Abstract
Antimicrobial resistance is one of the major health problems we face in the 21st century. Nowadays we cannot understand global health without the interdependence between the human, animal and environmental dimensions. It is therefore logical to adopt a “One Health” approach to address this problem. In this review we show why a collaboration of all sectors and all professions is necessary in order to achieve optimal health for people, animals, plants and our environment.
Keywords: One Health, antimicrobial resistance, last-resort antibiotics
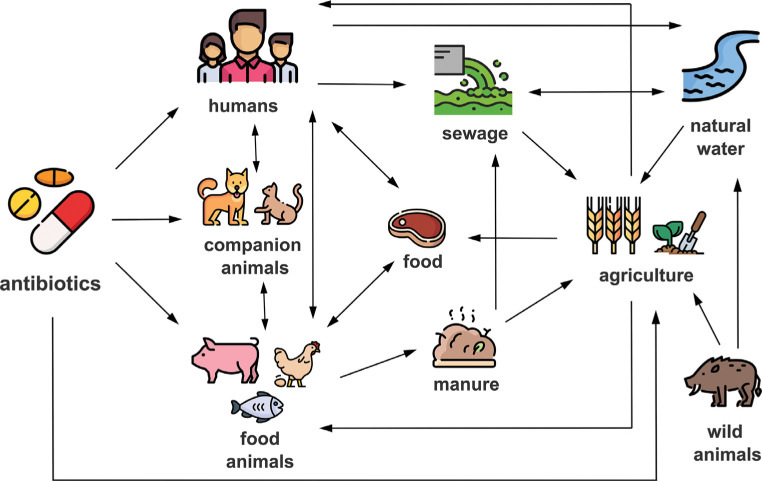
Since ancient times, infectious diseases have been one of the greatest health problems faced by humankind. The discovery of penicillin by Alexander Fleming in 1928 represented a worldwide revolution and one of the greatest achievements in medicine, since the use of antibiotics made it possible to deal with infectious diseases that had been fatal until then [1]. In the decades following the discovery of penicillin, numerous new molecules with antimicrobial activity were found, thus initiating the “golden age” of antibiotic discovery between the 1940s and 1960s. Antimicrobial agents were classified into different groups according to their mechanisms of action, e.g. molecules that target the cell wall (beta-lactam and glycopeptides), inhibitors of protein biosynthesis (aminoglycosides, tetracyclines, macrolides), inhibitors of DNA replication (quinolones) and folic acid metabolism inhibitors (sulfonamides and trimethoprim) [2]. From the earliest stages, antibiotics began to be used on a massive scale in both human and veterinary medicine, without full awareness of the implications that their indiscriminate use could entail. After the introduction of antibiotics, the development of resistance to them was assumed to be unlikely, based on the theory that the frequency of mutations that generated resistant bacteria was insignificant. Unfortunately, this was not the case, and almost simultaneously with the discovery and use of new antibiotics, it was observed that bacteria could develop a wide variety of mechanisms that made themselves resistant to them [3]. Nowadays it is well known that bacteria can develop resistance to antibiotics through mutation or acquisition of genetic material by conjugation, transformation and/or transduction, known as horizontal gene transfer (HGT) mechanisms. The rapid emergence and dissemination of these mechanisms of resistance to all antibiotics commonly used in the clinical setting, together with the scarce discovery or synthesis of new antibiotics by pharmaceutical companies in recent years, make antimicrobial resistance (AMR) one of the most serious threats to world health in the 21st century. According to The Review on Antimicrobial Resistance chaired by Jim O’Neill, approximately 700,000 people die each year globally as a result of antibiotic-resistant infections and AMR could kill 10 million people by 2050, surpassing other pathologies such as cancer [4]. For this reason, the World Health Organization (WHO) has recently reviewed the current pipeline and together with the Group of Eight (G8) declared this problem as a priority and are implementing action plans to address [5]. Wherever antimicrobials are used, reservoirs of AMR genes and drug-resistant pathogens emerge, including humans, companion and production animals, food and environment (Figure 1).
