Abstract
Background
Proteomic studies of the secretome of skeletal muscle cells can help us understand the processes that govern the synthesis, systemic interactions and organization of skeletal muscle and identify proteins that are involved in muscular adaptations to exercise, ageing and degeneration. In this systematic review, we aimed to summarize recent mass-spectrometry based proteomics discoveries on the secretome of skeletal muscle cells in response to disease, exercise or metabolic stress.
Methods
A literature search was performed in the Medline/Ovid and Scopus electronic bibliographic databases. Only papers reporting the analysis of the secretome by mass spectrometry were included.
Results
A total of 19 papers met the inclusion criteria for this systematic review. These papers included comparative analysis of differentially expressed proteins between healthy and unhealthy muscle cells and comparison of the secretome of skeletal muscle cells during myogenesis and after insulin stimulation or exercising. The proteins were separated into several categories and their differential secretion was compared. In total, 654 proteins were listed as being present in the secretome of muscle cells. Among them, 30 proteins were differentially regulated by physical exercise, 130 during myogenesis, 114 by dystrophin deficiency, 26 by muscle atrophy, 27 by insulin stimulation and finally 176 proteins secreted by insulin-resistant muscle cells.
Conclusions
This systematic review of the secretome of skeletal muscle cell in health and disease provides a comprehensive overview of the most regulated proteins in pathological or physiological conditions. These proteins might be therapeutic targets or biochemical markers of muscle diseases.
Keywords: Secretome, Skeletal muscle cells, Myokines, Proteomic, Mass spectrometry, Biomarkers
1. Introduction
Skeletal muscle accounts for approximately 40% of the total body weight and contains between 50 and 75% of all body proteins.1 This organ has many more functions than joint motion by contraction, it has fundamental participation in immunometabolic processes, as provider of substrates to immune system and as glucose buffering organ. It storages important molecules like amino acids and carbohydrates (i.e. glycogen), and provides the production of heat for body temperature regulation.1 Skeletal muscle architecture is characterized by the arrangement of the muscle fibers with the associated connective tissue. The main cells in mature muscles are myocytes. Myocytes are long, tubular cells that develop from myoblasts to form muscles in a process known as myogenesis. There are various specialized forms of myocytes with distinct properties: cardiac, skeletal, and smooth muscle cells. Skeletal muscle stem cells (also called skeletal muscle satellite cells) are located between the sarcolemma and the basal lamina. These quiescent cells are activated by muscle fiber degeneration or injury and proliferate in myoblasts. These last proliferate, differentiate and fuse to form multinucleated myofibers. The connective tissue surrounding the muscle is known as the epimysium. Another layer of connective tissue called perimysium surrounds the bundles of fibers. Finally, the sarcolemma envelops the single muscle fiber which has in mean 1 cm in length and 100 μm in diameter.1 The myofiber contains myofibrils which are composed of sarcomeres (the basic contractile units of skeletal muscle). The sarcomere is composed mainly by thick (myosin) and thin (actin, troponin and tropomyosin) filaments that can slide with each other's and lead to contraction of the muscle.1
Skeletal muscle is an active endocrine organ containing cells that may communicate in an auto-, para- or endocrine manner thanks to the secretion of mediators like myokines. Pedersen et al defined myokines as “cytokines and other peptides that are produced, expressed, and released by muscle fibers and exert either paracrine or endocrine effects”.2 Beside myokines, other molecules like proteins, lipids, amino acids, metabolites and small RNAs secreted by skeletal muscle cells are also involved in cells communication.3, 4, 5 Myokines mediate metabolic regulation, inflammatory processes, angiogenesis and myogenesis.6, 7, 8, 9 Myogenesis is the process during which the muscle stem cells, or satellite cells, proliferate and differentiate into mature muscle fibers, or myotubes. This process is crucial for maintenance and repair of muscle tissue.
Furthermore, myokines are likely to play important roles in the pathophysiology of diseases like sarcopenia, insulin resistance or type-2 diabetes. The secretory profile of the skeletal muscle cells is changed by strength and/or endurance exercising.10, 11, 12, 13,9 For example, in addition to its role in inflammation, immune responses and hematopoiesis, interleukin-6 (IL-6) released by muscles during contraction, influences lipid and glucose metabolism.14, 15, 16
The secretome has been defined by Makridakis et al as the “rich, complex set of molecules secreted from living cells”.17 The study of the secretome of skeletal muscle cells may help to understand the processes that govern synthesis and organization of the skeletal muscle, its relation with adipose tissue, immune system and could help to identify factors responsible for metabolic, structural and functional changing in muscle during aging. In addition, the secretome of skeletal muscle cells could be a source of biomarkers of muscle disease and source of therapeutic target for future therapy.
The types of cell cultures most commonly used for secretome analysis are rat L6 skeletal muscle cells, mouse C2C12 skeletal muscle cells or primary human skeletal muscle cells.9 Among techniques used to analyse the secretome of skeletal muscle cells, mass spectrometry is recognized as the most accurate, due to its specificity and robustness.18 There are two strategies for spectrometry-based global protein analysis: bottom-up and top-down proteomics.19, 20, 21 Top-down strategies directly analyze intact proteins while bottom-up proteomics include an enzymatic digestion step of proteins to analyze the resulting peptides. The bottom-up strategy is the most often used for global protein analysis, because top-down strategy is more complicated to handle with larger proteins due to their poor solubility.22 Furthermore, the detection limits and sensitivity of the mass spectrometer are lower for proteins than for peptides.22 This review focus only on bottom-up strategies.
The objectives of this paper were to list the proteins identified by mass spectrometry in the secretome of skeletal muscle cells and to summarize knowledges on the modification of this secretome in different experimental and pathological conditions. To our knowledge, this is the first review covering systematically the literature published in this field.
2. Materials and methods
This review was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. A literature search was performed in two electronic bibliographic databases: Medline/Ovid and Scopus. Regarding the search done on Medline, a combination of Mesh terms as well as free language was used. Details of the search terms which included keywords like “secretome”, “secreted proteins”, “myoblasts”, “skeletal muscle cells”, “skeletal muscle fibers” or “skeletal muscle satellite cells” were used. The search strategies are available in the supplementary file 1. Only papers published in English and reporting the analysis of the secretome of isolated skeletal muscle cells or skeletal muscle explants of all species by mass spectrometry were included. Articles were screened independently by two authors. Supplementary files of all papers were analyzed and relevant data were included in this review.
3. Results
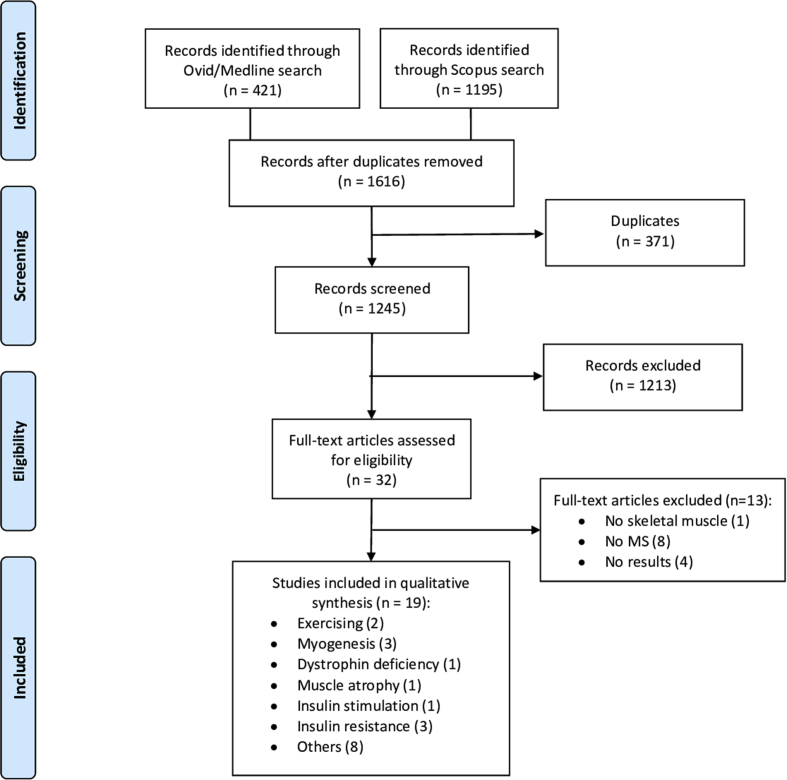
Using the search strategies given in supplementary file 1, 421 papers were found from Medline/Ovid database, and 1195 from Scopus. After eliminating 371 duplicates, 1245 articles were screened. Then, 32 full-texts were assessed for eligibility. Finally, 19 papers met the inclusion criteria for this review (fig. 1). In total, 654 proteins were listed in this systematic review as being present in the secretome of muscle cells (Table 2). Among them, 30 proteins were differentially regulated by physical exercise (all upregulated), 130 during myogenesis (90 up- and 40 downregulated), 114 by dystrophin deficiency (107 up- and 7 downregulated), 26 by muscle atrophy (15 up- and 11 downregulated), 27 by insulin stimulation (14 up- and 13 downregulated) and finally 176 proteins secreted by insulin-resistant muscle cells (26 up- and 150 downregulated). Comparative analysis of differentially expressed proteins between healthy and unhealthy (Duchenne muscular dystrophy, muscle atrophy, insulin resistant cells, etc.) skeletal muscle cells were conducted by different research groups. Comparison of secretome of skeletal muscle cells after exercising or during myogenesis were also conducted. The main characteristics of the proteomic studies are listed in Table 1.
Fig.1.
Preferred reporting items for systematic reviews and meta-analysis (PRISMA) Flow diagram.23 No MS = No Mass Spectrometry.
Table 2.
Summary of recent mass spectrometry-based studies carried out on human skeletal muscle cells to identify secretome components. Classically secreted proteins contain a signal sequence, unlike non-classically secreted proteins.
| ECM Proteins | Function | Secretion | Exercise | Myogenesis | Dystrophin deficiency | Atrophy | Insulin stimulation | Insulin resistant cells | References |
|---|---|---|---|---|---|---|---|---|---|
| Basement membrane-specific heparan sulfate proteoglycan core protein (Perlecan) | component of basement membranes | Classical | ↑ | ↑ | ↓ | ↓ | 6,27,26,28,31,33,36,37,40 | ||
| Biglycan | collagen fiber assembly | Classical | ↑ | ↓ | ↓ | 25,40,27,28,26,6,35, 36, 37, 38 | |||
| Collagen I(α1, α2), II(α1), III(α1), IV(α1,α2,α3), V(α1,α2,α3), VI(α1,α2,α3), VII(α1), VIII(α1), IX(α3), XI(α1,α2), XII(α1), XIII(α1), XIV(α1), XV(α1), XVIII(α1) | cell adhesion | Classical | III(α1), XVIII(α1) ↓ I (α1), II(α1), V (α1, α3) VI(α1), XI (α1) ↑ |
IV(α2)↑ | VI(α1)↑ V (α2)↓ III(α1)↓ |
I (α1, α2)↓ III(α1)↓ IV(α1, α2)↓ V (α1, α2)↓ VI(α1,α2)↓ VIII(α1,α2)↓ XI (α1)↓ XII(α1)↓ XV (α1)↓ |
30,29,31,27,6,33,32,34, 35, 36, 37, 38, 39,26 | ||
| Decorin | affects the rate of fibril formation | Classical | ↑ | ↓ | 27,28,26,6,35,36 | ||||
| Dystroglycan | laminin and basement membrane assembly, cell survival and migration | Classical | ↑ | ↓ | 27,28,26,33,35,36,38,39 | ||||
| Extracellular matrix protein 1, 2 | Angiogenesis, Biomineralization, Mineral balance, Osteogenesis | Classical | 2 ↓ | 29,28,6,34, 35, 36,39 | |||||
