Abstract
Objectives
A narrative review on recent published studies of metabolomics in osteoarthritis (OA) with the focus on how the metabolomic findings help stratify OA patients in precision medicine.
Design
A narrative review based on selected population-based metabolomics studies in OA.
Results
PubMed search resulted in a total 139 articles on metabolomics of OA. One hundred and seven articles were excluded because they were either reviews, in vitro studies, animal models. Thirty-two population-based metabolomic studies using either plasma/serum, synovial fluid, cartilage, or subchondral bone samples were reviewed. The most reported metabolic pathways to be involved in OA included energy metabolic pathways, arginine and proline metabolism, taurine and hypotaurine metabolism, and glycerophospholipid metabolism.
Conclusions
While metabolomics of OA research is still in its infancy, the published data showed that metabolomics is a promising tool to help better understanding of pathogenesis of OA, classify OA patients into different endotypes, and develop precision medicine tools for OA management.
Keywords: Osteoarthritis, Metabolomics, Biomarkers, Endotypes, Precision medicine
1. Introduction
Osteoarthritis (OA) is the most common form of arthritis and a leading cause of chronic disability, affecting about 10% of the world's population aged 60 years or older [1]. There is still no cure for it yet. Therapeutic interventions for OA are currently palliative and primarily focus on alleviating pain; however, current treatments for OA pain remain unsatisfactory because of the ‘one-size-fits-all’ approach, despite the fact that OA is a nebula of several diseases for which we still lack markers that would allow us to differentiate them [2]. Patient stratification and precision medicine tools are urgently needed to better stratify patients in order to conduct clinical trials effectively to detect efficacy of disease modifying OA drugs (DMOADs) and develop tailored treatments for OA patients [3].
Efforts are being made to identify subtypes of OA patients mostly based on epidemiological risk factors or joint structural changes seen on the magnetic resonance imaging (MRI). Roemer FW et al. [4] proposed five phenotypes for knee OA based on MRI features, namely: atrophic, hypertrophic, meniscal, inflammatory, and bone phenotypes. However, these features seen on MRI may just reflect different stages of the disease and most people have multiple MRI features, limiting its clinical utility. Dell’Isola A et al. [5] proposed six distinct knee OA phenotypes, including: chronic pain, inflammatory, metabolic syndrome, bone and cartilage metabolism, mechanical overload, and minimal joint disease phenotypes. Karsdal MA et al. [6] proposed five potential phenotypes: mechano-transduction, hormonal, metabolic, auto-inflammation, and genetic subtypes. The problems with these classifications are the overlapping between these phenotypes because many people have multiple risk factors and undefined causal relationships between OA and the risk factors used for these classifications. For example, the metabolic or metabolic syndrome OA subtype was based on the observational studies in which the association was found between OA and metabolic related diseases such as diabetes and obesity, but the causal relationship between metabolic related disease and OA is not yet established. Our metabolomics analysis showed that the observed association between diabetes and OA was most likely due to the shared alteration of the phosphatidylcholine metabolism [7] which is likely caused by different molecular mechanisms in OA [8] and diabetes [9]; thus, classifying patients into metabolic OA based on whether a patient has diabetes or other metabolic related diseases is too simplistic, and may be useless. Thus, while these classifications improved our knowledge in understanding of the pathogenesis of OA, their clinical utility for stratifying patients is limited.
The metabolome represents an integrated readout of upstream genetic variation and downstream environmental influence and each individual has their own metabolomic fingerprints. The advance of the metabolomics provides novel avenue to better understanding systems biology, detect altered metabolic pathways, identify biomarkers and endotypes for disease stratification, early diagnosis, and treatment monitoring, ultimately achieve the goal of precision medicine. The application of metabolomics in OA in recent years have produced robust and promising findings. In this review, I summarized and compared the findings of the selected population-based metabolomic studies of OA published in recent years and provided directions for future research in OA. This review is an update of our previous reviews [10,11] with focusing mainly on population-based metabolomic studies of OA and including much larger number of publications than the previous reviews [10,11].
2. Methods
PubMed search was conducted to identify publications on metabolomics of OA. The search resulted in 139 articles. One hundred and seven articles were excluded because they were reviews, in vitro studies, or animal models. Full texts of the 32 original population-based metabolomic studies of OA were obtained and reviewed. Nineteen of them studied serum/plasma samples, thirteen of them studied joint tissues including synovial fluid, cartilage, subchondral bone. The findings were summarized and compared, and the consistent results across studies were highlighted, and the relevant metabolic pathways were discussed. Table 1 lists the 32 original articles included in this review.
Table 1.
List of the publications reviewed.