Figure 1.
Diagram of the transmission routes of antimicrobial resistance between humans, animals and environment.
ANTIOTICS, ANIMALS AND THE FOOD CHAIN
It is widely known that antimicrobials can be used as growth promoters, e.g., given low doses to animals, giving rise to a 30-40% higher weight gain. This practice is however forbidden in January 2006 in Europe, and strictly controlled. In January 2017, the USA stopped using antimicrobials used in human medicine, as growth promoters. In some parts of the world, however, this is still a common practice. Further, a wide number of people in the society still believe, that antimicrobials used in animals flow from the animals to the food chain. However, antibiotics and all medicines used in food-producing animals are associated with a withdrawal period. That means that when an animal is treated, depending on the drug, there is a period of time in which none of the animal’s food products can be used for human consumption [6]. This is, especially in developed countries, a strictly controlled process, that includes analysis of farms and food throughout the complete food chain in order to ensure that no residue levels of any antibiotic are exceeded. Thus, the use of antibiotics in food-producing animals is a necessity as animals eventually get ill. However, the routine use of antibiotics in animals needs to be controlled because antimicrobial resistant bacteria emerge and may eventually reach humans or accelerate the emergence of resistance [3]. In the One health concept, all actors involved in or with antimicrobials could contribute to delay AMR. In the same line, in many countries worldwide antibiotics can be purchased without prescription, being a major driver of the emergence of antimicrobial resistance in the community.
ANTIBIOTIC-RESISTANT PATHOGENS: A CRITICAL ONE HEALTH ISSUE
Among the drug-resistant pathogens, one of the main concerns nowadays is multidrug-resistant (MDR), extensively drug-resistant (XDR) or even pandrug-resistant (PDR) Enterobacteriaceae. Apart from Enterobacteriaceae, the WHO has identified bacteria critically important for their resistance and clinical relevance (Table 1).
Table 1.
WHO list of AMR priority pathogens for research and development of new antibiotics.
| Species | Type of Antibiotic Resistance |
|---|---|
| Critical | |
| Acinetobacter baumannii | Carbapenem-resistant |
| Pseudomonas aeruginosa | Carbapenem-resistant |
| Enterobacteriaceae | Carbapenem-resistant, ESBL-producing |
| High | |
| Enterococcus faecium | Vancomycin-resistant |
| Staphylococcus aureus | Methicillin-resistant, vancomycin-intermediate and resistant |
| Helicobacter pylori | Clarithromycin-resistant |
| Campylobacter spp. | Fluoroquinolone-resistant |
| Salmonellae | Fluoroquinolone-resistant |
| Neisseria gonorrhoeae | Cephalosporin-resistant, fluoroquinolone-resistant |
| Medium | |
| Streptococcus pneumoniae | Penicillin-non-susceptible |
| Haemophilus influenzae | Ampicillin-resistant |
| Shigella spp. | Fluoroquinolone-resistant |
To illustrate AMR and One Health, over the last 15 years, 16S rRNA methylases (as armA gene) that possess the capacity to completely nullify the efficacy of aminoglycosides in clinical practice have been identified worldwide. Although the armA gene was initially isolated in a Klebsiella pneumoniae from a urinary tract infection of human origin [7], it was subsequently identified in an Escherichia coli pig isolate [8]. The importance of coordinated surveillance of human and animal isolates was recognized immediately. Over the following years, the presence of ArmA methylase was reported in Salmonella enterica isolates from food [9], in K. pneumoniae isolates from companion animals [10] and in members of the Enterobacteriaceae family from aquatic environments (wastewater treatment plants) [11]. A real concern is that most of these methylases are often part of mobile genetic elements (MGEs) such as plasmid-mediated transposons, increasing the possibility of horizontal transfer of these elements and their dissemination between bacterial species, genera and families, usually associated with genes encoding resistance to other classes of antibiotics. MGEs (such as phages and plasmids) play a key role in the development and dissemination of AMR. A recent study has shown that phages, viruses that are widespread in all environments and infect and replicate within bacteria, represent a novel high-efficiency transmission route of AMR genes [12]. Phages are capable to encapsidate and transfer AMR genes, and they do this in a highly efficient way when these genes are carried on small plasmids [12]. These elements, plasmids, are the most relevant MGEs in the evolution of bacterial resistance to antibiotics in clinical settings. Plasmids are small circular DNA molecules that replicate independently of the bacterial chromosome and are transferred through a protein tunnel that directly connects to bacteria in a process known as conjugation. In a One Health context, plasmids bearing AMR genes can be thus selected in bacterial species adapted to a given environment, e.g., river, dog, hospital sink, and serve as a reservoir for genes that can be mobilized through the plasmids to pathogenic bacteria for humans. For this reason, it is not enough to try to isolate disease-causing bacteria in humans in other ecological niches to identify their reservoirs. We need genomic approaches to find mobile genetic platforms, often, but not only, plasmids, in different environments to assess their role in AMR in humans.
