1 was purified from B. dermatitidis ATCC 60636, a virulent isolate associated with an outbreak of human disease (10). B. dermatitidis was maintained in the yeast form by growth on Middlebrook 7H10 agar medium containing oleic acid-albumin complex (OADC) (Sigma Chemical Co., St. Louis, Mo.). Liquid cultures of yeasts were grown in Histoplasma macrophage medium (HMM) (24).
For large-scale growth of yeast, Roux bottles of 7H10-OADC agar were seeded with 5 × 108 yeast cells in a final volume of 4 ml of HMM and the yeasts were grown at 37°C in a humidified incubator for 7 days. Yeasts were harvested and inoculated into 500 ml of HMM at a final concentration of 2.5 × 105 yeasts/ml. Cultures were grown for 14 days at 37°C with shaking at 250 rpm. Supernatants enriched for secreted WI-1 were collected and frozen at −20°C until WI-1 purification, which was performed using a two-step method (1).
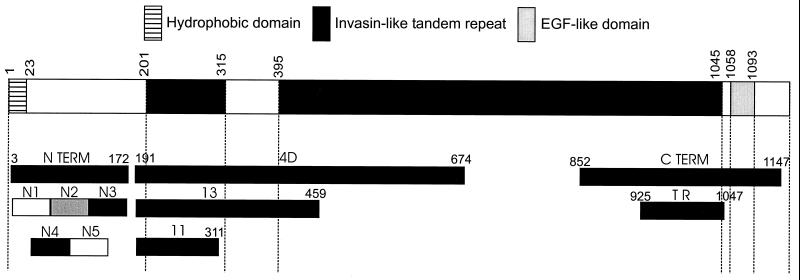
(ii) Recombinant WI-1 and its fragments (Fig. 1).
FIG. 1.
Full-length WI-1 and its three chief domains (top bar). Below are recombinant fragments and subfragments of WI-1 domains investigated in this study. Amino acid positions are shown above full-length WI-1. Each N-terminal segment extends about 50 residues: N1, amino acids (aa) 8 to 58; N2, aa 58 to 115; N3, aa 115 to 172; N4, aa 29 to 85; N5, aa 85 to 143. EGF, epidermal growth factor; TR, tandem repeat.
The full coding sequence of WI-1 was derived by BspI (New England Biolabs, Beverly, Mass.) digestion of a plasmid containing the complete WI-1 genomic sequence from B. dermatitidis ATCC 26199 (9). The 5.5-kb WI-1 genomic fragment, which is free of introns, was purified and ligated in frame 3′ to nucleotides coding for a six-histidine tag for affinity purification in the expression plasmid pQE32 (Qiagen, Valencia, Calif.). Plasmid DNA was electroporated into Escherichia coli strain XL1-Blue containing the repressor plasmid pRep4. Transformed E. coli cells were grown on Luria-Bertani agar containing 50 μg of ampicillin and 25 μg of kanamycin/ml. Plasmids with either full or partial coding sequences of WI-1 were sequenced to verify that inserts were in frame. Recombinant proteins were expressed in E. coli strain XL1-Blue, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Coomassie blue staining.
(iii) Carboxy-terminus and tandem repeat.
The coding sequence for the carboxy terminus (C terminus), a 942-bp cDNA fragment, was identified initially through immunologic screening of a B. dermatitidis (ATCC 60636) cDNA expression library using rabbit antiserum specific for WI-1 and colony hybridization (13). Sequence analysis indicated 4.5 copies of the 25-amino-acid tandem repeat encoded by the sequence at the 5′ end of the cDNA. Restriction sites flanking sequence encoding the repeats were used to subclone a 372-bp fragment encoding only the 4.5 copies of the tandem repeat. The C terminus cDNA and tandem repeat cDNA were purified and ligated in frame into the expression plasmid pQE32 as described above.
(iv) Subclones 4D, 11, and 13.
Fragment 4D spans a major portion of the invasin-like tandem repeat domain (Fig. 1). Nested deletions of genomic WI-1 DNA (strain 26199) were made with exonuclease III (Erase-a-base; Promega, Madison, Wis.). Coding sequences for subclones 4D, 11, and 13 were selected by size from minipreps. They were isolated, purified, and cloned in frame into the expression vector pQE32 as described above.
(v) N terminus.
The N terminus coding sequence (538 bp) was generated by restricting the WI-1 (strain 26199) genomic sequence with BsgI and KpnI (New England Biolabs). BsgI cuts 51 bp (encoding 17 amino acids) 5′ of the tandem repeat coding sequence; KpnI cuts 3′ of the WI-1 stop codon, thereby “dropping out” from the plasmid all WI-1 coding sequences except for that for the N terminus. This plasmid was purified, treated with Klenow fragment, religated, and cloned as described above.
(vi) Overlapping segments of the N terminus.
The coding sequences for five overlapping, 50-amino-acid fragments of the N terminus were generated by PCR amplification. The WI-1 genomic sequence from B. dermatitidis ATCC 26199 was used as the template, and primers were designed with a BglII restriction site at the 5′ end and a HindIII restriction site at the 3′ end (Operon Technologies, Alameda, Calif.). N segment coding sequences were cloned into expression plasmid pQE40 (Qiagen) to create a fusion protein with a six-histidine tag for affinity purification and with mouse dihydrofolate reductase (DHFR) to stabilize expression of small peptides.
Expression and purification of recombinant proteins. (i) rWI-1, C terminus, tandem repeat, and clones 4D, 11, and 13.
