Abstract
The rapid expansion of Next Generation Sequencing (NGS) data availability has made exploration of appropriate bioinformatics analysis pipelines a timely issue. Since there are multiple tools and combinations thereof to analyze any dataset, there can be uncertainty in how to best perform an analysis in a robust and reproducible manner. This is especially true for newer omics applications, such as miRNomics, or microRNA-sequencing (miRNA-sequencing). As compared to transcriptomics, there have been far fewer miRNA-sequencing studies performed to date, and those that are reported seldom provide detailed description of the bioinformatics analysis, including aspects such as Unique Molecular Identifiers (UMIs). In this article, we attempt to fill the gap and help researchers understand their miRNA-sequencing data and its analysis. This article will specifically discuss a customizable miRNA bioinformatics pipeline that was developed using miRNA-sequencing datasets generated from human osteoarthritis plasma samples. We describe quality assessment of raw sequencing data files, reference-based alignment, counts generation for miRNA expression levels, and novel miRNA discovery. This report is expected to improve clarity and reproducibility of the bioinformatics portion of miRNA-sequencing analysis, applicable across any sample type, to promote sharing of detailed protocols in the NGS field.
Keywords: High-throughput nucleotide sequencing, Computational biology, MicroRNAs, Bioinformatics, Osteoarthritis
1. Introduction
Next Generation Sequencing (NGS) technology has revolutionized the study of human genetic code, enabling a fast, reliable, and cost-effect method for reading the genome. Whereas “first generation” sequencing involved sequencing one molecule at a time, NGS involves sequencing multiple molecules in parallel [[1], [2], [3]]. This advance has reduced the time and cost per base that is sequenced, and has expanded sequencing applications which now includes microRNAs [[4], [5], [6], [7]]. MicroRNAs (miRNAs) are small RNAs of 22–25 base length, regulating gene expression through degradation of mRNA transcripts and inhibition of translation [8]. MiRNAs have emerged as critical regulators of health and disease, and when found in circulation, represent promising biomarkers given their stability, specificity, and ease of detection and quantification [9].
By providing a quantitative readout of all molecules of interest in a sample without relying on endogenous controls or pre-selected probes as do real-time PCR and microarray approaches, NGS has emerged as the gold standard approach for profiling nucleic acid, including miRNAs. Detecting single-nucleotide sequence changes or altogether novel sequences are added advantages of sequencing [10]. As a result, sequencing has the capacity to identify molecules with greater sensitivity, specificity, and predictive ability for detecting disease [11]. For these reasons, sequencing has been applied to biomarker discovery for a variety of diseases, but not without limitations. There are several sources of error that can be introduced during a sequencing experiment. Among these, the patient cohort may be underpowered [12]; sample extraction, library preparation, and sequencing may create bias that leads to over- or under-estimation of the expression level of a molecule or subset of molecules [13]; or a one-size-fits-all approach may be inappropriately applied to data analysis. To harness the potential of NGS to identify miRNAs as biomarkers – including novel miRNAs – a rigorous approach that overcomes existing limitations is needed [14]. This report focuses on the data analysis aspect, where a rigorous methodology for bioinformatics analysis of miRNA-sequencing data has been developed and applied to identify miRNAs in plasma samples from osteoarthritis patients [15].
Here we focus on two major advantages of miRNA-sequencing, the discovery of novel miRNAs, and the use of unique molecular identifiers (UMIs). A novel miRNA is predicted based on secondary structure and lack of homology with miRNAs in other species [16]. Novel miRNAs represent promise in precision medicine approaches given their potential specificity to disease states. Given this potential biological importance, we have developed and tested a method for discovery of novel miRNA sequences that are present in miRNA-sequencing data. In addition to novel miRNA discovery, our pipeline includes analysis of UMIs. During library preparation prior to amplification and sequencing, UMIs are added to each miRNA transcript. Following sequencing, UMI reads are collapsed such that the counts per miRNA remaining are more representative of the original starting sample prior to amplification. This is an internal control for managing library amplification bias, enabling accurate miRNA quantitation. While previous studies reporting miRNA-sequencing analysis may have incorporated UMI analysis, this level of detail is often not reported, nor is the method used to execute UMI analysis. Examples of available software which enable miRNA-sequencing analysis, but not UMI processing, include CAP-miRSeq and miRge [17,18]. Other software, such as TRUmiCount, handles UMI processing, and integrates the same UMI-tools software as we describe in our pipeline [19]. Yet other software, like sRNABench and sRNAtoolbox, provide a similar pipeline but the UMI processing is available only on the web-server mode and not standalone version, which is not secure for analyzing data generated from patient samples [20]. To overcome these limitations in the field, we put forth a detailed protocol for analysis of miRNA-sequencing data, including quality control, alignment, demultiplexing, UMI analysis, and novel miRNA analysis.
