Abstract
Dysbiosis of the intestinal microbiota has been shown to result in altered immune responses and increased susceptibility to infection; as such, the state of the intestinal microbiome may have profound implications in the perioperative setting. In this first-in-class study, we used 16s ribosomal RNA sequencing and analysis in a mouse model of general anesthesia to investigate the effects of volatile anesthetics on the diversity and composition of the intestinal microbiome. After 4-hour exposure to isoflurane, we observed a decrease in bacterial diversity. Taxonomic alterations included depletion of several commensal bacteria including Clostridiales. These data identify volatile anesthetics as potential contributors to microbial dysbiosis in the postoperative patient.
Commensal microbes protect host organisms from intestinal pathogens directly by interactions with pathologic species and indirectly via effects on local immune cells. They can also mediate systemic immune responses by influencing T-cell development, neutrophil activation, and cytokine production.1 The involvement of the intestinal microbiota in these responses has profound implications in the perioperative period, when immune function is critical. More than 25 million patients undergo surgery annually in the United States, and an estimated 5%–9% will develop a postoperative infectious complication.2 Surgical site infections alone account for 20% of all hospital-acquired infections and cost an estimated $5 billion dollars annually,3 and identifying ways to reduce infectious complications after surgery has become a major public safety concern.
With the advent of technologies like high-throughput sequencing of 16s ribosomal RNA, a gene marker shared by all bacterial micro-organisms, researchers are now able to characterize complex microbial communities. Using these techniques, numerous animal and human studies have demonstrated that a variety of factors including diet, antibiotic administration, and ongoing systemic illness can result in decreased bacterial diversity and derangements in microbial composition and/or function, a state referred to as microbial dysbiosis.4 In turn, dysbiosis has been associated with, and in some cases found to directly cause, an increased susceptibility to infection.5 In animal models, increased rates of systemic microbial translocation6 and disseminated infection7 were found to be the direct result of antibiotic-induced depletion of specific commensal microbes, such as taxa from the Clostridiales order. Clinical studies have shown that in patients with weakened immune function—those receiving immunosuppressive therapies5 or with infection-related hospitalizations8—dysbiosis is associated with a greater risk of sepsis. Surgery, particularly of the gastrointestinal tract, has also been cited as a cause of dysbiosis, which is thought to contribute to the development of adverse postoperative outcomes including surgical site infections and bacteremia.9 In fact, in a study of patients undergoing hepatectomy, preoperative administration of synbiotics attenuated intestinal microbial imbalances and reduced postoperative infectious complications.10 These findings underscore the need for further investigation into how all aspects of perioperative medicine, particularly the factors that may be modifiable, exert potentially beneficial or harmful effects on the microbiota.
In the studies described in this report, we tested the hypothesis that exposure to the volatile anesthetic isoflurane alters the microbiota. To address this question, we used a mouse model of isoflurane general anesthesia and used 16s ribosomal RNA sequencing and analysis of fecal samples to assess for changes in the intestinal microbiota. Our primary outcome was bacterial diversity, as measured by alpha diversity metrics that quantify the number of distinct taxa present (richness).11 Our secondary outcomes included taxonomic profiling and quantification of relative abundances at the phylum, order, and family level to determine the populations most affected by exposure.
METHODS
All study protocols were approved by the Animal Care and Use Committee at the Johns Hopkins University and conducted in accordance with the National Institutes of Health guidelines for care and use of animals. C57BL/6 mice (n = 13) were used in all experiments (20–22 weeks old). Both sexes were equally represented. Mice were housed in a temperature- and humidity-controlled room and fed a standard diet ad libitum, with free access to water.