| Fibrillin 1, 2 | Structural component of microfibrils | Classical | 1 ↑ | 1↓ | 25,27,28,26,6,35,36 | ||||
| Fibromodulin | affects the rate of fibril formation | Classical | ↑ | ↓ | 36,27 | ||||
| Fibronectin | Cell adhesion, cell shape | Classical | ↑ | ↓ | 32,29,27,28,6,34,36, 37, 38, 39,24 | ||||
| Fibulin 1, 2, 5, 7 | cell–cell interaction, cell migration, ECM remodelling | Classical | 1 (isoform C) ↓ 1 (isoform D), 2 (isoform B), 5↑ |
1, 2, 5 ↓ | 31,27,28,26,6,33,32,35,36,39 | ||||
| Glypican 1, 6 | Role in skeletal muscle differentiation | Classical | 1↑ | 1↓ | 29,27,28,6,33,35,37,39,40 | ||||
| Laminin α (1, 2, 3, 4, 5), β (1, 2), γ (1,2) | Classical | α5, α2, β2 ↓ | α(4,5), β1, γ1↓ | 6,27,28,30,33,35, 36, 37 | |||||
| Latent-transforming growth factor beta-binding protein 1(S, L), 2, 3, 4 | growth factor binding | Classical | 3↑ | 1, 2, 3, 4↓ | 6,26, 27, 28,35,36 | ||||
| Lumican | Collagen binding | Classical | ↓ | 6,28,29,36 | |||||
| Matrilin 2, 3 | involved in matrix assembly | Classical | 2↓ | 26, 27, 28,35,36 | |||||
| Matrix Gla Protein | Classical | 27,33,35,36 | |||||||
| Mimecan | Induces bone formation in conjunction with TGF-β-1 or TGF-β-2. | Classical | ↓ | ↓ | 26,27,31,32,35,36 | ||||
| Moesin | connection cytoskeletal structure - plasma membrane | ↑ | 6,27,40,28,29,31,34, 35, 36,38,39 | ||||||
| Nidogen 1, 2 | Classical | 1,2↓ | 6,26, 27, 28,33,35,36,38, 39, 40 | ||||||
| Periostin | cell adhesion | Classical | ↑ | ↑ | ↓ | 6,25, 26, 27, 28,35,36 | |||
| Prolargin | Classical | ↑ | ↓ | 26,27,36 | |||||
| Protein S100 (A4, A6 (calcyclin), A11, A13, A16) | Classical | A6↓ | 6,28,30,33, 34, 35, 36,38,39 | ||||||
| Proteoglycan 4 | Role in joint lubrication | ↑ | 6,27,35 | ||||||
| Secreted protein acidic and rich in cysteine (SPARC) | Regulates cell growth, binds calcium and copper | Classical | ↑ and ↓ | ↑ | ↓ | 6,26,36, 37, 38, 39, 40,27, 28, 29, 30, 31, 32,34,35 | |||
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | cell attachment process | Classical | ↓ | ↓ | 6,35,37 | ||||
| Syndecan 2, 4 | proteoglycan that bears heparan sulfate | Classical | 27,28,31, 32, 33,36 | ||||||
| Tenascin C, XB, X | Cell adhesion | Classical | C ↑ | ↓ | 6,26, 27, 28,30,35,36 | ||||
|
Vitronectin |
Cell adhesion and spreading factor |
Classical |
6,36 |
||||||
|
Cytokines and growth factors |
Specificity |
Secretion |
Exercise |
Myogenesis |
Dystrophin deficiency |
Atrophy |
Insulin stimulation |
Insulin resistant cells |
References |
| Anamorsin (Ciapin 1) | Apoptosis, metal binding | Non-classical | ↑ | 28,31,32,35,36 | |||||
| Angiopoietin 1, 4 | angiogenesis, cell proliferation, adhesion and migration | Classical | 1↑ | 4↓ | 27,28,35,36 | ||||
| Bone morphogenetic protein 1, 4 | Cytokine, growth factor, protease, metalloprotease | Classical | 1 ↓ | 1 ↓ | 26, 27, 28,30,35,36 | ||||
| Brain derived neurotrophic factor | regulates skeletal muscle metabolism | Classical | 28,36 | ||||||
| C–C motif chemokine 2, 3, 5, 7, 8, 9 | cytokine, inflammatory response, chemotaxis | Classical | 2↑ | 2, 7, 8↑ | 2↑ 9↓ |
6,27,28,30,35,36 | |||
| Cell adhesion molecule-related/down-regulated by oncogenes | Promotes differentiation of myogenic cells | Classical | ↑ | ↓ | 26,27,35,36 | ||||
| Complement C1q tumor necrosis factor-related protein 1, 3, 5 | Classical | 3, 5↑ | 1, 3 ↓ | 26, 27, 28,33,35, 36, 37 | |||||
| Connective tissue growth factor | Cell adhesion, DNA synthesis, growth factor activity | Classical | ↑ | ↓ | ↓ | 6,26, 27, 28,35,36,38,39 | |||
| C-X-C motif chemokine (CXCL) 1, 2, 5, 6, 10, 12, 16 | Cytokine, growth factor | Classical | 5↓ | 1, 5↑ | 6,27,28,32,35,36 | ||||
| Fibroblast growth factor 17, 21 | Mitogenic and cell survival activities | Classical | 21↑ | 27,30, 31, 32,35,36 | |||||
| Follistatin-related protein 1, 3 | Heparin binding, modulate action of some growth factors | Classical | 1 ↓, 3 ↑ | 1↓ | 1↓ | 6,26,37, 38, 39,27, 28, 29,31,32,34, 35, 36 | |||
| Galectin 1, 3, 9 | regulation of cell proliferation and migration + bind galactose | Classical | 1↑ | 1, 3↑ | 1↑ | 9↑ | 6,26,39,40,27,28,30,34, 35, 36, 37, 38 | ||
| Glucose-6-phosphate isomerase | Cytokine, growth factor, isomerase | Classical | ↑ | 6,24,39,26, 27, 28, 29,34, 35, 36,38 | |||||
| Granulins | Cytokine, role in inflammation and tissue remodeling | Classical | ↓ | 26, 27, 28,35,36 | |||||
| Granulocyte colony-stimulating factor | cytokine, growth factor, role in hematopoiesis | 6 | |||||||
| Gremlin 1 | Cytokine | Classical | 28,33,35,36 | ||||||
| Growth/differentiation factor 8 (myostatin), 11, 15 | Regulates food intake, energy expenditure and body weight | Classical | 11↓ | 27,28,30,35,36 | |||||
| Hepatocyte growth factor | Mitogen for hepatocytes cells, growth factor | Classical | ↓ | 27,35,36 | |||||
| Hepatoma-derived growth factor | Growth factor, repressor | Non-classical | ↓ | ↑ | 37,39 | ||||
| High mobility group protein B1, B2, B3 | Immunity, autophagy, chemotaxis | Non-classical | B1 ↑ and ↓ | 27,32,35, 36, 37 | |||||
| Insulin-like growth factor 1, 1A, 2 | Classical | 1,2↑ | 2↑ | 2↓ | 1↓ | 26,27,30,34, 35, 36 | |||
| Insulin-like growth factor binding protein(IGFBP) 2, 3, 4, 5, 6, 7 | regulation of cell proliferation | Classical | 2↓ 4↑ 5↑ and ↓ |
6↓ | 2, 5, 6, 7↓ 4 ↑ and ↓ |
6,26,39,40,27,28,31,32,35, 36, 37, 38 | |||
| Interleukin 1β, 2, 4, 6, 7, 8, 10, 13, 17A, 25, 34 | Classical | 6,28,35,36 | |||||||
| Macrophage colony-stimulating factor 1 | Cytokine, growth factor | Classical | ↓ | 6,25,26,28,29,35,36 | |||||
| Macrophage migration inhibitory factor | cytokine, isomerase, immunity | Classical | 6,27,38,39,28,29,31, 32, 33,35, 36, 37 | ||||||
| Osteoclast-stimulating factor 1 | induces bone resorption | 35,36 | |||||||
| Placenta growth factor | angiogenesis, differentiation | Classical | 28,3536 | ||||||
| Platelet-derived growth factor A, C | Developmental protein, mitogen, growth factor | Classical | A, C↑ | C ↓ | 27,35,36 | ||||
| Prosaposin (sulfated glycoprotein 1) | Lipid metabolism, growth factor activity | ↑ | ↑ | 6,26, 27, 28,31,35, 36, 37 | |||||
| Secreted frizzled-related protein 2, 4 | Developmental protein, regulation of cell growth and differentiation | Classical | 2 ↓ | 6,27,35,36 | |||||
| Stromal cell-derived factor 1, 2 | cytokine, growth factor, chemotaxis | Classical | 27,28,35,36 | ||||||
| Transforming growth factor β (1, 2, 3) | Multifunctional proteins | Classical | 1, 2, 3↑ | 2↓ | 26, 27, 28,35,36 | ||||
| Tumor necrosis factor α (TNF-α) | Cytokine | 6,28 | |||||||
|
Vascular endothelial growth factor A, C, D |
growth factor, angiogenesis, mitogen, differentiation |
Classical |
A, D↓ |
35,36 |
|||||
|
Enzymes |
Specificity |
Secretion |
Exercise |
Myogenesis |
Dystrophin deficiency |
Atrophy |
Insulin stimulation |
Insulin resistant cells |
References |
| 6-phosphogluconate dehydrogenase, decarboxylating | Oxydoreductase | ↑ | 6,26,29,32,34, 35, 36,38,39 | ||||||
| 6-phosphogluconolactonase | Hydrolase | Classical | ↑ | 6,29,31,32,34, 35, 36,38 | |||||
| A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) 1, 2, 4, 5, 6, 7, 9, 10, 12, 19 | cell migration, inflammation, development | Classical | 1↑ | 1, 2, 5, 7, 12↓ | 27,28,32,35,36 | ||||
| Adenosylhomocysteinase | Hydrolase | ↓ | 6,29,33, 34, 35, 36,39 | ||||||
| Alcohol dehydrogenase [NADP( + )] | Oxidoreductase | ↑ | ↑ | 6,25,26,29,34, 35, 36 | |||||
| Aldo-keto reductase family 1 member A4, C1, C2, C3 | Oxidoreductase | Non-classical | A4 ↓ | 6,29,32,35,36 | |||||
| Aldose reductase | Oxidoreductase | ↑ | 6,11,39,26,28,29,33, 34, 35, 36,38 | ||||||
| Aminoacylase-1 | Hydrolase | 6,26, 27, 28, 29,36 | |||||||
| Aminopeptidase-B,-N | Aminopeptidase, hydrolase, metalloprotease, protease | Classical | B↑ | 6,26,28,29,33, 34, 35, 36 | |||||
| Angiogenin | angiogenesis, differentiation, stress response | Classical | 28,36 | ||||||
| Arylsulfatase A, B, K | Hydrolase | Classical | A↓ | 28,29,36 | |||||
| ATP synthase subunit beta, mitochondrial | Production of ATP from ADP | ↑ | ↑ | 31 | |||||
| Carboxypeptidase A4, D, E, Q | Carboxypeptidase, Hydrolase, Metalloprotease, Protease | Classical | E↑ | E, Q↓ | 26,28,33,35,36,38 | ||||
| Cathepsin A, B, D, H, L1, Z, O | Hydrolase, Protease, Thiol protease | Classical | D, L1, O, Z, ↑ | D↑ B, Z, L1↓ |
L1, O ↓ | 6,26,38,39,27, 28, 29,31,32,34, 35, 36 | |||
| Chymotrypsinogen B | Hydrolase, Protease, Serine protease | Classical | ↑ | ↑ | 25,27,35,36 | ||||
| Creatine kinase B-type, M-type | Kinase, Transferase | B↑ | B↑ | 6,11,26, 27, 28, 29,34, 35, 36 | |||||
| Delta-aminolevulinic acid dehydratase | Allosteric enzyme, Lyase | Non-classical | ↑ | ↓ | 6,25,29,31,32,34, 35, 36 | ||||
| Dipeptidyl peptidase 1, 2, 3, 4, 8, 9 | Aminopeptidase, Hydrolase, Protease | Classical | 2↑ | 6,26, 27, 28, 29,35,36,38,39 | |||||
| Disintegrin and metalloproteinase domain-containing protein (ADAM) 9, 10, 12, 15 17, 19 | Role in tumorigenesis and angiogenesis | Classical | 10, 12, 19 ↓ | 28,35,36,39 | |||||
| Extracellular superoxide dismutase [Cu–Zn] | Antioxidant, Oxidoreductase | Classical | ↑ | ↓ | 6,26,27,35, 36, 37 | ||||
| Farnesyl pyrophosphate synthetase | Transferase | ↑ | 6,29,32, 33, 34, 35, 36 | ||||||
| Ferritin heavy chain, light chain | Stores iron in a soluble and non-toxic form | Non-classical | LC↓ | 6,11,36,26,28,29,31, 32, 33, 34, 35 | |||||
| Fibroblast growth factor receptor 1, 4 | Kinase, receptor, transferase | Classical | 4↑ | 28,36 | |||||
| Fructose-biphosphate aldolase A, B, C | Lyase involved in glycolysis | A↑ | A↑ | A↑ | 6,11,27,29,32, 33, 34,37, 38, 39 | ||||
| Glutathione S-transferase A4, MU1, MU2, ω-1, P1, alpha 3 | Oxidoreductase, transferase | Classical | P1, ω-1↑ | 6,11,36,38,39,26,28,29,31, 32, 33, 34, 35 | |||||
| Glutathione synthetase | Ligase | Non-classical | ↓ | 6,26,29,34,36 | |||||
| Glyceraldehyde-3-phosphate dehydrogenase | inhibit cell spreading, glycolysis | Non-classical | ↓ | ↑ | 6,11,30, 31, 32,34, 35, 36,40 | ||||
| Glycerol 3 phosphate dehydrogenase | 11,36 | ||||||||
| Glycogen phosphorylase, liver form, muscle form | Allosteric enzyme, Glycosyltransferase, Transferase | Muscle↑ | 6,11,26,29,34,36 | ||||||
| Histidine triad nucleotide-binding protein 1 | Hydrolase, role in apoptosis and transcription | ↓ | 28,35,36,38 | ||||||
| Hypoxanthine-guanine phosphoribosyltransferase | Glycosyltransferase, transferase | ↓ | ↑ | 29,31,32,35,36 | |||||
| Inosine triphosphate pyrophosphatase | hydrolase, magnesium binding | Non-classical | 26,31,35,36 | ||||||
| Inositol monophosphatase 1, 2 | Hydrolase | Classical | ↑ | 6,26,29,34, 35, 36 | |||||
| Inositol-3-phosphate synthase 1 | Involved in myo-inositol biosynthesis | ↑ | 26,34,36 | ||||||
| Lipoprotein lipase | Lipid metabolism, hydrolase | Classical | ↓ | 27,35,36 | |||||
| Mannan-binding lectin serine protease 1 | Role in immunity | Classical | ↑ | 25,26,35,36 | |||||
