Abstract
Objective
Sjögren’s disease (SjD) is an autoimmune disease characterised by inflammatory destruction of exocrine glands. Patients with autoantibodies to Ro/SSA (SjDRo+) exhibit more severe disease. Long non-coding RNAs (lncRNAs) are a functionally diverse class of non-protein-coding RNAs whose role in autoimmune disease pathology has not been well characterised.
Methods
Whole blood RNA-sequencing (RNA-seq) was performed on SjD cases (n=23 Ro/SSA negative (SjDRo−); n=27 Ro/SSA positive (SjDRo+) and healthy controls (HCs; n=27). Bioinformatics and pathway analyses of differentially expressed (DE) transcripts (log2 fold change ≥2 or ≤0.5; padj<0.05) were used to predict lncRNA function. LINC01871 was characterised by RNA-seq analyses of HSB-2 cells with CRISPR-targeted LINC01871 deletion (LINC01871−/−) and in vitro stimulation assays.
Results
Whole blood RNA-seq revealed autoantibody-specific transcription profiles and disproportionate downregulation of DE transcripts in SjD cases relative to HCs. Sixteen DE lncRNAs exhibited correlated expression with the interferon (IFN)-regulated gene, RSAD2, in SjDRo+ (r≥0.65 or ≤−0.6); four antisense lncRNAs exhibited IFN-regulated expression in immune cell lines. LINC01871 was upregulated in all SjD cases. RNA-seq and pathway analyses of LINC01871−/− cells implicated roles in cytotoxic function, differentiation and IFNγ induction. LINC01871 was induced by IFNγ in a myeloid cell line and regulated by calcineurin/NFAT pathway and T cell receptor (TCR) signalling in primary human T cells.
Conclusion
LINC01871 influences expression of many immune cell genes and growth factors, is IFNγ inducible, and regulated by calcineurin signalling and TCR ligand engagement. Altered LINC01871 expression may influence the dysregulated T cell inflammatory pathways implicated in SjD.
Keywords: Sjogren's Syndrome; Polymorphism, Genetic; Autoimmune Diseases; Autoimmunity; Autoantibodies
WHAT IS ALREADY KNOWN ON THIS TOPIC
Sjögren’s disease (SjD) is an understudied heterogeneous autoimmune disease generally characterised as extreme dryness of the eyes and mouth caused by immune-mediated destruction of exocrine glands.
Identifying transcripts that are differentially expressed in SjD subphenotypes and have functional implications in disease pathology (especially undercharacterised long non-coding RNAs (lncRNAs)) provides important new insights into the disease mechanisms that drive SjD and identify potential biomarkers of different subphenotypes.
WHAT THIS STUDY ADDS
Pathway analyses of whole blood transcriptomes from patients with SjD that are either positive or negative for the hallmark autoantibody, anti-Ro (SjDRo+ vs SjDRo−), and healthy controls suggest these two SjD subphenotypes may have different mechanisms of disease pathology: interferon-mediated disease in SjDRo+ patients and T cell-driven disease in SjDRo− and SjDRo+ patients.
Identified the lncRNA, LINC01871, as differentially expressed in SjD and important for T cell function.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Study clearly implicates dysregulation of lncRNAs as important mechanisms driving both interferon-mediated and T cell-driven pathology of SjD. As such, it provides critical insights for future studies aimed at identifying biomarkers of SjD for clinical diagnosis. The approaches described herein also provide a roadmap to guide future studies in the discovery and functional characterisation of novel lncRNAs with potential disease implications.
Introduction
Sjögren’s disease (SjD) is a heterogeneous autoimmune disease that affects ~0.7% of the worldwide population with a female-to-male disparity of greater than 9:1.1–3 Extreme dry eyes and mouth are hallmark symptoms of SjD caused by chronic inflammation, lymphocytic infiltration, and subsequent destruction of lacrimal and salivary glands.4 5 Many patients with SjD also experience extraglandular complications including pulmonary and kidney dysfunction, neuropathy, debilitating fatigue, arthritis/arthralgia, leucocytoclastic vasculitis and an increased risk of 9-fold to 20-fold of non-Hodgkin’s lymphoma.5 Although the aetiology and pathological mechanisms remain unknown, it is widely hypothesised that genetic susceptibility likely influences disease onset, progression and heterogeneity in the context of specific, but not yet defined, environmental conditions.6 7
Research classification criteria for SjD require focal lymphocyte sialoadenitis of a biopsied salivary gland and/or circulating autoantibodies to the RNA-binding protein, Ro60/SSA/TROVE-2 and/or the ubiquitin ligase protein, Ro52/TRIM21.8–11 Circulating anti-Ro/SSA antibodies are common in SjD (~60%–70%; SjDRo+) and have been correlated with the upregulation of interferon (IFN)-stimulated genes (ie, IFN signature), increased lymphocyte infiltration of glands, more severe salivary gland involvement, increased prevalence of systemic extraglandular disease and higher risk of developing non-Hodgkin’s lymphoma.12–14 In contrast, patients with anti-Ro autoantibody-negative SjD (SjDRo−) lack the IFN signature and have a lower risk of lymphoma, but exhibit increased dryness measures, peripheral nervous system involvement and evidence of T cell-driven pathologies.15–17
Long non-coding RNAs (lncRNAs) are a diverse group of non-protein-coding RNAs greater than 200 nucleotides in length that modulate chromatin remodelling, transcription, and/or post-transcriptional modifications.18 Transcriptome profiling studies have identified numerous differentially expressed (DE) lncRNAs in autoimmune diseases, including SjD.19–25 These and similar studies applied expression correlation analyses with protein-coding RNAs (pcRNAs) to gain insights into the molecular roles of DE lncRNAs, but precise functional mechanisms remain undefined for many of the ~150 000 reported lncRNAs in the NONCODE database (noncode.org). For example, the lncRNA, negative regulator of the IFN response (NRIR), was shown to impair hepatocyte responses to IFN, but was also reportedly upregulated and functioned to positively regulate IFN responses in monocytes from patients with systemic sclerosis.26 27 Nuclear enriched abundant transcript 1 (NEAT1) positively regulates BAFF-mediated type I IFN activation of B cells in lupus-prone mice, mitogen-activated protein kinase (MAPK) signalling in cells from patients with SjD and inflammation signalling through TLR4.22 28 29 IFNG-AS1 (TMEVPG1 or NeST) increases Th1 responses in patients with SjD.30 Together, these studies suggest previously uncharacterised DE lncRNAs may yield important mechanistic implications and/or serve as novel biomarkers for SjD and other human diseases.
This study leveraged whole blood RNA-sequencing (RNA-seq) analyses coupled with bioinformatics pathway and co-expression correlation analyses to identify and differentiate important regulators of the IFN signature-dominant SjDRo+ subphenotype from the T cell-driven pathologies shared by SjDRo+ and SjDRo−. To this end, we identified four antisense lncRNAs that were overexpressed in the SjDRo+ and co-expressed with IFN-regulated transcripts. LINC01871, a previously uncharacterised lncRNA, was found to be overexpressed in both SjD subphenotypes and regulated through both IFNγ and calcineurin/NFAT signalling. Collectively, this study provides insights into the pathways that influence SjDRo+ and SjDRo− subphenotypes and mechanistic roles of several lncRNAs implicated in immune cell regulation and SjD pathogenesis.
Methods
See online supplemental materials and methods.
rmdopen-2022-002672supp001.pdf (32.3MB, pdf)
Results
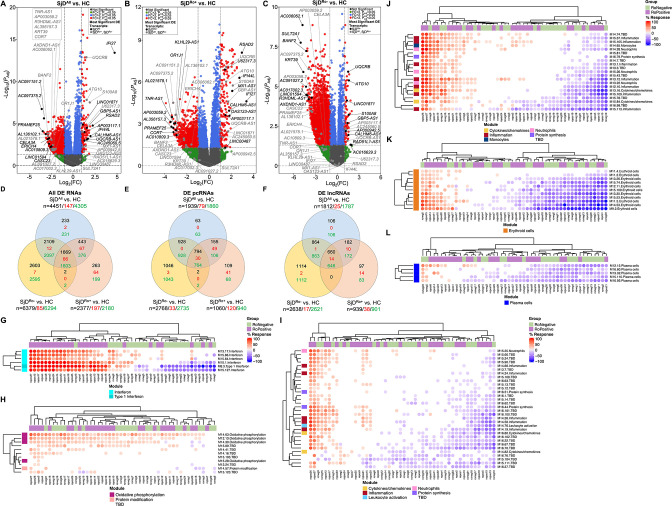
RNA-seq was performed using RNA from globin-depleted whole blood from 50 patients with SjD (27 SjDRo+, 23 SjDRo−) and 27 healthy controls (HCs) (online supplemental table 1). After quality control, a total of 37 821 transcripts were analysed for DE (log2 fold change (log2FC) ≥1 or ≤−1; false discovery rate-adjusted p value (padj)≤0.05). Four analyses were performed: (1) all SjD cases (SjDAll) versus HCs (figure 1A; online supplemental table 2); (2) SjDRo+ versus HCs (figure 1B; online supplemental table 3); (3) SjDRo− versus HCs (figure 1C; online supplemental table 4); (4) SjDRo+ versus SjDRo− (online supplemental table 5). The SjDAll versus HC, SjDRo+ versus HC and SjDRo− versus HC analyses identified 4451, 2377, and 6379 DE transcripts, respectively, with nearly equal representation of pcRNAs and lncRNAs (figure 1D–F; online supplemental table 6). The majority of genes were downregulated in all three SjD DE transcript sets relative to HCs (figure 1D–F), indicating repression of gene expression, lack of appropriate upregulation of gene expression or substantially different blood cellular composition in SjD. Differential blood cellularity, mostly involving leucocytopenia, has been described in SjD.16 31 Deconvolution was performed using quantiseqr32 to assess whether differences in cell type composition between the three datasets may confound interpretations of the RNA-seq results. Significant differences between SjDRo+ or SjDRo−, and HCs were only observed in macrophages and monocytes (online supplemental figure 1).
Figure 1.
Protein-coding (pc)RNAs and long non-coding (lnc)RNAs are differentially expressed in the whole blood of SjD cases compared with healthy controls (HCs). (A–C) Differentially expressed (DE) transcripts from whole blood RNA-sequencing analysis of (A) all SjD cases (SjDAll; n=50), (B) anti-Ro positive SjD cases (SjDRo+; n=27) or (C) anti-Ro negative SjD cases (SjDRo−; n=23) compared with HCs (n=27). Y-axis shows the −log10 of the FDR-adjusted p value (padj); x-axis shows the log2 of the fold change (FC). Black dots indicate the top three upregulated and downregulated pcRNAs and lncRNAs in the analysis. Grey dots indicate the top three upregulated and downregulated pcRNAs and lncRNAs in the other analyses. (D–F) Distribution of (D) all DE transcripts, (E) DE pcRNAs or (F) DE lncRNAs across the three analyses. Black text indicates total DE transcripts; red text indicates upregulated transcripts; green text indicates downregulated transcripts. (G–L) Hierarchical clustering of annotated module aggregates across individual SjDRo+ (purple) or SjDRo− (green) cases. Displayed individual modules had DE of >20% of constitutive transcripts in at least one case. Colour gradient indicates the proportion of DE transcripts ranging from 100% increased (red) to 100% decreased (blue), respective to HCs. FDR, false discovery rate; SjD, Sjögren’s disease.