Clones were grown in super broth (25 g of Bacto tryptone, 15 g of Bacto yeast extract, 5 g of NaCl/liter) to an optical density at 600 nm (OD600) of 0.6 and then induced with 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 to 5 h at 30 or 37°C. Cell pellets were harvested and stored at −20°C overnight. Pellets were thawed in sonication buffer (50 mM sodium phosphate, 300 mM NaCl, pH 7.8) and sonicated for 2.5 min on ice. After centrifugation for 20 min at 10,000 × g and 4°C, the pellet was resuspended in buffer A (6 M guanidine-HCl, 0.1 M NaH2PO4, 0.01 M Tris adjusted to pH 8.0 with NaOH) and stirred for 1.5 h at room temperature. This material was centrifuged at 10,000 × g and 15°C to pellet cell debris. The supernatant was batch adsorbed with Ni-nitrilotriacetic acid (NTA) resin (Qiagen) for 30 to 45 min and washed three times with buffer A and then once each with buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 8.0), buffer C (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 6.3) and 20 mM imidazole until the OD280 was <0.02. Resin was poured into a column and washed again with buffer C. Proteins were refolded on the column using 20 column volumes of a stepwise urea gradient, from 6 M to 0 M in 20 mM Tris, pH 7.5. The column was washed with 20 mM Tris, pH 7.5, and recombinant proteins were eluted with 0.25 M imidazole in 20 mM Tris, pH 7.5. Eluted proteins were dialyzed against 4 liters of 20 mM Tris, pH 7.5, and then quantified by measurement of OD280.
(ii) N terminus, DHFR, and N1 to N5 segments.
Clones were grown and induced, and recombinant proteins were bound to Ni-NTA resin and placed into columns as described above. After being washed with buffer C until the OD280 was <0.02, the proteins were eluted using 8 M urea with 0.25 M imidazole and then dialyzed against 4 liter of deionized water. Recombinant proteins were quantified by measuring OD280, lyophilized, and resuspended in dimethyl sulfoxide at 4 mg/ml.
Synthetic peptides. (i) N3/N5 segments.
Four 24-amino-acid peptides, overlapping by 12 amino acids were synthesized (Research Genetics, Huntsville, Ala.) to span the N3 fragment and extend beyond it (Table 1). The sequence from the N terminus of B. dermatitidis ATCC 60636 was used to generate the synthetic peptides.
TABLE 1.
Synthetic peptides used to stimulate WI-1-specific T-cells in blastomycosis patients
| WI-1 domain | Amino acid sequence | Residue locationb |
|---|---|---|
| N3/N5a | TGHGKHFYDCDWDPSHGDYSWYLW | 113–136 |
| N3/N5 | DPSHGDYSWYLWDYLCGNGHHPYD | 125–148 |
| N3/N5 | DYLCGNGHHPYDCELDNSHEDYSW | 137–160 |
| N3 | CELDNSHEDYNWNLWFKWCSGHGR | 149–172 |
| Tandem repeat | DPYNCDWDPYHEKYDWDLWNKWCN | NA (PSN 265)c |
| Tandem repeat | KYDWDLWNKWCNKDPYNCDWDPYH | NA (PSN 266) |
(ii) Tandem repeat.
Two 24-amino-acid peptides, overlapping by 12 amino acids, were synthesized from a conserved and nondegenerate area of the tandem repeat by the University of Wisconsin Biotechnology Center (Table 1).
Antigen-presenting cells. (i) EBV B-lymphoblastoid cell lines (LCL).
Autologous B cells from patients were transformed by EBV with supernatants from an EBV-infected B95-8 cell line (12).
(ii) HLA loss mutant LCLs and transferents.
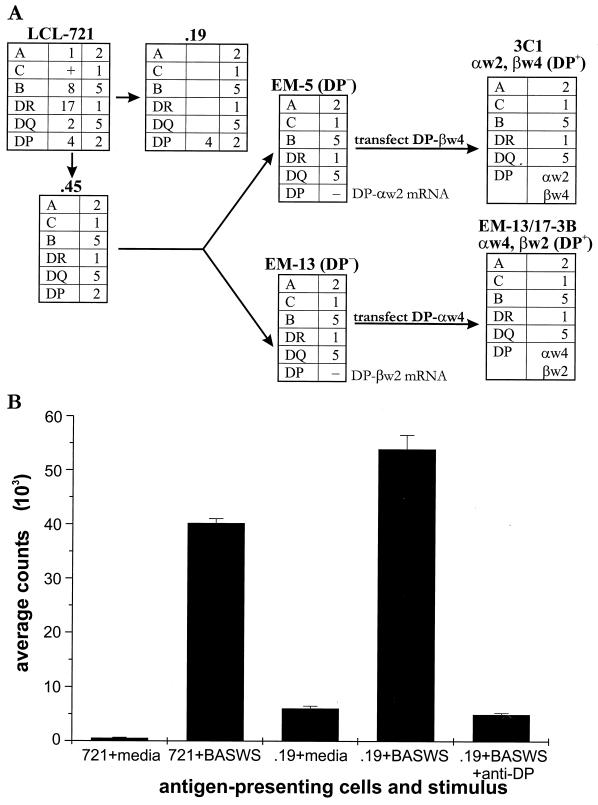
Some of these cell lines have been described previously (15). HLA loss mutant LCLs were derived by γ-irradiation and selected by antibody and complement depletion from the parental 721 LCL. HLA-DR transferents expressing selected HLA-DRβ alleles were derived from mutant .174, which lacks the major histocompatibility complex (MHC) class II region. HLA-DR expression was restored to .174 by transferring HLA-DM A and B genes to confer antigen processing and presentation ability. The monomorphic HLA-DR A gene was then introduced to create a “founder” cell line. Introduction of specific HLA-DR B genes into the founder cell line yielded transferents, each of which expressed just one or another kind of HLA-DRβ molecule (18). HLA-DP transferents were created here by means of gene transfer with episomal vectors into HLA-DP null mutants that had been derived originally (22) from the parental 721 cell line heterozygous at the DP locus (haplotype DPw4/2). cDNAs encoding either the DPβw4 chain or DPαw4 chain were isolated from a cDNA library constructed with mRNA from the parental LCL 721 cell line using standard screens and cloned into pRSV.5 (neo) plus ori P. Two independent HLA-DP loss mutants, devoid of surface HLA-DP due to defects in either the DPα or DPβ chain gene (22), served as the recipients for gene transfer. Thus, mutant EM5, retaining DPαw2 mRNA as determined by Northern analysis, received DPβw4 (Fig. 5A). Mutant EM13, retaining DPβw2 mRNA as determined by Northern analysis, received DPαw4 (Fig. 5A). Surface HLA-DR or -DP expression on the transferents was monitored by flow cytometric analysis using monoclonal antibodies (MAbs) L243 (14) and B7/21 (23), respectively. HLA-DR transferents were cultured in fetal calf serum (FCS)-RPMI 1640 (RPMI 1640, 10% FCS, penicillin-streptomycin, l-glutamine) under selection with 0.5 mg of G418 (Gibco)/ml–0.036 M xanthine (Sigma)–0.03 mM mycophenolic acid (Sigma) as described previously (18). HLA-DP transferents were cultured in FCS-RPMI 1640 with 0.5 mg of G418/ml.