| Matrix metalloproteinase 2, 9, 14, 19 | Regulation of cell migration, metalloproteinase, breakdown of ECM | Classical | 2↑ | 2, 9, 19↓ | 27,28,31,35,36,39,40 | ||||
| NEDD8-conjugating enzyme Ubc 12 | Transferase, involved in cell proliferation | Non-classical | 6,31,35,36 | ||||||
| Obg-like ATPase 1 | ATP hydrolysis | ↑ | ↓ | 25, 26, 27, 28,34,36 | |||||
| Peptidyl-prolyl cis–trans isomerase A, B, C, D, H, like 1, CWC27 homolog, FKBP (1, 3, 4, 9) | accelerate the folding of proteins | Classical | A, B, C↑ | FKBP9 ↓ | 6,11,36, 37, 38, 39,26, 27, 28,31, 32, 33, 34, 35 | ||||
| Peroxidasin homolog | Peroxidase activity and role in ECM formation | Classical | ↓ | 6,25, 26, 27,35,36 | |||||
| Peroxiredoxin 1, 2, 4, 5, 6 | Protecting cells from free-radical damage | Classical | 1, 2 ↓ | 1, 5, 6↑ | 4 ↑ and ↓ 6↑ | 6,11,38,39,27,29,31, 32, 33, 34, 35, 36 | |||
| Phosphoglycerate mutase 1, 2 | Hydrolase, isomerase | 2↑ | 2↑ | 6,11,39,28, 29, 30,32,34, 35, 36,38 | |||||
| Phospholipase A1 member A, A2 activating protein, D3 | Hydrolase, Protease, Serine protease | Classical | A1A↓ | 26,27,33,35,36 | |||||
| Procollagen C-endopeptidase enhancer 1 | collagenase enhancer | Classical | ↓ | 6,27,31,32,35,38, 39, 40 | |||||
| Prolyl endopeptidase | Role in tissue remodeling, fibrosis, inflammation | 26,29,33,35,36 | |||||||
| Proteasome subunit β (type 1, 2, 3, 4, 5, 6, 8), α (1, 2, 3, 4, 5, 6, 7) | Hydrolase, protease, threonine protease | Non-classical | β type-6 ↑ | β type (4, 5) ↓ | β6, β8↑ | 6,11,27,31,32,34, 35, 36,38 | |||
| Protein disulfide isomerase | Classical | ↑ | ↓ | 11,35,36,38,39 | |||||
| Protein disulfide-isomerase A (3, 4, 5, 6) | chaperone, isomerase, | Classical | A3, A6↑ | A3↑ | A3↑ | A6↓, A3, A4↑ |
6,11,37, 38, 39,25,27,28,31,32,34, 35, 36 | ||
| Protein/nucleic acid deglycase DJ-1 | Chaperone, hydrolase, protease, RNA binding | Non-classical | ↑ | ↓ | 6,11,31,32,34, 35, 36,38,39 | ||||
| Puromycin-sensitive aminopeptidase | involved in cell growth and viability | ↑ | 6,34,36,38,39 | ||||||
| Pyridoxal phosphate phosphatase | involved in mitosis and cytokinesis | 27,35,36 | |||||||
| Pyruvate kinase (isozymes R/L, M1/M2, PKM) | Plays a key role in glycolysis, marker of myonecrotic conditions | PKM↑ | M1/M2↑ | 11,28,29,33, 34, 35, 36,38,39 | |||||
| Secretory leucocyte protease inhibitor | 36 | ||||||||
| Serine protease HTRA1, 2, 23 | growth factor binding, hydrolase, protease, serine protease | Classical | 2↑ 1↓ |
6,26, 27, 28,35,36 | |||||
| Spermidine synthase | Transferase | Non-classical | ↑ | 31,32,34, 35, 36 | |||||
| Sulfhydryl oxidase 1, 2 | Classical | 6,26, 27, 28,35,36 | |||||||
| Superoxide dismutase [Cu–Zn] | Antioxidant, Oxidoreductase | Non-classical | ↑ | ↓ | ↑ | 6,11,27,28,31,32,34,35,38,39 | |||
| Tissue-type plasminogen activator | Tissue remodeling and degradation | Classical | ↑ | 26,27,35,36 | |||||
| Transketolase | Transferase, calcium and magnesium binding | 6,28,29,34, 35, 36,38,39 | |||||||
| Triosephosphate isomerase | Isomerase involved in gluconeogenesis and glycolysis | ↑ | 6,11,36,38,39,26,28, 29, 30,32, 33, 34, 35 | ||||||
| Ubiquitin carboxyl-terminal hydrolase 17-like protein D | Protease, hydrolase, thiol protease | ↓ | 25 | ||||||
| Ubiquitin-conjugating enzyme E1-like 2 (isoform CRA_a), E2 (D1, D2, L3, K, N, Z, variant 1 et 2) | Transferase | Non-classical | E1-like 2 ↓ | E2 variant 1, K, N↑ | 26,27,31,32,34, 35, 36 | ||||
| α, β, γ enolase | lyase | α, γ ↑ | α, β, γ↑ | α↑ | α ↑ | 6,11,38,39,26,28,29,32, 33, 34, 35, 36 | |||
|
α, β-mannosidase |
Classical |
β↓ |
6,26,36 |
||||||
|
Enzymatic inhibitors |
Specificity |
Secretion |
Exercise |
Myogenesis |
Dystrophin deficiency |
Atrophy |
Insulin stimulation |
Insulin resistant cells |
References |
| Amyloid-β A4 protein | Apoptosis, cell adhesion, endocytosis, Notch signaling pathways | Classical | ↓ | 26 | |||||
| Cystatin B, C | cysteine protease inhibitor | Classical | C↑ | C↓ | 6,26,39,40,27,28,31,32,35, 36, 37, 38 | ||||
| Metalloproteinase inhibitor (TIMP) 1, 2 | Cytokine, growth factor, metalloendopeptidase inhibitor | Classical | 1, 2↑ | 1↓ | 1↓ | 2↓ | 2↓ | 6,25,35, 36, 37, 38, 39,26, 27, 28, 29, 30, 31, 32,34 | |
| Phosphatidylethanolamine-binding protein 1 | Protease inhibitor, serine protease inhibitor | Non-classical | ↑ | ↑ and ↓ | ↑ | ↑ | 6,11,28,29,35, 36, 37, 38, 39 | ||
| Serpin A6, A8 (angiotensinogen), A11, A3N | Regulator of blood pressure, body fluid and electrolytes homeostasis | Classical | A3N↑ | A6↓ | 25,28,36 | ||||
| Serpin B1 (Leukocyte elastase inhibitor), B2 (Plasminogen activator inhibitor 2), B6, B9 | Inhibitor of serine protease, protease inhibitor | Non-classical | B2↓ | B1, B6↑ | 6,27, 28, 29,32, 33, 34, 35, 36,39 | ||||
| Serpin C1 (Antithrombin-III) | Inhibitor of serine protease, regulation of cell migration | Classical | ↑ | ↓ | 6,27,29,31,32,35 | ||||
| Serpin E1 (Plasminogen activator inhibitor 1), E2 | Inhibitor of serine protease, regulation of cell migration | Classical | E1, E2↑ | E1↑ | E1↑ E2↓ |
6,26, 27, 28, 29,35,36,40 | |||
| Serpin F1 (pigment epithelium derived factor (PEDF)) | Inhibitor of serine protease, regulation of cell migration | Classical | ↓ | 6,26, 27, 28, 29,35, 36, 37,40 | |||||
| Serpin G1 (Plasma protease C1 inhibitor) | Inhibitor of serine protease, regulation of cell migration | ↓ | 6,26,27,30,34 | ||||||
| Serpin H1 | Inhibitor of serine protease, regulation of cell migration | Classical | ↑ | ↑ and ↓ | 6,28,31,32,35,36,38,39 | ||||
| Tissue factor pathway inhibitor | Anti-thrombic action and associates lipoproteins in plasma | Classical | ↑ | ↓ | 27,35,36 | ||||
|
α(1,2)-macroglobulin |
Protease inhibitor, serine protease inhibitor |
Classical |
1, 2↑ |
↓ |
6,27, 28, 29,34,35 |
||||
|
Cytoskeletal proteins |
Specificity |
Secretion |
Exercise |
Myogenesis |
Dystrophin deficiency |
Atrophy |
Insulin stimulation |
Insulin resistant cells |
References |
| Actin, aortic smooth muscle | 6,11,28,29,35,36 | ||||||||
| Actin, cytoplasmic 1, 2 | 6,28,29,32,36,39 | ||||||||
| Actin, α cardiac muscle 1 | ↑ | 6,11,28,29 | |||||||
| Actin, α skeletal muscle | ↑ | ↑ | 6,11,28,29,34, 35, 36,39 | ||||||
| Actin-related protein 2, 3 | 2↑ | 6,28,29,34, 35, 36 | |||||||
| Calponin 2, 3 | regulation and modulation of muscle contraction | non-classical | 3↑ | 28,31,32,34, 35, 36 | |||||
| Cell division control protein 42 homolog | Differentiation, neurogenesis | non-classical | 31,32,35,36 | ||||||
| Cofilin 1, 2 | Regulates actin cytoskeleton dynamics | non-classical | 1 ↓ | 2↑ | 1 ↑ | 6,11,38,39,26,28,31, 32, 33, 34, 35, 36 | |||
| Desmin | Muscle-specific type III intermediate filament | non-classical | ↑ | ↑ | 6,11,36,26,28, 29, 30, 31, 32,34,35 | ||||
| Destrin | Actin-depolymerizing protein | non-classical | 6,31,32,35,36 | ||||||
| Ezrin | Cell shape | 6,28,29,33,35,36 | |||||||
| F-actin-capping protein subunit α (1, 2), β | actin capping, actin binding | 6,29,35,36 | |||||||
| Filamin A, B, C | Actin binding | C↑ | 6,11,28,29,33, 34, 35, 36,38,39 | ||||||
| Gelsolin | actin capping, actin binding | Classical | ↑ | 6,24,38,26, 27, 28, 29,33, 34, 35, 36 | |||||
| Myomesin 1, 3 | Major component of the vertebrate myofibrillar M band | 11,36 | |||||||
| Myosin 1, 3, 4, 6, 7, 8, 9, 10 | Role in cytokinesis, cell shape, secretion and capping | 3↑ | 6,11,39,28, 29, 30,33, 34, 35, 36,38 | ||||||
| Myosin binding protein C, fast type | cell adhesion | 11,30 | |||||||
| Myosin light chain 1, 2, 3, 4, 6B, 12B, 1/3 | Muscle contraction, motor activity, calcium binding | 1, 4, 6B, 12B↑ | 6,13,28,29,31,35, 36, 37,40,29 | ||||||
| Myosin regulatory light chain 2, skeletal muscle isoform, 4, 12B | Contractile protein | 2↑ | 6,11,28,34, 35, 36,38,39 | ||||||
| Myotilin | Involved in a complex of actin cross-linking proteins | 11 | |||||||
| Palladin | Role in cell morphology, motility, cell adhesion | 28,29,35,36 | |||||||
| Peripherin | Neuronal intermediate filament protein | Non-classical | 6,31,32 | ||||||
| Plectin | actin binding | ↑ | 6,11,28,33,35,36 | ||||||
| Profilin 1, 2 | Actin binding | Non-classical | 1 ↓ | 1↑ | 1↑ | 6,11,28,31,32,34,36,38,39 | |||
| Titin | Key component in the functioning of striated muscle | ↑ | 11,29,30,33, 34, 35, 36 | ||||||
| Transgelin 1, 2 | Non-classical | 2 ↓ | 1, 2↑ | 6,28,31, 32, 33, 34, 35,38,39 | |||||
| Tropomyosin α (1, 3, 4), β (1, 2) | actin-binding | Non-classical | α1, β ↑ | β ↓ | β↑ | β 2 ↑ | 6,11,30, 31, 32,34,35,38,39 | ||
| Troponin I (slow skeletal muscle), C (slow, skeletal and cardiac muscle), T | inhibitory subunit of troponin, muscle protein, actin binding | I, C, T↑ | 6,11,34, 35, 36,39 | ||||||
| Tubulin α (A,4A, B), β (2A, 3, 4B, 5, 6) | Non-classical | α4A↑ | α1A↑ | 6,26,32, 33, 34, 35, 36,39 | |||||
| Vimentin | cytoskeletal protein | Non-classical | ↑ | ↑ | ↑ | 6,11,36,38, 39, 40,28, 29, 30, 31, 32, 33, 34, 35 | |||
| Vinculin | Actin binding, cell adhesion | ↑ | ↑ | 6,28, 29, 30,34, 35, 36,38,39 | |||||
|
α-actinin 1, 2, 3, 4 |
actin binding |
3, 4↑ |
6,24,28,29,34, 35, 36,38,39 |
||||||
|
Miscellaneous |
Specificity |
Secretion |
Exercise |
Myogenesis |
Dystrophin deficiency |
Atrophy |
Insulin stimulation |
Insulin resistant cells |
References |
| 14-3-3 protein β/α, γ, ζ/δ, ε, η, θ | Non-classical | ε↑ | 6,28,29,32, 33, 34, 35, 36,38,39 | ||||||
| ADAMTS-like protein 1, 2, 3, 4 | Hydrolase | Classical | 3↑ | 2↓ | 27,28,35,36 | ||||
| Adapter molecule crk | cell adhesion, spreading, migration | 6,26,28,35,36 | |||||||
| Adiponectin | hormone involved in fat metabolism and insulin sensitivity | 26 | |||||||
| Agrin | Developmental protein | Non-classical | ↓ | 27,28,35,36 | |||||
| Alpha-1-acid glycoprotein 1 | Transport | Classical | ↑ | 25 | |||||
| Angiopoietin-related protein 1, 2, 4 | Induces sprouting in endothelial cells | Classical | 2↑ | 2↓ 4↑ |
26,28,31,32,35,36 | ||||
| Annexin A (1, 2, 4, 5, 6) | involved in exocytosis, regulation of cell migration and fusion | 1, 5, Non-classical 2 Classical |
1, 2 ↓ | A1, A2↓ | 6,26,38, 39, 40,27,28,31, 32, 33, 34, 35, 36 | ||||
| AP-1 complex subunit β1, γ1, σ (1, 2), μ1 | Transport, protein transport | Non-classical | 28,31,35,36 | ||||||
| Apolipoprotein B-100, D, E | Lipid metabolism and transport | Classical | B-100↑ | E↑ | 6,27,28,35,36 | ||||
| Basal cell adhesion molecule | Blood group antigen, receptor | 6,26,29 | |||||||
| Brain acid soluble protein | ↑ | 28,33,38,39 | |||||||
| Cadherin 2, 5, 11, 13, 15 (M-cadherin), 23 | cell adhesion | Classical | 23 ↑ | 11, 13, 15↑ | 2, 15↑ | 15↑ | 2, 11, 13, 15↓ | 6,26,39,27, 28, 29,33, 34, 35, 36,38 | |
| Calcium/calmodulin-dependent protein kinase type 1 | Cell cycle, differentiation, neurogenesis | 27,35,36 | |||||||
| Calmodulin | mediates a large number of enzymes | ↑ | 6,11,28,33,34,36,39 | ||||||