Differential expression of pcRNAs
A total of 3097 unique pcRNAs were DE across all three analyses (figure 1E). In the SjDAll versus HC and SjDRo+ versus HC analyses, the most upregulated DE pcRNAs were IFN-stimulated genes: RSAD2, IFI44L and IFI27 (figure 1A, B; table 1; online supplemental tables 2 and 3).33 The IFN-stimulated gene, S100A8, was upregulated in all three analyses. In contrast, many of the upregulated DE pcRNAs in the SjDRo− versus HC analysis, including ATG10 and UQCRB, were involved in protein processing and cell metabolism (figure 1C; table 1; online supplemental table 4).
Table 1.
RNA-seq analysis of whole blood from SjD cases reveals common and subphenotype-specific differently expressed pcRNAs and lncRNAs
| ENSEMBL ID | Gene symbol | Cytoband | Strand | Chromosome position (hg19) | SjDAll | SjDRo+ | SjDRo− | ||||
| Log2FC | Padj | Log2FC | Padj | Log2FC | Padj | ||||||
| pcRNA | ENSG00000143546 | S100A8 | 1q21.3 | – | chr1:153,362,508–153,363,664 | 1.91 | 3.03E-08 | 2.13 | 1.75E-06 | 1.60 | 6.17E-04 |
| ENSG00000137959 | IFI44L | 1p31.1 | + | chr1:78,619,922–78,646,145 | 1.99 | 3.09E-04 | 2.72 | 4.98E-07 | 0.02 | 9.60E-01 | |
| ENSG00000134321 | RSAD2 | 2p25.2 | + | chr2:6,865,806–6,898,239 | 2.25 | 1.69E-05 | 2.95 | 7.82E-09 | 0.24 | 5.79E-01 | |
| ENSG00000152348 | ATG10 | 5q14.1 | + | chr5:81,972,023–82,276,857 | 1.56 | 1.52E-08 | 1.53 | 9.35E-07 | 1.60 | 4.07E-06 | |
| ENSG00000156467 | UQCRB | 8q22.1 | – | chr8:96,222,947–96,235,546 | 1.52 | 1.35E-13 | 1.54 | 8.08E-08 | 1.51 | 1.14E-06 | |
| ENSG00000165949 | IFI27 | 14q32.12 | + | chr14:94,104,836–94,116,698 | 4.74 | 3.90E-14 | 3.36 | 2.39E-05 | −0.40 | 3.76E-01 | |
| ENSG00000241563 | CORT | 1p36.22 | + | chr1:10,450,031–10,451,998 | −2.59 | 8.40E-03 | −3.22 | 3.95E-03 | −2.07 | 5.69E-02 | |
| ENSG00000229571 | PRAMEF25 | 1p36.21 | + | chr1:13,068,677–13,077,884 | −3.46 | 8.10E-05 | −3.24 | 1.95E-03 | −3.71 | 2.87E-04 | |
| ENSG00000142789 | CELA3A | 1p36.12 | + | chr1:22,001,657–22,012,542 | −2.87 | 5.61E-04 | −2.16 | 1.32E-02 | −1.57 | 7.87E-02 | |
| ENSG00000136834 | OR1J1 | 9q33.2 | – | chr9:122,476,958–122,477,926 | −2.35 | 8.86E-03 | −3.06 | 8.80E-04 | −1.77 | 6.25E-02 | |
| ENSG00000196859 | KRT39 | 17q21.2 | – | chr17:40,958,417–40,966,948 | −2.43 | 1.51E-02 | −1.72 | 9.07E-02 | −4.65 | 1.20E-06 | |
| ENSG00000204978 | ERICH4 | 19q13.2 | + | chr19:41,443,156–41,444,765 | −3.02 | 6.33E-04 | −2.97 | 5.99E-03 | −3.06 | 3.75E-03 | |
| ENSG00000105398 | SULT2A1 | 19q13.33 | – | chr19:47,870,467–47,886,315 | −1.66 | 7.15E-02 | −0.79 | 3.96E-01 | −5.02 | 1.70E-08 | |
| ENSG00000125888 | BANF2 | 20p12.1 | + | chr20:17,693,672–17,735,871 | −2.83 | 1.05E-04 | −2.18 | 7.37E-03 | −4.23 | 5.65E-08 | |
| Antisense lncRNA | ENSG00000237568 | GBP5-AS1 | 1p22.2 | + | chr1:89,260,582–89,269,754 | 1.58 | 3.20E-05 | 1.72 | 2.54E-05 | 1.42 | 1.99E-03 |
| ENSG00000244158 | CALHM6-AS1 | 6q22.1 | – | chr6:116,460,739–116,463,692 | 1.68 | 6.12E-04 | 1.80 | 1.13E-04 | 1.54 | 1.12E-02 | |
| ENSG00000254224 | UQCRB-AS1 | 8q22.1 | + | chr8:96,235,427–96,239,149 | 1.57 | 8.02E-04 | 1.70 | 3.39E-03 | 1.41 | 2.64E-02 | |
| ENSG00000257452 | OAS123-AS1 | 12q24.13 | – | chr12:112,907,628–113,017,751 | 1.24 | 1.83E-02 | 1.83 | 2.61E-04 | 0.00 | 9.98E-01 | |
| ENSG00000259038 | RAD51L1-AS1 | 14q24.1 | – | chr14:68,627,166–68,628,445 | 1.64 | 5.64E-02 | 1.28 | NR | 1.98 | 4.80E-02 | |
| ENSG00000228318 | MX1-AS1 | 21q22.3 | – | chr21:42,813,321–42,814,669 | 1.43 | 4.44E-03 | 2.07 | 4.36E-06 | −0.09 | 8.84E-01 | |
| ENSG00000260990 | TNR-AS1 | 1q25.1 | + | chr1:175,307,218–175,335,459 | −2.52 | 6.47E-03 | −3.16 | 9.46E-04 | −1.99 | 3.62E-02 | |
| ENSG00000261250 | AXDND1-AS1 | 1q25.2 | – | chr1:179,543,201–179,548,922 | −2.43 | 3.35E-03 | −1.91 | 2.86E-02 | −3.68 | 5.32E-05 | |
| ENSG00000224361 | KLHL29-AS1 | 2p24.1 | – | chr2:23,507,043–23,524,344 | −1.68 | 5.51E-02 | −3.09 | 2.49E-03 | −0.82 | 3.93E-01 | |
| ENSG00000225328 | LINC01594 | 2q12.3 | – | chr2:108,167,125–108,217,886 | −2.57 | 4.91E-03 | −1.87 | 4.83E-02 | −4.03 | 2.94E-05 | |
| ENSG00000255420 | CASC23 | 11p15.4 | – | chr11:8,011,278–8,016,520 | −2.53 | 4.46E-03 | −1.96 | 4.61E-02 | −3.59 | 1.97E-04 | |
| ENSG00000276702 | AC010809.3 | 15q14 | – | chr15:33,850,538–33,851,178 | −2.80 | 2.14E-03 | −3.08 | 5.63E-03 | −2.52 | 1.72E-02 | |
| ENSG00000226812 | R3HDML-AS1 | 20q13.12 | – | chr20:44,347,552–44,355,185 | −2.40 | 8.07E-03 | −1.65 | 9.74E-02 | −3.92 | 6.23E-05 | |
| lncRNA | ENSG00000235576 | LINC01871 | 2p25.1 | + | chr2:7,725,801–7,730,705 | 1.16 | 1.78E-05 | 1.01 | 3.26E-03 | 1.33 | 7.67E-05 |
| ENSG00000205837 | LINC00487 | 2p25.2 | – | chr2:6,728,177–6,770,311 | 0.91 | 1.75E-01 | 1.49 | 4.04E-02 | −0.31 | 7.12E-01 | |
| ENSG00000270077 | AP003117.1 | 8p23.1 | + | chr8:97,144,170–97,144,723 | 1.33 | 1.02E-04 | 1.47 | 2.31E-04 | 1.15 | 3.79E-03 | |
| ENSG00000288528 | AP000942.5 | 11q22.2 | – | chr11:102,316,173–102,328,586 | 1.13 | 5.14E-03 | 1.04 | 4.99E-02 | 1.25 | 5.54E-03 | |
| ENSG00000286104 | AC016629.3 | 19q13.43 | + | chr19:58,575,445–58,601,316 | 1.25 | 1.33E-01 | 0.47 | NR | 1.84 | 4.71E-02 | |
| ENSG00000274422 | AC245060.5 | 22q11.22 | – | chr22:22,283,928–22,287,220 | 1.22 | 6.99E-04 | 1.22 | 9.18E-03 | 1.22 | 5.39E-03 | |
| ENSG00000273272 | U62317.3 | 22q13.33 | + | chr22:50,541,108–50,543,011 | 1.03 | 8.18E-05 | 1.44 | 1.67E-07 | 0.36 | 1.83E-01 | |
| ENSG00000285016 | AC017002.3 | 2q13 | – | chr2:111,429,324–111,699,033 | −2.01 | 3.03E-02 | −1.21 | 2.16E-01 | −4.24 | 3.27E-05 | |
| ENSG00000249818 | AC097375.2 | 4q31.3 | – | chr4:151,904,932–151,928,710 | −3.56 | 1.87E-07 | −3.25 | 3.00E-05 | −3.94 | 7.68E-07 | |
| ENSG00000287173 | AL356157.3 | 10q11.21 | + | chr10:44,509,708–44,515,421 | −2.48 | 1.26E-02 | −3.41 | 1.56E-03 | −1.73 | 1.01E-01 | |
| ENSG00000255250 | AP003059.2 | 11q14.2 | + | chr11:86,727,355–86,765,467 | −2.40 | 7.75E-03 | −3.49 | 7.12E-04 | −1.64 | 8.11E-02 | |
| ENSG00000267354 | AC091151.2 | 18q12.3 | + | chr18:45,507,202–45,550,183 | −3.31 | 2.36E-06 | −3.03 | 2.24E-04 | −3.64 | 1.93E-05 | |
| ENSG00000266586 | AC091027.2 | 18q12.3 | – | chr18:78,908,629–78,918,993 | −2.04 | 3.21E-02 | −1.20 | 2.28E-01 | −4.28 | 1.01E-05 | |
| ENSG00000275894 | AL021578.1 | 20q13.12 | – | chr20:45,345,115–45,345,823 | −2.87 | 2.91E-04 | −3.35 | 3.18E-04 | −2.38 | 8.14E-03 | |
| ENSG00000287107 | AL136102.1 | 20q13.13 | + | chr20:48,120,269–48,136,844 | −3.12 | 1.75E-04 | −2.86 | 3.48E-03 | −3.50 | 3.28E-04 | |
| ENSG00000287171 | AC006062.1 | Xq21.1 | + | chrX:8,487,165–8,504,564 | −2.43 | 7.94E-04 | −1.61 | 2.62E-02 | −4.76 | 1.32E-09 | |
Bold indicates top three upregulated and downregulated pcRNAs and lncRNAs in each analysis.
FDR, false discovery rate; lncRNAs, long non-coding RNAs; Log2FC, log2 fold change; Padj, FDR-adjusted p value; pcRNAs, protein-coding RNAs; RNA-seq, RNA sequencing; SjD, Sjögren’s disease; SjDAll, all SjD cases; SjDRo−, anti-Ro negative SjD; SjDRo+, anti-Ro positive SjD.