| Phenotypes | Biospecimen | Methodology | Sample size | References |
|---|---|---|---|---|
| Knee OA, RA, and postmortem controls | SF | UPLC Q-TOF MS | OA 5, RA 3, Controls 5. | Carlson A et al. [16] |
| Early and later knee OA and controls (all postmortem) | SF | UPLC Q-TOF MS | Early OA 55; late OA 17; controls 7. | Carlson A et al. [18] |
| Knee OA and cadaveric controls | SF | 1H NMR and GC-MS | Knee OA 55; controls 13. | Mickiewicz B et al. [13] |
| Knee OA, RA, postmortem controls | SF | ESI-MS/MS | Early OA 17; late OA 13; RA 18; controls 9. | Kosinska M et al. [14] |
| Knee OA vs. controls | SF | GC-TOF/MS | OA 49; controls 21. | Zheng K et al. [17] |
| Knee OA, gout, calcium pyrophosphate disease (CPPD), spondylarthritis, septic arthritis, and RA | SF | 1H NMR | OA 15; gout 18; CPPD 11; septic arthritis 4; RA 4; reactive arthritis 3; Crohn's disease 2; ankylosing spondylitis 1; psoriasis arthritis 1. | Hügle T et al. [22] |
| Reactive arthritis and undifferentiated spondyloarthropathy; RA, and OA | SF | 1H NMR | OA 21; RA 25; and reactive arthritis 30. | Muhammed H et al. [23] |
| Knee OA severity | SF | GC/TOF MS | OA 15. | Kim S et al. [12] |
| Knee and hip OA | SF | 1H NMR | Hip 12; knee 12. | Akhbari P et al. [19] |
| Classification of OA | SF | hip and knee OA 80. | Zhang W. et al. | |
| Knee OA | Cartilage | MS/MS | Knee OA 34. | Xu Z et al. [15] |
| Knee OA | Subchondral bone | UPLC/Q-TOF-MS | Knee OA 42. | Yang G. et al. [20] |
| Knee OA with and without metabolic syndrome | SF and plasma | HPLC-MS/MS | 54 paired SF and plasma samples and 30 plasma controls in the discovery; 143 plasma samples in the replication. | Zhang W. et al. [7] |
| Knee OA vs. controls and other forms of arthritis | Serum | GC-TOF MS/UPLC-QTOF MS | OA 27; RA 27; AS 27; gout 33, and controls 60. | Jiang M et al. [25] |
| OA, RA, and FM | Bloodspot | IRMS | OA 12; RA 15; FM14. | Hackshaw KV et al. [26] |
| Knee OA vs. controls | Plasma | GC/Q-TOF-MS | OA 12; controls 29. | Huang Z et al. [27] |
| OA vs. controls | Serum | LC/MS | Knee and hip OA 70; controls 82. | Tootsi K. et al. [28] |
| OA vs. controls | Serum | 1H NMR | OA 1556; controls 2125. | Meessen, J. et al. [29] |
| OA vs. controls | Serum | UPLC-TQ-MS | OA 32 and controls 35 in discovery cohort; OA 30 and controls 30 in replication cohort. | Chen R. et al. [30] |
| Obesity and non-obesity knee OA vs. controls | Serum | LC/Q-TOF/MS/MS | Obesity knee OA 14; non-obesity knee OA 14, and controls 15. | Senol O et al. [31] |
| Knee OA and risk for TKR | Plasma and serum | HPLC-MS/MS | Knee OA 64 and control 45 in the discovery cohort; knee OA 72 and controls 76 in the replication cohort; 158 subjects in the longitudinal study. | Zhang W. et al. [8] |
| Knee cartilage volume loss over 2 years | Serum | HPLC-MS/MS | Knee OA 139. | Zhai G et al. [33] |
| Drug response in knee OA | Serum | HPLC-MS/MS | Knee OA 158. | Zhai G et al. [34] |
| Knee OA | Plasma | HPLC-MS/MS | Knee OA 64 and controls 45 in the discovery cohort; knee OA 72 and controls 76 in the replication cohort. | Zhang W. et al. [35] |
| Knee OA | Serum | HPLC-MS/MS | Knee OA 123 and controls 299 in the discovery cohort; knee OA 76 and controls 100 in the replication cohort. | Zhai G. et al. [36] |
| Knee OA progression in 5 years | Serum | HPLC-MS/MS | Knee OA progressor 234; nonprogressor 322. | Zhai G et al. [39] |
| Non-responders to TJR | Plasma | HPLC-MS/MS | 461 TJR. | Costello C et al. [40] |
| Non-responders to TJR | Plasma | HPLC-MS/MS | 461 TJR. | Costello C et al. [41] |
| Endotypes of OA | Plasma | HPLC-MS/MS | OA 615; controls 237. | Werdyani S et al. [42] |
| Knee and hip OA | Plasma | HPLC-MS/MS | Knee and hip OA 153; controls 236. | Hu T et al. [37] |
| Knee OA | Plasma | HPLC-MS/MS | Knee OA 152; controls 194. | Rockel J et al. [32] |
| Knee OA | Plasma | HPLC-MS/MS | Knee OA 136; controls 121. | Hu T et al. [38] |
∗MS – mass spectrometry; ESI – electrospray ionization; Q-TOF – quadrupole time-of-flight; HPLC – high performance liquid chromatography; GC – gas chromatography; UPLC – ultra performance liquid chromatography; IRMS – infrared microspectroscopy; 1H NMR - proton nuclear magnetic resonance.
3. Results and discussion
3.1. Metabolomics of joint tissues
Joint tissues are the most proper samples to study for OA because they can provide information on local tissue metabolism and metabolic alteration relevant to the disease. The most obtainable joint tissue is synovial fluid (SF). Among 13 published studies with joint tissues, eleven of them studied SF samples.
Owing to the challenge to obtain human joint tissues, sample size was small in all these published papers, ranging from 13 to 80 subjects in total [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]] and the controls were mostly from postmortem samples which were usually much younger than OA cases [13,14,16,18] and may introduce misclassification bias. These studies applied either MS-based or NMR-based methods for metabolomic profiling and covered different numbers of metabolites. Data analyses were mostly done with multivariate analytic methods such as Principal Component Analysis (PCA), Orthogonal Projections to Latent Structures - Discriminative Analysis (OPLS-DA), or Hierarchical Clustering Analysis (HCA). These methods provided means to assess the overall discrimination power of all the metabolites collectively for OA but had challenges to identify the specific metabolites and pathways. Thus, authors also performed univariable analyses afterwards for those potential OA-associated metabolites without taking into account of multiple testing correction. As a result, large number of OA-associated metabolites and related metabolic pathways were reported, and some of them were most likely to be false positive purely due to chance because of large number of statistical testing performed.