FIG. 5.
Analysis of the role of HLA-DP in presentation of WI-1 to T cells. (A) The “family tree” of parental cell line LCL 721, its HLA loss mutants, and the HLA-DP transferents used in this study. HLA-DP transferents 3C1 and EM13/17-3B were generated by gene transfer into HLA-DP loss mutants EM5 and EM13, respectively. As illustrated, mutant EM5, which retains DPαw2 mRNA as determined by Northern analysis, received DPβw4. Conversely, mutant EM13, which retains DPβw2 mRNA as determined by Northern analysis, received DPαw4. (B) Antigen presentation by LCL cells and HLA loss mutants. WI-1-reactive T cells (clone K20B) are from a patient homozygous for HLA-DPw4. BASWS was used to stimulate T cells as it shares epitopes with WI-1. An antibody against HLA-DP (MAb B7/21) was added to wells (2 μg/well) to block proliferation evoked by .19. MAbs against HLA-DR (L243) and HLA-DQ (G.2B.2) (21) were similarly added to separate wells as controls, without effect.
Responder T-cells. (i) WI-1-specific T-cell clones.
WI-1-specific T-cell clones were derived by limiting dilution from a 22-year-old patient with disseminated blastomycosis as previously described (12). The clones, which had been stored at −135°C, were thawed and cultured in 96-well round-bottom microtiter plates as described previously (12). Clones were restimulated every 10 to 14 days.
(ii) PBMCs from patients.
Thirty-five patients with a history of blastomycosis confirmed by histopathology or culture were identified and contacted for blood donation by the Health Department in Eagle River, Wis., an area of hyperendemicity for blastomycosis. Two other patients were residents of Madison, Wis. A volume of 50 to 60 ml of heparinized blood was obtained from a peripheral vein. Blood was transported to the laboratory where PBMCs were isolated by Lymphoprep (Gibco) sedimentation. PBMCs were washed with phosphate-buffered saline and resuspended in 10% pooled human serum in RPMI-1640 supplemented with l-glutamine, penicillin, streptomycin, and HEPES.
Proliferation assays. (i) T-cell clones.
WI-1-specific T-cell clones were plated at 2 × 104 to 5 × 104/well in a volume of 100 μl. Irradiated autologous LCL (11,400 cGy) as antigen-presenting cells were plated at 5 × 104/well in a volume of 50 μl. Antigens were plated at 50 μl/well with concentrations that were determined in preliminary titration experiments to be optimal. Concentrations were as follows: Candida antigen (Hollister-Stier Laboratories, Spokane, Wash.), 1:100; tetanus toxoid (Wyeth-Ayerst Laboratories, Paoli, Pa.), 1:10; BASWS, 100 μg/ml; native WI-1, 10 μg/ml; recombinant WI-1 (rWI-1), C terminus, N terminus, tandem repeat, 4D, 11, 13, N fragments, DHFR, and N fragment and tandem repeat synthetic peptides, 20 μg/ml each. Concanavalin A (ConA; Boehringer Mannheim, Indianapolis, Ind.) at 10 μg/ml and recombinant interleukin-2 (rIL-2) at 200 U/ml served as positive controls. Cells and antigens were plated in flat-bottomed 96-well plates. Proliferation was measured after 72 h by incorporation of [3H]thymidine as described previously (12). A stimulation index (SI) was calculated from the counts of [3H]thymidine incorporation per minute induced by an antigen divided by the baseline uptake in media.
(ii) PBMCs.
Freshly isolated PBMCs from patients with a history of blastomycosis were used at 105/well. Cells were incubated with antigen for 6 days. [3H]thymidine was added to wells, and cells were harvested and counted as described above.
Flow cytometry.
Hybridoma supernatants of MAbs specific for HLA-DR (L243) and HLA-DP (B7/21) were used in flow cytometry. Each MAb recognizes the respective class II molecule, regardless of allele (14, 23). Cells were labeled using a standard protocol for indirect staining (4). The secondary antibody was goat anti-mouse fluorescein isothiocyanate (Immunotech, Westbrook, Maine).
RESULTS
Response of PBMCs to WI-1 in patients with blastomycosis.
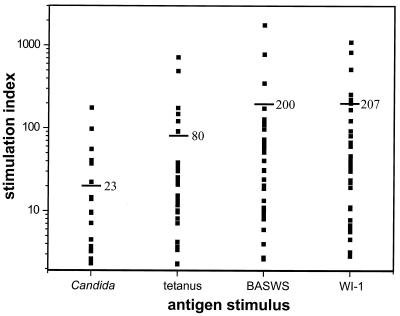
A prior, small study showed that PBMCs from 10 patients with blastomycosis responded to both WI-1 and a crude yeast cell wall antigen, BASWS (which shares determinants with WI-1) (12). To extend those findings, we tested PBMCs from 37 patients who had resolved cases of blastomycosis and no underlying impairment of immunity. Patients had had infection for from 1 to 15 years (median, 4 years) before they donated a blood specimen for this study. Of these subjects, 35 (95%) demonstrated a significant response to both WI-1 and BASWS, as defined by an SI greater than or equal to 3 (Fig. 2). The PBMC responses to WI-1 (mean SI = 208; mean cpm ± standard error of the mean [SEM] = 22,500 ± 3,570) were slightly greater than the responses to BASWS (mean SI = 200; mean cpm ± SEM = 21,300 ± 3,930). PBMCs of two patients who did not respond to WI-1 also did not respond to Candida and tetanus control antigens and thus were unable to respond to either test or control antigens in the assay.