Many of the most downregulated DE pcRNAs from the SjDRo− versus HC analysis were also DE in the SjDAll versus HC and SjDRo+ versus HC analyses (figure 1A–C; table 1), indicative of common pathways across disease subphenotypes. Further, several of these transcripts, including CORT, PRAMEF25, KRT39, ERICH4 and BANF2, have not been previously reported as DE in SjD.
Pathway analyses of DE transcripts
Gene Set Enrichment Analysis34 35 (online supplemental tables 7 and 8), Ingenuity Pathway Analysis (IPA) (online supplemental figure 2A and tables 9–11) and the BloodGen3 module approach previously developed for whole blood lupus studies36–38 (figure 1G–L; online supplemental figure 2B–D) were performed using the DE transcript lists to gain insights into the functional similarities and differences between SjDRo+ and SjDRo−. Consistent with previous reports, the pathway analyses of the DE transcripts from the SjDRo+ versus HC analysis strongly indicated upregulated type I IFN signalling and other pro-inflammatory pathways (figure 1G, I, J; online supplemental figure 2A–C and table 12). Pathways of significance for the SjDRo− versus HC analysis included antigen receptor, cytokine and calcium/cAMP signalling (online supplemental figure 2A, B, D and table 12). Examination of all gene module sets from the BloodGen3 approach revealed significant heterogeneity among individual cases and, apart from the type I IFN signature, no clear segregation based on autoantibody status. The majority of subjects with SjD did exhibit increased expression of genes in the oxidative phosphorylation module set (figure 1H). Further, a subset of subjects with SjD, irrespective of autoantibody status, showed increased expression of genes in the erythroid module set (figure 1K), perhaps consistent with recent work showing the presence of erythrocyte mitochondria in juvenile systemic lupus erythematosus.39 Collectively, these analyses suggest that the disease pathologies of SjDRo+ involve a prominent IFN signature that is absent from, and less likely to influence disease pathology of, SjDRo−. However, the heterogeneous stratification of SjDRo+ and SjDRo− subjects with respect to other gene module sets suggests that multiple mechanisms likely contribute to SjD, with shared mechanisms of cellular metabolism and immune cell dysfunction.
Differential expression of lncRNAs
A total of 660 unique DE lncRNAs were shared across all three whole blood RNA-seq analyses; 97 lncRNAs were unique in the SjDRo+ versus HCs; 1114 lncRNAs were unique in the SjDRo− versus HCs (figure 1F; online supplemental table 6). The most upregulated DE antisense lncRNAs in the SjDRo+ versus HCs were also upregulated in the SjDAll versus HC analysis and are antisense to pcRNAs implicated in type I IFN signalling: AC004551.1 (antisense spanning the genes OAS1, OAS2 and OAS3, therefore, named OAS123-AS1; ENSG00000257452) and AP001610.1 (directly overlapping antisense to MX1, therefore, named MX1-AS1; ENSG00000228318) (figure 1A, B; table 1; online supplemental tables 2 and 3).33 The IFN-inducible lncRNA, NRIR,26 27 was also upregulated in both SjDAll versus HCs and SjDRo+ versus HCs (online supplemental tables 2 and 3). Significant upregulation of NRIR, OAS123-AS1 and MX1-AS1 in SjDAll and SjDRo+ was validated by quantitative reverse transcription PCR (RT-qPCR) in an independent cohort of 22 cases with SjD (14 SjDRo+, 8 SjDRo−) and 24 HCs (online supplemental figure 3A–C). AC099063.1 (directly overlapping antisense to the type II IFN responsive gene, GBP5; therefore, named GBP5-AS1; ENSG00000237568), CALHM6-AS1 and UQCRB-AS1 were upregulated DE antisense lncRNAs in all three RNA-seq analyses (figure 1A–C; table 1; online supplemental tables 2–4). GBP5-AS1 upregulation was validated by RT-qPCR in the independent SjDRo+and trended in SjDAll (online supplemental figure 3E). As observed with the pcRNAs, many of the downregulated antisense lncRNAs were shared among the three analyses, such as TNR-AS1, AXDND1-AS1 and CASC23 (figure 1A–C; table 1; online supplemental tables 2–4).
Several lncRNAs, including LINC01871 and AC245060.5, were also DE across all three RNA-seq analyses (figure 1A–C; table 1; online supplemental tables 2–4). LINC01871 upregulation was validated by RT-qPCR in the independent cohort (online supplemental figure 3D). Two DE lncRNAs were uniquely upregulated in the SjDRo+ versus HCs: LINC00487 and U62317.3 (figure 1B; table 1; online supplemental table 3). AC016629.3 was unique to and the most upregulated DE lncRNA in the SjDRo− versus HCs (figure 1C; table 1; online supplemental table 4). Lastly, the SjDRo− versus HC analysis exhibited the most downregulated lncRNAs with two subphenotype-specific transcripts: AC017002.3 and AC091027.2 (figure 1C; table 1; online supplemental table 4).
Identification and cell type-specific expression of IFN-related lncRNAs
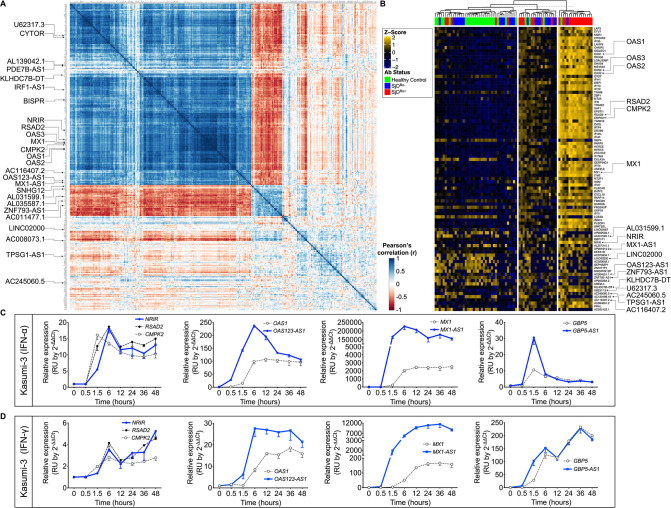
Pearson’s correlation analyses were performed using the highly DE IFN-stimulated pcRNA, RSAD2 and DE transcript lists from the SjDAll, SjDRo+, SjDRo−, and HC datasets to identify RSAD2-correlated transcripts (r>0.65 or <−0.6; p<0.05) (figure 2A; online supplemental table 13). Numerous IFN-responsive pcRNAs and 16 lncRNAs from the SjDRo+ DE transcript list correlated with RSAD2 expression, including MX1-AS1, NRIR, BISPR, OAS123-AS1 and IRF1-AS1 (figure 2A; online supplemental tables 13 and 14).33 40 CYTOR, an lncRNA implicated in cancer and autophagy,41–43 was also correlated. Five anti-correlated lncRNAs were also identified, including ZNF793-AS1, which was DE in SjDAll versus HC and SjDRo− versus HC RNA-seq analyses.
Figure 2.
Differentially expressed (DE) interferon (IFN)-responsive lncRNAs were coordinately modulated with associated pcRNAs and correlated with antibody status of SjD cases. (A) RSAD2 correlation analysis of the normalised RNA-seq data from the SjDRo+-only primary expression matrix (r≥0.65 or ≤−0.6; p<0.05). Type I IFN-responsive pcRNAs and other pcRNAs and lncRNAs of interest are indicated. (B) Individual-level expression of transcripts that were correlated with RSAD2 expression and DE in any of the whole blood RNA-seq analyses. Z-scores were computed using scale function in R, after calculating fold change of expression relative to healthy controls. K-means clustering (K=3) segregated cases by high, moderate and low IFN status, and are shown relative to patient antibody status. pcRNAs and lncRNAs of interest are indicated. (C, D) RT-qPCR analysis of indicated DE transcripts in Kasumi-3 cells stimulated with (C) universal type I IFN (IFNα; 150 U/mL) or (D) IFNγ (2800 U/mL) from 0 to 48 hours. Target lncRNAs are shown in blue and respective pcRNAs in grey (n>3). lncRNAs, long non-coding RNAs; pcRNAs, protein-coding RNAs; RNA-seq, RNA sequencing; RT-qPCR, quantitative reverse transcription PCR; SjD, Sjögren’s disease; SjDRo+, anti-Ro positive SjD; SjDRo−, anti-Ro negative SjD.
Individual-level expression of transcripts that were both DE in the whole blood RNA-seq analyses and correlated with RSAD2 expression segregated into IFN high, moderate and low clusters that corresponded with anti-Ro antibody status (figure 2B). Correlated expression of RSAD2 and IFN-responsive lncRNAs in SjDRo+ subjects further implicated these lncRNAs as potential regulators of IFN responses. Interestingly, Blueprint Epigenome cell type-specific RNA-seq data from 62 cell types revealed lower and more cell type-restricted expression of these lncRNAs, relative to IFN-responsive pcRNAs (online supplemental figure 3G).
Modulation of lncRNAs in response to IFN in vitro
RT-qPCR was performed with RNA from Kasumi-3 (early myeloid) cell lines stimulated with IFNα or IFNγ over a 48-hour time course to determine if the RSAD2 correlated and DE lncRNAs were modulated by IFN. NRIR and the IFN-stimulated pcRNAs, RSAD2 and CMPK2 were coordinately upregulated in response to IFNα in Kasumi-3 cells (figure 2C). OAS123-AS1, MX1-AS1 and GBP5-AS1 were also upregulated, but their induction preceded upregulation of respective pcRNAs, OAS1, MX1 or GBP5 (figure 2C). IFNγ stimulation also upregulated NRIR expression by 6 hours, and OAS123-AS1, MX1-AS1 and GBP5-AS1 expression by 1.5 hours (figure 2D). Observed kinetics of the IFN response suggests that these antisense lncRNAs may positively regulate the spatially linked pcRNAs.
Identification of dysregulated lncRNAs common across SjD subphenotypes
Identifying and characterising lncRNAs that are common among different SjD subphenotypes may provide new insights into common disease mechanisms. Fourteen of the 660 shared DE lncRNAs identified in the whole blood RNA-seq analyses were overexpressed (figure 1F; online supplemental table 6). Twelve were antisense or sense-intronic transcripts, including GBP5-AS1, CALHM6-AS1, LARP7-AS1 (miR302CHG) and UQCRB-AS1, and two were long intergenic non-coding RNAs, LINC01871 and AC245060.5 (table 1; online supplemental tables 2–4). Minimal information was available on AC245060.5, but LINC01871 was reported as DE in multiple cancer studies, autoinflammatory diseases, T cells and monocytes (data derived from Gene Expression Atlas; online supplemental figure 3F),44–49 making it a transcript of interest in human disease.
LINC01871, located at the 2p25.1 locus, has low exonic conservation among vertebrates, multiple isoforms found only in humans and low protein-coding potential (online supplemental note).50 The surrounding genomic interval has an enrichment of H3K27ac epigenetic marks, DNaseI hypersensitivity and transcription factor binding indicative of active transcription. Consistently, LINC01871 is strongly expressed in mature CD4+ and CD8+ T cells, class switched memory B cells, plasma cells, natural killer (NK) cells and haematopoietic progenitor cells (data derived from Blueprint Epigenome RNA-seq data; online supplemental figure 3H). Conversely, regulatory T cells and most B cells have low basal expression of LINC01871 and early lineage T cells (CD3− thymocytes) do not express LINC01871. Analysis of publicly available single-cell RNA-seq data of minor salivary gland tissue revealed that LINC01871 is most highly expressed in tissue-resident T cell subtypes and NK cells (online supplemental figure 3I, J).51 52 These data confirm the expression of this lncRNA in immune cells present in SjD-affected tissues, indicating potential relevance to disease pathology.