In a very small sample size including OA, rheumatoid arthritis (RA), and postmortem controls, Carlson A et al. [16] detected a total of 1233 metabolites in the SF samples and found that 58 metabolites were significantly different between OA and controls. The enrichment analysis identified nitric oxide production, chondroitin sulfate degradation, and arginine and proline metabolism were upregulated in OA. While they found that RA SF samples contained expected metabolites consistent with the inflammatory nature of the disease, they did not find any significant metabolic dissimilarities between OA and RA SF samples, most likely due to the small sample size. Their second study had a relative larger sample size [18]. Based on the Outerbridge scoring system for OA, they classified the samples into three groups - healthy controls (n = 7), early OA (n = 55), and late OA (n = 17). A total of 9903 metabolite features were detected in SF samples and 188 features were significantly different between healthy and early OA with 162 lower and 26 higher in early OA. Sixty-four metabolite features were significantly different between healthy and late OA with 39 decreased and 25 increased in late OA. Within OA, they found 191 metabolite features were significantly different between early and late-stage diseases, with 9 lower and 182 higher in late-stage disease. Pathway analysis showed that OA was associated with altered extracellular matrix component metabolism (glucosamine and galactosamine biosynthesis, ascorbate metabolism, keratin sulfate metabolism, and N-glycan metabolism), amino acid metabolism, fatty acid and lipid metabolism (glycosphingolipid and glycerophospholipid metabolism, the carnitine shuttle), inflammation (leukotriene metabolism), central energy metabolism (glycolysis and gluconeogenesis, the TCA cycle), oxidative stress (vitamin E, glutathione metabolism), and vitamin metabolism (C, E, B1, B3, B6, and B9). Further, the unsupervised HCA identified four SF phenotypes in OA patients – two for early OA and two for late OA, characterized by increased inflammation (early and late OA), oxidative stress (late OA), and structural deterioration (early and late OA). While these studies could not point out more specific OA-associated metabolites, they provided potential altered metabolic pathways for further investigation.
Using the similar study design, Mickiewicz B et al. [13] found that fructose and citrate were increased whereas O-acetyl-carnitine, N-phenylacetylglycine, methionine, ethanol, creatine, malate, ethanolamine, 3-hydroxbutyrate and hexanoylcarnitine were decreased in OA SF samples compared to the controls, suggesting an increase energy demand and extended fatty acids and lipids metabolism in OA affected knee joints. Kosinska M et al. [14] found that sphingomyelin (SM) molecular species were the most abundant in SF and the concentration of the total SMs was increased 2.4-fold in early OA, 4.4-fold in late OA, and 3.2-fold in RA compared to the controls. SM 34:1 was the predominant species among SMs, accounting for 38%–44% of total SMs in all cohorts. Of note, all SM species has risen approximately 2-fold in synovial fluid from early OA to late OA. The second most prominent lipid molecule in the synovial fluid was ceramide molecular species. The concentration of total ceramides was increased 2-fold in early OA, 3.9-fold in late OA, and 3-fold in RA compared to controls. The most predominant species was ceramide d18:0/24:0.
The study done by Zheng K et al. [17] was the only one that included an independent replication cohort; thus, the results would be more robust. They found that six metabolites were significantly different between OA and controls and three of them, namely glutamine, 1,5-anhydroglucitol, and gluconic lactone were replicated in the replication cohort. While glutamine was significantly lower in OA patients, the other two metabolites 1,5-anhydroglucitol and gluconic lactone were significantly higher in OA patients than controls. The receiver operating curve (ROC) analysis showed that all three different metabolites had an area under the curve (AUC) of 0.85–0.93 to distinguish OA from controls. Further, the study showed that gluconic lactone was 2.76-fold higher in OA patients than in RA patients, suggesting it could be a marker for differential diagnosis of OA and RA. However, both Hügle T et al. [22] and Muhammed H et al. [23] did not find any metabolic difference in SF samples between OA and RA as well as other forms of arthritis including gout, calcium pyrophosphate disease, and spondylarthritis. The authors acknowledged the small sample size may be the blame.
Using affected and unaffected samples from the same individual would solve the problems of the difficulty to obtain control samples and minimize the potential confounding effects. Xu Z et al. [15]. studied cartilage samples obtained from osteophyte affected site and no osteophyte site (lateral posterior femoral condyle (LPC) from 34 postmenopausal women who underwent total knee joint replacement (TKR) due to OA. They identified 25 metabolites were elevated and 3 were decreased in osteophyte group. Eight metabolites had good power to distinguish cartilage samples of osteophyte site from that of no osteophyte site, which included leucine, phenylalanine, proline, taurine, tyrosine, cis,cis-muconic acid, Lyso PC (16:0), and pyroglutamic acid. The pathway analysis showed phenylalanine metabolism, taurine and hypotaurine metabolism, and arginine and proline metabolism were the major metabolic pathways involved in OA. Yang G. et al. [20] studied the metabolomics of subchondral bone samples obtained from 42 primary knee OA patients. All the patients had the Kellgren-Lawrence (KL) score = 3 and the centric subarticular spongiosa of the medial femoral condyle was sampled as the experimental group and the centric subarticular spongiosa of the lateral medial femoral condyle was samples as the control group. They found 68 out of 469 metabolites were significant between the two groups with p < 0.05, and metabolic pathway analysis showed that these metabolites were involved in taurine and hypotaurine metabolism, beta-alanine metabolism, phenylalanine metabolism, tyrosine metabolism, lysine degradation, pyrimidine metabolism purine metabolism, and sphingolipid metabolism.
Based on the radiographic assessment, Kim S et al. [12] investigated the metabolic alterations in SF for knee OA severity defined by the KL score and found that 28 out of the 114 metabolites were different between early and late radiographic knee OA. Three metabolites – squalene, palmitoleic acid, and pentadecanoic acid had changes more than 2 folds between early and late disease stages. Pathway analysis indicated fatty acid biosynthesis, glycerophospholipid metabolism, and glycolipid metabolism were the major metabolic pathways to be associated with increasing degree of the severity of knee OA.