It is our aim to establish a standardized protocol in the field such that subsequent miRNA-sequencing studies will have a pipeline for guidance in bioinformatics analysis. This will enable biologists who may not have sufficient expertise in bioinformatics methods to understand the steps that need to be taken when analyzing a miRNA-sequencing dataset. It will also benefit bioinformaticians who have not previously worked with miRNA-sequencing data, given that this approach is relatively new as compared to more established sequencing approaches such as DNA-sequencing and RNA-sequencing. Furthermore, having a standardized protocol will promote integration of research findings from different groups, consistent with the efforts of established guidelines such as ‘Minimum Information about a high-throughput Nucleotide SEQuencing Experiment’ (MINSEQE - http://fged.org/projects/minseqe/) and ‘Encyclopedia of DNA Elements’ (ENCODE) pipelines - https://www.encodeproject.org/microrna/microrna-seq/). We leverage only open source software in our pipeline, offering customizable scripts for more advanced users. Having applied this pipeline and identified a unique signature of 11 circulating miRNAs in early knee osteoarthritis, we present the pipeline in sufficient detail to be replicated and widely used by others for the bioinformatics analysis of miRNA-sequencing data [15].
2. Overview of miRNA NGS analysis pipeline
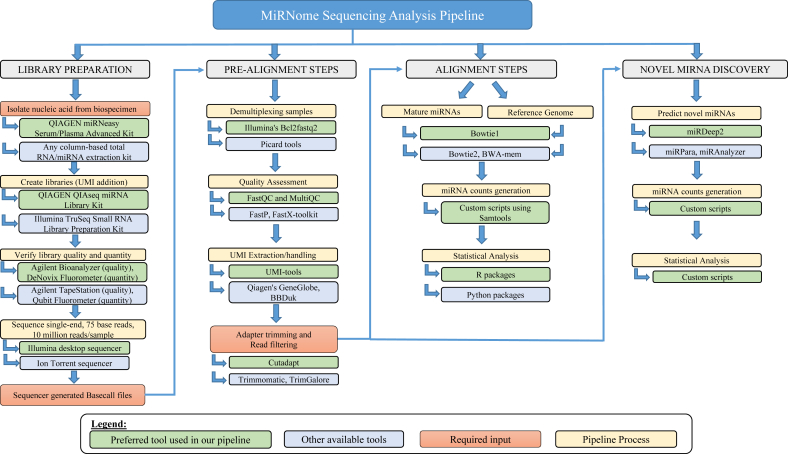
There is more than one way to analyze miRNA-sequencing data so here we present the approach we determined to be most suitable for bioinformatics analysis of miRNA-sequencing data generated from human plasma samples. Fig. 1 depicts an overview of the pipeline in its entirety, including: Prerequisite sequencing quality checks, Alignment steps, and Novel miRNA analysis. The first section begins with assessing the quality of the raw sequencing data, which is crucial to defining the path of downstream data processing. The second section involves read mapping and populating the UMI-based miRNA expression table for all samples in an experiment. This section represents the core of analysis. The third and final section describes the steps involved in novel miRNA prediction analysis. For those who are interested in applying this analysis pipeline, a detailed method is provided in Supplementary File 1 (miRNA analysis protocol) and Supplementary File 2 (novel miRNA analysis pipeline), along with details about software and databases used in Table 1.
Fig. 1.
MiRNome sequencing analysis pipeline schematic. An outline of the bioinformatics software used in the analysis, where the blue linear arrows depict the order of the software to be executed in the pipeline and the blue bent arrows depict the options of software available for each process. Abbreviations used: miRNA (microRNA), UMI (Unique Molecular Identifier)
Table 1.