FIG. 2.
PBMC responses in 37 patients with a history of confirmed blastomycosis. The cells were tested in a primary proliferation assay for response to B. dermatitidis antigens, BASWS (100 μg/ml), and WI-1 (10 μg/ml) and to control antigens of tetanus (1:10) and Candida (1:100). Response to the antigen is expressed as SI, which is the average counts of [3H]thymidine uptake induced by the antigen/average counts induced by medium. The average SI for each antigen is depicted by a horizontal bar.
Domains of WI-1 recognized by PBMCs of patients with blastomycosis.
Six patients who demonstrated strong responses to WI-1 (SI ≥ 50) in the initial assay were selected for further study to identify regions of WI-1 chiefly recognized by responding T cells. Fresh PBMCs from these patients were tested in a primary proliferation assay for the response to native WI-1, rWI-1, and fragments of WI-1 (Fig. 1 and Table 2). These patients responded most strongly to native WI-1, but they also responded significantly to rWI-1. All patients except for one (patient 11) responded to each of the WI-1 fragments, including the C terminus, the tandem repeat, derivatives 11 and 13 of 4D, and the N terminus. In the test of responses to the N segments, three of six patients, patients 11, 20, and 32, responded chiefly to the N3 segment and each of them showed a greater response to the N terminus than to the tandem repeat represented by the 4D fragment (Table 1). Two healthy control subjects without a history of blastomycosis responded to control stimuli of ConA and IL-2 but did not respond to WI-1 or its recombinant fragments. The apparent focusing of patient T-cell responses toward a particular WI-1 domain and N segment led us to investigate further which epitopes are recognized in the N terminus and which HLAs select and display them to the T cells.
TABLE 2.
PBMC responses to WI-1 and its domains and fragments in blastomycosis patients and healthy control subjectsa
| Patient or control no. | SIb for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| nWI-1 | rWI-1 | C-term | T-repeat | 4D | N-term | N1 | N2 | N3 | N4 | N5 | |
| Patients | |||||||||||
| 1 | 170 | 70 | 47 | 121 | 88 | 14 | 0.3 | 0.9 | 0.3 | 0.4 | 0.5 |
| 11 | 15 | 6.6 | 2.5 | 5.4 | 1.0 | 3.3 | 1.4 | 2.2 | 6.6 | 0.6 | 1.4 |
| 18 | 110 | 80 | 64 | 86 | 75 | 13 | 0.3 | 0.9 | 0.2 | 0.2 | 0.3 |
| 20 | 44 | 35 | 24 | 21 | 19 | 20 | 0.8 | 0.5 | 4.7 | 0.3 | 0.2 |
| 29 | 270 | 110 | 82 | 190 | 96 | 39 | 0.1 | 1.0 | 0.1 | 0.1 | 0.3 |
| 32 | 48 | 22 | 15 | 24 | 15 | 19 | 0.5 | 1.1 | 6.3 | 0.9 | 0.5 |
| Controlsc | |||||||||||
| 1 | 2.2 | 2.5 | 1.1 | 0.8 | 2.9 | 0.7 | 0.8 | 1.2 | 0.7 | 0.8 | 0.7 |
| 2 | 0.1 | 0.5 | 0.3 | 1.2 | 1.7 | 0.4 | 0.6 | 1.0 | 1.4 | 0.6 | 1.0 |
WI-1, native WI-1; C-term, C terminus; T-repeat, tandem repeat; N-term, N terminus.
An SI of 3 or greater is considered positive.
PBMCs from control subjects responded to positive control stimuli of conA (SI = 24 and 38) and IL-2 (SI = 77 and 130).
WI-1 domains and epitopes recognized by cloned T cells.
WI-1-specific T-cell clones used here were derived from a 22-year-old Phillipino man with a case of pulmonary and cutaneous blastomycosis and have been characterized previously with regard to their phenotype and function (12). Briefly, all of the clones were CD3+, CD4+, and T-cell receptor (TCR) αβ+ and most were Vβ8+ with respect to TCR β-chain usage. In functional assays, all clones responded to WI-1 in vitro by proliferating in the presence of antigen and autologous antigen-presenting cells; however, a small number of them also were cytolytic and lysed targets that displayed the antigen. The response of noncytolytic T-cell clones was inhibited with a MAb directed against HLA-DR, whereas the response of cytolytic clones was inhibited by a MAbs directed against HLA-DP. These findings pointed to a relationship between the restricting element and the in vitro function of clones and suggested that HLA selection of WI-1 epitopes might influence T-cell function.