The above-mentioned studies were all for knee OA. When comparing between knee and hip OA, Akhbari P et al. [19] found that knee OA patients had greater quantities of metabolites including N-acetylated molecules, glycosaminoglycans, citrate, and glutamine than hip OA, and these metabolites play roles in collagen degradation, the TCA cycle and oxidative metabolism. However, we [21] did not find any metabolic difference in SF between hip and knee OA patients but found that the SF metabolic profiles can clearly classify the patients into two groups, A and B. The group A had a significantly higher concentration of all acylcarnitines but lower free carnitine than the group B that can be further subdivided into two subgroups characterized by different levels of glycerophospholipids, sphingolipids, and biogenic amine. While it was not statistically different, we found that the group B patients had a higher prevalence of metabolic-related diseases including hypertension, high cholesterol and diabetes than the group A patients. Following these findings, we examined the difference in metabolic profiles of the SF samples between knee OA patients with and without metabolic syndrome [7] and found that the two groups can be clearly separated based on their metabolic profiles in the SF samples. The clear separation appeared to be driven by knee OA patients with diabetes but no other metabolic syndrome components. The same pattern was seen when we examined the metabolic profiles in the plasma sample of these patients, indicating that blood samples could be used as surrogate for joint tissues in OA research which was also supported by another study in which we demonstrated particularly that metabolite ratios as proxies for enzymatic reaction had a high correlation between blood and SF samples [24]. These findings were important and provided justification for using blood samples in metabolomics of OA.
3.2. Metabolomics of plasma/serum in OA
Blood samples are the most obtainable biospecimen from patients, thus the studies of metabolomics of plasma or serum samples in OA generally had a larger sample size than those studies with SF samples and mostly they included healthy control samples, thus making the results more robust.
Jiang M et al. [25] studies four forms of arthritis including OA, RA, ankylosing spondylitis (AS), gout, and healthy control subjects. Both OA and RA patients were all females whereas both AS and gout patients were all males. The healthy controls included 30 males and 30 females. They identified 196 metabolites in the serum samples and 6 of them were identified as common metabolites shared by four common arthritis whose levels were different from the healthy controls. These six metabolites included homoserine, 4,8-dimethylnonanoyl carnitine, glyceraldehyde, lactic acid, dihydroxyfumatic acid and aspartic acid. All of them except for 4,8-dimethylnonanoyl carnitine were upregulated in arthritis patients compared to the controls. Together, these six metabolites can distinguish arthritis patients from controls with an AUC of 0.91, sensitivity of 0.81 and specificity of 0.88. These metabolites indicated metabolic pathways related joint lesion and inflammation. The authors did not take into account of multiple testing. Further, they found 13 metabolites can be used to distinguish OA from RA. These metabolites included alanine, tryptophan, 5-oxoproline, sarcosine, tyrosine, threonine, citric acid, lysine, actylornithine, histamine, 24-hydroxycalcitriol, cic-aconitic acid, and pyroglutamic acid. Together these metabolites had an AUC of 0.86 with a sensitivity of 85% and specificity of 81%. Among them, the most significant ones were tryptophan, sarcosine, and alanine. No specific metabolites were identified for separating OA from gout and AS. However, Hackshaw KV et al. [26] found that OA and RA were metabolically similar, but biochemical differences were identified in the fibromyalgia syndrome (FM) patients that were quite distinctive from OA and RA. Both mid-infrared microspectroscopy (IRMS) and metabolomic analysis identified changes in tryptophan metabolism pathway was the main alteration in FM compared to OA and RA.
Huang Z et al. [27] studied 12 knee OA patients and 20 healthy controls and identified three metabolites – succinic acid, xanthurenic acid, and tryptophan, were the potential markers for distinguishing OA from controls. All three metabolites were lower in OA than controls.
Tootsi K et al. [28] studied 70 knee and hip OA patients and 82 controls and found that glycine and arginine were independently associated with OA radiographic severity. They also found significantly higher levels of arginine, asparagine, leucine, serine, asymmetric dimethylarginine (ADMA), phenylalanine and spermidine, and lower levels of serotonin and spermine/spermidine rationin the OA groups after adjusting for BMI. The OA group also had significantly lower serum levels of lysoPC a C14:0, PC aa C30:0; PC aa C32:2; PC aa C32:3, PC aa C34:3, PC aa C34:4, PC ae C30:0, PC ae C34:2 and PC ae C34:3, and. Higher levels of lysoPC a C20:4, PC aa C38:6, PC aa C40:6 and SM C20:2 compared with the control group. The significance level was defined at p < 0.05.
Meessen, J et al. [29] studied a total 227 metabolites assessed by NMR platform in a total 2125 controls and 1556 OA cases from the Dutch population. Factor analysis was conducted on the 227 metabolites and 23 factors were identified which accounted for 91.4% of the total variance. Further adjustment for age, sex, BMI and fasting status removed four factors which accounted for 28.06% of the variance. They identified two factors that were most significantly associated with overall OA. One factor represented mainly the medium and large triglycerides to total lipids ratio and another factor represented amino acids glutamine and histidine. The most significant metabolic factor was fatty acids chain length (FALen) was associated with overall OA and total joint replacement (TJR).
Chen R et al. [30] studied 32 OA patients and 35 controls as discovery cohort and 30 OA patients and 30 controls as validation cohort. They focused on serum amino acids and nicotinamide and identified and validated four metabolites – alanine, γ-aminobutyric acid, 4-hydroxyproline, and taurine, to be associated with OA. Each of these four metabolites had an AUC of at least 0.91 with sensitivity of >0.8 and specificity of >0.83 to distinguish OA from controls.