List of software and databases used in both miRNA analysis and novel miRNA prediction pipeline.
| Name | URL/link | Utility | Date of Access |
|---|---|---|---|
| Bcl2fastq2 | https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html | Software: Conversion of.bcl files to.fastq files | 12-Feb-2019 |
| Bowtie1 | http://bowtie-bio.sourceforge.net/index.shtml | Software: Read alignment | 13-Mar-2019 |
| miRDeep2 | https://github.com/rajewsky-lab/mirdeep2 | Software: Novel miRNA prediction | 20-Jun-2019 |
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | Software: Single sample Quality Control (QC) report creation | 21-Mar-2019 |
| MultiQC | https://multiqc.info | Software: Creation of multi-sample QC report | 22-Mar-2019 |
| UMI-tools | https://github.com/CGATOxford/UMI-tools | Software: Handling of UMI-tagged reads | 31-Mar-2019 |
| Cutadapt | https://cutadapt.readthedocs.io/en/stable/guide.html | Software: Adapter trimming | 25-Feb-2019 |
| Samtools | http://www.htslib.org | Software: Processing aligned files (.SAM) | 24-Jun-2019 |
| miRBase | http://www.mirbase.org | Database: Mature and hairpin miRNA sequences | 05-Jan-2019 |
| vGRCh38 | http://hgdownload.soe.ucsc.edu/goldenPath/hg38/chromosomes/ | Database: Human reference genome sequence | 06-Jan-2019 |
3. Detailed NGS analysis pipeline
3.1. Pre-requisite sequencing quality checks
A critical issue to consider in NGS experiments is accurate preparation of the sample sheet. A sample sheet is a comma-separated values (.csv) file, outlining the information required for a text editor (e.g. Illumina Experiment Manager) to interpret the data generated from a sequencing run. Without the sample sheet, demultiplexing of samples is not possible, and therefore it is essential that the sample sheet accurately identifies the specific index that was used for each sample. During library preparation prior to sequencing, a specific barcode (8 nucleotides [nt] in length) is assigned to each sample, therefore enabling the individual libraries to be pooled and sequenced in the same run. The unique barcode (8 nt in length) is not to be confused with the UMI (12 nt in length). Following sequencing, the sample sheet is used to ‘decode’ and separate the samples from each other based on these unique barcodes. In the event barcodes are mismatched, demultiplexing is not possible. In other words, the sequencing reads for individual samples cannot be separated from each other and those data are lost. Further detail on demultiplexing following sequencing is provided below.
3.2. Demultiplexing
Once sequencing is complete the result is base call files (.bcl) that are generated for all the samples in a run, consisting of a series of images captured and converted into base calls. These files need to be separated into the individual biological samples based on aforementioned barcodes added during library preparation. This process is known as demultiplexing. These bcl files (binary format and not viewable) are converted into Fastq files using the bcl2fastq tool provided by Illumina, which allows users to open and view the file. At the end of demultiplexing, millions of reads for each sample are obtained in Fastq file format. For example, in our previous study, we obtained an average of 10 million reads per sample [15].
3.3. Quality assessment
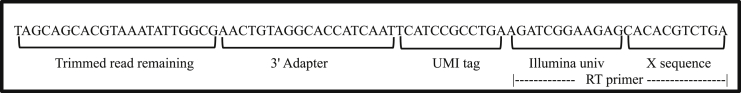
It is essential to ensure the reads are of sufficient quality for subsequent analyses. Quality control of reads avoids ‘Garbage In Garbage Out (GIGO)’ during downstream analysis, which is a computer science abbreviation implying poor quality input (i.e. sequencing reads) will result in poor quality data (i.e. expression values). There are two types of read quality checks we perform for every sample. The first is to check the quality of raw sequencing reads and the second is to check the number of reads post-alignment to the reference database. Read quality is measured using a metric called Q score or Phred score, which is logarithmically related to base calling error probabilities [21]. A Q score of 30 (99.9% accuracy) is considered acceptable across the field. For miRNA-sequencing data, a higher percentage (>40%) of reads having 3′ adapter and universal adapter contamination (e.g. Illumina Universal adapter - ‘AGATCGGAAGAG’) is often observed. This is attributable to the short length of mature miRNAs, where in a 75-base single-end read sequencing run, the sequencer will read into the adapters in addition to reading the entire length of the mature miRNA as shown in Fig. 2. In cases where <40% reads have 3′ adapter contamination, this may be an indication of degraded RNA or other small RNA molecules such as snoRNA, piRNA, etc. This can be confirmed by carefully noting the genomic coordinates of these RNA molecules to the reference genome and determining whether they have a disproportionate amount of aligned reads [22].