Interrogation of publicly available GeneFriends,53 FuncPred54 and lncRNA2function55 databases found that LINC01871 expression was correlated with pcRNAs implicated in IFNγ stimulation, T cell activation and differentiation, myeloid cells and dendritic cells (online supplemental tables 15–17). In this SjD cohort, LINC01871 expression was highest in the SjDRo− subjects (online supplemental figure 3D).
Pearson’s correlation analyses were performed using the DE transcript list from the SjDRo− versus HC analysis to identify transcripts of specific interest to immune cell activation and regulation correlated with LINC01871 (r>0.7 or <−0.6; p<0.05) (figure 3A; online supplemental table 18). Of specific interest, CAMK2N1 and NFAT5 are both directly regulated by calcium signalling,56 while EOMES, GZMA and FGFBP2 are important for cytotoxic immune responses.33 Consistently, Gene Ontology (GO) Enrichment Analysis,57 performed using the LINC01871-correlated DE transcripts list, identified GO terms implicating LINC01871 in immune system function, leucocyte and lymphocyte activation, and IFNγ production (online supplemental table 19).
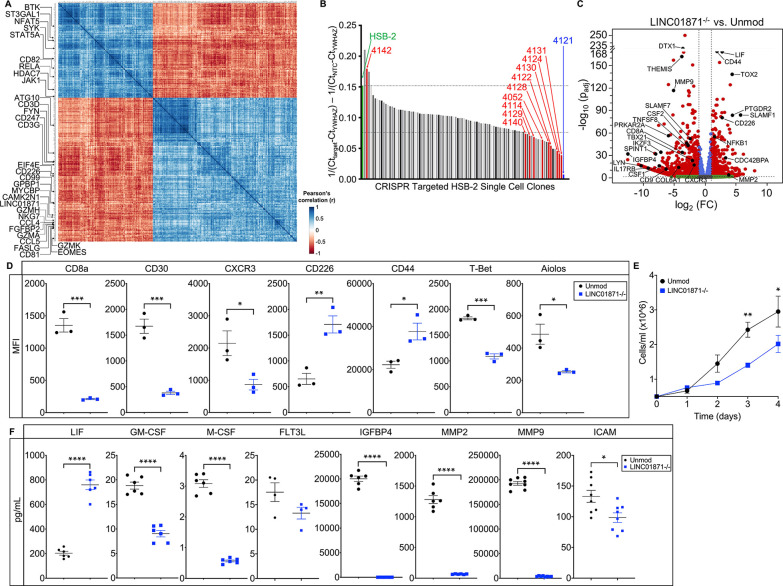
Figure 3.
Loss of LINC01871 disrupts basal expression of genes involved in immune cell regulation. (A) LINC01871 correlation analysis of normalised RNA-seq data from the SjDRo−-only primary expression matrix (r≥0.7 or ≤−0.6; p<0.05). Transcripts implicated in immune function or SjD pathology are indicated. (B) Quantitative PCR screen of LINC01871 expression in HSB-2 single-cell clones after CRISPR-targeted deletion of LINC01871. (C) Differentially expressed transcripts from the RNA-seq analysis of HSB-2 clone 4121 (hereafter LINC01871−/−) relative to HSB-2 parental cell line. Y-axis shows the −log10 of the FDR-adjusted p value (padj); x-axis shows the log2 of the fold change (FC). Black dots indicate independently replicated transcripts of interest. (D) Levels of indicated surface or intracellular proteins, reported as mean fluorescence intensity (MFI), in HSB-2 parental cells (black) and LINC01871−/− cells (blue); n=3; unpaired t-test where *p<0.05, **p<0.01 or ***p<0.001. (E) Growth curve analysis of HSB-2 parental cells (black) and LINC01871−/− cells (blue) from 0 to 96 hours; n=3; unpaired t-test where *p<0.05 or **p<0.01. (F) Concentration of indicated secreted protein in supernatant collected from HSB-2 parental cells (black) and LINC01871−/− cells (blue) at 96 hours; n>6; unpaired t-test where *p<0.05 or ****p<0.0001. FDR, false discovery rate; RNA-seq, RNA sequencing; SjD, Sjögren’s disease; SjDRo−, anti-Ro negative SjD.
Functional analysis of LINC01871 by RNA-seq analysis of a clonal CRISPR-targeted LINC01871 deletion
To explore the function of LINC01871, CRISPR-guided RNAs were designed to target the LINC01871 interval in early T cell lineage HSB-2 cell line, which constitutively express LINC01871 (figure 3B; online supplemental figure 4A and table 20). Clones of the CRISPR-edited HSB-2 cells were screened for LINC01871 deletion by RT-qPCR, identifying clone 4121 (hereafter referred to as LINC01871−/−) as having complete deletion of the targeted LINC01871 interval confirmed by Nanopore sequencing (figure 3B; online supplemental figure 4B, C and tables 21 and 22).
RNA-seq analysis of LINC01871−/− cells relative to unedited HSB-2 parental cells identified 1166 DE transcripts (log2FC≥1 or ≤−1; padj≤0.05) (figure 3C; table 2; online supplemental table 23). Loss of LINC01871 disproportionately decreased the expression of pcRNAs (~3:1); perturbations of ncRNAs were more equally distributed (online supplemental table 24). Notably, several transcripts that were positively correlated with LINC01871 in the whole blood RNA-seq analyses, such as CCL5 (log2FC=−3.46; padj=9.82e-12) and FGFBP2 (log2FC=−3.54; padj=1.62e-17) (online supplemental tables 2–4,18), were significantly downregulated in LINC01871−/− cells (figure 3C; table 2; online supplemental table 23). In addition, loss of LINC01871 completely abrogated the expression of 127 transcripts, including adaptive immunity genes TNFSF8 (CD30L), IL22, CD27 and LYN (table 2; online supplemental table 23).
Table 2.
CRISPR deletion of LINC01871 alters the expression of several immune regulatory transcripts
| Gene (protein) symbol | Log2FC | Padj | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| Immune regulation and function | LYN | −10.392 | 1.24E-16 | ||||||||||||
| CD109 | −9.707 | 2.05E-14 | |||||||||||||
| TNFSF15 | −9.598 | 4.64E-14 | |||||||||||||
| IL17RB | −9.324 | 1.87E-13 | |||||||||||||
| IL22 | −8.080 | 1.70E-09 | |||||||||||||
| IKZF3 (Aiolos) | −7.158 | 1.62E-34 | |||||||||||||
| PDCD1 (PD-1) | −6.243 | 1.07E-06 | |||||||||||||
| CD27 | −6.128 | 1.20E-04 | |||||||||||||
| TNFSF8 (CD30L) | −5.727 | 8.29E-04 | |||||||||||||
| LAG3 | −4.684 | 1.04E-14 | |||||||||||||
| CD79A | −4.091 | 2.66E-08 | |||||||||||||
| EGR2 | −3.925 | 5.74E-62 | |||||||||||||
| THEMIS | −3.770 | 5.85E-162 | |||||||||||||
| CD8A | −3.683 | 4.03E-34 | |||||||||||||
| CCL5 | −3.456 | 9.82E-12 | |||||||||||||
| FOXP3 | −3.064 | 1.78E-03 | |||||||||||||
| IRF5 | −2.959 | 1.19E-10 | |||||||||||||
| TNFRSF8 (CD30) | −2.537 | 3.47E-44 | |||||||||||||
| SLAMF7 | −2.398 | 1.90E-52 | |||||||||||||
| TBX21 (T-Bet) | −2.066 | 2.45E-23 | |||||||||||||
| IL6ST | −1.820 | 2.16E-21 | |||||||||||||
| CXCR3 | −1.728 | 2.68E-18 | |||||||||||||
| TNFRSF4 | 1.910 | 1.37E-09 | |||||||||||||
| CD44 | 1.924 | 1.68E-174 | |||||||||||||
| TGFBR3 | 2.069 | 8.65E-16 | |||||||||||||
| CD226 | 2.750 | 1.96E-80 | |||||||||||||
| SELL (CD62L) | 3.061 | 1.82E-15 | |||||||||||||
| HAVCR2 | 3.139 | 1.95E-87 | |||||||||||||
| CCR4 | 3.816 | 4.47E-08 | |||||||||||||
| KLF2 | 4.043 | 4.13E-60 | |||||||||||||
| TOX2 | 4.353 | 2.58E-138 | |||||||||||||
| CCR2 | 4.893 | 9.69E-13 | |||||||||||||
| TIGIT | 5.531 | 2.31E-08 | |||||||||||||
| SLAMF1 (CD150) | 5.726 | 4.86E-84 | |||||||||||||
| Growth signalling/cancer regulation | IGFBP4 | −12.476 | 3.61E-32 | ||||||||||||
| CSF1 (M-CSF) | −7.356 | 5.58E-16 | |||||||||||||
| PTPN13 | −5.998 | 4.19E-05 | |||||||||||||
| TLR2 | −5.914 | 7.51E-05 | |||||||||||||
| PRKAR2A | −5.899 | 2.08E-56 | |||||||||||||
| MMP9 | −5.096 | 9.32E-117 | |||||||||||||
| FSCN1 | −5.003 | 0.00E+00 | |||||||||||||
| PDGFD | −4.730 | 7.54E-03 | |||||||||||||
| GZMB | −4.688 | 2.93E-02 | |||||||||||||
| P2R×7 | −4.190 | 1.17E-08 | |||||||||||||
| ADCY6 | −4.163 | 5.86E-21 | |||||||||||||
| FGFBP2 | −3.543 | 1.62E-17 | |||||||||||||
| CSF2 (GM-CSF) | −3.052 | 6.62E-48 | |||||||||||||
| TNF | −1.631 | 1.22E-88 | |||||||||||||
| CD28 | 1.614 | 7.59E-30 | |||||||||||||
| XCL1 | 1.636 | 1.92E-09 | |||||||||||||
| TLR1 | 1.674 | 4.87E-02 | |||||||||||||
| LIF | 2.613 | 2.15E-171 | |||||||||||||
| ITGA6 | 3.072 | 0.00E+00 | |||||||||||||
| HPGD | 3.517 | 1.88E-37 | |||||||||||||
| MMP2 | 3.851 | 1.17E-02 | |||||||||||||
| ITGAM | 4.198 | 4.15E-03 | |||||||||||||
| IL7R | 4.393 | 1.41E-02 | |||||||||||||
| PTGDR2 | 4.399 | 1.27E-83 | |||||||||||||
| ALOX15B | 5.189 | 5.42E-03 | |||||||||||||
| PTGDR | 5.460 | 1.02E-10 | |||||||||||||
| HPGDS | 7.992 | 1.99E-09 |
Genes in bold denote correlation with LINC01871 in SjDRo− (online supplemental table 18).