ISOFLURANE EXPOSURE
Mice were induced with 3% isoflurane in an induction chamber until righting reflect was lost and then transferred to exposure via rodent nose cones for maintenance at 1.5% isoflurane for 4 hours. An isoflurane-specific calibrated vaporizer was used to deliver agent mixed with 100% oxygen at 5 L/min as described previously.12 No other medications were given before, during, or after the anesthetic exposure, and all mice had unrestricted access to food and water during this period. Oral intake was assessed hourly, and all mice were noted to have resumed normal feeding by 6 hours following general anesthesia. Fecal samples were collected from each mouse at 3 time points: 24 hours before general anesthesia (“pre” group), then 24 hours and 7 days after exposure. These time points were chosen based on mouse gastrointestinal transit time, which is 22 hours for complete elimination of an oral tracer.13 To assess the stability of the normal mouse microbiota, samples were also collected from 3 naive mice at the same times. Naive mice were littermates of experimental mice, and cagemates in the months before and following the study, but were not placed in the induction chamber or exposed to isoflurane directly. All samples were sterilely collected in E-swab tubes (Copan Diagnostics Inc, Murrieta, CA) and stored at −80°C according to manufacturer’s protocols until delivery to American Type Culture Collection Center for Translational Microbiology for 16s ribosomal RNA profiling.
16S RIBOSOMAL RNA GENE SEQUENCING AND ANALYSIS
The Figure, panel A, depicts steps involved in workflow. Please see Supplemental Digital Content 1, Section S1, http://links.lww.com/AA/C656, for detailed methods and Supplemental Digital Content 2, Table S1, http://links.lww.com/AA/C657, for raw 16s ribosomal RNA data per mouse.
Figure.
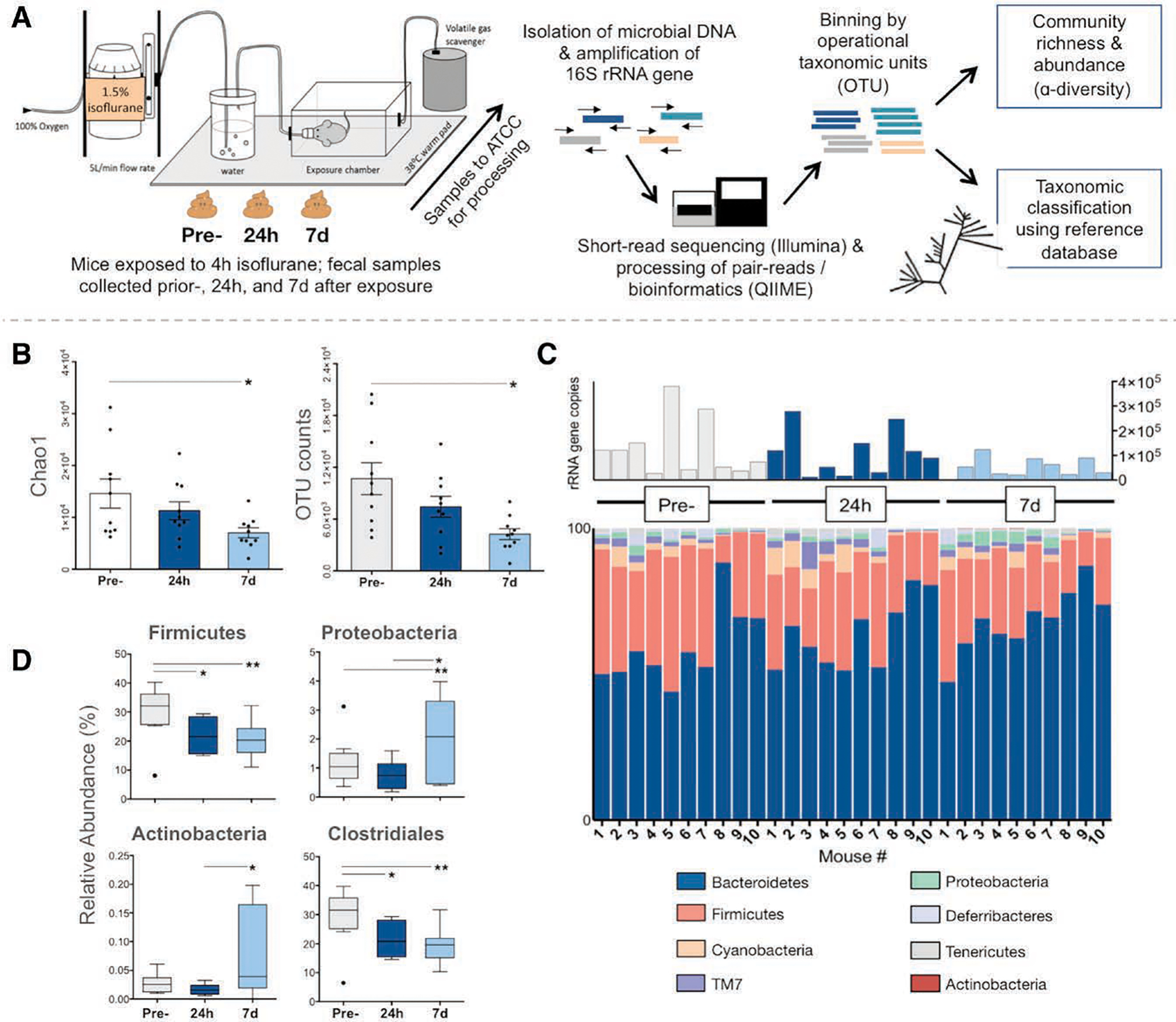
Changes in the mouse gut microbiota after exposure to isoflurane. A, Experiment workflow: fecal samples were collected from mice at 3 time points. Samples were sent to ATCC for processing where DNA was extracted. PCR primers complementary to the V3 and V4 conserved regions of the 16S ribosomal RNA gene were used to amplify the intervening variable sequence using