| Calpastatin | inhibition of calpain, may be involved in protein degradation in muscle | 28,35,36,39 | |||||||
| Calreticulin | calcium binding chaperone | Classical | ↓ | 6,11,27,28,34, 35, 36, 37, 38 | |||||
| Calsequestrin 1, 2 | calcium storage | Classical | 2↑ | 6,26,27,33,36,38 | |||||
| Calsyntenin 1, 2 | Classical | 1↓ | 6,26, 27, 28,35,36,38 | ||||||
| Calumenin | Metal-binding, calcium binding | Classical | 6,27, 28, 29,34, 35, 36,38,39 | ||||||
| Cartilage intermediate layer protein 1, 2 | Classical | 1, 2↑ | ↓ | 26,27,36 | |||||
| Catenin α1 | Association with cadherin | ↑ | 25,28,35,36 | ||||||
| Cellular nucleic acid-binding protein | Single-stranded DNA-binding protein | ↓ | 31 | ||||||
| Chloride intracellular channel protein 1, 4 | transport, chloride transport | Non-classical | 4 <-> | 4↑ | 6,26,31, 32, 33, 34, 35, 36 | ||||
| Chordin | Developmental protein | Classical | ↓ | 27,35,36 | |||||
| Clusterin | apoptosis, immunity | Classical | ↑ | ↑ and ↓ | 38,39 | ||||
| Coatomer subunit α, β, δ, ε | Transport | Classical | 6,28,30,32,34, 35, 36 | ||||||
| Coiled-coil domain-containing protein 80, 126 | Promotes cell adhesion and matrix assembly | Classical | 80↑ | 6,27,28,36,38,39 | |||||
| Complement component 1, C3, C4 (A, B) | Immune response | C3,C4B↑ | 6,28,30,35,39 | ||||||
| Complement factor B, D, H | Immune response | Classical | H ↑ | ↑ | H↓ | 6,25, 26, 27, 28,30,35,36 | |||
| Cytochrome C | 6,11,27,28,35,36 | ||||||||
| Drebrin | Actin binding, differentiation, neurogenesis | 6,28,35,36,39 | |||||||
| Elongation factor 1 (α1, β, δ, γ) 2 | Factor elongation | 2↑ | 1α1, 1γ, 1,β, 2↑ | 6,11,36,38,39,26, 27, 28, 29,32, 33, 34, 35 | |||||
| Endoplasmin | Chaperone | Classical | ↑ | 6,27,28,31,34, 35, 36,38 | |||||
| Ephrin type-A receptor 1, 2, 4, 7 | cell adhesion, neurogenesis | Classical | 2↓ | 6,27,28,33,36 | |||||
| Exostosin 1, 2 | protein glycosylation | 26, 27, 28,35,36 | |||||||
| Fascin | actin binding | ↑ | 6,32,34, 35, 36,38 | ||||||
| Fatty acid-binding protein, heart | Transport of lipids | ↑ | 11,28,35,36 | ||||||
| Follistatin | activin antagonist, inhibitor of FSH biosynthesis | Classical | ↑ | ↑ | ↓ | 25,26,35,36 | |||
| Four and a half LIM domains protein 1, 2, 3 | Metal binding | Non-classical | 6,11,31,32,35,36,38 | ||||||
| Galectin-3-binding protein | Cell adhesion | Classical | ↓ | 25, 26, 27,35,36 | |||||
| Glucosidase 2 subunit β | Metal binding, calcium binding | Classical | ↑ | 6,28,29,34,36 | |||||
| Growth factor receptor-bound protein 2 | link between growth factor and RAS signaling pathway | 6,26,35,36 | |||||||
| Heat shock 70 kDa protein 1A/1B, 1 like,2, 4 | ATP binding, nucleotide binding | 2↑ | 6,11,26,29,34, 35, 36,38,39 | ||||||
| Heat shock protein beta(1, 2, 3, 6, 7) | Chaperone | 2↑ | 1 ↑ and ↓ | 6,11,28,29,33, 34, 35, 36,38,39 | |||||
| Heat shock protein HSP 90 (α, β) | Chaperone | α↑ | α↑ | 6,36, 37, 38 | |||||
| Heme-binding protein 1, 2 | 6,28,29,35,36 | ||||||||
| High mobility group protein HMGI-C | Cell cycle, cell division, growth regulation | 28,35,36 | |||||||
| Hypoxia up-regulated protein 1 | role in cytoprotection triggered by oxygen deprivation | Classical | 27,28,36 | ||||||
| Importin 5, 7, 9 | transport nucléaire | 6,33,35,36 | |||||||
| Integrin α (1, 3, 5, 6, 7), β (1, 5) | cell adhesion, integrin, receptor, cell adhesion | Classical | α3, 5, 7↓ β1, 5 ↓ |
27,28,30,33,35,36 | |||||
| Lactadherin | angiogenesis, cell adhesion | Classical | 27,28,35,36,38 | ||||||
| Leukemia inhibitory factor receptor | Signal transducing molecule | Classical | ↓ | 27,36 | |||||
| l-lactate dehydrogenase A chain, B chain | Pyruvate fermentation to lactate | Non-classical | A ↓ | A↑ | A↑ | 6,11,38,39,24,26,28,31,32,34, 35, 36 | |||
| Lipopolysaccharide-binding protein | Innate immune response | Classical | ↑ | 25,36 | |||||
| Low-density lipoprotein receptor | Binds and transport LDL | Classical | 26,36,38 | ||||||
| Lysosome-associated membrane protein 1, 2 | 1↑ 2↓ |
26,36 | |||||||
| Lysyl oxidase homolog 1, 2, 3, 4 | Active on elastin and collagen substrates | Classical | 1↑ | ↑ | 1, 3, 4↓ | 25, 26, 27, 28,35,36 | |||
| Macrophage-capping protein | actin capping and binding | Classical | 6,27,28,33,34,36 | ||||||
| Meteorin-like protein | Hormone induced by exercising and promotes energy expenditure | Classical | 27,35,36 | ||||||
| Microtubule-associated proteins 1A/1B light chain 3B, 1S, 4 | Mitophagy | Non-classical | 26,31,32,35,36,39 | ||||||
| Musculoskeletal embryonic nuclear protein 1 | May be involved in the development of the musculoskeletal system | ↓ | 31 | ||||||
| Myc box-dependent-interacting protein 1 | Differentiation, endocytosis | ↑ | 6,27,28,33, 34, 35, 36,38 | ||||||
| Myoferlin | involved in muscle contraction | ↑ | 33, 34, 35, 36 | ||||||
| Myoglobin | oxygen transport | Non-classical | 11,26,30,34, 35, 36 | ||||||
| Myotrophin | regulation of the growth of actin filaments | 6,28,35,36,38 | |||||||
| NAC α domain containing | Protein transport | Classical | ↑ | 32,40 | |||||
| Nascent polypeptide-associated complex subunit alpha, muscle-specific form | Muscle -specific transcription factor | ↑ | 34, 35, 36 | ||||||
| Neogenin | cell adhesion in myogenesis | Classical | ↓ | 27,28,36 | |||||
| Nephronectin | Cell adhesion, spreading and survival | Classical | ↓ | 27,33,35,36 | |||||
| Neudesin | Promotes cell proliferation and neurogenesis | Classical | 27,35,36 | ||||||
| Neural cell adhesion molecule 1 | Cell adhesion | Classical | ↑ | ↓ | 6,26,39,27, 28, 29,33, 34, 35, 36,38 | ||||
| Neuropilin 1, 2 | Angiogenesis, differentiation, neurogenesis | Classical | 1, 2↓ | 26, 27, 28,33,36 | |||||
| Nuclease-sensitive element-binding protein 1 | mitogen, activator, repressor | Classical | 28,35, 36, 37 | ||||||
| Nucleobindin 1, 2 | Calcium binding, may have a role in calcium homeostasis | Classical | 1↑ | 1, 2↓ | 1 ↓, 2 ↑ | 24,26, 27, 28,34, 35, 36,38,39 | |||
| Nucleophosmin | cell proliferation, regulation of tumor suppresors | ↑ | 11,32,35,36 | ||||||
| Olfactomedin-like protein 2A, 2B, 3 | extracellular matrix organization | Classical | 2A↑ | 2B, 3↓ | 26, 27, 28,35,36 | ||||
| Parvalbumin alpha | involved in relaxation after contraction | 11,27,35 | |||||||
| Phospholipid transfer protein | Lipid transport | Classical | ↓ | 6,26, 27, 28,33,36 | |||||
| Plastin 3 | Actin binding | 26,28,33,35,36,39 | |||||||
| Plexin A (1, 2, 3, 4), B2 | Classical | B2↓ | 27,33,35,36 | ||||||
| Poly(rC)-binding protein 1, 2, 3 | DNA binding, RNA binding | Non-classical | 6,31,32,35,36 | ||||||
| Programmed cell death 5, 6-interacting protein | apoptosis, cell cyle, cell division, transport | 5↓ | 26, 27, 28,33,35,36,39 | ||||||
| Proliferating cell nuclear antigen | DNA binding | Non-classical | 26,28,31,32,35,36 | ||||||
| Prostaglandin F2 receptor negative regulator | Regulation of myoblast fusion | ↓ | 25,27,28,33,36,38 | ||||||
| Protein jagged-1 | Inhibits myoblast differenciation | Classical | 28,36 | ||||||
| Protein NOV homolog | Role in proliferation, adhesion, migration, differentiation, survival | Classical | ↑ | 26,36 | |||||
| Protein RCC2 | mitosis and cytokinesis | 35,36 | |||||||
| Protein VAC14 homolog | phosphatidylinositol biosynthetic process and signal transduction | ↑ | 31 | ||||||
| Protocadherin-1, gamma C3, 18, 19, beta 14, Fat 4 | cell to cell adhesion and interaction processes | Classical | 19, Fat 4↓ | 27,35,36 | |||||
| Ras suppressor protein 1 | regulation of cell substrate adhesion | Non-classical | ↑ | 34, 35, 36 | |||||
| Ras-related protein Rab 5A, 14, 35 | Protein transport, transport | Non-classical | 29,33,35,36 | ||||||
| Reticulocalbin 1, 2, 3 | Calcium, metal binding | Classical | ↓ | 6,26,28,34, 35, 36,39 | |||||
| Reticulon 2, 4 | Neurogenesis | 4↑ | 33, 34, 35, 36 | ||||||
| Rho GDP dissociation inhibitor (GDI) α | Controls rho proteins homeostasis | Non-classical | ↓ | ↑ | 28,35 | ||||
| Semaphorin 3(A,B, C, D, E), 4(B, C), 5A, 6(A, B), 7A | Role in differenciation, inflammation and neurogenesis | Classical | 3A, 3D, 3E, 6A (isoform 1)↑ 7A↑ and↓ |
3 (A, B), 4B, 4C, 6A↓ | 26, 27, 28, 29,35, 36, 37 | ||||
| Septin 2, 7, 9, 11 | Cell cycle, cell division, differentiation, mitosis | 11↑ | 26,28,34, 35, 36,38 | ||||||
| Serotransferrin | Iron transport | ↑ | 11 | ||||||
| SH3 domain-binding glutamic acid-rich protein | 6,30,31,35,36 | ||||||||
| Slit homolog 2 protein | Differentiation, neurogenesis | Classical | ↓ | 27,36 | |||||
| Spondin 2 | cell adhesion, immunity | Classical | ↓ | 6,26,27,31,32,35 | |||||
| Stanniocalcin 1, 2 | Stimulates renal phosphate reabsorption | Classical | 6,28,31,32,35,36,38 | ||||||
| Stathmin 1, 2 | Regulation of the microtubule filament | 1 ↑ | 6,30, 31, 32,35,36,39 | ||||||
| Sterile α motif domain-containing protein 3 | ↓ | 32 | |||||||
| Sushi repeat-containing protein SRPX2 | angiogenesis, cell adhesion | ↓ | 6,26, 27, 28,36 | ||||||
| Syntenin-1 | Tumorigenesis, exosome biosynthesis | 6,27,28,32,35,36 | |||||||
| Talin 1, 2 | connections in the cytoskelettal structure | 28,33,35,36,38 | |||||||
| Telethonin | Muscle assembly regulating factor | 11 | |||||||
| Testican 1, 2 | Neurogenesis | Classical | 2↑ | 6,25, 26, 27, 28,36 | |||||
| Tetraspanin 4, 6, 7, 9, 14 | may be involved in cell proliferation and motility | Non-classical | 33,35,36 | ||||||
| Thrombospondin 1, 2 | mediates cell to cell and cell to matrix interactions | Classical | ↑ | 1↑ | 6,26, 27, 28,31,32,35, 36, 37 | ||||
| Transcobalamin 2 | Vitamin B12 binding, transport protein | Classical | ↓ | 26,27,35,36,39 | |||||
| Transcription elongation factor B polypeptide 2 (Elongin-B) | Transcription factor | ↑ | 25, 26, 27, 28,35,36 | ||||||
| Translationally-controlled tumor protein | involved in microtubule stabilization and calcium binding | Non-classical | ↑ | ↑ | 6,27,28,31,32,34, 35, 36, 37, 38 | ||||
| Ubiquilin 1, 2, 3 | Regulation of protein degradation | 28,35,36 | |||||||
| Vacuolar protein sorting-associated protein 26A, 29, 35, 37C VTA1 homolog | protein transporter | ↑ | 26A, 35↑ | 6,26, 27, 28,33, 34, 35, 36 | |||||
| Vascular cell adhesion protein 1 | Cell–cell recognition | Classical | Isoform 1↑ | 27,28,36 | |||||
| Vasorin | Classical | 6,27,28,36 | |||||||
| Vigilin | Lipid metabolism | Non-classical | ↑ | 28,35,36 | |||||
| Vitamin K-dependent protein S | Blood coagulation, fibrinolysis, hemostasis | Classical | ↓ | 6,26,27,36 | |||||
| WNT1-inducible-signaling pathway protein 1 | Associated with cell survival | Classical | 27,35,36 | ||||||
| α-crystallin A chain (HSP4), B chain | chaperone, eye-lens protection | B↑ | 6,11,26,30,33, 34, 35, 36,39 | ||||||
| β-2-microglobulin | component of the class I MHC | Classical | ↑ | ↓ | 26,38,39 | ||||
Table 1.