1. Th1 pathway; 2. Th2 pathway; 3. Th1/Th2 activation; 4. CD8 cytotoxic/cytokines; 5. Treg/Th17; 6. T cell development; 7. T cell exhaustion; 8. associated with autoimmune disease; 9. molecular mechanisms of cancer; 10. proliferation/crosstalk; 11. extravasation/migration; 12. prostaglandin pathway.
FDR, false discovery rate; log2FC, log2 fold change; Padj, FDR-adjusted p value; SjD, Sjögren’s disease; SjDRo−, anti-Ro negative SjD; Treg, regulatory T cell.
Consistent with the whole blood SjD RNA-seq correlation analyses and database functional predictions, GO Enrichment Analysis and IPA of the LINC01871−/− DE transcripts identified several pathways involved in immune cell development and signalling, including Th1/Th2 cells, T/B cell signalling and leucocyte extravasation, as well as pathways impacting growth and cell survival (table 2; online supplemental figure 4D–J and tables 19 and 25). Predictions of such wide-ranging impacts suggest that LINC01871 may act as a broad regulator impacting many immune cell pathways.
Validation of transcript expression and characterisation of growth and protein expression in LINC01871−/− cells
RT-qPCR was used to validate the DE of 32 transcripts using independent biological replicates of the LINC01871−/− cells compared with unmodified HSB-2 cells. Twenty-nine of the selected genes, including LINC01871, exhibited significant changes in gene expression (online supplemental figure 5A–D). Two of the remaining three transcripts trended but did not reach significance: SLAMF7 and CSF2 (online supplemental figure 5A and D). LINC01871−/− cells also displayed significant and stable changes in the expression of five cell surface proteins (CD8a, CD30, CXCR3, CD226 and CD44) and two intracellular transcription factors (T-bet and Aiolos) important in immune cell function (figure 3D; online supplemental figure 5E–K).58 59
Given the observed DE of transcripts involved in cell proliferation, the growth characteristics of the LINC01871−/− cells were also examined. While early growth rates of the LINC01871−/− cells appeared comparable with the unmodified HSB-2 cells, the growth rate of LINC01871−/− cells slowed with time in culture, suggesting that deletion of LINC01871 may perturb the secretion of growth factors (figure 3E). Consistently, several key secreted proteins/growth factors, including CSF1 (M-CSF protein) and CSF2 (GM-CSF protein), were reduced in the supernatants from the LINC01871−/− cells compared with the unmodified HSB-2 cells (figure 3F; online supplemental figure 5D). IGFBP4, a secreted inhibitor of insulin-like growth factor (IGF) with functions in inflammation and growth regulation,60 was also depleted. FLT3L, which encodes the protein Flt3-L, a critical dendritic cell cytokine that acts in concert with GM-CSF,61 62 trended toward a decrease. Interestingly, the pleiotropic cytokine, LIF (leukaemia inhibitory factor), was markedly increased, implicating LINC01871 as a negative regulator of this gene.
Perturbations in the concentrations of several proteins commonly associated with cell−cell interactions and extravasation were also observed. The extracellular adhesion and endoprotease proteins, MMP2 and MMP9,63 64 were both significantly reduced in the media of LINC01871−/− cells (figure 3F). However, MMP2 gene expression was increased in the RNA-seq analysis (log2FC=3.85; online supplemental figure 5D), suggesting some dysregulation of MMP2 protein secretion in the LINC01871−/− cells. Changes in the expression of MMP9 could not be replicated by qPCR, perhaps due to differential isoform detection by qPCR. Soluble ICAM, a marker of vascular inflammation,65 was also significantly decreased. Overall, deletion of LINC01871 in a T cell line caused widespread disruption of diverse cellular proteins, including the disruption of basal level expression of many proteins involved in adaptive immune responses, cell migration, adhesion and extravasation.
LINC01871 expression is modulated by IFNγ in a myeloid cell line
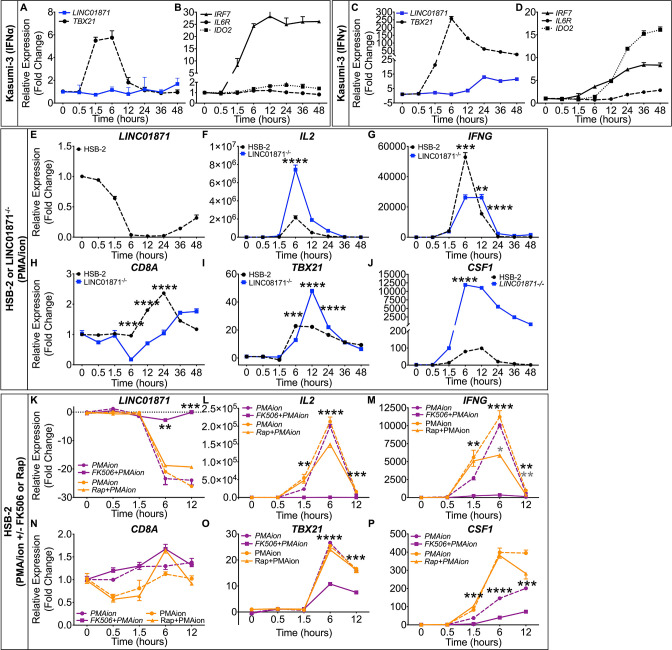
LINC01871 correlation analyses, bioinformatics data and CRISPR-edited LINC01871−/− in vitro studies suggested that LINC01871 may play a role in both IFN and immune cell activation pathways, specifically IFNγ signalling. Because HSB-2 cells do not respond to in vitro IFNγ stimulations and LINC01871 was reported to be overexpressed in acute myeloid leukaemia,46 we used Kasumi-3 cells to determine if LINC01871 expression is regulated by universal type I IFN (IFNα) and/or IFNγ (type II IFN) stimulations. IFNα stimulation did not induce LINC01871 in Kasumi-3 cells, but robustly induced the expression of IFN-inducible genes, TBX21 and IRF7 (figure 4A, B). In contrast, IFNγ upregulated LINC01871 expression (peak 12-fold induction), as well as the IFN-stimulated genes, TBX21, IL6R, IDO2 and IRF7 (figure 4C, D), thus demonstrating that LINC01871 expression in myeloid cells is regulated by type II IFN signalling.
Figure 4.
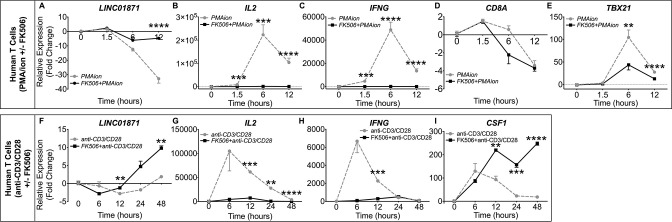
LINC01871 is modulated by interferon (IFN)γ and calcineurin/NFAT signalling in immune cell lines. (A−D) RT-qPCR analysis of LINC01871 (A and C), TBX21 (A and C), IRF7 (B and D), IL6R (B and D) and IDO2 (B and D) in Kasumi-3 cells stimulated with either (A, B) universal type I IFN (IFNα; 150 U/mL) or (C, D) IFNγ (2800 U/mL) from 0 to 48 hours. Expression is reported as fold change relative to unstimulated controls. (E−J) RT-qPCR analysis of LINC01871 (E), IL2 (F), IFNG (G), CD8A (H), TBX21 (I) and CSF1 (J) in LINC01871−/− cells (blue) and HSB-2 parental (black) cells stimulated from 0 to 48 hours with PMA/I (100 ng/mL; 1000 ng/mL). Expression is reported as fold change relative to unstimulated controls; n=3; unpaired t-test where **p<0.01, ***p<0.001 or ****p<0.0001. (K−P) RT-qPCR analysis of LINC01871 (K), IL2 (L), IFNG (M), CD8A (N), TBX21 (O) and CSF1 (P) in HSB-2 parental cells stimulated from 0 to 12 hours with PMA/I (100 ng/mL; 1000 ng/mL) with and without FK506 (2.5 µM; purple) or rapamycin (Rap; 2.5 µM; orange). Expression is reported as fold change relative to unstimulated inhibitor controls; n=3; paired t-test where *p<0.05, **p<0.01, ***p<0.001 or ****p<0.0001. Black * indicates FK506 with PMA/I relative to PMA/I alone and grey * indicates Rap with PMA/I relative to PMA/I alone. PMA/I, phorbol 12-myristate 13-acetate and ionomycin; RT-qPCR, quantitative reverse transcription PCR.
LINC01871 expression is regulated by calcineurin/NFAT signalling
To further test the hypothesis that LINC01871 may play a role in adaptive immune cell activation, specifically T cell activation, LINC01871−/− and parental HSB-2 cells were stimulated with phorbol 12-myristate 13-acetate and ionomycin (PMA/I) and the modulation of LINC01871, IL2, IFNG, CD8A, TBX21 and CSF1 expression was assessed by RT-qPCR. PMA/I was used to mimic T cell receptor (TCR) signalling because HSB-2 cells lack a functional TCR.66 67 PMA/I increases intracellular calcium without surface receptor engagement, leading to activation of the calmodulin-dependent protease, calcineurin, as well as protein kinase C (PKC), and subsequent downstream transcriptional activation through multiple pathways including NFAT and NFκB.66 67 LINC01871 expression was undetectable in LINC01871−/− cells. Parental HSB-2 cells showed marked LINC01871 downregulation after 6 hours of PMA/I stimulation that returned to near basal levels after ~36 hours (figure 4E). Targets of calcineurin/NFAT signalling, IL2 and IFNG, exhibited robust inductions in parental HSB-2 cells, indicating normal stimulation (figure 4F, G). LINC01871−/− cells showed an increased induction of IL2 expression, but an impaired induction of IFNG in response to PMA/I (figure 4F, G). This was not due to differences in basal expression as neither transcript was expressed in unstimulated cells. CD8A expression increased modestly in HSB-2 cells after PMA/I stimulation but decreased in LINC01871−/− cells (figure 4H). TBX21 and CSF1 both exhibited a higher magnitude of induction in LINC01871−/− cells compared with HSB-2 cells (figure 4I, J); however, this was likely due to lower basal expression in LINC01871−/− cells, resulting in a larger fold change induction.
Flow cytometry was used to determine if loss of LINC01871 also modulated PMA/I-mediated expression of cell surface proteins. PMA/I stimulation induced CD8A surface expression in parental HSB-2 cells but did not change CD8A expression in the LINC01871−/− cells, likely because of abrogated CD8A expression in LINC01871−/− cells (online supplemental figure 5L). Loss of LINC01871 prevented the PMA/I-mediated downregulation of CD30 and CXCR3 surface expression observed in parental HSB-2 cells (online supplemental figure 5M, N). In contrast, PMA/I upregulated the surface expression of CD226 and CD44 in both parental HSB-2 and LINC01871−/− cells (online supplemental figure 5O, P).