Fig. 2.
UMI read architecture. The raw sequenced read is divided into the following parts:1. Trimmed read remaining - is the first part of the read, which codes for the mature miRNA sequenc. 2. 3′ Adapter – fixed length (19 nucleotide [nt]) of Illumina adapter sequence. 3. UMI tag – stands for Unique Molecular Identifier (UMI) tag of fixed length of 12 nt. 4. Illumina univ – stands for Illumina Universal adapter of fixed length of 12 nt. 5. X seq – stands for a random sequence of nucleotides, when read in combination with Illumina universal adapter makes up for the Reverse Transcription (RT) primer.
The second major quality check is performed post-alignment to the mature miRNA reference database, where miRBase is most often used [23]. Poor quality samples with an insufficient number of aligned reads should be excluded from further analysis to avoid biasing the final results. On average, successful alignment will yield at least 3 million aligned reads per sample, provided that the sequencing depth was assigned 10 million reads per sample. Samples with low numbers of aligned reads can be identified based on having log-reads below the 0.025 normal quantile of the log-reads distribution across all samples. In our previous study, this resulted in samples with fewer than 400,000 aligned reads being excluded due to insufficient read depth [15]. An option to salvage such samples is to repeat library preparation and re-sequence to see if the number of reads can be increased. MiRNA-sequencing typically does not require great read depth or long read overlap since mature miRNAs are 22–25 bases in length, although sufficient read depth will provide greater confidence in identifying isomiRs (miRNA isoforms) and novel miRNA sequences. Quality Control (QC) can be visualized through plots based on individual base sequence quality scores, sequence length distribution, individual sequence GC content, duplicate sequences and many other criteria using tools like FastQC and FastX toolkit. To create multiple sample QC reports, MultiQC tool is recommended (Table 1; Supplementary File 1).
3.4. UMI analysis and read filtering
In our previous study, we used the QIAGEN QIAseq miRNA library kit which incorporates a UMI for each read. During bioinformatics analysis, UMI reads must be collapsed to produce counts for each miRNA that were present in the original biological sample prior to amplification. In our experience, UMI-tools is the best open source software to accurately process reads since it has multiple steps to extract, assign, deduplicate and count the UMI-based reads [24]. Other tools available for UMI analysis include bcl2fastq and FastP, but these tools do not provide the number of reads tagged with each UMI [25]. Even with a tool to assist, it is crucial to understand the structure of the entire read, identifying the location of the 3′ adapter, UMI, Reverse Transcription (RT) primer and 5′ adapter (Fig. 2). Based on this structure, we generated a regular expression (a string of characters which define a search pattern) with some leniency to accommodate both the maximum number of reads and also the most correct reads based on their UMI tag. Since it is imperative to retain and trim only those reads having the 3’ adapter the sequencer will read into the adapter in order to capture miRNAs. The utility of UMIs is only seen post-alignment as there is greater confidence in the genomic read location. Since the length of mature miRNAs is known to be around 22–25 bp, the final raw read filtering step is to trim the reads to retain only the expected miRNA read lengths with some leniency, to remove reads that are either too short (<18 bp) and too long (>30 bp). The result of UMI analysis and read filtering is a set of good quality raw sequences, ready to be processed for any analysis, such as alignment.
3.5. Reference-based read alignment
Given that miRNA-sequencing produces very short reads, accurate alignment requires stringent parameters to avoid multiple matches across the reference database. Our tool of choice is the original version of Bowtie, since it is better known for short read alignment and also performed better than tools like Bowtie2 and BWA. In our method, we perform two rounds of alignment, one against a database of mature miRNA sequences and the other against the reference genome sequence, both being organism-specific. The most used reference database for miRNA alignment is miRBase, currently in version 22.1 (Table 1) with 38,589 miRNA entries [23]. In the first round of alignment, reads are aligned to mature miRNA entries from miRBase with stringent criteria. Alignment to the reference genome is an important additional step since the matrix of counts generated from miRBase alignment can be further increased. Reads that do not align to miRBase are aligned to the genomic coordinates of mature miRNA from the reference genome (e.g. vGRCh38). Ideally, alignment to miRBase should account for the majority of the sequencing reads at roughly 60–80%. If this is the case, it is an indication that the preceding filtering steps were successful in retaining high quality data. This also lends confidence to the starting sample quality and proper library preparation. In cases where the percentage of aligned reads to miRBase is low (<40%), the problem could be an incorrect adapter sequence being used, regular expression being used in UMI extraction, or an inappropriate reference database.