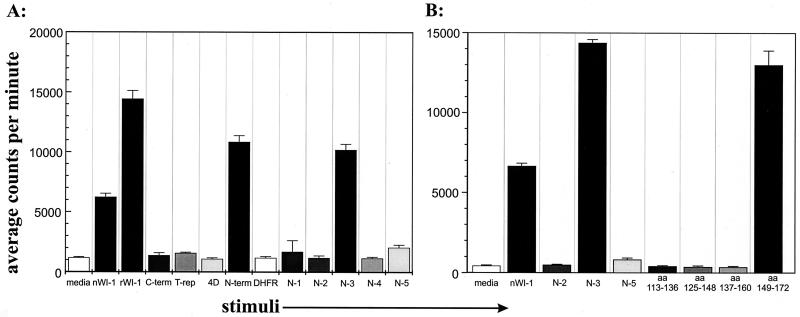
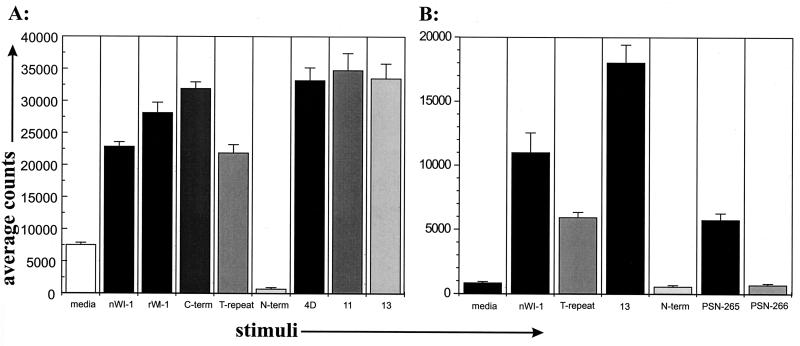
To identify WI-1 T-cell epitopes and assess whether T cells with disparate functions in vitro recognize different epitopes, clones were analyzed for the response to the purified native WI-1, rWI-1, and WI-1 fragments shown in Fig. 1. A total of eight T-cell clones were tested here: six noncytolytic clones and two cytolytic clones. Figures 3A and 4A illustrate representative responses of noncytolytic and cytolytic T-cell clones, respectively. Noncytolytic clone K20F responded to both native and rWI-1 and to the N terminus, but it did not respond to the C terminus, derivatives 11 and 13 of 4D, or the tandem repeat (Fig. 3A). The five other noncytolytic clones responded similarly (data not shown). Thus, noncytolytic clones recognized an epitope in the N terminus between amino acid residues 3 to 172. The epitope was narrowed further by detecting responses to the N3 segment, which spans residues 123 to 172 (Fig. 1 and 3A) but not to the other N segments. Peptides of 24 amino acids and overlapping by 12 residues were used to delineate the epitope in the N3 segment (Table 1; Fig. 3B). The peptide spanning amino acid residues 149 to 172 stimulated a response in clone K20F. None of the other peptides, including the one spanning residues 137 to 160, evoked a response. These findings indicate that the epitope is most likely located between amino acid residues 160 and 172.
FIG. 3.
Response of representative noncytolytic T-cell clone (K20F) to WI-1 fragments and peptides. (A) Response of clone K20F to N fragments within the N terminus. (B) Response of clone K20F to four overlapping 24-amino acid (aa) peptides spanning the N3 fragment. Residues spanning the peptide are indicated below histogram bars. C-term, C terminus; nWI-1, native WI-1; T-rep, T repeat; N-term, N terminus.
FIG. 4.
Response of a representative cytolytic T-cell clone (K20B) to WI-1 fragments and peptides. (A) Response of clone K20B to fragments that display the tandem repeat (T-repeat). (B) Response of clone K20B to 24-amino-acid peptides PSN 265 and 266, which span the tandem repeat (amino acid residues are in Materials and Methods). nWI-1, C-term, and N-term are as defined for Fig. 3.
T-cell clone K20B, which exhibited cytolytic activity, responded to native WI-1 and rWI-1, the C terminus, successive deletions 4D, 11, and 13, and the tandem repeat, but it did not respond to the N terminus (Fig. 4A). Other clones from this group responded similarly. The response in this group to derivatives 11 and 13 of 4D and to the tandem repeat indicated that the epitope resided somewhere within the 25-amino-acid tandem repeat (Fig. 1). The response to synthetic peptides PSN 265 and PSN 266 within the tandem repeat was analyzed to define the epitope further. K20B responded only to PSN 265 (Table 1; Fig. 4B). Inspection of the 24 residues in PSN 265 and in PSN 266 (Table 1) shows that the 12-amino-acid blocks of residues at the N terminus and the C terminus are simply transposed with each other from one peptide to the next. Thus, the blocks of sequence are in the order, for example, AB in PSN 265 and BA in PSN 266, but the sequences are essentially the same in each peptide. This suggests that the epitope recognized by this group of T-cell clones does not reside in either 12-amino-acid block of residues but rather spans the junction of the two blocks as they are arranged in PSN 265.
HLA restriction elements that display WI-1 to cloned T cells.
We sought to firmly establish for these two groups of clones the identity of the HLA restricting element and allele. In experiments below, HLA loss mutant LCL cells and HLA-DR and HLA-DP transferents were used for presentation of antigens to T-cell clones. The haplotype of the patient from whom T cells were cloned is as follows: HLA-DR, 15/6; HLA-DPw, 4/4; HLA-DQ, 1/3. The haplotype for the parental strain (LCL 721) of HLA loss mutants, which served as recipients for HLA-DR and -DP α- and β-chain gene transfers is as follows: HLA-DR, 17/1; HLA-DP, 4/2; HLA-DQ, 2/5 (1). The HLA transferents used in these assays are monospecific, expressing only one kind of class II MHC isotype and allele, matched with that in the patient who donated the T cells. We confirmed the surface expression of class II MHC before transferents were used in antigen presentation assays.
HLA loss mutants used to assess the role of HLA-DP are illustrated (Fig. 5A). LCL .19 displays both maternal and paternal alleles of HLA-DP on its surface. One allele (DPw4) is matched with the homozygous DPw4 alleles in the blastomycosis patient's T-cell clones. LCL .45, expressing only one parental HLA allele, does not have a HLA-DP allele that matches those of the T-cell clones. LCL EM5 and LCL EM13, HLA-DP loss mutants derived from .45 (22), lack surface HLA-DP (Fig. 5A). These mutants are otherwise similar to LCL .19 with respect to HLA-DR and -DQ. However, because both of these LCL express HLA-DR and -DQ alleles that do not match the patient's haplotype, they would not be expected to present antigen to these T-cell clones. In antigen presentation assays, LCL 721 and mutant .19 presented antigen (BASWS) to T-cell clone K20B (Fig. 5B), which specifically recognizes the tandem repeat. In addition, the response to LCL .19 plus antigen is abolished by treatment with an anti-HLA-DP MAb, suggesting that HLA-DP is required for presentation to K20B (Fig. 5B). Neither LCL .45 nor EM5 or EM13 presented BASWS to K20B (SI values of 1.04, 2.5, and 0.96, respectively), further supporting the importance of HLA-DPw4 in the recognition of antigen.