an Illumina (San Diego, CA) platform. Output sequences were then analyzed using the QIIME software package/pipeline.17 After a preprocessing step (primers and low-quality reads trimmed), reads were binned into observed clustered sequences (OTUs). Taxonomy was assigned using the 16S rRNA gene database Greengenes (http://greengenes.lbl.gov) and OTU tables were used for alpha diversity calculations. B–D, Samples were collected at 3 time points: before isoflurane exposure (pre-), and 24 hours and 7 days after (n = 10 mice). Repeated measures 1-way ANOVA was used to compare data from each animal across time points (within-subject effect), and multiple comparisons (comparisons—pre- versus 24 hours, pre- versus 7 days, and 24 hours versus 7 days) were adjusted for by Bonferroni post hoc analysis. Multiplicity-adjusted P values are reported (* = adjusted P < .05 and ** = adjusted P < .01); graphs show mean ± SEM. B, Alpha diversity, as measured by Chao1 scores estimating the predicted number of taxa accounting for rare organisms (left) and OTU count (right), is significantly reduced at 7 days following general anesthesia compared to preexposure (pre- versus 7 days adjusted P = .015 for Chao1 and adjusted P = .043 for OTUs). C, Stacked column graph showing relative abundance of bacterial species in the mouse gut microbiota at the phyla level. Total 16S ribosomal RNA gene copies per sample displayed above. D, Boxplots showing significant changes in relative abundances of taxa. Compared to preexposure, there is a decreased abundance of Firmicutes at 24 hours and 7 days following general anesthesia (pre- versus 24 hours adjusted P = .028; pre- versus 7 days adjusted P = .006), largely due to the decrease in taxa from the Clostridiales order (pre- versus 24 hours adjusted P = .036; pre- versus 7 days adjusted P = .004), whereas abundance of Proteobacteria has increased compared to preexposure (pre- versus 7 days adjusted P = .0293). Boxplots indicate median with edges at 25th and 75th percentiles, and whiskers determined by the Tukey method. ATCC indicates American Type Culture Collection; Chao1, predicted number of taxa after accounting for rare organisms; OTUs, operational taxonomic units; PCR, polymerase chain reaction.