Senol O et al. [31] studied 14 obesity knee OA, 14 non-obesity knee OA, and 15 controls. Untargeted metabolic profiling was performed on the collected serum samples. By using p ≤ 0.01 and fold change >1.5, they identified 21 different metabolites for patients vs controls and 15 metabolites for obesity knee OA vs non-obesity knee OA. Twelve of these metabolites were found in both comparisons. However, when multiple testing was considered, benzoic acid would be the only metabolite that associated with knee OA regardless of the obesity status and all the 12 metabolites for obesity knee OA would not be significant. Benzoic acid is a fungistatic compound that is widely used as a food preservative and is also produced when gut bacteria process polyphenols. Benzoic acid can be also produced by phenylalanine metabolism in bacteria but not clear yet whether this is the case in human body. We demonstrated that knee OA patients with metabolic syndrome can be clearly separated from knee OA patients without metabolic syndrome by their metabolic profiles in SF and plasma, and the separation was mostly driven by patients with diabetes but no other metabolic syndrome components such as obesity, hypertension, or high cholesterol [7]. Further, we demonstrated that two specific metabolites - phosphatidylcholine acyl-alkyl C34:3 (PC ae C34:3) and phosphatidylcholine acyl-alkyl C36:3 (PC ae C36:3), were the key driver for the separation of knee OA patients with and without diabetes. The two metabolites had a clear linear relationship with the disease status – knee OA patients with diabetes had the lowest plasma concentrations of these two metabolites and healthy controls had the highest concentrations. Knee OA without diabetes had higher concentrations than knee OA with diabetes but lower than those affected by only diabetes whereas diabetic patients had higher than both knee OA with and without diabetes but lower concentration than healthy controls [7]. The results suggested that phosphatidylcholine metabolism was altered in both knee OA and diabetic patients. The alteration has additive effects, but the potential mechanisms are different between knee OA and diabetes.
In a subsequent study [9], we found that diabetic patients had an elevated level of advanced glycation end-products (AGEs) including methylglyoxal (MG) and methylglyoxal-derived hydroimidazolone (MG-H1) in SF but not plasma samples and both MG and MG-H1 were negatively and significantly correlated with PC ae C34:3 and PC ae C36:3, suggesting that hyperglycemia-related AGEs might be responsible for the altered phosphatidylcholine metabolism in diabetic patients. However, in knee OA patients, we found that the conversion pathway of phosphatidylcholines (PCs) to lysophosphatidylcholines (lysoPCs) was likely responsible for the altered phosphatidylcholine metabolism in OA patients as the ratio of lysoPCs to PCs were found to be significantly higher in knee OA patients than healthy controls in both discovery and validation cohorts [8]. Further, in the 10 years follow-up study, we found that people with the ratio≥0.09 were 2.3 times more likely to undergo TKR than those with the ratio<0.098, indicating the ratio could be a good biomarker for predicting knee OA progression. When applying a classification modeling approach [32] to the same dataset [8], we confirmed that lysoPCs and PCs are dominant indicators of OA, particularly in males [32].
In a separate study, we demonstrated that a specific lysoPC to PC ratio – lysoPC C18:2 to PC aa C44:3, was associated with knee cartilage volume loss over two years measured by MRI [33], supporting the previous findings [8]. Further, in a clinical trial cohort, we demonstrated that symptomatic knee OA patients with serum lysoPCs to PCs ratio ≥0.088 had 2.93 times more likely to respond to licofelone and naproxen treatment than those with the ratio <0.088 [34]. These results are robust and generalizable as the samples used in these studies were from three different geographic locations including Newfoundland, Montreal, and Tasmania. Together, these data showed that lysoPCs to PCs ratio could be a promising blood marker that can be used for OA diagnosis, progression monitoring, and treatment response prediction.
In a study with utilizing a discovery cohort of 64 knee OA patients and 45 controls and a validation cohort of 72 knee OA patients and 76 age and sex matched controls collected in Newfoundland [35], we found that knee OA patients had their plasma arginine concentration reduced by 35% than the healthy controls. The ROC curve analysis showed that plasma arginine concentration had 98.3% sensitivity and 89% specificity at the optimal cut-off 57 μM to discriminate knee OA patients from controls. The findings suggested that arginine supplement might be a new nutraceutics for OA treatment. A clinical trial of the effect of arginine supplement on knee OA is currently underway (ClinicalTrials.gov Identifier: NCT03665116). In an UK cohort, we found that serum branched-chain amino acid to histidine ratio was associated with knee OA [36] which was replicated in the Newfoundland population [8].
When applying an evolutionary learning and network approach [37] and the differential correlation network approach [38] to the same datasets [8,35], we confirmed our previous findings [21,35,36] but also identified additional OA-associated metabolites including nitrotyrosine, taurine, threonine, and tyrosine [37,38].
When examining unilateral and bilateral knee OA radiographic progression in five year in the Multicenter Osteoarthritis Study (MOST) cohort, we found that baseline serum level of phenylalanine was significantly associated with both unilateral and bilateral knee OA progression, especially in women [39]. When examining responders to TJR, we found that two metabolite ratios were significantly associated with pain non-responders to TJR; acetylcarnitine (C2) to phosphatidylcholine acyl-alkyl C40:1 (PC ae C40:1) was five times higher in pain non-responders whereas phosphatidylcholine diacyl C36:4 (PC aa C36:4) to isoleucine was twenty-one times lower in pain non-responders than responders. one metabolite ratio, glutamine to isoleucine, was significantly lower in function non-responders than responders [40]. The differential correlation network analysis showed that these metabolite ratios suggested that inflammation, muscle breakdown, and wound healing may all play roles in TJR response [41].