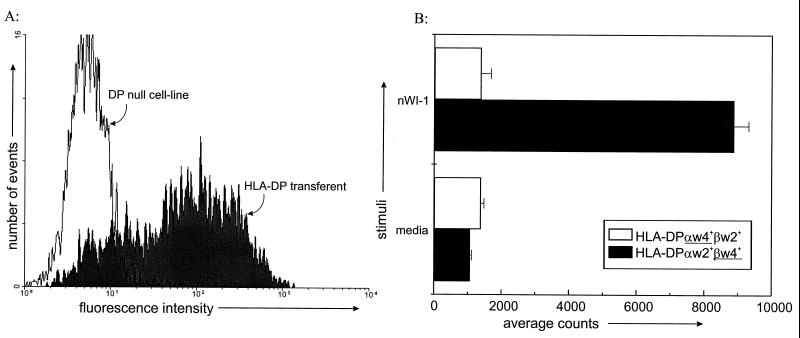
Two kinds of HLA-DP transferents were created and tested to confirm the role of HLA-DP in presentation. Initially, a DPα4 gene was transferred into EM13, which specifically lacks a DPα chain (22), giving rise to transferent EM13/17-3B, which displays a surface HLA-DP heterodimer of a DPw4α chain and the endogenous DPw2β chain (Fig. 5A). In antigen presentation assays, transferent EM13/17-3B did not present the WI-1 antigen to clone K20B (Fig. 6B). Subsequently, we transferred a DPβ4 gene into cell line EM5 to construct transferent 3C1, which displays surface HLA-DP heterodimers composed of the endogenous DPw2α chain (22) and a DPw4β chain (Fig. 5A). Quantitatively similar levels of surface expression of HLA-DP by both transferents were confirmed by labeling with MAb B7/21 and flow cytometry analysis (Fig. 6A), yet only transferent 3C1 was able to present WI-1 to K20B in antigen presentation assays (Fig. 6B). These data confirm the role of HLA-DP in WI-1 presentation and suggest the importance of the DPw4β chain in displaying the tandem repeat sequence to T cells. Further studies would be required to determine if this observation can be generalized to presentation of other peptide epitopes by HLA-DP.
FIG. 6.
HLA-DP surface expression on transferents and their presentation of WI-1 to T cells. (A) Flow cytometric analysis of HLA-DP expression on DP-null mutants and on the HLA-DP transferents described in Fig. 5A. Expression of HLA-DP is restored by the gene transfer, as demonstrated by MAb B7/21 staining of representative transferent 3C1. Transferent EM-13/17-3B expressed an amount of DP (not shown) similar to that on 3C1 despite its failure to present antigen (see below). (B) The HLA-DP mutants and transferents were tested for presentation of WI-1 to clone K20B. The HLA-DP transferent with the βw4 gene (3C1 in Fig. 5A) presented WI-1 to K20B, whereas the transferent with the DPw4α chain (EM-13/17-3B) did not present antigen. nWI-1, native WI-1.
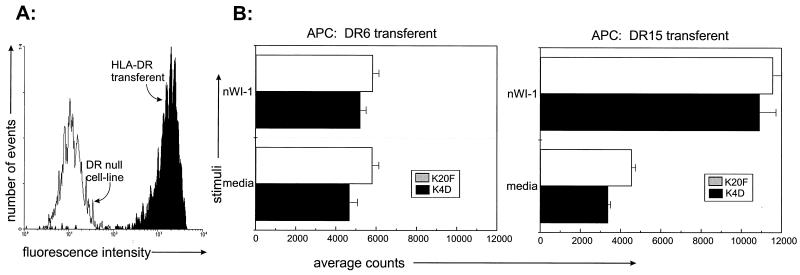
To establish the role of HLA-DR in WI-1 presentation and identify the allele involved, monospecific transferents displaying only HLA-DR 15 or only HLA-DR 6 were used for antigen presentation (see Materials and Methods). The transferents displayed substantial levels of HLA-DR on their surfaces when analyzed by flow cytometry (Fig. 7A). HLA-DR6 and HLA-DR 15 transferents each express an allele that matches those in the patient from whom the T cells were cloned. In antigen presentation assays, T-cell clones K4D and K20F responded to WI-1 displayed by HLA-DR 15 but not by HLA-DR 6 (Fig. 7B). These results indicate that epitopes in the N terminus of WI-1, spanning residues 160 to 172, are displayed to T cells by HLA-DR and that in this particular patient and clone, HLA-DR 15 serves as the restricting element.
FIG. 7.
HLA-DR surface expression on transferents and their presentation of WI-1 to T cells. (A) Flow cytometric analysis of HLA-DR expression on a DR-null mutant cell line and on HLA-DR transferents described in reference 18, as illustrated by MAb L243 staining of an HLA-DR 6 transferent. An HLA-DR 15 transferent expressed an amount of DR (not shown) similar to that on HLA-DR 6 and presented antigen (see below). (B) HLA-DR transferents matched with the haplotype of the patient T-cell clones were used in antigen presentation assays. Only the HLA-DR 15 transferent presented WI-1 to noncytolytic T-cell clones K4D and K20F. nWI-1, native WI-1; APC, antigen-presenting cell.
DISCUSSION
In this study, PBMCs from nearly all patients with a history of blastomycosis responded to purified native WI-1 and the crude cell wall antigen BASWS, confirming findings from a prior study in which WI-1 was demonstrated to be an immunodominant antigen of cell-mediated immunity (12). Prior work has demonstrated that PBMC responses to WI-1 are observed only in patients with blastomycosis and not in control subjects (12). Here, we show that recombinant WI-1 and its fragments also stimulate patient cells specifically, as might be expected, although only a small number of healthy control subjects were investigated.