Once both alignment steps are completed for each sample, counts for each miRNA are added to create a matrix of sums of UMI-based counts, where each row is a miRNA and each column corresponds to a sample. This step can be performed as per the user’s convenience and knowledge using custom scripts (Supplementary File 1). These raw counts can then be used for downstream Differential Expression Analysis (DEA) and statistical analysis. DEA and other downstream analyses are beyond the scope of this article but have been described elsewhere [26]. These analyses will depend on the research question and key variables such as the number of samples, groups, batch effects, metadata (e.g. age, sex, comorbidities), time points, and so on.
4. Novel miRNA analysis
Sequencing applications are increasingly used for detection of single nucleotide polymorphisms, gene isoforms, and other lowly-expressed genetic variants. With miRNA-sequencing, prediction of novel miRNAs is possible based on features such as alignment, secondary structure prediction, energy scores, and homology to other species. This prediction pipeline makes use of open source tools such as mirDeep2 [16]. However, to the best of our knowledge, there are no existing tools which incorporate UMI information into novel miRNA prediction. Therefore, it is critical to remove UMI tags from all reads using UMI-tools before processing reads for novel miRNAs to avoid incorrect alignment. When the first step of alignment occurs in miRDeep2.pl script, it is recommended to find a species, genetically similar to the species profiled in your sequencing data, so that homology between the observed and expected mature and hairpin miRNA sequences can be accounted for before moving ahead. The recommended goal of these prediction-based analyses is fewer novel miRNAs with greater confidence. In order to achieve this, we removed sequences showing homology with known human and mouse mature and hairpin miRNA sequences from miRBase. It is equally essential to only retain sequences that are predicted to form a hairpin-like structure (since this is the secondary structure of final mature miRNAs) based on nucleotide energy calculations using specific software like RNAfold within miRDeep2 [27].
Once a list of predicted novel miRNAs is generated for each sample, an appropriate filtering strategy must be applied. The predicted novel miRNAs are often not consistently found across all samples, therefore precluding typical differential expression analyses. To increase confidence that a predicted novel miRNA may be a true biological phenomenon rather than a computational artefact, one way to filter the list is to select novel miRNAs that are consistently found across biological samples in a pre-defined group. When applied in our previous study, we identified 4 novel miRNAs that were consistently found in >95% of samples in our group of interest [15]. Novel miRNAs that are consistently present in one group and absent in another may reflect a new biomarker of disease or a new regulator of disease processes. A step-by-step guide has been curated for scientists interested in novel miRNA prediction (Supplementary File 2).
5. Discussion
The recent surge in exploring bone and joint pathologies such as osteoarthritis using NGS continues to advance our understanding of underlying molecular processes [28,29]. MiRNA-sequencing offers the power to identify both known and novel miRNAs in any sample, free of specific probe-binding or endogenous references as required by older technologies, revealing the complete miRNome of the biological sample. Discovery of a panel of differentially expressed miRNAs can be utilized to understand disease mechanisms, identify novel disease biomarkers, or design therapeutic agents such as antisense oligonucleotides [30]. MiRNAs are implicated in a variety of disease processes and have been demonstrated to have cell- and tissue-specific expression and behavior [31]. Therefore, the pipeline we present for miRNA-sequencing represents an important advance in the omics field, with a key advantage being the level of customization offered to the user. While the upstream experimental design and downstream analyses (e.g. DEA) are beyond the scope of this article, here we provide a detailed method for the bioinformatics portion of miRNA-sequencing analysis. Given the complexity and importance of this step in obtaining high-quality sequencing data, greater attention to bioinformatics processing is needed in the omics field. Going forward, reports using sequencing would be strengthened by including detailed bioinformatics methodology such that analyses can be understood and replicated by other groups. Collaboration between academic labs and private companies can leverage different expertise to accelerate translation of research data.