Characteristics of the reviewed studies.
| Article title | Author | Year | Cell type | Culture | Technique | Objectives |
|---|---|---|---|---|---|---|
| Conditioned media from AICAR-treated skeletal muscle cells increases neuronal differentiation of adult neural progenitor cells | Youl et al.24 | 2019 | L6 (rat) | Culture | Tandem mass tag | Comparative analysis between AICAR-treated (AMPK agonist) vs. untreated skeletal muscle cells |
| Increased Serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. | Gueugneau et al.25 | 2018 | C2C12 (mouse) | Culture | MS and label-free quantification | Comparative analysis between glucocorticoid-induced muscle atrophy and healthy muscle cells |
| Mining the secretome of C2C12 Muscle cells: Data dependent experimental approach to analyze protein secretion using label-free quantification and peptide based analysis. | Grube et al.26 | 2018 | C2C12 (mouse) | Culture | MS and label-free quantification | |
| Dynamics of the Skeletal Muscle Secretome during myoblast differentiation. | Henningsen et al.27 | 2010 | C2C12 (mouse) | Culture | SILAC | |
| Secretome profiling of primary human skeletal muscle cells. | Hartwig et al.6 | 2014 | Human primary muscle cells | Culture | MS and label-free quantification | |
| In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. | Lebihan et al.28 | 2012 | Human primary muscle cells | Culture | MS and label-free quantification | Comparative analysis between nanovesicles vs microvesicles content. |
| Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. | Norheim et al.29 | 2011 | Human primary muscle cells | Culture | MS and label-free quantification | Comparative analysis between strength training patient biopsies vs. untrained individuals. |
| Increased Secretion and Expression of Myostatin in skeletal muscle from extremely obese women. | Hittel et al.30 | 2009 | Human primary muscle cells | Culture | SILAC | Comparative analysis between extremely obese vs. Healthy nonobese women. |
| Comparative proteomic analysis of the insulin-induced L6 myotube secretome. | Yoon et al.31 | 2009 | L6 (rat) | Culture | MS and label-free quantification | Comparative analysis of insulin treated vs non treated myotubes. |
| Identification of Differentially Regulated Secretome components during skeletal myogenesis. | Chan et al.32 | 2011 | C2C12 (mouse) | Culture | SILAC | |
| Proteomic Analysis of C2C12 Myoblast and Myotube exosome-like vesicles: A new paradigm for myoblast-myotube cross talk. | Forterre et al.33 | 2014 | C2C12 (mouse) | Culture | MS and label-free quantification | Comparative analysis of the proteins present in myotubes vesicles vs. Myoblasts vesicles. |
| Muscle tissue as an endocrine organ: Comparative secretome profiling of slow-oxidative and fast-glycolytic rat muscle explants and its variation with exercise. | Roca-Rivada et al.11 | 2012 | Rat muscles | Explants | MS and label-free quantification | |
| Dystrophin deficiency leads to disturbance of LAMP-1-vesicle-associated protein secretion. | Duguez et al.34 | 2013 | H–2K (mouse) | Culture | SILAC | Comparative analysis between wild-type vs. Dystrophin-deficient myotubes. |
| Quantitative analysis of the secretion of the MCP family of chemokines by muscle cells. | Henningsen et al.35 | 2011 | C2C12 (mouse) | Culture | SILAC | |
| Secretome Analysis of Lipid-Induced Insulin Resistance in skeletal muscle cells by a combined experimental and bioinformatics workflow. | Deshmukh et al.36 | 2015 | C2C12 (mouse) | Culture | MS and label-free quantification | Comparative analysis of insulin-resistant vs. healthy skeletal muscle cells. |
| Proteomic analysis of secreted proteins from skeletal muscle cells during differentiation. | Ojima et al.37 | 2014 | Mouse muscles biopsies | Culture | iTRAQ | |
| Proteomic Analysis of the Palmitate-induced myotube secretome reveals involvement of the annexin A1-formyl peptide receptor 2 (FPR2) pathway in insulin resistance. | Yoon et al.38 | 2015 | L6 (rat) | Culture | MS and label-free quantification | Comparative analysis of insulin-resistant vs. healthy skeletal muscle cells. |
| Proteomic Analysis of Tumor Necrosis Factor-Alpha (TNF-α)-induced L6 myotube secretome reveals novel TNF-α-dependent myokines in diabetic skeletal muscle. | Yoon et al.39 | 2011 | L6 (rat) | Culture | MS and label-free quantification | Comparative analysis of (TNF-α)-induced insulin resistant vs. wild type skeletal muscle cells. |
| Identification of Secreted Proteins during Skeletal Muscle development. | Chan et al.40 | 2007 | C2C12 (mouse) | Culture | MS and label-free quantification |
3.1. Exercise
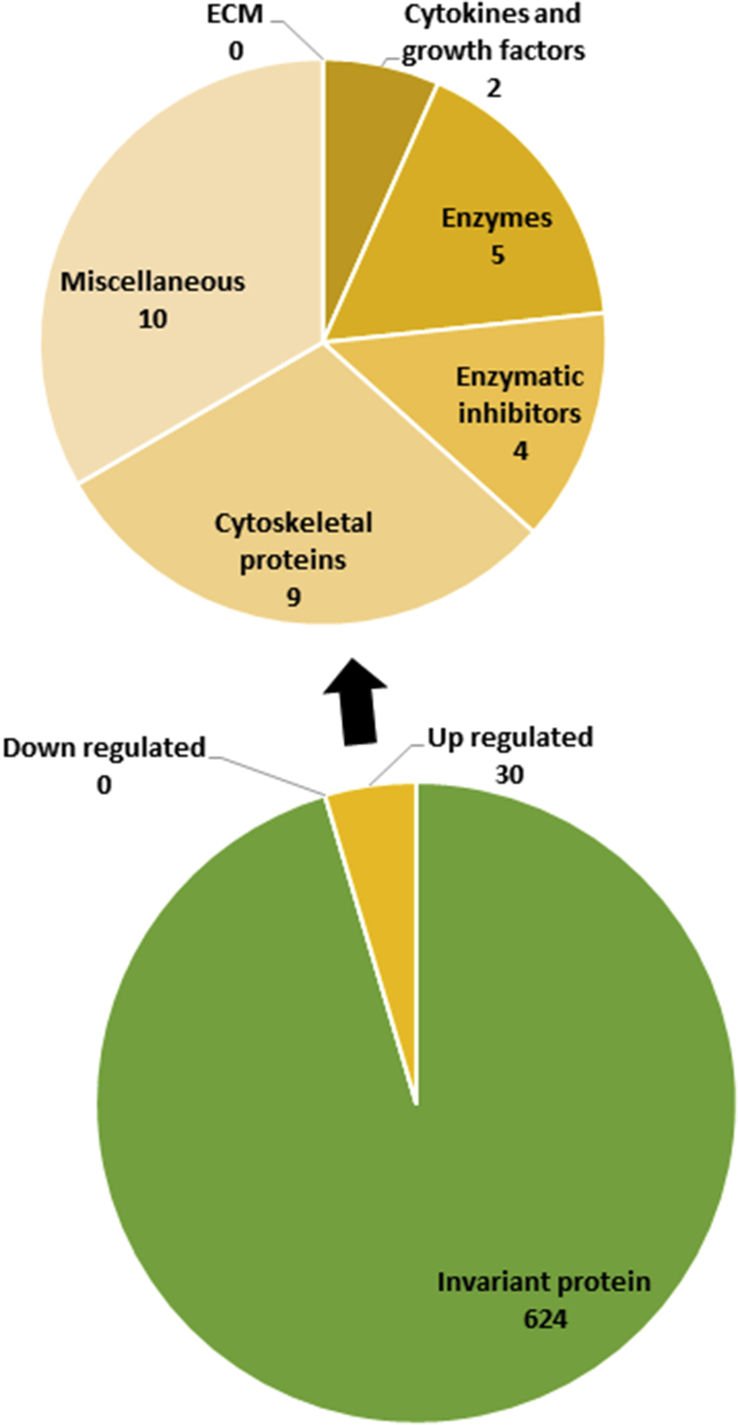
Analysis of secretome shows that no proteins are down-regulated and 30 are up-regulated by exercises (fig. 2). Galectin 1, CC motif chemokine 2, serpin C1, superoxide dismutase and cadherin 23 are oversecreted in exercise-mimicking AMP-kinase agonist 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR)-treated skeletal muscle cells. AICAR mimics some aspects of exercising. Indeed, AMPK activation blocks energy-consuming processes and promotes ATP synthesis from glucose uptake, glycosylation and fatty acid oxidation.
Fig.2.
The secretome analysis of skeletal muscle cells after exercising showed that no proteins were down-regulated and 30 proteins were up-regulated. Proteins were then classified and counted on the basis of their function.
Interestingly, a lot of cytoskeletal proteins are up-regulated during exercise (9 out 30 proteins). Exercising leads to a rise in actin, vimentin, vinculin and desmin secretion.29 The desmin is an intermediate filament protein of the cytoskeleton which stabilizes the sarcomere.41 This protein is used as a marker of myogenicity because it is prominent in activated myoblasts and developing or regenerating fibers.28
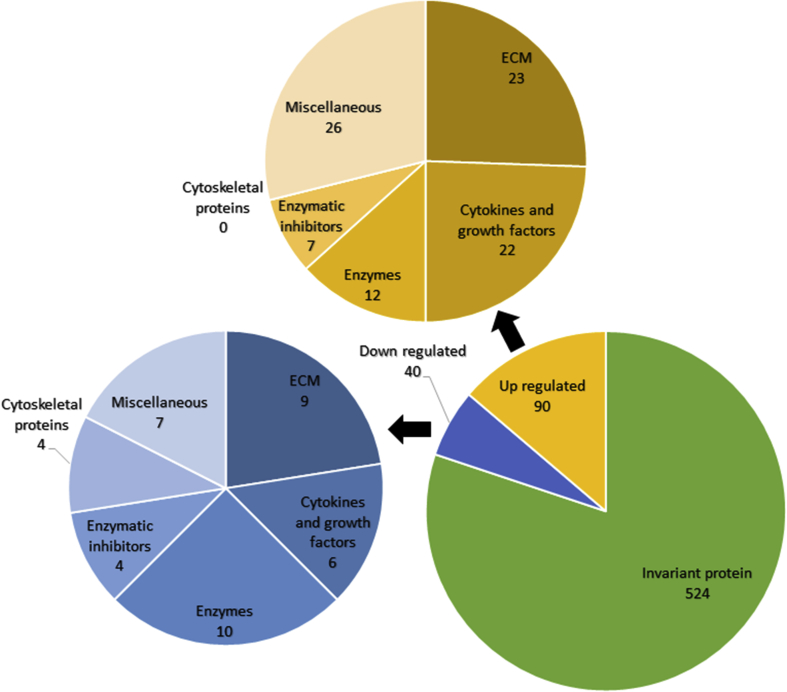
3.2. Myogenesis
The secretion of 130 proteins are regulated during myogenesis (90 up- and 40 downregulated) (fig. 3). Among those, decorin and biglycan, two major components of the extracellular matrix, are found to be elevated in the secretome of skeletal muscle cells. These two small proteoglycans reduce the bioavailability of TGF-β during myogenesis.27,42,43 Considering that TGF-β plays a role in promoting the synthesis of extracellular components, to reduce TGF-β availability leads to a decrease in extracellular matrix components synthesis. Mimecan (also called osteoglycin), a component of the extracellular matrix which is involved in collagen fibrillogenesis, was found to be decreased in the secretome of skeletal muscle cells during myogenesis.
Fig.3.
The secretome analysis of skeletal muscle cells during myogenesis showed that 40 proteins were down-regulated and 90 proteins were up-regulated. Proteins were then classified and counted on the basis of their function.
Regarding the collagens, most of them are upregulated during myogenesis (I (α1), II(α1), V (α1, α3) VI(α1), XI (α1)). Only two collagens are downregulated (III(α1), XVIII(α1)). This is the contrary for laminins, which are mostly downregulated during myogenesis (α2, α5, β2).
The Secreted Protein Acidic and Rich in Cysteine (SPARC) is a protein regulating cell growth through interactions with cytokines and the extracellular matrix. SPARC is highly expressed in injured muscles and in muscle cells undergoing development or regeneration.29,44 Studies contradict one another concerning the regulation of SPARC during myogenesis.
Beside these extracellular matrix components, numerous cytokines and growth factors are modulated during myogenesis. C–C motif chemokines 2, 7 and 8, the complement C1q or tumor necrosis factor-related protein 3 and 5 are upregulated. The only ones downregulated cytokines and growth factors during myogenesis are the follistatin related protein 1 and Insulin-like Growth Factor Binding Protein 2 (IGFBP2).
In addition to its role in ECM components synthesis, TGF-β is a complex regulator of skeletal muscle development. This protein is regulated in many different ways, for instance by sequestration via latent TGF-β-binding proteins (LTBPs).27 There is a pronounced increased in the secretion of TGF-β1, - β2, - β3 during myogenesis. In parallel, LTBP-3 is modestly enhanced on day 2 but markedly increase on day 5 of myogenesis.27 On the other hand, the secretion of Follistatin-like 1 (FSTL1) decreases during myogenesis.32 FSTL1 is secreted by skeletal muscle cells and is involved in muscle vascularization and metabolism.45,46,27,47 FSTL1 may be a positive regulator of myogenesis by counteracting TGF- β signaling pathway.32 This protein may be involved in the early phase of myogenesis (cell migration, cell-cycle exit, …) since it is more expressed by myoblasts than myotubes cultures.32
Insulin-like Growth Factors (IGFs) promote differentiation of skeletal muscle cells.27 They are found increased during myogenesis.27 The regulation of the IGFs is regulated by IGFBPs in the extracellular environment, inhibiting or enhancing the effect depending on cell type and context. The growth hormone (GH) stimulates the secretion of IGF-1 by muscles.48 IGF-1 plays essential roles in regeneration and hypertrophy of the muscles and is also an osteogenic factor.27,49,50 This protein may be involved in muscle-bone crosstalk.49 During myogenesis, the secretion of IGF-2 is markedly increased throughout differentiation. The highest level of IGF-1 is observed at day 2 of differentiation.27 The secretion of IGFBPs is also modulated during myogenesis. IGFBP2 secretion decreased during differentiation process whereas globally IGFBP4 levels increased.27 In contrast, IGFBP6 or -7 levels were not significantly modified.27 Finally, contradictory results have been published regarding IGFBP532,37.