More recently, we applied the machine learning method to the metabolomic data obtained from a large sample size of 615 OA patients and 237 controls and found that there were three possible clinically actionable OA endotypes existed in OA [42]. Each OA endotype is characterized by a specific metabolomic marker. Endotype I was characterized by an elevated butyrylcarnitine (C4) level in blood which is thought to be related to muscle weakness and wasting; endotype II was characterized by significantly reduced arginine levels; and endotype II was characterized by a significantly lower level of lysophosphatidylcholine with palmitic acid. Further, we found that endotype I had a higher BMI and prevalence of diabetes than other endotypes and also a higher prevalence of coronary heart disease than endotype III. Endotype II had a higher prevalence of coronary heart disease than endotype III whereas endotype III had a higher prevalence of osteoporosis. While these findings need to be confirmed in independent studies, it appears that the results are robust as both arginine and lysophosphatidylcholine were found to be associated with OA as mentioned above. Based on the potential function of these metabolites, OA patients can be classified into three subtypes: muscle weakness, arginine deficit, and low inflammatory OA.
4. Conclusions
Although metabolomics of OA research is still in its infancy, the published data pointed out several metabolic pathways are involved in OA risk and progression. Several studies particularly with SF samples [13,[17], [18], [19],21] showed an upregulation of energy metabolic pathway in OA, including glycolysis, TCA cycle, and fatty acid β-oxidation. These findings reflect the increased energy demand in OA to repair the damaged cartilage and other joint tissues. Thus, it is more likely that the upregulation of the energy metabolic pathways is due to the need for cartilage repair rather than causing joint tissue damage. However, this upregulation of the energy metabolic pathways in return produces more reactive oxygen species (ROS) which may lead to damage of articular cartilage [10]. Oxidative pathways have been found to be involved in OA in several studies [16,18].
Many amino acid metabolic pathways have been found to be associated with OA. Arginine and proline metabolism is the most reported one [15, 16, 18, 28, 30, 35, 42]. Arginine might be the key metabolite as it is the precursor that not only involves in the arginine and proline metabolism but also the nitric oxide production pathway which has also been found to be associated with OA [16]. Reduced arginine level in OA patients [35,42] may result in the limited availability of proline for collagen synthesis and reduce the nutrient supply to the joint. Taurine and hypotaurine metabolic pathways were found mostly in studies with OA-affected and unaffected samples obtained from the same patients, and those control samples were from non-weight bearing sites, thus these altered metabolic pathways might be due to biomechanical loading on the joint [15,20]. Other amino acid metabolic pathways including glutamine, phenylalanine, and branched-chain amino acids have also been reported to be associated with OA and further investigations are required to elaborate them in OA.
Lipid metabolism, especially glycerophospholipid and SM metabolisms, have been reported to be involved in OA. While SM and ceramides were examined in detail in one study [14], many studies have reported the involvement of glycerophospholipids in OA, particularly phosphatidylcholines (PCs) and lysophosphatidylcholines (lysoPCs) [7,8,12,15,18,21,28,29,33,34,42]. The most likely pathway is the conversion pathway of PCs to lysoPCs which can be quantified by the ratio of lysoPCs to PCs [8,33,34] and the upregulation of the pathway is likely due to the overexpression of PLA2 enzyme in the joint tissues [33]. Thus, this pathway could be a promising target for developing new OA drugs.
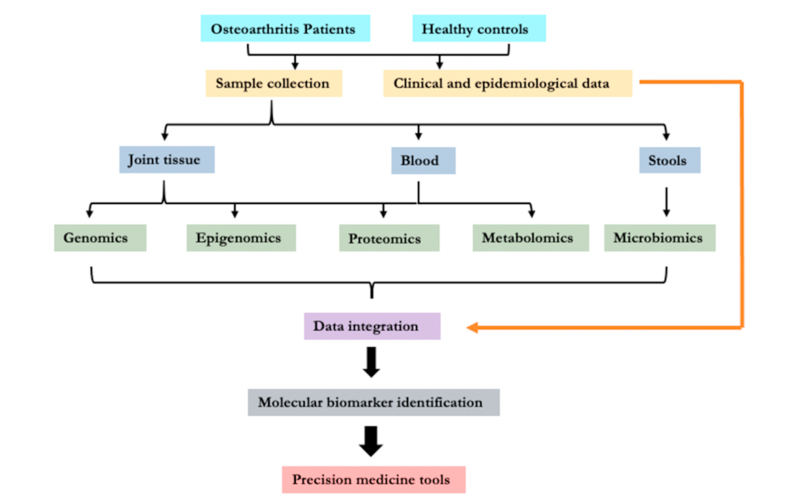
Overall, these data demonstrated the promise of the metabolomics in OA research and its utility to help develop precision medicine tools for OA. A recent study on endotypes of OA identified by metabolomics demonstrated its robustness [42]. However, data are still limited. While metabolome reflects both genetic and environmental influence, combining metabolomics with other OMICs [11,43] with a systems biology approach (Fig. 1) would greatly facilitate the efforts to achieve our goal in realizing the precision medicine of OA.
Fig. 1.
Schematic representation of the systems biology approach to develop precision medicine tools for OA.
Acknowledgement
This work was supported by Canadian Institutes of Health Research (CIHR) and Memorial University of Newfoundland.