PBMCs of six patients investigated for their response to WI-1 fragments recognized multiple regions and domains of WI-1, including the C terminus, the tandem repeat, and the N terminus. Half of these patients responded to the N terminus. Those who responded better to the tandem repeat sequence in fragment 4D than to the N terminus did not respond at all to the N segments, as would be expected, while those who responded better to the N terminus than to fragment 4D responded chiefly to the N3 segment. These observations using bulk populations of freshly isolated PBMCs indicate that multiple regions of WI-1 harbor epitopes recognized by T cells in patients with blastomycosis. It is possible that selection of six patients with particularly strong proliferative responses could have biased findings in this study, since Th1 cells typically respond with stronger proliferation than Th2 cells. However, the patterns of recognition observed in bulk populations foreshadowed and underscore the patterns observed with T-cell clones that were subsequently investigated in this study.
WI-1-reactive T cells cloned from the peripheral blood of a patient also recognized various regions of the antigen. Most of the clones recognized an epitope in the N terminus, spanning amino acid residues 160 to 172, and only a few recognized an epitope in the 25-amino-acid tandem repeat. This skew could reflect in vitro culture conditions that favored growth of certain clones over others. However, the data are consistent with frequent recognition of the N terminus by bulk populations of lymphocytes from individual patients. Thus, some patients' T cells may recognize certain regions of the antigen, particularly the N terminus, in favor of other regions. This finding may relate to the relative density or haplotype of appropriate restricting elements on the surfaces of the antigen-presenting cells in a given patient.
N-terminal epitopes of WI-1 were shown to be displayed to T cells by HLA-DR class II molecules, whereas tandem repeat epitopes were shown to be displayed by HLA-DP class II molecules. HLA-DR molecules are expressed abundantly on antigen-presenting cells and commonly display nominal antigens to the immune system, whereas HLA-DP is expressed in smaller amounts and only infrequently presents epitopes to the immune system (15). Participation of HLA-DP in the presentation of WI-1 may be explained by some unusual sequence properties of the antigen, particularly in the tandem repeat, which is rich in the aromatic amino acids that anchor peptides to the peptide-binding region of HLA-DP (15). Thus, more than one kind of restricting element participates in the presentation of WI-1 to T cells. The role of restricting elements is extended further in the present study through the use of HLA transferents. These cells were shown to express only one kind of restricting element on their surfaces after gene transfer into HLA-null mutants. When the correct HLA allele was present, these transferents bound WI-1 epitopes and stimulated a response from WI-1-specific T-cell clones. These results further define the role of HLA alleles in the presentation of WI-1 to patient T cells.
Not surprisingly, specific epitopes of WI-1 required specific HLA alleles for presentation to T cells. HLA-DR 15 presented N-terminal peptides of WI-1 to T cells, whereas HLA-DPw4 presented the tandem repeat. Dominant epitopes of WI-1 are likely to vary, depending on the haplotype of the patient and on the WI-1 sequence of the infecting B. dermatitidis strain. However, our study provides some new insight into how HLA-DP heterodimers interact with antigenic peptides. Different pairwise combinations of DPα and -β chains behaved differently with respect to antigen presentation, despite having equivalent levels of surface DP expression as assessed by flow cytometry. Our results suggest a pivotal role for the β chain because an α2-β4 chain combination presented antigen, whereas an α4-β2 chain heterodimer failed to present antigen to cells from a patient with an HLA-DPw4 haplotype.
What importance do these studies hold for understanding cell-mediated immunity in blastomycosis or other fungal diseases? First, it is possible that HLA may regulate the type of immune response observed in vivo by selection of epitopes and, thus, by stimulation of certain T-cell subsets or functions. Although our study did not address this issue directly, our results are consistent with that premise. We found that T-cell clones demonstrated previously to perform a cytolytic function in vitro recognized a different HLA-peptide combination (HLA-DP and tandem repeat) than clones demonstrated not to exhibit cytolytic functions (HLA-DR and N terminus). Other work has demonstrated that a variation in HLA-peptide combination can significantly influence T-cell functions, such as the proliferative capacity of T-cells, the T-helper phenotype, and secretion of IL-4 (8, 16). We did not investigate cytokine production, which could be germane to understanding how HLA-peptide combinations affect immunity. Nevertheless, we can speculate that epitope selection of WI-1 or other B. dermatitidis antigens might influence the immune response and manifestations of disease in a given patient. In support of this notion, patients with either HLA-DR 2 or -DR 5 haplotypes who respond to certain antigens of Aspergillus fumigatus are predisposed toward development of allergic bronchopulmonary aspergillosis (3). Proof of this principle for blastomycosis patients will require further study, as our study investigated T cells from only one patient in sufficient detail.
Finally, this work illustrates a method for systematically defining antigenic peptides and HLA restricting elements that display them for the purpose of developing a peptide-based vaccine. In situations where such vaccines are contemplated, the dominant epitopes together with their HLA restricting elements must be identified to ensure that individuals in the target population will respond to the vaccine. We investigated only one individual in detail using the approach outlined, whereas investigation of a larger number of representative individuals in the target population would be needed to design a rational and effective vaccine. Such an approach would be feasible with the use of HLA transferents described here, as illustrated by studies of Chlamydia trachomatis major outer membrane protein epitopes and HLA restricting elements that stimulate T cells in infected patients (18).
ACKNOWLEDGMENTS
This work was supported by a Mentored Clinical Scientist Development Award from NIH, K08 AI 01348-04 (W.L.C.), NIH grants AI-40996, AI-35681 (B.S.K.), and AI-15486 (R.I.D.). B.S.K. is the recipient of an NIH Research Career Development Award, AI-01308, and a Scholar Award in Molecular Mycology from the Burroughs Wellcome Fund.
We thank Delores Ludwig of the Vilas County, Wis., Health Department for assistance in coordinating the collection of peripheral blood specimens from 35 people with a history of blastomycosis, and we thank Hanna Filutowicz for technical assistance. We are especially grateful to the patients who participated in this study.