Semaphorins are important modulators of neurogenesis,51 organogenesis, tumor progression, angiogenesis and immune responses.52 Further, they play an important role in skeletal muscle development.53,54 Semaphorin 3A, 3D, 3E and 6A are secreted by skeletal muscle satellite cells and are up-regulated at the early phase of muscle differentiation.27,55 Concerning the secretion of semaphorin 7A, contradictory results exist.27 According the study, this protein was found up- or downregulated during myogenesis.18,37
MMP2 is up-regulated during myogenesis, and the highest level is reached during early phase of the differentiation process.27 The metalloproteinase inhibitor 2 (TIMP2) inhibits MMPs40 and also decreased myogenin expression, leading to an inhibition of myogenesis.40
3.3. Dystrophin deficiency
A total of 114 proteins were regulated by dystrophin deficiency (107 up- and 7 downregulated). Approximately twice as much proteins were secreted by dystrophin deficient (mdx) vs wild-type (WT) skeletal muscle cells and most of the oversecreted proteins were cytosolic. In addition, fibronectin was found in higher amount in the conditioned media of mdx muscle cells.34
3.4. Muscle atrophy
Fifteen proteins were oversecreted during muscle atrophy and 11 were under-secreted. Regarding the variation of extracellular matrix proteins, perlecan, fibrillin 1 and biglycan were downregulated on the contrary to periostin and collagen IV(α2). About the cytokines and growth factors, only macrophage colony-stimulating factor 1 was downregulated.25
The alcohol dehydrogenase [NADP (+)], chymotrypsinogen B, and protein disulfide isomerase A3 are 3 enzymes which were found to be more secreted by atrophied skeletal muscle cells. Delta-aminolevulinic acid dehydratase, Obg-like ATPase 1, peroxidasin homolog were found to be less secreted.25
TIMP2 was less secreted and serpin A3n was more secreted by atrophied skeletal muscle cells. This serpin is localized around the myofiber and is an extracellular inhibitor of proteases such as granzyme B, trypsin, chymotrypsin, cathepsins G/B/L and leucocyte elastase. Follistatin is another protein which is more expressed during skeletal atrophy.25
3.5. Insulin stimulation
A total of 27 proteins were regulated by insulin stimulation with 14 up- and 13 downregulated proteins. Four extracellular matrix protein are found to be regulated by insulin in the secretome of skeletal muscle cells. The collagens V (α2) and III(α1) and calcyclin (formerly known as Protein S100-A6) are downregulated while collagen VI(α1) is upregulated.31
Some cytokines and growth factors were also regulated by insulin. FSTL1, IGF-2, IGFBP6 and bone morphogenetic protein 1 (BMP1) were downregulated by insulin stimulation.31 Finally, only one matrix metalloproteinase (MMP 2) was upregulated by insulin. On the contrary, TIMP-2 is downregulated by insulin31 while plasminogen activator inhibitor (PAI1, also known as serpin E1) and serpin H are increased.56,31
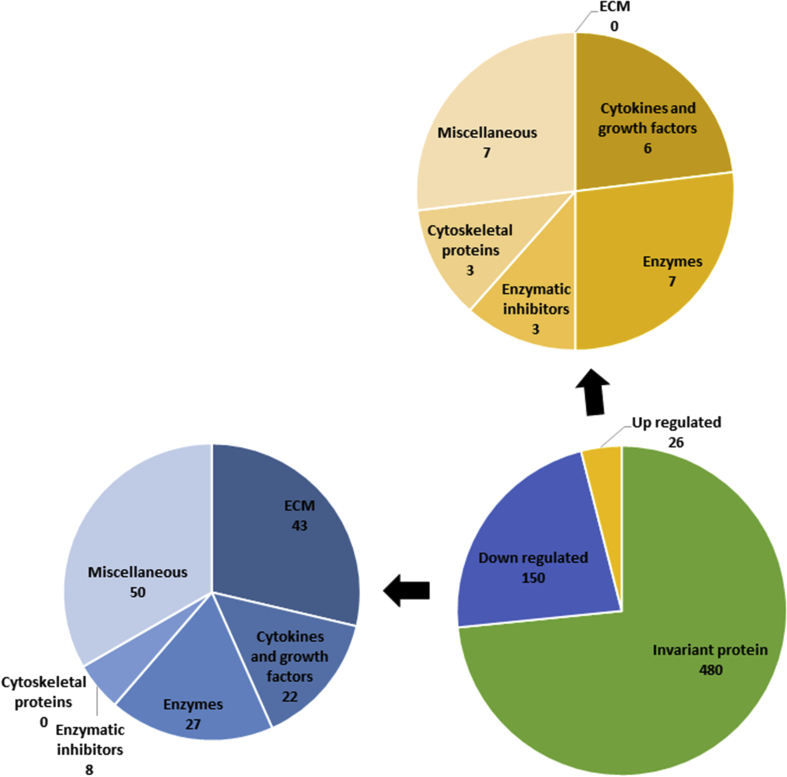
3.6. Insulin resistance
Twenty-six proteins were over-secreted and 150 down-secreted in the secretome of insulin-resistant muscle cells, for a total of 176 regulated proteins (fig. 4). Most of the ECM proteins were downregulated under insulin resistant conditions. Indeed, the expression of collagens I, III, IV, V, VI, XV and XVIII,41 but also laminin, perlecan, nidogen, fibronectin and periostin, by insulin resistant muscle cells was reduced compared to that of healthy muscle cells.36,38,39,41
Fig.4.
The secretome analysis of insulin resistant skeletal muscle cells showed that 150 proteins were down-regulated and 26 proteins were up-regulated. Proteins were then classified and counted on the basis of their function.
The cytokines and growth factors were also differently expressed in insulin-resistant muscle cells compared to healthy cells. For example, FSTL1 and C–C motif chemokine 9 were downregulated in insulin resistant muscle cells31 while monocyte chemotactic protein-1 (MCP-1) (also known as C–C motif chemokine 2) was upregulated. Interestingly, MCP-1 was involved in the recruitment of macrophages into the site of muscle damage. They also influence the migration of muscle cells during development.57 MCP-1 activates the extracellular signal-regulated kinase 1 and 2 (Erk ½) leading to myoblast proliferation in response to muscle injury.35,58,36
Concerning the Insulin-like growth factor, IGF-1 was 3 times less concentrated in the secretome of insulin resistant muscle cells.36 Furthermore, IGFBP2, IGFBP4, IGFBP5, IGFBP6 and IGFBP7 were all downregulated in insulin-resistant muscle cells and in patients with type 2 diabetes.36,59 Furthermore, low plasma levels of IGFBP7 was correlated with the incidence of type 2 diabetes in humans.59
The concentration of BMP1 in the secretome of insulin resistant skeletal muscle cells was also decreased compared to healthy skeletal muscle cells.36 Granulin was found in the secretome of skeletal muscle cells and has many roles including cell growth and anti-inflammatory effects. The secretion of this cytokine was decreased in insulin-resistant skeletal muscle cells, leading to an increase in the inflammatory response under insulin-resistant conditions.60,36
In the family of Semaphorins, the Semaphorins 3A, 3B, 4B, 4C and 6A were all less concentrated in the secretome of insulin resistant muscle cells compared to healthy muscle cells.36,38,39
The growth/differentiation factor 11 (GDF11) was significantly decreased in insulin-resistant muscle cells.36 Hepatocyte growth factor and platelet derived growth factor C are involved in muscle proliferation and differentiation and are also decreased in the secretome of insulin resistant muscle cells.61,62 Follistatin, an inhibitor of myostatin49 was found to be decreased in the secretome of insulin resistant skeletal muscle cells.
Concerning the enzymes, the secretion of MMP2, MMP9, MMP19 but also ADAMTS -1, −2, −5, −7 and −12 are downregulated in insulin resistant muscle cells. On the contrary, the secretion of TIMP-2 is up-regulated in insulin resistant compared to healthy muscle cells.38,39,36
Regarding the regulation of the enzymatic inhibitors, the secretion of Serpin A6, E2 and F1 are downregulated in insulin resistant skeletal muscle cells while Serpin E1 is upregulated.39,38,36
3.7. Others
The secretion of certain proteins can also be modified according to the muscle. Roca-Rivada et al compared a slow-oxidative muscle with a fast-oxidative muscle to highlight differential expression of proteins. One of the regulated proteins was the heart fatty acid binding protein (FAPB-3) which was 3 fold more abundant in the secretome of soleus versus gastrocnemius muscle.11 This protein is thought to be involved in intracellular transport of long-chain fatty acids which is consistent with its upregulation in oxidative muscle. Indeed, type I fibers are predominant in slow oxidative muscle and uses fatty acids and glucose as fuel to produce ATP.
4. Discussion
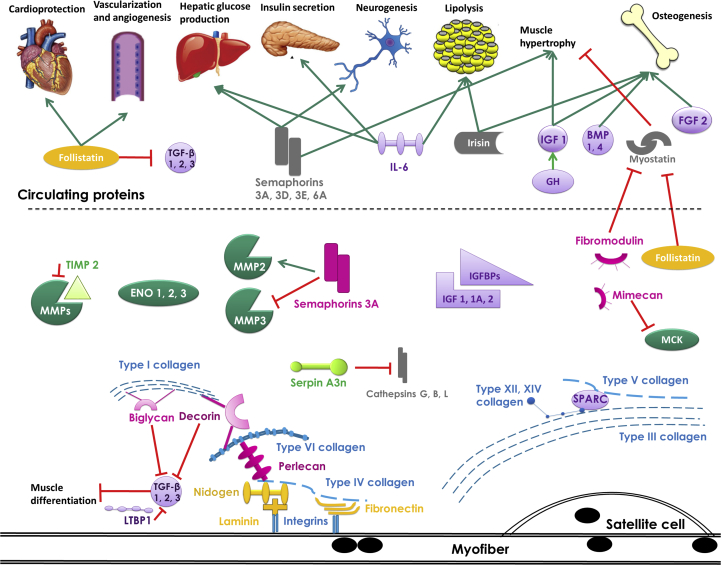
Skeletal muscle is known to secrete proteins and especially myokines that allow communication in an autocrine, paracrine and endocrine manner (fig. 5). Some of these secreted proteins and myokines can be found in the blood or in the urine and can reflect a change in the pathophysiological state of an individual. The study of the secretome of skeletal muscle cells is of great interest to identify a range of candidate biomarkers from muscle diseases like sarcopenia, allowing early diagnosis and appropriate interventions. This review summarizes the data obtained by proteomic studies of the secretome of skeletal muscle cells or explants. The advantage of this method is the control of the key parameters and the live conditions of cells or tissues. For instance, only 6 conditions have been reported: exercising, myogenesis, dystrophin deficiency, muscle atrophy, insulin-treated and insulin-resistant. Clearly, insulin-resistance and myogenesis are the two most documented conditions. There is a paucity of published data on the secretome of sarcopenic muscle.
Fig.5.
Schematic representation of the secretome of skeletal muscle cells. The stimulation is represented by green arrows and the inhibition is represented by red crossed arrows. Collagens are represented in blue; proteoglycans and glycosaminoglycans are represented in pink; glycoproteins are represented in orange; growth factors and cytokines are represented in purple; enzymes are represented in dark green; enzymatic inhibitors are represented in light green and other proteins are represented in dark gray. MMP: Matrix Metalloproteinase; TIMP: Metalloproteinase inhibitor; IL: Interleukin; IGF: Insulin-like growth factor; FGF: Fibroblast Growth Factor; GDF: Growth/Differentiation factor; GH: Growth hormone; BMP: Bone morphogenetic protein; ENO: Enolase; IGFBP: Insulin-like growth factor binding protein; LTBP: Latent-transforming growth factor beta-binding protein; MCK: Muscle creatine kinase; TGF: Transforming growth factor.
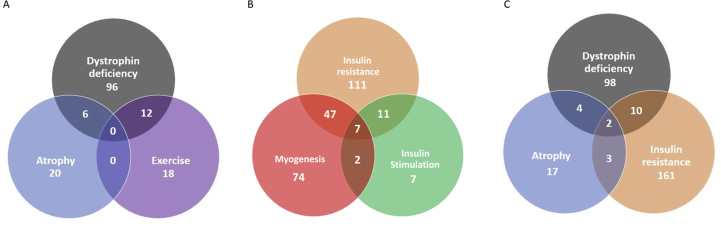
The interest in insulin expression and signalling within muscle comes from the fact that skeletal muscle energy supply depends on up to 75% of the insulin dependent glucose uptake and storage as glycogen. Furthermore, insulin resistance leads to the development of type 2 diabetes, which is associated with muscle dysfunction and weakness. Several methods have been used to induce insulin resistance in muscle cells. Yoon et al induced resistance by addition of TNF-α39 or palmitate38 while Deshmuk et al used palmitic acid.36 These models drastically influence the secretome of skeletal muscle cells. Eighteen proteins were regulated under both conditions (fig. 6). The type 1 collagen III and the type 2 collagen V are the 2 only collagens which are downregulated by these two treatments. The other proteins down-regulated in these two models were BMP1, FSTL1, IGFBP6, TIMP2 and NUCB1 while PDIA3, ENO1 and Serpin E1 (PAI-1) were up-regulated. FSTL1 and IGFBP6 regulate the action of some growth factors and their decrease may lead to a reduced regenerative capacity of the muscle. The reduction of TIMP2 secretion leads to a greater matrix cellular degradation. PDIA3 is also upregulated by dystrophin deficient cells and during atrophy. This protein has recently been shown to be a marker of muscle aerobic capacity.65 ENO1 is a key glycolytic enzyme and its expression is increased by insulin suggesting that ENO1 increase could be a compensatory mechanism to metabolize glucose in insulin-resistant muscle cells. Serpin E1 is increased in the plasma of obese and insulin-resistant patient,56 this is also the case in the secretome of insulin resistant skeletal muscle cells or after insulin stimulation.31,36 In addition to its role in regulating growth and metabolism, IGF-1 has also insulin-sensitizing and glucose lowering actions.36 IGF-1 decreases in insulin-resistant skeletal muscle cells' secretome leading to this insulin resistant state. Many chemokines (CCL2, CXCL1 and CXCL5) were up-regulated in insulin-resistant muscle cells, this may lead to a low-grade chronic inflammation which is a hallmark of type 2 diabetes.66 Furthermore, patient with type 2 diabetes have MMP-2 levels significantly higher than healthy patient,66 this is in accordance with the increase of MMP-2 and the decrease of TIMP2 in insulin-treated muscle cells’ secretome. Interestingly, MMP-2 secretion decrease in the conditioned media of insulin-resistant skeletal muscle cells leading to a reduced extracellular matrix component degradation and subsequent cell migration and tissue remodeling.30,40 Annexin A1 was found to be down-regulated in insulin-stimulated skeletal muscle cells.38 The Annexin A1-FPR2 axis plays a protective role in insulin resistance by mediating leukocyte recruitment and anti-inflammatory effects. This pathway might be a new therapeutic target for insulin resistance.38
Fig.6.
Venn diagrams showing the number of differentially regulated proteins between different conditions/pathologies. A. Proteins differentially regulated between the conditions dystrophin deficiency, atrophy and exercise. B. Proteins differentially regulated between the conditions insulin resistance, myogenesis and insulin stimulation. C. Proteins differentially regulated between the conditions dystrophin deficiency, atrophy and insulin resistance.