To determine if the PMA/I-mediated downregulation of LINC01871 resulted from calcineurin activation, parental HSB-2 cells were treated with the calcineurin-specific inhibitor, FK506 (tacrolimus),68 or rapamycin (mTOR signalling inhibitor in the same molecular class as FK506), prior to PMA/I stimulation. Treatment with FK506 alone did not alter LINC01871 expression, but presence of FK506 during PMA/I stimulation restored LINC01871 expression nearly to that of unstimulated cells (figure 4K). Rapamycin had no effect on LINC01871 expression. In addition, FK506 mitigated PMA/I-mediated induction of NFAT-regulated genes, IL2 and IFNG (figure 4L, M), and partially inhibited induction of two genes modulated in LINC01871−/− cells, TBX21 and CSF1 (figure 4O, P). Rapamycin also partially impaired PMA/I-mediated induction of IL2, but had no effect on TBX21 and CSF1. Both FK506 and rapamycin caused significant impairment of IFNG expression (figure 4M), indicating this pathway is regulated by both calcineurin and mTOR signalling. Neither inhibitor affected PMA/I-induced expression of CD8A (figure 4N). Overall, these data show that LINC01871 expression is downregulated in T cells in response to direct calcineurin/NFAT activation.
Human T cells regulate LINC01871 expression by calcineurin/NFAT signalling
To further examine the role of LINC01871 in T cells, the expression of LINC01871 and other transcripts of interest were measured in human T cells purified from healthy donor PBMCs treated with FK506 or DMSO and stimulated with PMA/I using RT-qPCR. As in the HSB-2 cells, PMA/I repressed LINC01871 expression in purified human T cells, beginning at 1.5 hours, and maintained repression 12 hours post-treatment (figure 5A), an effect that was abrogated by FK506. PMA/I-treated human donor T cells also exhibited similar transcriptional regulation of IL2, IFNG, CD8A and TBX21, relative to HSB-2 cells (figure 5B–E).
Figure 5.
LINC01871 is modulated by IFNγ and calcineurin/NFAT signalling in purified human T cells. (A–E) Negatively selected human donor T cells were treated with DMSO or 2.5 µM FK506 overnight, then stimulated with PMA/I (100 ng/mL; 1000 ng/mL) for 12 hours. Expression of LINC01871 (A), IL2 (B), IFNG (C), CD8A (D) and TBX21 (E) was assessed by RT-qPCR and reported as fold change relative to DMSO-treated cells; n=4; paired t-test where *p<0.05, **p<0.01, ***p<0.001 or ****p<0.0001. Grey lines indicate PMA/I. Black lines indicate PMA/I with FK506 treatment. (F–I) Negatively selected human donor T cells were treated with either DMSO or 2.5 µM FK506 overnight, then stimulated with anti-CD3/CD28 for 48 hours. Expression of LINC01871 (F), IL2 (G), IFNG (H) and CSF1 (I) was assessed by RT-qPCR and reported as fold change relative to DMSO-treated cells at each time point; n=4; paired t-test where *p<0.05, **p<0.01, ***p<0.001 or ****p<0.0001. Grey lines indicate anti-CD3/CD28. Black lines indicate anti-CD3/CD28 with FK506 treatment. IFN, interferon; PMA/I, phorbol 12-myristate 13-acetate and ionomycin; RT-qPCR, quantitative reverse transcription PCR.
Engagement of the TCR triggers a complex signalling cascade, including the activation of multiple signalling effectors including MAPKs, PKC and calcineurin.66 In contrast to our findings using PMA/I, LINC01871 expression was previously shown to increase in response to anti-CD3/CD28-activated TCR signalling in human T cells (online supplemental figure 3F; unpublished data).69 To address this, LINC01871 expression was assessed in purified human T cells stimulated with anti-CD3/CD28 antibodies. TCR signalling decreased LINC01871 expression at early time points (figure 5F), although to a lower, but significant, degree compared with PMA/I stimulation. However, LINC01871 expression increased twofold to threefold with longer anti-CD3/CD28 stimulation (48 hours), perhaps mimicking chronic TCR stimulation in vitro. FK506 treatment of anti-CD3/CD28-stimulated T cells shifted the kinetics of downregulation of LINC10871 at early time points, but augmented expression at late time points. As expected, anti-CD3/CD28 stimulation induced expression of IL2 and IFNG, which was nearly abrogated by FK506 treatment (figure 5G, H). CSF1 expression also exhibited increased expression with prolonged TCR stimulation in the presence of FK506 treatment (figure 5I). In conclusion, LINC01871 expression is modulated in human T cells in response to calcineurin/NFAT and TCR signalling. The overexpression of LINC01871 in SjD, coupled with the alterations observed in LINC01871−/− cells and its regulation by both IFNγ and calcineurin/TCR signalling, suggest that this lncRNA plays a role in immune cells that may be relevant to autoimmune disease.
Discussion
Dysregulated immune and salivary gland functions, including a prominent IFN signature, are implicated in the pathogenesis of SjD.12 70 Consistently, the whole blood RNA-seq from this study revealed IFN signalling dysregulation in the SjDRo+. In contrast, pathway analyses were indicative of T cell-driven disease mechanisms in the SjDRo−. Interestingly, the DE transcripts from both the SjDRo+ and SjDRo− exhibited overall repression of transcript expression relative to HCs, including downregulation of many lncRNAs. Further, direct comparison of SjDRo+ and SjDRo− yielded surprisingly few DE transcripts (online supplemental table 5), the majority of which were IFN-induced genes. Given previously reported differences in the absolute numbers of specific immune cell types in patients with SjD,16 the observed minor alterations in the abundance of immune cell types by deconvolution in this study (online supplemental figure 1), and the significant differences in the mean ages of the SjD and HC subjects from this study (online supplemental table 1), it is possible that these and other observations from this study may be influenced by the slight differences in the percentages of different immune cell types in the limited peripheral blood samples available from SjD cases subphenotyped based on Ro autoantibody status. Future studies using carefully matched case–control samples would be needed to resolve this limitation, as well as further improve our understanding of SjD heterogeneity.
Although recent studies have revealed important regulatory roles of several lncRNAs in immune responses and autoimmunity (reviewed in 71 72), few studies have investigated the potential roles of lncRNAs in SjD pathogenesis. The IFN signalling pathway is tightly regulated, requiring genetic ‘on/off’ switches to rapidly activate or suppress transcription in response to changing cellular environments.33 More recent studies have identified lncRNAs that function as important regulators of IFN signalling, autoimmunity and cancer. Conflicting reports of NRIR functioning as a positive regulator of IFN-inducible gene expression in monocytes,26 27 but a negative regulator of expression in hepatocellular carcinomas,27 suggest that NRIR expression is subject to cellular and context-specific regulatory effects. In this study, NRIR was overexpressed in whole blood of SjDRo+ and induced by both type I and type II IFN stimulation in vitro. We also demonstrated that three previously uncharacterised antisense lncRNAs were IFN responsive: OAS123-AS1, MX1-AS1 and GBP5-AS1. Our kinetic data suggest that these antisense lncRNAs are rapidly induced in response to IFNs and their induction may act as a massive upregulation signal to induce their respective pcRNAs. The precise mechanisms of action for these novel antisense lncRNAs will require further studies.
We identified 15 other lncRNAs (online supplemental table 14) that were also correlated with IFN-inducible gene expression, including BISPR (BST2 IFN-stimulated positive regulator), which is implicated in the regulation of tetherin function,40 and CYTOR, which interacts with microRNAs to modulate numerous signalling systems implicated in cancer.41–43 CYTOR, BISPR, NRIR73 and LINC0048774 (table 1) were previously reported as upregulated in patients with SjD. Interestingly, LINC00487 resides ~50 kb upstream of NRIR, suggesting the entire genomic region containing LINC00487/NRIR/CMPK2/RSAD2 is subject to IFN regulation. However, other lncRNAs reportedly overexpressed in SjD, such as IFNG-AS1,30 NEAT122 and PTV1,23 were not DE in our analyses (online supplemental tables 2–4). Further studies will be required to elucidate the mechanisms of action of undefined lncRNAs.
In this study, we identified LINC01871 as overexpressed in all patients with SjD. Deep bioinformatics data mining, expression correlation analyses in the whole blood SjDRo− RNA-seq dataset, and pathway analyses all implicated LINC01871 as a potential regulator of T cell activation and differentiation. Loss of LINC01871 in a T cell line altered the expression of numerous transcripts implicated in innate and adaptive immune function. Dysregulation of LINC01871 in SjD may play a role in the T cell-driven pathogenesis of SjD by impacting leucocyte migration and extravasation (CD44, MMP2, MMP9, ITGAM, SELL), IFNγ signalling (CXCR3) and cellular growth pathways (CSF1, CSF2).58 59 75 Notably, many of the pcRNAs perturbed due to LINC01871 deletion have been previously implicated in the pathogenesis of SjD and/or autoimmunity. MMP9 is reportedly overexpressed in the salivary gland of patients with SjD,63 64 and CXCR3 is strongly induced by IFNγ and highly expressed in the T cells of patients with SjD.75 CSF2 is also reportedly elevated in the plasma of patients with SjD.76 Further, LINC01871 is overexpressed in several human cancers, highlighting the importance of this transcript in the regulation of multiple cellular processes.44–47
Finally, we observed that LINC01871 was negatively regulated by both short-term calcineurin/NFAT and TCR signalling in both a T cell line and primary human T cells, but positively regulated during long-term TCR stimulation. While it is tempting to assume an overall downregulation of LINC01871 based on the use of PMA/I (figure 5A), this pharmacological stimulus is much more powerful and broadly acting than TCR signalling, modelled herein by cross-linking with anti-CD3/CD28 beads (figure 5F). The extent and duration of calcineurin activation after TCR signalling are distinct from that of PMA/I treatment, and this difference is readily appreciated in the bimodal regulation of LINC01871 through longer stimulation and the effect of FK506 as an augmenter, rather than an inhibitor, of expression at late time points. These seemingly contradictory outcomes point to complex regulation, likely by more than one pathway (eg, IFNγ, figure 4C). Further, we speculate that the upregulation of LINC01871 during chronic or continual TCR stimulation is consistent with LINC01871 dysregulation in response to dysregulated T cell function in autoimmune diseases. Dutta et al recently showed that calcineurin also has NFAT-independent activity by recruitment to the TCR and regulation of ICAM-1-dependent cell adhesion.77 Our observed kinetic-dependent differences in LINC01871 regulation in the primary human T cells emphasise the need for careful dissection of the involved molecular pathways in the future.
Our data clearly implicate the dysregulation of lncRNAs as an important mechanism driving the IFN-mediated and/or T cell-driven pathology of SjD. Understanding the underlying differential regulation of immunomodulatory gene expression in clinical subphenotypes of SjD will provide critical insights for future identification of biomarkers and/or development of novel therapeutic treatments. Further, the approaches described in this study provide a roadmap to guide future studies in the discovery and functional characterisation of novel lncRNAs with potential disease implications.
Data availability
Whole blood RNA-seq datasets generated during this study will be made available through the Databases of Genotypes and Phenotypes controlled-access study phs002723.v1.p1 after publication of this manuscript. RNA-seq datasets generated from the CRISPR-edited HSB-2 cell lines will be made available through the NCBI Sequence Read Archive after publication of this manuscript. Alternatively, access to the RNA-seq data will be made available in accordance with consent and data transfer/use agreement from the Oklahoma Medical Research Foundation by request to the corresponding author, Dr Christopher J Lessard (chris-lessard@omrf.org). All other data associated with this study were publicly available and cited in the manuscript and URLs provided as an online supplemental note.