REFERENCES
- 1.Audet R, Brandhorst T T, Klein B. Purification in quantity of the secreted form of WI-1: a major adhesin on Blastomyces dermatitidis yeasts. Protein Expr Purif. 1997;11:219–226. doi: 10.1006/prep.1997.0783. [DOI] [PubMed] [Google Scholar]
- 2.Brandhorst T T, Wuthrich M, Warner T, Klein B. Targeted disruption reveals an adhesin indispensible for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauhan B, Knutsen A P, Hutcheson P S, Slavin R G, Bellone C L. T cell subsets, epitope mapping, and HLA-restriction in patients with allergic bronchopulmonary aspergillosis. J Clin Investig. 1996;97:2324–2331. doi: 10.1172/JCI118675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current protocols in immunology. I. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Cozad G C, Chang C-T. Cell-mediated immunoprotection in blastomycosis. Infect Immun. 1980;28:398–403. doi: 10.1128/iai.28.2.398-403.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deepe G S, Gibbons R, Brunner G D, Gomez F J. A protective domain of heat-shock protein 60 from Histoplasma capsulatum. J Infect Dis. 1996;174:828–834. doi: 10.1093/infdis/174.4.828. [DOI] [PubMed] [Google Scholar]
- 7.Deepe G S. Prospects for the development of fungal vaccines. Clin Microbiol Rev. 1997;10:585–596. doi: 10.1128/cmr.10.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evavold B D, Allen P M. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- 9.Hogan L H, Josvai S, Klein B S. Genomic cloning, characterization, and functional analysis of the major surface adhesin WI-1 on Blastomyces dermatitidis yeasts. J Biol Chem. 1995;270:30725–30732. doi: 10.1074/jbc.270.51.30725. [DOI] [PubMed] [Google Scholar]
- 10.Klein B S, Vergeront J M, Weeks R J, Kumar U N, Mathai G, Varkey B, Kaufman L, Bradsher R W, Stoebig J F, Davis J P. Isolation of Blastomyces dermatitidis in soil associated with a large outbreak of blastomycosis in Wisconsin. N Engl J Med. 1986;314:529–534. doi: 10.1056/NEJM198602273140901. [DOI] [PubMed] [Google Scholar]
- 11.Klein B S, Jones J M. Isolation, purification, and radiolabeling of a novel 120-kD surface protein on Blastomyces dermatitidis yeasts to detect antibody in infected patients. J Clin Investig. 1990;85:152–161. doi: 10.1172/JCI114406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein B S, Sondel P M, Jones J M. WI-1, a novel 120-kilodalton surface protein on Blastomyces dermatitidis yeast cells, is a target antigen of cell-mediated immunity in human blastomycosis. Infect Immun. 1992;60:4291–4300. doi: 10.1128/iai.60.10.4291-4300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein B S, Hogan L H, Jones J M. Immunologic recognition of a 25-amino acid repeat arrayed in tandem on a major antigen of Blastomyces dermatitidis. J Clin Investig. 1993;92:330–337. doi: 10.1172/JCI116571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lampson L A, Levy R. Two populations of IA-like molecules on a human B-cell line. J Immunol. 1980;125:293. [PubMed] [Google Scholar]
- 15.Meyer C G, Schnittger M J. HLA-DP—part of the concert. Immunol Today. 1997;18:58–61. doi: 10.1016/s0167-5699(96)30071-6. [DOI] [PubMed] [Google Scholar]
- 16.Murray J S. How MHC selects Th1/Th2 immunity. Immunol Today. 1998;19:157–163. doi: 10.1016/s0167-5699(97)01237-1. [DOI] [PubMed] [Google Scholar]
- 17.Newman S L, Chaturvedi S, Klein B S. The WI-1 antigen of Blastomyces dermatitidis yeasts mediates binding to human macrophage CD11b/CD18 (CR3) and CD14. J Immunol. 1995;154:753–761. [PubMed] [Google Scholar]
- 18.Ortiz L, Demick K P, Petersen J W, Polka M, Rudersdorf A R, Van der Pol B, Jones R, Angevine M, DeMars R. Chlamydia trachomatis major outer membrane protein (MOMP) epitopes that activate HLA class II-restricted T cells from infected humans. J Immunol. 1996;157:4554–4567. [PubMed] [Google Scholar]
- 19.Pappas P G, Pottage J C, Powderly W G, Fraser V J, Stratton C W, McKenzie S, Tapper M L, Chmel H, Bonebrake F C, Blum R, et al. Blastomycosis in patients with the acquired immunodeficiency syndrome. Ann Intern Med. 1992;116:847–853. doi: 10.7326/0003-4819-116-10-847. [DOI] [PubMed] [Google Scholar]
- 20.Pappas P G, Threlkeld M G, Bedsole G D, Cleveland K O, Gelfand M S, Dismukes W E. Blastomycosis in immunocompromised patients. Medicine. 1993;72:311–325. doi: 10.1097/00005792-199309000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Parham P, Kipps T J, Ward F E, Herzenberg L A. Isolation of heavy chain class I switch variants of a monoclonal anti-DC1 hybridoma cell line: effective conversion of noncytotoxic IgG1 to cytotoxic IgG2 antibodies. Hum Immunol. 1983;8:141. doi: 10.1016/0198-8859(83)90009-5. [DOI] [PubMed] [Google Scholar]
- 22.Perez M S, Orejas R D, Petersen J W, DeMars R I, Shaw S. Differences in specificity of DPw2-specific cytotoxic T-cell clones revealed with HLA mutant lines: evidence that non-DP HLA genes influence recognition by some clones. Eur J Immunol. 1990;20:673–681. doi: 10.1002/eji.1830200331. [DOI] [PubMed] [Google Scholar]
- 23.Watson A J, DeMars R, Trowbridge I S, Bach F H. Detection of a novel human class II HLA antigen. Nature. 1983;304:358. doi: 10.1038/304358a0. [DOI] [PubMed] [Google Scholar]
- 24.Worsham P L, Goldman W E. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;26:137–143. [PubMed] [Google Scholar]
- 25.Wuthrich M, Chang W L, Klein B S. Immunogenicity and protective efficacy of the WI-1 adhesin of Blastomyces dermatitidis. Infect Immun. 1998;66:5443–5449. doi: 10.1128/iai.66.11.5443-5449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]