The second most investigated condition is myogenesis. The formation of myotubes involves multiple steps such as cell migration, recognition, alignment, adhesion, cell fusion and ECM reorganization. Three published papers have been devoted to the study of proteins secretion during the skeletal muscle development. Interestingly, these studies investigated the secretome at different stages of muscle development.27 At day 0, 2 and 5 during differentiation for Henningsen et al.27, before differentiation and after 30 h, 72 h and 120 h of differentiation for Ojima et al37 and finally after 24 h and 120 h of differentiation in the Chang's study.32 Table 2 highlights the proteins that are regulated (up or down) in the secretome of myotubes (after 120 h of differentiation) and myoblasts (before differentiation). Interestingly, the secretion of some proteins resulting from different studies appear to be contradictory. This is the case for the proteins high mobility group B1, semaphorin 7A, phosphatidylethanolamine binding protein 1, IGFBP5 and SPARC. These differences may be due to the different culture media used in these in vitro studies to induce the differentiation of myoblasts. These media may lead to a differential secretion and a variable advance in differentiation of myoblasts into myotubes. Henningsen et al27 studied the dynamics of the skeletal muscle cell secretome during differentiation, this method is useful for understanding which factors are secreted during myogenesis. They observed that ECM reorganization is critical during differentiation and is particularly controlled by the TGF-β proteins by promoting the synthesis of ECM components.27 For example, mimecan (also called osteoglycin) was downregulated by more than 6 fold during myogenesis and appeared to decrease the transcriptional activity of muscle creatine kinase (MCK), which suggests that mimecan may only influence the early stages of myogenesis.32,67
Furthermore, TGF-β proteins have a role in inhibiting differentiation of skeletal muscle cells, which is in accordance with the continuous increase of these proteins during the course of differentiation.27 The growth factors IGF-1 and IGF-2 play also a crucial role during differentiation. IGF-1 is important in the beginning of the differentiation whereas IGF-2 is essential throughout the differentiation process. They are regulated by IGFBPs. The interactions and regulations of IGFs by IGFBPs are complex. This is highlighted by variation of IGFBPs production during differentiation. IGFBP2 decreases and IGFBP4 increases during differentiation whereas IGFBP5, -6 and -7 were not significantly regulated.27 About semaphorins, the dynamics of expression suggest that they mainly regulate the early phase of differentiation in an autocrine way, except for semaphorin 7A which increases throughout the course of differentiation. Furthermore, SEMA3A activates MMP-2 which has a crucial role for ECM remodeling during differentiation. Indeed, MMP-2 degrades extracellular matrix, leading to a release of growth factors and signaling molecules that play a role in activation and proliferation of satellite cells.68 This is in accordance with the up-regulation of MMP-2, especially during the early phase of myogenesis. On the other hand, SEMA3A decreases both the expression and activity of MMP-3. Interestingly, they are a lot of proteins (54) that are regulated both by myogenesis and insulin resistance (fig. 6). Insulin resistance leads to an accumulation of adipose tissue resulting in a rise in circulating adipokines. Some adipokines like resistin leads to lipid accumulation in skeletal muscle and subsequently to insulin resistance through activation of protein kinase C.69 Furthermore, the adipokine resistin were found to impair myogenesis through activation of NF-κB.70 Insulin resistance also decrease skeletal muscle anabolism and so induce loss of skeletal muscle mass.71 On the contrary, insulin induce myogenesis through inhibition of glycogen synthase kinase 3 beta (GSK-3β) and is mediated by AKT.72 Furthermore, enzymatic inhibitor Serpin E1 and the glycoprotein FSTL1 were downregulated both during myogenesis, insulin resistance and insulin stimulation (fig. 6).73
The third condition studied is dystrophin deficiency. Dystrophin deficient H–2K skeletal muscle cells were extracted from mdx-H2Kb-ts58 (CBA/ca X C57B1/10ScSn-mdx) mice at 1 month of age. The secretome of the mdx myotubes were compared to wild-type myotubes.34 Interestingly, mdx myotubes secreted twice as much total protein and most of the secreted proteins were cytoplasmic.34 In addition, fibronectin was found in higher amount in the conditioned media of mdx muscle cells. This study demonstrates that dystrophin deficiency impairs vesicle trafficking leading to excess protein secretion and fibrotic deposition.34
Another condition studied in this review was the glucocorticoid-mediated muscle atrophy. Muscle cell atrophy was induced by adding 10−6 M dexamethasone in the cell medium for 24 h in serum free conditions. Chronic glucocorticoid (GC) exposure in humans is well known to result in whole-body insulin resistance and obesity. This occur mainly through the suppression of osteoblasts’ osteocalcin secretion rather than GC signaling in the liver or the skeletal muscle.74 Insulin resistance impairs glucose metabolism and high-glucose levels are known to inhibit myogenesis.71 Interestingly, some proteins are modulated both in the insulin resistance and in the muscle atrophy models. A decrease in secretion of perlecan and biglycan were observed in both models.25 The protein disulfide isomerase A3 increased in the conditioned media of atrophied skeletal muscle25 and during insulin-stimulated and insulin resistant conditions. Furthermore, this protein is expressed more by low capacity compared to high capacity running rats and may be a biomarker of muscle aerobic capacity. This may be due to differences in insulin muscle sensitivity.65 It would be interesting to study the mechanism of action of this protein and its role in these different conditions. Serpin A3n was found to rise in the serum of mice treated with dexamethasone.25 Serpin A3n may have a protective role towards skeletal muscle through his antiprotease activity against extracellular proteases.25 This protein may be a potential biomarker of glucocorticoid-induced muscle atrophy.25
The last condition reported in this review was secretome modification by exercise. The first study compared the secretome of slow-oxidative (soleus) and fast-oxidative (gastrocnemius) muscle explant and its variation with endurance exercising.11 The rats where housed in rodent cages with a running wheel coupled to a turn counter. Rats running 8 h a day were selected for the secretome study. The control group (non-exercising) was compared to the exercising group who performed this endurance exercise during 6 days. As anticipated, the two muscle types showed different secretome. For example, IL-6 and FAPB-3 were more abundant in soleus muscle compared to gastrocnemius muscle. On the contrary, the protein DJ-1 was secreted in higher amount in the gastrocnemius muscle compared to the soleus. This means that exercises can modify the fiber-type of the muscle and so influence the secretion pattern. The second article mimicked the effects of exercising by using the AMP-kinase agonist 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR). Indeed, AMPK activation blocks energy-consuming processes and promotes ATP synthesis from glucose uptake, glycosylation and fatty acid oxidation. AICAR treated mice show decreased fat mass, increase running endurance and muscle mass.24 With these experiments, Youl et al observed that secretory proteins from AICAR treated skeletal muscle cells could influence in vitro expression of neuronal differentiation markers, suggesting that exercise-induced secreted proteins from skeletal muscle may play a role in neurogenesis.24 Interestingly, no proteins are regulated both by atrophy and by exercise. This is not the case for dystrophin deficiency and exercise, because 12 proteins were regulated by these two conditions (fig. 6). This may be a consequence of the damage or the inflammation induced in the first case by the dystrophin deficiency and in the second case by exercising.75 It is interesting to notice that ENO1 was upregulated in most of the conditions (exercising, dystrophin deficiency, insulin stimulation and insulin resistance). The upregulation of ENO1 may be an adaptation to better metabolize glucose during exercising or in the case of exercise intolerance leading dystrophy.75,76
Another important cytokine with myokine properties is IL-6. IL-6 is a crucial exercise-induced cytokine that probably mediates the immunoregulatory and anti-inflammatory effects of physical exercise.49,77,78 Unfortunately, the detection of IL-6 in the conditioned media of skeletal muscle cells has not been possible. Indeed, due to the intolerance of myotubes to serum-free conditions, the incubation was limited to 6 h to avoid leakage of intracellular proteins, which may result in the inability to detect low-abundance proteins. It may also be due to the small size of this protein which is a limitation of the analysis by mass spectrometry. In conclusion, the up- or downregulation of IL-6 by exercise has not yet been demonstrated by an in vitro proteomic approach. Currently, no regulation of myostatin (GDF8) in the secretome of skeletal muscle cell has been demonstrated by mass spectrometric analysis. This protein is a crucial negative regulator of skeletal muscle growth, modulate adipose tissue mass and function and is involved in the maintenance of metabolic homeostasis.
This systematic review has some limitations due to the significant difference in the methodologies used to study the skeletal muscle secretome. One main concern is that each research group has employed a different type of skeletal muscle cell preparation, or different culture media and different protocols to analyze the secreted proteins. These differences in methodology complicate objective comparisons between the studies. For example, in explant cultures, a large part of the proteins secreted are entrapped in the extracellular matrix, modifying the secretome analysis.79 Other cell types are present within skeletal muscle in vivo (for instance, fibroblasts and adipocytes) that could influence the secretome of the skeletal muscle. This is particularly important because the secretome of infiltrating adipocytes may be largely catabolic and inflammatory, leading to low grade inflammation within skeletal muscle. Another drawback of using skeletal muscle cells in vitro is that they are non-functional. They are not exposed to physiological mechanical strains that exist in vivo during muscle contraction. Several studies have shown that contraction influence skeletal muscle cells’ secretome.80 It is possible to overcome this issue by studying mRNA expression in skeletal muscle biopsies29 or by exploring the secretome of skeletal muscle explants.11 Manabe et al developed an in vitro muscle contraction model based on mouse primary cultured myotubes and this model may enable the discovery of novel myokines secreted by muscle contraction.81 Furthermore, the quantitative assessment of proteins remains complex due to their different solubility, binding to other components, size, length, amino acid sequence and post-translational modifications. Finally, during the process of proteomic analysis, at least 2 independent peptides are needed for the unambiguous identification of proteins, but very small proteins (like cytokines) generate few peptides after trypsinization, leading to a possible exclusion of key proteins from the final analysis.
In conclusion, the study of the secretome of skeletal muscle cells highlights novel proteins and myokines that could be candidate biomarkers for the diagnosis, prognosis or follow-up of interventions of in muscular diseases like sarcopenia. Particular attention should be given to proteins like TNF-α, β-aminoisobutyric acid (BAIBA),82 irisin,83 interleukin 1583, decorin, myostatin, myonectin,84 meteorin-like 1,85 adiponectin, C-reactive protein CRP, HSP72 and C-terminal fragments of agrin86,87 which may be involved in ageing and inflammatory processes. However, very limited studies associated with the role of myokines are currently available and more researches need to first determine and address the expression profile in muscle development and regeneration. The underlying signaling pathways also need to be studied in details. Secretome analysis has generated a lot of data that could be implemented in big data system and learning machine, a way currently used in other research field to identify patient phenotypes, leading to a more personalized treatment.
Declaration of Competing Interest
Yves Henrotin has received consulting fees from Artialis SA, Tilman, Laboratoires Expanscience, Nestle.
Acknowledgements
The authors thank the Belgian Walloon Region for the financial support provided to this research project in the form of two subsidies (N° 1320131 & N°7781).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ocarto.2019.100019.
Contributions
Authors’ roles: Study design: AF, CL, CS, JZ, ND and YH. Data collection: AF, CL and ND. Data analysis: AF, CL. Data interpretation: AF, CL, CS, JZ, AMT, AM and YH. Drafting manuscript: AF and YH. Revising manuscript content: CL, CS, JZ, ND, AMT and AM. Approving final version of the manuscript: AF, CL, CS, JZ, ND, AMT, AM and YH. AF takes the responsibility for the integrity of the data analysis.