STATISTICAL ANALYSIS
Data were analyzed using the statistical software GraphPad Prism 6.0 (GraphPad Software, Inc, La Jolla, CA). In this time series design, repeated measures 1-way ANOVA was used to assess for changes in the intestinal microbiota over time (within-subject effect). Multiple comparisons (comparisons—pre- versus 24 hours, pre- versus 7 days, and 24 hours versus 7 days) were adjusted for by Bonferroni post hoc analysis; multiplicity-adjusted P values are reported herein. The criteria of significance were set a priori as P < .05 (adjusted), such that with the planned sample size of 10, we had 90% power to detect an effect size of ≥1.5. To assess the stability of the normal mouse microbiota, samples from naive cagemates taken at the same time points were compared using nonrepeated measures (ordinary) 1-way ANOVA with Bonferroni analysis to correct for multiple comparisons, as previous. In addition, to determine the effects of time and isoflurane exposure on the results, experimental and naive groups were analyzed using ordinary 2-way ANOVA. Naive and experimental means were compared at each time point using Bonferroni correction (naive pre- versus general anesthesia pre-, naive 24 hours versus general anesthesia 24 hours, and naive 7 days versus general anesthesia 7 days).
RESULTS
In the experimental group, 3 samples were collected from each of 10 mice (1 per time point—preexposure, 24 hours, and 7 days, as previous). Bacterial diversity was assessed by 2 measures of alpha diversity: (1) operational taxonomic units, or the actual number of different taxa observed, and (2) Chao1 index, or the predicted number of taxa after accounting for rare organisms that may have been missed due to undersampling.14 Naive and experimental groups exhibited similar bacterial diversity at baseline, and richness remained stable over time in naive cagemates (Supplemental Digital Content 3, Figure S1, http://links.lww.com/AA/C658). In contrast, mice exposed to isoflurane demonstrated a significant reduction in diversity at 7 days (adjusted P = .015 and adjusted P = .043, respectively; Figure, panel B). Comparison between experimental and naive groups confirmed that isoflurane exposure, but not time, has a significant effect on diversity (Supplemental Digital Content 3, Figure S1, http://links.lww.com/AA/C658).
Taxonomy analysis was performed to assess the specific communities affected by general anesthesia. At the phylum level, >90% of all samples belonged to 1 of 8 phyla. Taxa from the Bacteroidetes and Firmicutes phyla comprised the majority, with smaller contributions from Cyanobacteria, TM7, Proteobacteria, Deferribacteres, Tenericutes, and Actinobacteria (Figure, panel C). In naive mice, relative abundances of all major taxa did not change over time (Supplemental Digital Content 3, Figure S2, http://links.lww.com/AA/C658). After exposure to general anesthesia, we observed a contraction in the Firmicutes phyla at 24 hours and 7 days, which was found to be due almost entirely to a decreased representation of taxa belonging to the Clostridiales order (Figure, panel D). The reduction in these populations after general anesthesia exposure was also significant when compared to naive samples at the same time points (Supplemental Digital Content 3, Figure S3, http://links.lww.com/AA/C658). General anesthesia exposure also resulted in increased representation of Proteobacteria at 7 days compared to preexposure and 24 hours and in Actinobacteria at 7 days when compared to 24 hours (Figure, panel D). At the family level, there was a transient reduction in Rikenellaceae at 24 hours (adjusted P = .001), whereas the abundance of Bacteroidales S24-7 increased at 24 hours and 7 days after general anesthesia (adjusted P = .026 and adjusted P = .048, respectively).
DISCUSSION
In this first-in-kind study, we used 16s ribosomal RNA profiling to assess the effects of general anesthesia on the intestinal microbiota. By comparing pre- and postexposure microbial communities, we obtained evidence that isoflurane general anesthesia is associated with decreased bacterial diversity and alterations in specific taxa. The mechanism(s) driving these changes remain unknown. Direct inhibition via binding of isoflurane to bacterial receptors is not impossible, particularly in light of a recent study that demonstrated that nearly all classes of chemically diverse antipsychotics have anticommensal activity in vitro.15 Alternatively, isoflurane effects may be indirectly mediated, through changes enacted on intestinal integrity or availability of local nutrients and/or circulating factors, thereby inhibiting the growth of some bacterial species and favoring others. While further studies are also needed to delineate the clinical significance of isoflurane’s effects on the microbiota, it is interesting to note that similar changes in bacterial composition have been reported in other models and linked to deleterious consequences. In several chronic diseases, a reduced population of Firmicutes relative to Bacteroides has been associated with impaired immune function and worsened disease progression.9 Depletion of Clostridiales from the intestinal microbiota has been associated with increased susceptibility to infections, particularly in immunocompromised hosts.7 Last, a pattern of decreased Clostridiales and increased Proteobacteria has been described in aging mice and humans and is thought to explain the increased numbers of opportunistic pathogens in these populations.16 Our study has some limitations that should be noted. Small rodent anesthesia using the techniques described previously involves the use of 100% oxygen and results in some modest physiologic perturbation, which we have quantified in previous work.12 Thus, the possibility that the changes we observed are in some way influenced by these factors cannot be excluded, although of course neither supplemental oxygen nor physiologic perturbation is absent from clinical anesthesia practice. To address this question and others raised by our study, we are currently investigating how isoflurane exerts effects on the gut microbiota, the immune consequences of these changes, and whether other anesthetic agents enact similar derangements. These and other studies will be crucial to establishing how various aspects of perioperative care influence infectious complications via effects on the microbiota.