References
- 1.The Burden of Musculoskeletal Conditions at the Start of the New Millennium: Report of a WHO Scientific Group. World Health Organisation; Geneva: 2003. [PubMed] [Google Scholar]
- 2.Berenbaum F. Deep phenotyping of osteoarthritis: a step forward. Ann. Rheum. Dis. 2019;78:3–5. doi: 10.1136/annrheumdis-2018-213864. [DOI] [PubMed] [Google Scholar]
- 3.Mobasheri A., Saarakkala S., Finnila M., Karsdal M.A., Bay-Jensen A.C., van Spil W.E. 2019. Recent Advances in Understanding the Phenotypes of Osteoarthritis. F1000Res 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roemer F.W., Kwoh C.K., Hayashi D., Felson D.T., Guermazi A. The role of radiography and MRI for eligibility assessment in DMOAD trials of knee OA. Nat. Rev. Rheumatol. 2018;14:372–380. doi: 10.1038/s41584-018-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dell'Isola A., Allan R., Smith S.L., Marreiros S.S., Steultjens M. Identification of clinical phenotypes in knee osteoarthritis: a systematic review of the literature. BMC Muscoskel. Disord. 2016;17:425. doi: 10.1186/s12891-016-1286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karsdal M.A., Christiansen C., Ladel C., Henriksen K., Kraus V.B., Bay-Jensen A.C. Osteoarthritis--a case for personalized health care? Osteoarthritis Cartilage. 2014;22:7–16. doi: 10.1016/j.joca.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Sun G., Likhodii S., Aref-Eshghi E., Harper P.E., Randell E., et al. Metabolomic analysis of human synovial fluid and plasma reveals that phosphatidylcholine metabolism is associated with both osteoarthritis and diabetes mellitus. Metabolomics. 2015;12:24. [Google Scholar]
- 8.Zhang W., Sun G., Aitken D., Likhodii S., Liu M., Martin G., et al. Lysophosphatidylcholines to phosphatidylcholines ratio predicts advanced knee osteoarthritis. Rheumatology. 2016;55:1566–1574. doi: 10.1093/rheumatology/kew207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Randell E.W., Sun G., Likhodii S., Liu M., Furey A., et al. Hyperglycemia-related advanced glycation end-products is associated with the altered phosphatidylcholine metabolism in osteoarthritis patients with diabetes. PloS One. 2017;12 doi: 10.1371/journal.pone.0184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai G. Alteration of metabolic pathways in osteoarthritis. Metabolites. 2019;9 doi: 10.3390/metabo9010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai G., Randell E.W., Rahman P. Metabolomics of osteoarthritis: emerging novel markers and their potential clinical utility. Rheumatology. 2018;57:2087–2095. doi: 10.1093/rheumatology/kex497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S., Hwang J., Kim J., Ahn J.K., Cha H.S., Kim K.H. Metabolite profiles of synovial fluid change with the radiographic severity of knee osteoarthritis. Joint Bone Spine. 2017;84:605–610. doi: 10.1016/j.jbspin.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Mickiewicz B., Kelly J.J., Ludwig T.E., Weljie A.M., Wiley J.P., Schmidt T.A., et al. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J. Orthop. Res. 2015;33:1631–1638. doi: 10.1002/jor.22949. [DOI] [PubMed] [Google Scholar]
- 14.Kosinska M.K., Liebisch G., Lochnit G., Wilhelm J., Klein H., Kaesser U., et al. Sphingolipids in human synovial fluid--a lipidomic study. PloS One. 2014;9 doi: 10.1371/journal.pone.0091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z., Chen T., Luo J., Ding S., Gao S., Zhang J. Cartilaginous metabolomic study reveals potential mechanisms of osteophyte formation in osteoarthritis. J. Proteome Res. 2017;16:1425–1435. doi: 10.1021/acs.jproteome.6b00676. [DOI] [PubMed] [Google Scholar]
- 16.Carlson A.K., Rawle R.A., Adams E., Greenwood M.C., Bothner B., June R.K. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochem. Biophys. Res. Commun. 2018;499:182–188. doi: 10.1016/j.bbrc.2018.03.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng K., Shen N., Chen H., Ni S., Zhang T., Hu M., et al. Global and targeted metabolomics of synovial fluid discovers special osteoarthritis metabolites. J. Orthop. Res. 2017;35:1973–1981. doi: 10.1002/jor.23482. [DOI] [PubMed] [Google Scholar]
- 18.Carlson A.K., Rawle R.A., Wallace C.W., Brooks E.G., Adams E., Greenwood M.C., et al. Characterization of synovial fluid metabolomic phenotypes of cartilage morphological changes associated with osteoarthritis. Osteoarthritis Cartilage. 2019;27:1174–1184. doi: 10.1016/j.joca.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhbari P., Jaggard M.K., Boulange C.L., Vaghela U., Graca G., Bhattacharya R., et al. Differences in the composition of hip and knee synovial fluid in osteoarthritis: a nuclear magnetic resonance (NMR) spectroscopy study of metabolic profiles. Osteoarthritis Cartilage. 2019;27:1768–1777. doi: 10.1016/j.joca.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Yang G., Zhang H., Chen T., Zhu W., Ding S., Xu K., et al. Metabolic analysis of osteoarthritis subchondral bone based on UPLC/Q-TOF-MS. Anal. Bioanal. Chem. 2016;408:4275–4286. doi: 10.1007/s00216-016-9524-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Likhodii S., Zhang Y., Aref-Eshghi E., Harper P.E., Randell E., et al. Classification of osteoarthritis phenotypes by metabolomics analysis. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2014-006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugle T., Kovacs H., Heijnen I.A., Daikeler T., Baisch U., Hicks J.M., et al. Synovial fluid metabolomics in different forms of arthritis assessed by nuclear magnetic resonance spectroscopy. Clin. Exp. Rheumatol. 2012;30:240–245. [PubMed] [Google Scholar]