Author contributions
PP and SAA contributed in analysis and interpretation of the data, drafting of the article and critical revision of the article for intellectual content. SAA also contributed in final approval of the article and provision of patient samples. MK contributed in conception and design of the article, critical revision of the article, final approval of the article and obtaining of funding.
Funding source
This work is supported by grants to MK by the Canadian Institute of Health Research, Canada (# 156299), the Krembil Foundation (Canada) and the Schroeder Arthritis Institute, University Health Network via the Toronto General and Western Hospital Foundation, Toronto, Canada
Declaration of competing interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ocarto.2020.100131.
Contributor Information
Shabana Amanda Ali, Email: sali14@hfhs.org.
Mohit Kapoor, Email: mkapoor@uhnresearch.ca.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sanger F., Coulson A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maxam A.M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 4.Slatko B.E., Gardner A.F., Ausubel F.M. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. 2018;122:e59. doi: 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., et al. DNA sequencing at 40: past, present and future. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy S.E., Myers R.M. Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 2016;17:95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church G.M., Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraus V.B., Karsdal M.A. Osteoarthritis: current molecular biomarkers and the way forward. Calcif. Tissue Int. 2020 doi: 10.1007/s00223-020-00701-7. [DOI] [PubMed] [Google Scholar]
- 12.Kok M.G.M., de Ronde Mwj, Moerland P.D., Ruijter J.M., Creemers E.E., Pinto-Sietsma S.J. Small sample sizes in high-throughput miRNA screens: a common pitfall for the identification of miRNA biomarkers. Biomol Detect Quantif. 2018;15:1–5. doi: 10.1016/j.bdq.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dijk E.L., Jaszczyszyn Y., Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali S.A., Gandhi R., Potla P., Keshavarzi S., Espin-Garcia O., Shestopaloff K., et al. Osteoarthritis Cartilage; 2020. Sequencing Identifies a Distinct Signature of Circulating microRNAs in Early Radiographic Knee Osteoarthritis. [DOI] [PubMed] [Google Scholar]
- 16.Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Z., Evans J., Bhagwate A., Middha S., Bockol M., Yan H., et al. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genom. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y., Baras A.S., Halushka M.K. miRge 2.0 for comprehensive analysis of microRNA sequencing data. BMC Bioinf. 2018;19:275. doi: 10.1186/s12859-018-2287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pflug F.G., von Haeseler A. TRUmiCount: correctly counting absolute numbers of molecules using unique molecular identifiers. Bioinformatics. 2018;34:3137–3144. doi: 10.1093/bioinformatics/bty283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aparicio-Puerta E., Lebron R., Rueda A., Gomez-Martin C., Giannoukakos S., Jaspez D., et al. sRNAbench and sRNAtoolbox 2019: intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019;47:W530–W535. doi: 10.1093/nar/gkz415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 22.Zhong X., Heinicke F., Lie B.A., Rayner S. Accurate adapter information is crucial for reproducibility and reusability in small RNA seq studies. Noncoding RNA. 2019;5 doi: 10.3390/ncrna5040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May S.M., Abbott T.E.F., Del Arroyo A.G., Reyes A., Martir G., Stephens R.C.M., et al. MicroRNA signatures of perioperative myocardial injury after elective noncardiac surgery: prospective observational mechanistic cohort study. Br. J. Anaesth. 2020;125(5):661–671. doi: 10.1016/j.bja.2020.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofacker I.L., Stadler P.F. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- 28.Trajanoska K., Rivadeneira F. The genetic architecture of osteoporosis and fracture risk. Bone. 2019;126:2–10. doi: 10.1016/j.bone.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Reynard L.N., Barter M.J. Osteoarthritis year in review 2019: genetics, genomics and epigenetics. Osteoarthritis Cartilage. 2020;28:275–284. doi: 10.1016/j.joca.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura A., Ali S.A., Kapoor M. Antisense oligonucleotide-based therapies for the treatment of osteoarthritis: opportunities and roadblocks. Bone. 2020;138:115461. doi: 10.1016/j.bone.2020.115461. [DOI] [PubMed] [Google Scholar]
- 31.Tahamtan A., Teymoori-Rad M., Nakstad B., Salimi V. Anti-inflammatory MicroRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 2018;9:1377. doi: 10.3389/fimmu.2018.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.