Supplementary Material
Funding:
This work was supported by grants from the National Institutes of Health (R01 GM120519-01).
Footnotes
The authors declare no conflicts of interest.
Reprints will not be available from the authors.
DISCLOSURES
Name: Mara A. Serbanescu, MD.
Contribution: This author helped design the project, analyze and interpret the data, and write the manuscript.
Name: Reilley P. Mathena, BS.
Contribution: This author helped complete the experiments and review the manuscript.
Name: Jing Xu, MD.
Contribution: This author helped design the experiment, collect the data, and review the manuscript.
Name: Tasha Santiago-Rodriguez, PhD.
Contribution: This author helped analyze the data (16S ribosomal RNA sequencing) and review the manuscript.
Name: Theresa L. Hartsell, MD, PhD.
Contribution: This author helped design the project and review the manuscript.
Name: Raul J. Cano, PhD.
Contribution: This author helped design the project, analyze and interpret the data, and review the manuscript.
Name: Cyrus D. Mintz, MD, PhD.
Contribution: This author helped design the project, supervise the experiments, analyze and interpret the data, and prepare and review the manuscript.
This manuscript was handled by: Alexander Zarbock, MD.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.anesthesia-analgesia.org).
REFERENCES
- 1.Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology. 2017;6:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuri SF, Henderson WG, DePalma RG, Mosca C, Healey NA, Kumbhani DJ; Participants in the VA National Surgical Quality Improvement Program. Determinants of long-term survival after major surgery and the adverse effect of postoperative complications. Ann Surg. 2005;242:326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban KA, Minei JP, Laronga C, et al. American College of Surgeons and Surgical Infection Society: Surgical Site Infection Guidelines, 2016 Update. J Am Coll Surg. 2017;224:59–74. [DOI] [PubMed] [Google Scholar]
- 4.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. [DOI] [PubMed] [Google Scholar]
- 5.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis. 2013;26:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu LC, Shih YA, Wu LL, et al. Enteric dysbiosis promotes anti-biotic-resistant bacterial infection: systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G824–G835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becattini S, Littmann ER, Carter RA, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med. 2017;214:1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescott HC, Dickson RP, Rogers MA, Langa KM, Iwashyna TJ. Hospitalization type and subsequent severe sepsis. Am J Respir Crit Care Med. 2015;192:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyton K, Alverdy JC. The gut microbiota and gastrointestinal surgery. Nat Rev Gastroenterol Hepatol. 2017;14:43–54. [DOI] [PubMed] [Google Scholar]
- 10.Sugawara G, Nagino M, Nishio H, et al. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery: a randomized controlled trial. Ann Surg. 2006;244:706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang E, Jiang D, Ryu YK, et al. Early postnatal exposure to isoflurane causes cognitive deficits and disrupts development of newborn hippocampal neurons via activation of the mTOR pathway. PLoS Biol. 2017;15:e2001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padmanabhan P, Grosse J, Asad AB, Radda GK, Golay X. Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res. 2013;3:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao A Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 15.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langille MG, Meehan CJ, Koenig JE, et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