- 23.Muhammed H., Kumar D., Dubey D., Kumar S., Chaurasia S., Guleria A., et al. Metabolomics analysis revealed significantly higher synovial Phe/Tyr ratio in reactive arthritis and undifferentiated spondyloarthropathy. Rheumatology. 2020;59:1587–1590. doi: 10.1093/rheumatology/kez493. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., Likhodii S., Aref-Eshghi E., Zhang Y., Harper P.E., Randell E., et al. Relationship between blood plasma and synovial fluid metabolite concentrations in patients with osteoarthritis. J. Rheumatol. 2015;42:859–865. doi: 10.3899/jrheum.141252. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M., Chen T., Feng H., Zhang Y., Li L., Zhao A., et al. Serum metabolic signatures of four types of human arthritis. J. Proteome Res. 2013;12:3769–3779. doi: 10.1021/pr400415a. [DOI] [PubMed] [Google Scholar]
- 26.Hackshaw K.V., Rodriguez-Saona L., Plans M., Bell L.N., Buffington C.A. A bloodspot-based diagnostic test for fibromyalgia syndrome and related disorders. Analyst. 2013;138:4453–4462. doi: 10.1039/c3an36615d. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z., He Z., Kong Y., Liu Z., Gong L. Insight into osteoarthritis through integrative analysis of metabolomics and transcriptomics. Clin. Chim. Acta. 2020;510:323–329. doi: 10.1016/j.cca.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Tootsi K., Vilba K., Martson A., Kals J., Paapstel K., Zilmer M. Metabolomic signature of amino acids, biogenic amines and lipids in blood serum of patients with severe osteoarthritis. Metabolites. 2020;10 doi: 10.3390/metabo10080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meessen J., Saberi-Hosnijeh F., Bomer N., den Hollander W., van der Bom J.G., van Hilten J.A., et al. Serum fatty acid chain length associates with prevalent symptomatic end-stage osteoarthritis, independent of BMI. Sci. Rep. 2020;10:15459. doi: 10.1038/s41598-020-71811-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen R., Han S., Liu X., Wang K., Zhou Y., Yang C., et al. Perturbations in amino acids and metabolic pathways in osteoarthritis patients determined by targeted metabolomics analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018;1085:54–62. doi: 10.1016/j.jchromb.2018.03.047. [DOI] [PubMed] [Google Scholar]
- 31.Senol O., Gundogdu G., Gundogdu K., Miloglu F.D. Investigation of the relationships between knee osteoarthritis and obesity via untargeted metabolomics analysis. Clin. Rheumatol. 2019;38:1351–1360. doi: 10.1007/s10067-019-04428-1. [DOI] [PubMed] [Google Scholar]
- 32.Rockel J.S., Zhang W., Shestopaloff K., Likhodii S., Sun G., Furey A., et al. A classification modeling approach for determining metabolite signatures in osteoarthritis. PloS One. 2018;13 doi: 10.1371/journal.pone.0199618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai G., Pelletier J.P., Liu M., Aitken D., Randell E., Rahman P., et al. Activation of the phosphatidylcholine to lysophosphatidylcholine pathway is associated with osteoarthritis knee cartilage volume loss over time. Sci. Rep. 2019;9:9648. doi: 10.1038/s41598-019-46185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai G., Pelletier J.P., Liu M., Randell E.W., Rahman P., Martel-Pelletier J. Serum lysophosphatidylcholines to phosphatidylcholines ratio is associated with symptomatic responders to symptomatic drugs in knee osteoarthritis patients. Arthritis Res. Ther. 2019;21:224. doi: 10.1186/s13075-019-2006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., Sun G., Likhodii S., Liu M., Aref-Eshghi E., Harper P.E., et al. Metabolomic analysis of human plasma reveals that arginine is depleted in knee osteoarthritis patients. Osteoarthritis Cartilage. 2016;24:827–834. doi: 10.1016/j.joca.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Zhai G., Wang-Sattler R., Hart D.J., Arden N.K., Hakim A.J., Illig T., et al. Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Ann. Rheum. Dis. 2010;69:1227–1231. doi: 10.1136/ard.2009.120857. [DOI] [PubMed] [Google Scholar]
- 37.Hu T., Oksanen K., Zhang W., Randell E., Furey A., Sun G., et al. An evolutionary learning and network approach to identifying key metabolites for osteoarthritis. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu T., Zhang W., Fan Z., Sun G., Likhodi S., Randell E., et al. Metabolomics differential correlation network analysis of osteoarthritis. Pac. Symp. Biocomput. 2016;21:120–131. [PubMed] [Google Scholar]
- 39.Zhai G., Sun X., Randel E., Liu M., Wang N., Tolstykh I., et al. Phenylalanine is a novel marker for radiographic knee osteoarthritis progression: the MOST study. J. Rheumatol. 2020;48:123–128. doi: 10.3899/jrheum.200054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello C.A., Hu T., Liu M., Zhang W., Furey A., Fan Z., et al. Metabolomics signature for non-responders to total joint replacement surgery in primary osteoarthritis patients: the Newfoundland osteoarthritis study. J. Orthop. Res. 2020;38:793–802. doi: 10.1002/jor.24529. [DOI] [PubMed] [Google Scholar]
- 41.Costello C.A., Hu T., Liu M., Zhang W., Furey A., Fan Z., et al. Differential correlation network analysis identified novel metabolomics signatures for non-responders to total joint replacement in primary osteoarthritis patients. Metabolomics. 2020;16:61. doi: 10.1007/s11306-020-01683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werdyani S., Liu M., Zhang H., Sun G., Furey A., Randell E.W., et al. Endotypes of primary osteoarthritis identified by plasma metabolomics analysis. Rheumatology. 2020 doi: 10.1093/rheumatology/keaa693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menni C., Zierer J., Valdes A.M., Spector T.D. Mixing omics: combining genetics and metabolomics to study rheumatic diseases. Nat. Rev. Rheumatol. 2017;13:174–181. doi: 10.1038/nrrheum.2017.5. [DOI] [PubMed] [Google Scholar]