Material availability
Participant whole blood RNAs used in this study were obtained from the Oklahoma Medical Research Foundation Sjögren’s Research Clinic (OMRF-SRC) with Institutional Review Board (IRB) approval. Request for samples must be submitted to Dr A Darise Farris (principal investigator of OMRF-SRC; Darise-Farris@omrf.org). Access will depend on limited sample availability and will require IRB approval and completion of a material transfer agreement. LINC01871−/− CRISPR-edited HSB-2 cell line generated in this study will be made available by request to the corresponding author (Dr Christopher Lessard; chris-lessard@omrf.org) after a material transfer agreement is completed. Sequences of the primers used to create and/or evaluate this cell line are provided in the online supplemental tables and can be ordered through commercial sources. All other materials used in this study are commercially available.
Patient and public involvement
This study leveraged pre-existing banked whole blood RNA. Therefore, it was not appropriate or possible to involve patients or the public in the design, conduct, reporting or dissemination plans of our research.
Acknowledgments
We thank the study participants who made this study possible. We also thank the investigators and funding organisations of dbGaP Study phs0002446.v1.p1, supported by NIDCR Z01DE000704 (Warner BM) through the National Institutes of Health, National Institute of Dental and Craniofacial Research, Division of Intramural Research. All patients seen at NIDCR provided informed consent prior to participation in IRB-approved research protocols (NIH IRB: 15-D-0051, NCT02327884).
Footnotes
MLJ, BK, CL, KLT and JAI contributed equally.
Contributors: CJL supervised the research and had final responsibility for the decision to submit for publication. MLJ, BK, CL, JAI, AMS, NM, GBW, JDW and IA conceived, designed and/or performed the experiments and/or analyses under the supervision of CJL. KMG, JAK, DML, LR, DUS, JMG, JJ, RHAS, PMG, KS, AR, ADF and CJL collected and characterised the SjD cases and healthy controls used in this study. KMG manages the human sample and data curation. SBG manages the technology required for processing and analysis of large-scale sequencing data. JMG, JJ, PMG, CGM, KS, ADF, IA and CJL acquired funding that supported the research reported in this manuscript. All authors contributed intellectual content during the drafting and revision of the work and approved the final version to be published.
Funding: This work was financially supported by the National Institutes of Health (NIH) (R01AR065953 to CJL, R01AR074310 to ADF, P50AR060804 to KS, UM1AI144292 and P30AR073750 to JJ and JMG, U54GM104938 to JJ, JMG and CGM, R01HL113326 to CGM, P20GM103456 to PMG, and R33AR076803 and R21AR079089 to IA), the Presbyterian Health Foundation (CJL), and the Sjögren’s Syndrome Foundation (CJL, KS).
Disclaimer: The content of this publication is solely the responsibility of the authors and does not represent the official views of the listed funding agencies.
Competing interests: KS is a current employee of Janssen. ADF and CJL have an active collaborative research agreement with Janssen. All other authors have reported that they have no competing interests to report.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Data generated during this study are available in accordance with consent and data transfer/use agreement from the Oklahoma Medical Research Foundation (OMRF) by request to the corresponding author, Dr. Christopher J. Lessard (chris-lessard@omrf.org). Participant whole blood RNAs used in this study were obtained from the OMRF Sjogren’s Research Clinic (OMRF-SRC) with institutional IRB Approval. Request for samples must be submitted to Dr. A. Darise Farris (PI of OMRF-SRC; darise-farris@omrf.org). Access will depend on limited sample availability and will require IRB Approval and completion of a material transfer agreement. LINC01871-/- CRISPR-edited HSB-2 cell lines generated in this study will be made available pending material transfer agreement upon request to Dr. Christopher J. Lessard. All other data associated with this study were publicly available and cited in this manuscript.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All research received prior approval from the Institutional Review Boards at the Oklahoma Medical Research Foundation (OMRF IRB 12-03, 20-08). Informed consent was obtained prior to clinical evaluation at the University of Minnesota or OMRF Sjögren’s Research Clinics.8
References
- 1.Qin B, Wang J, Yang Z, et al. Epidemiology of primary Sjögren's syndrome: a systematic review and meta-analysis. Ann Rheum Dis 2015;74:1983–9. 10.1136/annrheumdis-2014-205375 [DOI] [PubMed] [Google Scholar]
- 2.Helmick CG, Felson DT, Lawrence RC, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum 2008;58:15–25. 10.1002/art.23177 [DOI] [PubMed] [Google Scholar]
- 3.Gøransson LG, Haldorsen K, Brun JG, et al. The point prevalence of clinically relevant primary Sjögren's syndrome in two Norwegian counties. Scand J Rheumatol 2011;40:221–4. 10.3109/03009742.2010.536164 [DOI] [PubMed] [Google Scholar]
- 4.Goules AV, Kapsogeorgou EK, Tzioufas AG. Insight into pathogenesis of Sjögren's syndrome: dissection on autoimmune infiltrates and epithelial cells. Clin Immunol 2017;182:30–40. 10.1016/j.clim.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 5.Mariette X, Criswell LA. Primary Sjögren's syndrome. N Engl J Med 2018;378:931–9. 10.1056/NEJMcp1702514 [DOI] [PubMed] [Google Scholar]
- 6.Imgenberg-Kreuz J, Rasmussen A, Sivils K, et al. Genetics and epigenetics in primary Sjögren’s syndrome. Rheumatology 2021;60:2085–98. 10.1093/rheumatology/key330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björk A, Mofors J, Wahren-Herlenius M. Environmental factors in the pathogenesis of primary Sjögren's syndrome. J Intern Med 2020;287:475–92. 10.1111/joim.13032 [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen A, Ice JA, Li H, et al. Comparison of the American-European consensus group Sjogren's syndrome classification criteria to newly proposed American College of rheumatology criteria in a large, carefully characterised sicca cohort. Ann Rheum Dis 2014;73:31–8. 10.1136/annrheumdis-2013-203845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiboski SC, Shiboski CH, Criswell LA, et al. American College of rheumatology classification criteria for Sjögren's syndrome: a data-driven, expert consensus approach in the Sjögren's international collaborative clinical alliance cohort. Arthritis Care Res 2012;64:475–87. 10.1002/acr.21591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitali C, Bombardieri S, Jonsson R, et al. Classification criteria for Sjögren's syndrome: a revised version of the European criteria proposed by the American-European consensus group. Ann Rheum Dis 2002;61:554. 10.1136/ard.61.6.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitali C, Bootsma H, Bowman SJ, et al. Classification criteria for Sjogren's syndrome: we actually need to definitively resolve the long debate on the issue. Ann Rheum Dis 2013;72:476–8. 10.1136/annrheumdis-2012-202565 [DOI] [PubMed] [Google Scholar]
- 12.Emamian ES, Leon JM, Lessard CJ, et al. Peripheral blood gene expression profiling in Sjögren's syndrome. Genes Immun 2009;10:285–96. 10.1038/gene.2009.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brito-Zerón P, Acar-Denizli N, Wan-Fai N, et al. How immunological profile drives clinical phenotype of primary Sjögren’s syndrome at diagnosis: Analysis of 10,500 patients (Sjögren Big Data Project). Clin Exp Rheumatol 2018;36:S102–11. [PubMed] [Google Scholar]
- 14.Soret P, Le Dantec C, Desvaux E, et al. A new molecular classification to drive precision treatment strategies in primary Sjögren’s syndrome. Nat Commun 2021;12:1–18. 10.1038/s41467-021-23472-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joachims ML, Leehan KM, Dozmorov MG, et al. Sjögren’s syndrome minor Salivary Gland CD4+ memory T Cells associate with glandular disease features and have a germinal center T follicular helper transcriptional profile. J Clin Med 2020;9:2164–17. 10.3390/jcm9072164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mingueneau M, Boudaoud S, Haskett S, et al. Cytometry by time-of-flight immunophenotyping identifies a blood Sjögren's signature correlating with disease activity and glandular inflammation. J Allergy Clin Immunol 2016;137:e12:1809–21. 10.1016/j.jaci.2016.01.024 [DOI] [PubMed] [Google Scholar]
- 17.de Vita S, Gandolfo S, Zandonella Callegher S, et al. The evaluation of disease activity in Sjögren’s syndrome based on the degree of MALT involvement: Glandular swelling and cryoglobulinaemia compared to ESSDAI in a cohort study. Clin Exp Rheumatol 2018;36:S150–6. [PubMed] [Google Scholar]
- 18.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756–70. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 19.Dolcino M, Tinazzi E, Vitali C, et al. Long non-coding RNAs modulate Sjögren’s syndrome associated gene expression and are involved in the pathogenesis of the disease. J Clin Med 2019;8:1349. 10.3390/jcm8091349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Cao N, Pu Y, et al. Long non-coding RNA expression profile in minor salivary gland of primary Sjögren’s syndrome. Arthritis Res Ther 2016;18:1–14. 10.1186/s13075-016-1005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandhya P, Joshi K, Scaria V. Long noncoding RNAs could be potential key players in the pathophysiology of Sjögren's syndrome. Int J Rheum Dis 2015;18:898–905. 10.1111/1756-185X.12752 [DOI] [PubMed] [Google Scholar]
- 22.Ye L, Shi H, Yu C, et al. LncRNA NEAT1 positively regulates MAPK signaling and is involved in the pathogenesis of Sjögren's syndrome. Int Immunopharmacol 2020;88:106992. 10.1016/j.intimp.2020.106992 [DOI] [PubMed] [Google Scholar]
- 23.Fu J, Shi H, Wang B, et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4+ T cell activation and Sjögren's syndrome-like autoimmune response. J Autoimmun 2020;107:102358. 10.1016/j.jaut.2019.102358 [DOI] [PubMed] [Google Scholar]
- 24.Cheng C, Zhou J, Chen R, et al. Predicted disease-specific immune infiltration patterns decode the potential mechanisms of long non-coding RNAs in primary Sjogren’s syndrome. Front Immunol 2021;12. 10.3389/fimmu.2021.624614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Cheng Q, Du Y, et al. Differential long non-coding RNA expression profile and function analysis in primary Sjogren’s syndrome. BMC Immunol 2021;22. 10.1186/s12865-021-00439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariotti B, Servaas NH, Rossato M, et al. The long non-coding RNA NRIR drives IFN-Response in monocytes: implication for systemic sclerosis. Front Immunol 2019;10:100. 10.3389/fimmu.2019.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 2014;42:10668–80. 10.1093/nar/gku713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong G, Yang Y, Li X, et al. Granulocytic myeloid-derived suppressor cells contribute to IFN-I signaling activation of B cells and disease progression through the lncRNA NEAT1-BAFF axis in systemic lupus erythematosus. Biochim Biophys Acta Mol Basis Dis 2020;1866:165554. 10.1016/j.bbadis.2019.165554 [DOI] [PubMed] [Google Scholar]
- 29.Pan Y, Wang T, Zhao Z, et al. Novel insights into the emerging role of NEAT1 and its effects downstream in the regulation of inflammation. J Inflamm Res 2022;15:557–71. 10.2147/JIR.S338162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Peng H, Tian J, et al. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjögren syndrome. Immunol Res 2016;64:489–96. 10.1007/s12026-015-8715-4 [DOI] [PubMed] [Google Scholar]
- 31.Loureiro-Amigo J, Palacio-García C, Martínez-Gallo M, et al. Utility of lymphocyte phenotype profile to differentiate primary Sjögren's syndrome from sicca syndrome. Rheumatology 2021;60:5647–58. 10.1093/rheumatology/keab170 [DOI] [PubMed] [Google Scholar]
- 32.Plattner C, Finotello F, Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-Seq data using quanTIseq. Methods Enzymol 2020;636:261–85. 10.1016/bs.mie.2019.05.056 [DOI] [PubMed] [Google Scholar]