In 2015, a plasmid-mediated colistin resistance (mcr-1) gene was reported for the first time in food animals, food and humans in China [13]. Why did this report have a big impact? Because colistin is a crucial last-resort option. Colistin has been used, both in human and veterinary medicine, for more than 50 years. The drug was stopped for use in humans because of its side effects (nephrotoxicity and neurotoxicity). However, colistin has recently and increasingly been used to treat patients with infections caused by multidrug-resistant bacteria against which colistin remains effective, despite its side effects [13]. The use of colistin in veterinary medicine has been quite different, where colistin is widely used, especially for controlling diarrheal diseases in pig and poultry production. In 2012, it was estimated that colistin consumption was on average more than 600 times higher in food animals than in humans in the European Union. Data from other parts of the world are scarcer, however, China was reported to be the largest user with an expected 12,000 tons in 2015 [13]. But it doesn’t stop there, following the identification of this new gene spread worldwide, bacteria in which mcr-1 coexists with genes that have the ability to hydrolyze penicillins, cephalosporins, monobactams and carbapenems (carbapenemase genes) were reported [14]. This combination is of particular concern, as carbapenems are also last-resort antibiotics, and these bacteria could cause truly untreatable infections. In Spain, the Spanish Agency of Medicines and Medical Devices (AEMPS) has coordinated efforts from a One Health perspective involving professionals from different sectors (human, animal and environmental health) to fight against AMR. In 2014, AEMPS implemented and approved the Spanish Action Plan on Antimicrobial Resistance (PRAN). Among other achievements, the REDUCE program successfully reduced the use of colistin in swine production by 98.88% (1015-2020) and 97% in poultry production (2015-1019) [15].
AMR AND AQUATIC ENVIRONMENTS
Finally, an important (and sometimes somewhat overlooked) component of One Health concept is the environment. We know, that when antimicrobials are used clinically, most active molecules are secreted by the patient, human or animal, through urine or feces. These active molecules, together with the bacteria and mobile genetic elements flow then into the wastewater, mix, and eventually are shared between bacteria, including environmental bacteria. Thus, we can find antimicrobial resistant genes in clinically important bacteria when we analyze the hosts, but within environmental bacteria when we analyze wastewater and environmental reservoirs [11]. Identifying and characterizing the sources of AMR emissions to the environment is crucial and, for this reason, a great emphasis has been placed in recent years on this issue. This situation is especially notorious in wastewater treatment plants (WWTPs) since they collect residual waters from diverse origins and populations where distinct anthropogenic activities occur and where processes often do not sufficiently neutralize antibiotic resistant bacteria and genes. A recent work demonstrated that wastewater environments promoted the expansion of conserved E. coli sequence types (STs), bacteria that share a specific allelic profile, and resistance gene flow through highly disseminated plasmids, leading to specific associations between plasmids and STs [11]. WWTPs allowed the exchange of a diverse genetic repertoire, and therefore, continued close monitoring of these hotspots is needed.
AMR is an increasingly worrisome phenomenon. In this short review, we have shown how this problem involves several actors. Regarding human health, AMR jeopardizes the use of so-called last-resort antibiotics (such as colistin and carbapenems). In recent years, action plans are being implemented involving and coordinating both sectors. We cannot fully understand the problem without considering the environment, which serves as the common link to global health. Therefore, efficient collaboration between all actors involved (humans, animals and environment) is crucial to combat this uncontrolled pandemic.
CONFLICT OF INTEREST
Authors declare no conflict of interest
References
- 1.Fleming A. The development and use of penicillin. Chic Med Sch Q. 1946;7(2):20-8. PMid: . [PubMed] [Google Scholar]
- 2.Kapoor G, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J Anaesthesiol Clin Pharmacol. 2017;33(3):300-5. doi: 10.4103/joacp.JOACP_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.González-Zorn B, Escudero JA. Ecology of antimicrobial resistance: humans, animals, food and environment. Int Microbiol. 2012;15(3):101-9. doi: 10.2436/20.1501.01.163. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill J. Two Years On: An update on achievement towards the recommendations of the Antimicrobial Resistance Report. J Appl Microbiol. 2018. doi: 10.1111/jam.13933. [DOI] [PubMed] [Google Scholar]
- 5.Wernli D, Harbarth S, Levrat N, Pittet D. A ‘whole of United Nations approach’ to tackle antimicrobial resistance? A mapping of the mandate and activities of international organisations. BMJ Glob Health. 2022;7(5):e008181. Doi: 10.1136/bmjgh-2021-008181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay MK. Use of antibiotics as feed additives: a burning question. Front Microbiol. 2014. Jul 2;5:334. doi: 10.3389/fmicb.2014.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003;47(8):2565–71. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Zorn B, Teshager T, Casas M, Porrero MC, Moreno MA, Courvalin P, et al. armA and aminoglycoside resistance in Escherichia coli. Emerg Infect Dis. 2005;11(6):954–6. doi: 10.3201/eid1106.040553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granier SA, Hidalgo L, San Millan A, Escudero JA, Gutierrez B, Brisabois A, et al. ArmA methyltransferase in a monophasic Salmonella enterica isolate from food. Antimicrob Agents Chemother. 2011;55(11):5262–6. doi: 10.1128/AAC.00308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo L, Gutierrez B, Ovejero CM, Carrilero L, Matrat S, Saba CKS, et al. Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob Agents Chemother. 2013;57(9):4532–4. doi: 10.1128/AAC.00491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado-Blas JF, Valenzuela Agüi C, Marin Rodriguez E, Serna C, Montero N, Saba CKS, et al. Dissemination Routes of Carbapenem and Pan-Aminoglycoside Resistance Mechanisms in Hospital and Urban Wastewater Canalizations of Ghana. mSystems. 2022;7(1):e0101921. doi: 10.1128/msystems.01019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Rubio L, Serna C, Ares-Arroyo M, Matamoros BR, Delgado-Blas JF, Montero N, et al. Extensive antimicrobial resistance mobilization via multicopy plasmid encapsidation mediated by temperate phages. Journal of Antimicrobial Chemotherapy. 2020;75(11):3173–80. doi: 10.1093/jac/dkaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Eurosurveillance. 2016;21(9):30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 14.Delgado-Blas JF, Ovejero CM, Abadia-Patiño L, Gonzalez-Zorn B. Coexistence of mcr-1 and bla NDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60(10):6356–8. doi: 10.1128/AAC.01319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.PRAN . PRAN [Internet]. Plan Nacional de Resistencia a Antibióticos (PRAN). 2022-2024. [accessed: 20 May 2022]. Available from: https://resistenciaantibioticos.es/es