- 33.Muskardin TLW, Niewold TB. Type I interferon in rheumatic diseases. Nat Rev Rheumatol 2018;14:214–28. 10.1038/nrrheum.2018.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 2007;23:3251–3. 10.1093/bioinformatics/btm369 [DOI] [PubMed] [Google Scholar]
- 36.Rinchai D, Roelands J, Toufiq M, et al. BloodGen3Module: blood transcriptional module repertoire analysis and visualization using R. Bioinformatics 2021;37:2382–9. 10.1093/bioinformatics/btab121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Rouphael N, Duraisingham S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014;15:195–204. 10.1038/ni.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 2008;29:150–64. 10.1016/j.immuni.2008.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caielli S, Cardenas J, de Jesus AA, et al. Erythroid mitochondrial retention triggers myeloid-dependent type I interferon in human SLE. Cell 2021;184:e19:4464–79. 10.1016/j.cell.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barriocanal M, Carnero E, Segura V, et al. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 2014;5:655. 10.3389/fimmu.2014.00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang J, Wei X, Liu Z, et al. Long noncoding RNA CYTOR in cancer: a TCGA data review. Clin Chim Acta 2018;483:227–33. 10.1016/j.cca.2018.05.010 [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Ma Q, Zhang M, et al. LncRNA CYTOR drives L-OHP resistance and facilitates the epithelial-mesenchymal transition of colon carcinoma cells via modulating miR-378a-5p/SERPINE1. Cell Cycle 2021;20:1415–30. 10.1080/15384101.2021.1934626 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Chen W, Du M, Hu X, et al. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR-613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer Med 2020;9:1209–19. 10.1002/cam4.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Q, Hu L, Huang D, et al. Six-lncRNA immune prognostic signature for cervical cancer. Front Genet 2020;11:1252. 10.3389/fgene.2020.533628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Y, Wang X, Biomedical W. Identification of molecular features correlating with tumor immunity in gastric cancer by multi-omics data analysis. Ann Transl Med 2020;8:1050. 10.21037/atm-20-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, Shen Y, Chen H, et al. Expression profile analysis of long non-coding RNA in acute myeloid leukemia by microarray and bioinformatics. Cancer Sci 2018;109:340–53. 10.1111/cas.13465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma W, Zhao F, Yu X, et al. Immune-related lncRNAs as predictors of survival in breast cancer: a prognostic signature. J Transl Med 2020;18:1–13. 10.1186/s12967-020-02522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu M, Zhang R, Qiu J. A four immune-related long noncoding RNAs signature as predictors for cervical cancer. Hum Cell 2022;35:348–59. 10.1007/s13577-021-00654-5 [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Zhang J, Liu Y, et al. An integrated autophagy-related long noncoding RNA signature as a prognostic biomarker for human endometrial cancer: a bioinformatics-based approach. Biomed Res Int 2020;2020:5717498. 10.1155/2020/5717498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volders P-J, Anckaert J, Verheggen K, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res 2019;47:D135–9. 10.1093/nar/gky1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang N, Pérez P, Kato T, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med 2021;27:892–903. 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin H, Pranzatelli TJF, French BN, et al. Sclerosing Sialadenitis is associated with salivary gland hypofunction and a unique gene expression profile in Sjögren’s syndrome. Front Immunol 2021;12. 10.3389/fimmu.2021.699722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dam S, Craig T, de Magalhães JP. GeneFriends: a human RNA-seq-based gene and transcript co-expression database. Nucleic Acids Res 2015;43:D1124–32. 10.1093/nar/gku1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perron U, Provero P, Molineris I. In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinformatics 2017;18:144. 10.1186/s12859-017-1535-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Q, Ma R, Wang J, et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-Seq data. BMC Genomics 2015;16 Suppl 3:S2. 10.1186/1471-2164-16-S3-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Y-J, Yoo S-A, Kim M, et al. The role of calcium-calcineurin-NFAT signaling pathway in health and autoimmune diseases. Front Immunol 2020;11:195. 10.3389/fimmu.2020.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. CarbonS, DouglassE, DunnN, et al. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res 2019;47:D330–8. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuhrman CA, Yeh W-I, Seay HR, et al. Divergent phenotypes of human regulatory T cells expressing the receptors TIGIT and CD226. J Immunol 2015;195:145–55. 10.4049/jimmunol.1402381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng C, Chen Y, Li W, et al. Alteration of CD226/TIGIT immune checkpoint on T cells in the pathogenesis of primary Sjögren's syndrome. J Autoimmun 2020;113:102485. 10.1016/j.jaut.2020.102485 [DOI] [PubMed] [Google Scholar]
- 60.Miyagawa I, Nakayamada S, Nakano K, et al. Induction of regulatory T cells and its regulation with insulin-like growth factor/insulin-like growth factor binding protein-4 by human mesenchymal stem cells. J Immunol 2017;199:1616–25. 10.4049/jimmunol.1600230 [DOI] [PubMed] [Google Scholar]
- 61.Merad M, Sathe P, Helft J, et al. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 2013;31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Satpathy AT, Wu X, Albring JC, et al. Re(de)fining the dendritic cell lineage. Nat Immunol 2012;13:1145–54. 10.1038/ni.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez P, Kwon Y-J, Alliende C, et al. Increased acinar damage of salivary glands of patients with Sjögren's syndrome is paralleled by simultaneous imbalance of matrix metalloproteinase 3/tissue inhibitor of metalloproteinases 1 and matrix metalloproteinase 9/tissue inhibitor of metalloproteinases 1 ratios. Arthritis Rheum 2005;52:2751–60. 10.1002/art.21265 [DOI] [PubMed] [Google Scholar]
- 64.Asatsuma M, Ito S, Watanabe M, et al. Increase in the ratio of matrix metalloproteinase-9 to tissue inhibitor of metalloproteinase-1 in saliva from patients with primary Sjögren’s syndrome. Clinica Chimica Acta 2004;345:99–104. 10.1016/j.cccn.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 65.Witkowska AM. Soluble ICAM-1: a marker of vascular inflammation and lifestyle. Cytokine 2005;31:127–34. 10.1016/j.cyto.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 66.Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol 2018;18:485–97. 10.1038/s41577-018-0020-8 [DOI] [PubMed] [Google Scholar]
- 67.Burnett RC, Thirman MJ, Rowley JD, et al. Molecular analysis of the T-cell acute lymphoblastic leukemia-associated t(1;7)(p34;q34) that fuses LCK and TCRB. Blood 1994;84:1232–6. [PubMed] [Google Scholar]
- 68.Annett S, Moore G, Robson T. FK506 binding proteins and inflammation related signalling pathways; basic biology, current status and future prospects for pharmacological intervention. Pharmacol Ther 2020;215:107623. 10.1016/j.pharmthera.2020.107623 [DOI] [PubMed] [Google Scholar]
- 69.Ehx G, Somja J, Warnatz H-J, et al. Xenogeneic graft-versus-host disease in humanized NSG and NSG-HLA-A2/HHD mice. Front Immunol 2018;9:1943. 10.3389/fimmu.2018.01943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hjelmervik TOR, Petersen K, Jonassen I, et al. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren's syndrome patients from healthy control subjects. Arthritis Rheum 2005;52:1534–44. 10.1002/art.21006 [DOI] [PubMed] [Google Scholar]
- 71.Zou Y, Xu H. Involvement of long noncoding RNAs in the pathogenesis of autoimmune diseases. J Transl Autoimmun 2020;3:100044. 10.1016/j.jtauto.2020.100044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Benedittis G, Ciccacci C, Latini A, et al. Emerging role of microRNAs and long non-coding RNAs in Sjögren's syndrome. Genes 2021;12. doi: 10.3390/genes12060903. [Epub ahead of print: 11 06 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng Y, Luo X, Chen Y, et al. LncRNA and mRNA expression profile of peripheral blood mononuclear cells in primary Sjögren’s syndrome patients. Sci Rep 2020;10. 10.1038/s41598-020-76701-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inamo J, Suzuki K, Takeshita M, et al. Identification of novel genes associated with dysregulation of B cells in patients with primary Sjögren's syndrome. Arthritis Res Ther 2020;22:153. 10.1186/s13075-020-02248-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao Y, Ma J-F, Chang C, et al. Immunobiology of T cells in Sjögren's syndrome. Clin Rev Allergy Immunol 2021;60:111–31. 10.1007/s12016-020-08793-7 [DOI] [PubMed] [Google Scholar]
- 76.Greenwell-Wild T, Moutsopoulos NM, Gliozzi M, et al. Chitinases in the salivary glands and circulation of patients with Sjögren's syndrome: macrophage harbingers of disease severity. Arthritis Rheum 2011;63:3103–15. 10.1002/art.30465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dutta D, Barr VA, Akpan I, et al. Recruitment of calcineurin to the TCR positively regulates T cell activation. Nat Immunol 2017;18:196–204. 10.1038/ni.3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rmdopen-2022-002672supp001.pdf (32.3MB, pdf)
Data Availability Statement
Whole blood RNA-seq datasets generated during this study will be made available through the Databases of Genotypes and Phenotypes controlled-access study phs002723.v1.p1 after publication of this manuscript. RNA-seq datasets generated from the CRISPR-edited HSB-2 cell lines will be made available through the NCBI Sequence Read Archive after publication of this manuscript. Alternatively, access to the RNA-seq data will be made available in accordance with consent and data transfer/use agreement from the Oklahoma Medical Research Foundation by request to the corresponding author, Dr Christopher J Lessard (chris-lessard@omrf.org). All other data associated with this study were publicly available and cited in the manuscript and URLs provided as an online supplemental note.
Material availability
Participant whole blood RNAs used in this study were obtained from the Oklahoma Medical Research Foundation Sjögren’s Research Clinic (OMRF-SRC) with Institutional Review Board (IRB) approval. Request for samples must be submitted to Dr A Darise Farris (principal investigator of OMRF-SRC; Darise-Farris@omrf.org). Access will depend on limited sample availability and will require IRB approval and completion of a material transfer agreement. LINC01871−/− CRISPR-edited HSB-2 cell line generated in this study will be made available by request to the corresponding author (Dr Christopher Lessard; chris-lessard@omrf.org) after a material transfer agreement is completed. Sequences of the primers used to create and/or evaluate this cell line are provided in the online supplemental tables and can be ordered through commercial sources. All other materials used in this study are commercially available.
Data are available upon reasonable request. Data generated during this study are available in accordance with consent and data transfer/use agreement from the Oklahoma Medical Research Foundation (OMRF) by request to the corresponding author, Dr. Christopher J. Lessard (chris-lessard@omrf.org). Participant whole blood RNAs used in this study were obtained from the OMRF Sjogren’s Research Clinic (OMRF-SRC) with institutional IRB Approval. Request for samples must be submitted to Dr. A. Darise Farris (PI of OMRF-SRC; darise-farris@omrf.org). Access will depend on limited sample availability and will require IRB Approval and completion of a material transfer agreement. LINC01871-/- CRISPR-edited HSB-2 cell lines generated in this study will be made available pending material transfer agreement upon request to Dr. Christopher J. Lessard. All other data associated with this study were publicly available and cited in this manuscript.