Role of the funding source
The funding source had no role in the writing of the manuscript or in the decision to submit the manuscript for publication.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Frontera W.R., Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen B.K., Akerström T.C.A., Nielsen A.R., Fischer C.P. Role of myokines in exercise and metabolism. J. Appl. Physiol. 2007;103(3):1093–1098. doi: 10.1152/japplphysiol.00080.2007. [DOI] [PubMed] [Google Scholar]
- 3.Furuichi Y., Fujii N.L. Mechanism of satellite cell regulation by myokines. J. Phys. Fit. Sport. Med. 2017;6(5):311–316. doi: 10.7600/jpfsm.6.311. [DOI] [Google Scholar]
- 4.Baggish A.L., Hale A., Weiner R.B., et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011;589(16):3983–3994. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guescini M., Canonico B., Lucertini F., et al. Muscle releases alpha-sarcoglycan positive extracellular vesicles carrying miRNAs in the bloodstream. PLoS One. 2015;10(5):1–19. doi: 10.1371/journal.pone.0125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwig S., Raschke S., Knebel B., et al. Secretome profiling of primary human skeletal muscle cells. Biochim. Biophys. Acta. 2014;1844:1011–1017. doi: 10.1016/j.bbapap.2013.08.004. (5 PG-1011-7) [DOI] [PubMed] [Google Scholar]
- 7.Eckardt K., Görgens S.W., Raschke S., Eckel J. Myokines in insulin resistance and type 2 diabetes. Diabetologia. 2014;57(6):1087–1099. doi: 10.1007/s00125-014-3224-x. [DOI] [PubMed] [Google Scholar]
- 8.Evers-Van Gogh I.J.A., Alex S., Stienstra R., Brenkman A.B., Kersten S., Kalkhoven E. Electric pulse stimulation of myotubes as an in vitro exercise model: cell-mediated and non-cell-mediated effects. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep10944. (April) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon J.H., Kim J., Song P., Lee T.G., Suh P.G., Ryu S.H. Secretomics for skeletal muscle cells: a discovery of novel regulators? Adv. Biol. Regul. 2012;52(2):340–350. doi: 10.1016/j.jbior.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Hojman P., Dethlefsen C., Brandt C., Hansen J., Pedersen L., Pedersen B.K. Exercise-induced muscle-derived cytokines inhibit mammary cancer cell growth. Am. J. Physiol. Metab. 2011;301(3):E504–E510. doi: 10.1152/ajpendo.00520.2010. [DOI] [PubMed] [Google Scholar]
- 11.Roca-Rivada A., Al-Massadi O., Castelao C., et al. Muscle tissue as an endocrine organ: comparative secretome profiling of slow-oxidative and fast-glycolytic rat muscle explants and its variation with exercise. J. Proteom. 2012;75(17):5414–5425. doi: 10.1016/j.jprot.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 12.Scheler M., Irmler M., Lehr S., et al. Cytokine response of primary human myotubes in an in vitro exercise model. AJP Cell Physiol. 2013;305(8):C877–C886. doi: 10.1152/ajpcell.00043.2013. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman N.J., Parker B.L., Chaudhuri R., et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metabol. 2015;22:922–935. doi: 10.1016/j.cmet.2015.10.004. (5 PG-922-935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. 2014;6:1–16. doi: 10.1101/cshperspect.a016295. (Kishimoto 1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Febbraio M.A., Hiscock N., Sacchetti M., Fischer C.P., Pedersen B.K. Interleukin-6 is a novel factor mediating glucose homeostasis during skeletal muscle contraction. Diabetes. 2004;53(7):1643–1648. doi: 10.2337/diabetes.53.7.1643. [DOI] [PubMed] [Google Scholar]
- 16.Carey A.L., Steinberg G.R., Macaulay S.L., et al. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes. 2006;55(10):2688–2697. doi: 10.2337/db05-1404. [DOI] [PubMed] [Google Scholar]
- 17.Makridakis M., Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J. Proteom. 2010;73(12):2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 19.Chen D., Landers-ramos R.Q., Mázala D.A.G. Advances in the Application of Protein Mass Spectrometry to Skeletal Muscle Biology Open Research Article Advances in the Application of Protein Mass Spectrometry to Skeletal Muscle Biology. J. Med. Discov. 2017 doi: 10.24262/jmd.2.2.17026. [DOI] [Google Scholar]
- 20.Zhang Y., Fonslow B.R., Shan B., Baek M.C., Yates J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalski A., Damoc E., Lange O., et al. Ultra high resolution linear ion trap orbitrap mass spectrometer (orbitrap elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteom. 2012;11(3) doi: 10.1074/mcp.O111.013698. O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Ge Y. Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ. Cardiovasc. Genet. 2012;4(6):1–21. doi: 10.1161/CIRCGENETICS.110.957829.Comprehensive. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moher D., Liberati A., Tetzlaff J., Altman D.G., Prisma Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement (reprinted from annals of internal medicine) Ann. Intern. Med. 2009;151(4):264–269. doi: 10.1371/journal.pmed.1000097. [DOI] [PubMed] [Google Scholar]
- 24.Youl H., Javadi S., Stremlau M., et al. Neuropharmacology Conditioned media from AICAR-treated skeletal muscle cells increases neuronal differentiation of adult neural progenitor cells. Neuropharmacology. 2019;145:123–130. doi: 10.1016/j.neuropharm.2018.10.041. (November 2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gueugneau M., Hose D., Barbé C., et al. Increased Serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. J. Cachexia Sarcopenia Muscle. 2018;9:929–946. doi: 10.1002/jcsm.12315. (July) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grube L., Dellen R., Kruse F., Schwender H., Stühler K., Poschmann G. Mining the secretome of C2C12 muscle cells: data dependent experimental approach to analyze protein secretion using label-free quantification and peptide based analysis. J. Proteome Res. 2018;17:879–890. doi: 10.1021/acs.jproteome.7b00684. (2 PG-879-890) [DOI] [PubMed] [Google Scholar]
- 27.Henningsen J., Rigbolt K.T.G., Blagoev B., Pedersen B.K., Kratchmarova I. Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol. Cell. Proteom. 2010;9(11):2482–2496. doi: 10.1074/mcp.M110.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bihan M.C., Bigot A., Jensen S.S., et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J. Proteom. 2012;77:344–356. doi: 10.1016/j.jprot.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Norheim F., Raastad T., Thiede B., Rustan A.C., Drevon C a, Haugen F. Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am. J. Physiol. Endocrinol. Metab. 2011:1013–1021. doi: 10.1152/ajpendo.00326.2011. [DOI] [PubMed] [Google Scholar]
- 30.Hittel D.S., Berggren J.R., Shearer J., et al. Increased secretion and expression of myostatin in skeletal muscle from extremely obese women. Diabetes. 2009;58:30–38. doi: 10.2337/db08-0943. (1 PG-30-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon J.H., Yea K., Kim J., et al. Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics. 2009;9(1):51–60. doi: 10.1002/pmic.200800187. [DOI] [PubMed] [Google Scholar]
- 32.Chan C.Y.X., Masui O., Krakovska O., et al. Identification of differentially regulated secretome components during skeletal myogenesis. Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.M110.004804. (5 PG-M110.004804):M110.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forterre A., Jalabert A., Berger E., et al. Correction: proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: a new paradigm for myoblast-myotube cross talk? (PLoS ONE) PLoS One. 2014;9 doi: 10.1371/annotation/ecd1e074-2618-4ad0-95c0-efdb467c714b. (1 PG-e84153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duguez S., Duddy W., Johnston H., et al. Dystrophin deficiency leads to disturbance of LAMP1-vesicle-associated protein secretion. Cell Mol. Life Sci. 2013;70(12):2159–2174. doi: 10.1007/s00018-012-1248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henningsen J., Pedersen B.K., Kratchmarova I. Quantitative analysis of the secretion of the MCP family of chemokines by muscle cells. Mol. Biosyst. 2011;7(2):311–321. doi: 10.1039/C0MB00209G. [DOI] [PubMed] [Google Scholar]
- 36.Deshmukh A.S., Cox J., Jensen L.J., et al. Secretome analysis of lipid-induced insulin resistance in skeletal muscle cells by a combined experimental and bioinformatics workflow. J. Proteome Res. 2015;14:4885–4895. doi: 10.1021/acs.jproteome.5b00720. (11 PG-4885-95) [DOI] [PubMed] [Google Scholar]
- 37.Ojima K., Oe M., Nakajima I., et al. Proteomic analysis of secreted proteins from skeletal muscle cells during differentiation. EuPA Open Proteom. 2014;5:1–9. doi: 10.1016/j.euprot.2014.08.001. [DOI] [Google Scholar]
- 38.Yoon J.H., Kim D., Jang J., et al. Proteomic analysis of the palmitate-induced myotube secretome reveals involvement of the annexin A1-formyl peptide receptor 2 (FPR2) pathway in insulin resistance. Mol. Cell. Proteom. 2015;14:882–892. doi: 10.1074/mcp.M114.039651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon J.H., Song P., Jang J.H., et al. Proteomic analysis of tumor necrosis factor-alpha (TNF-α)-induced L6 myotube secretome reveals novel TNF-α-dependent myokines in diabetic skeletal muscle. J. Proteome Res. 2011;10(12):5315–5325. doi: 10.1021/pr200573b. [DOI] [PubMed] [Google Scholar]
- 40.Chan X.C.Y., McDermott J.C., Siu K.W.M. Identification of secreted proteins during skeletal muscle development. J. Proteome Res. 2007;6(2):698–710. doi: 10.1021/pr060448k. [DOI] [PubMed] [Google Scholar]
- 41.Dubowitz V., Sewry A.C., Oldfors A. A Practical Approach.; 2013. Muscle Biopsy. [Google Scholar]
- 42.Brandan E., Cabello-Verrugio C., Vial C. Novel regulatory mechanisms for the proteoglycans decorin and biglycan during muscle formation and muscular dystrophy. Matrix Biol. 2008;27(8):700–708. doi: 10.1016/j.matbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Droguett R., Cabello-Verrugio C., Riquelme C., Brandan E. Extracellular proteoglycans modify TGF-β bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biol. 2006;25(6):332–341. doi: 10.1016/j.matbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Jørgensen L.H., Petersson S.J., Sellathurai J., et al. Secreted protein acidic and rich in cysteine (SPARC) in human skeletal muscle. J. Histochem. Cytochem. 2009;57(1):29–39. doi: 10.1369/jhc.2008.951954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hojman P., Pedersen M., Nielsen A.R., et al. Fibroblast growth factor-21 is induced in human skeletal muscles by hyperinsulinemia. Diabetes. 2009;58(12):2797–2801. doi: 10.2337/db09-0713. db09-0713 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh K. Adipokines, myokines and cardiovascular disease. Circ. J. 2009;73(1):13–18. doi: 10.1253/circj.cj-08-0961. doi:JST.JSTAGE/circj/CJ-08-0961. [DOI] [PubMed] [Google Scholar]
- 47.Giudice J., Taylor J.M. Muscle as a paracrine and endocrine organ. Curr. Opin. Pharmacol. 2017;34:49–55. doi: 10.1016/j.coph.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sattler F.R. Growth hormone in the aging male. Best Pract. Res. Clin. Endocrinol. Metabol. 2013;27(4):541–555. doi: 10.1016/j.beem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen B.K., Febbraio M.A. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012;8(8):457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 50.Glass D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 2005;5:87–90. doi: 10.1038/ncb0203-87. (February) [DOI] [PubMed] [Google Scholar]
- 51.De Wit J., Verhaagen J. Role of semaphorins in the adult nervous system. Prog. Neurobiol. 2003;71(2-3):249–267. doi: 10.1016/j.pneurobio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 52.Roth L., Koncina E., Satkauskas S., Crémel G., Aunis D., Bagnard D. The many faces of semaphorins: from development to pathology. Cell Mol. Life Sci. 2009;66(4):649–666. doi: 10.1007/s00018-008-8518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H., Wang X., Liu S., et al. Sema4C participates in myogenic differentiation in vivo and in vitro through the p38 MAPK pathway. Eur. J. Cell Biol. 2007;86(6):331–344. doi: 10.1016/j.ejcb.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Tatsumi R., Sankoda Y., Anderson J.E., et al. Possible implication of satellite cells in regenerative motoneuritogenesis: HGF upregulates neural chemorepellent Sema3A during myogenic differentiation. Am. J. Physiol. Cell Physiol. 2009;297(2):C238–C252. doi: 10.1152/ajpcell.00161.2009. [DOI] [PubMed] [Google Scholar]
- 55.Sato Y., Do M.K.Q., Suzuki T., et al. Satellite cells produce neural chemorepellent semaphorin 3A upon muscle injury. Anim. Sci. J. 2013;84(2):185–189. doi: 10.1111/asj.12014. [DOI] [PubMed] [Google Scholar]
- 56.Ma L.J., Mao S.L., Taylor K.L., et al. Prevention of obesity and insulin resistance in mice lacking plasminogen activator inhibitor 1. Diabetes. 2004;53(2):336–346. doi: 10.2337/diabetes.53.2.336. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14747283 [DOI] [PubMed] [Google Scholar]
- 57.McLennan I.S. Degenerating and regenerating skeletal muscles contain several subpopulations of macrophages with distinct spatial and temporal distributions. J. Anat. 1996;188:17–28. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1167629&tool=pmcentrez&rendertype=abstract (Pt 1) [PMC free article] [PubMed] [Google Scholar]
- 58.Yahiaoui L., Gvozdic D., Danialou G., Mack M., Petrof B.J. CC family chemokines directly regulate myoblast responses to skeletal muscle injury. J. Physiol. 2008;586(16):3991–4004. doi: 10.1113/jphysiol.2008.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu H.F., Gu T., Hilding A., et al. Evaluation of IGFBP-7 DNA methylation changes and serum protein variation in Swedish subjects with and without type 2 diabetes. Clin. Epigenet. 2013;5(1):20. doi: 10.1186/1868-7083-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kessenbrock K., Fröhlich L., Sixt M., et al. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Investig. 2008;118(7):2438–2447. doi: 10.1172/JC134694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen R.E., Sheehan S.M., Taylor R.G., Kendall T.L., Rice G.M. Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J. Cell Physiol. 1995;165(2):307–312. doi: 10.1002/jcp.1041650211. [DOI] [PubMed] [Google Scholar]
- 62.Doumit M.E., Cook D.R., Merkel R.A. Fibroblast growth factor, epidermal growth factor, insulin-like growth factors, and platelet-derived growth factor-BB stimulate proliferation of clonally derived porcine myogenic satellite cells. J. Cell Physiol. 1993;157(2):326–332. doi: 10.1002/jcp.1041570216. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8227164 [DOI] [PubMed] [Google Scholar]
- 65.Burniston J.G., Kenyani J., Gray D., et al. Conditional independence mapping of DIGE data reveals PDIA3 protein species as key nodes associated with muscle aerobic capacity. J. Proteom. 2014;(106):230–245. doi: 10.1016/j.jprot.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Derosa G., D'Angelo A., Scalise F., et al. Comparison between metalloproteinases-2 and -9 in healthy subjects, diabetics, and subjects with acute coronary syndrome. Heart Vessel. 2007;22(6):361–370. doi: 10.1007/s00380-007-0989-6. [DOI] [PubMed] [Google Scholar]
- 67.Funderburgh J.L., Corpuz L.M., Roth M.R., et al. Nucleic acids , protein synthesis , and molecular GENETICS : mimecan , the 25-kDa corneal keratan sulfate proteoglycan , is a product of the gene producing osteoglycin mimecan , the 25-kDa corneal keratan sulfate proteoglycan , is a product of the. Gen. Pract. 1997;272(44):28089–28095. doi: 10.1074/jbc.272.44.28089. [DOI] [PubMed] [Google Scholar]
- 68.Thomas K., Engler A.J., Meyer G.A., et al. Extracellular Matrix Regulation in the Muscle Satellite Cell Niche Extracellular Matrix Regulation in the Muscle Satellite Cell Niche. Connect. Tissue Res. 2015:8207. doi: 10.3109/03008207.2014.947369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meex R.C.R., Blaak E.E. Lipotoxicity plays a key role in the development of both insulin resistance and muscle atrophy in patients with type 2 diabetes. Obes. Rev. 2019;20:1205–1217. doi: 10.1111/obr.12862. (March): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leary M.F.O., Wallace G.R., Davis E.T., et al. Obese subcutaneous adipose tissue impairs human myogenesis , particularly in old skeletal muscle , via resistin-mediated activation of NF κ B. Sci. Rep. 2018:1–13. doi: 10.1038/s41598-018-33840-x. (October) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo W., Ai L., Wang B., Zhou Y. High glucose inhibits myogenesis and induces insulin resistance by down- regulating AKT signaling. Biomed. Pharmacother. 2019;120:109498. doi: 10.1016/j.biopha.2019.109498. (September) [DOI] [PubMed] [Google Scholar]
- 72.Litwiniuk A., Pijet B., Pijet-kucicka M. FOXO1 and GSK-3 β are main targets of insulin-mediated myogenesis in C2C12 muscle cells. PLoS One. 2016:1–25. doi: 10.1371/journal.pone.0146726. (Cox Iv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattiotti A., Prakash S., Barnett P., Van Den Hoff M.J. Follistatin - like 1 in development and human diseases. Cell Mol. Life Sci. 2018;75(13):2339–2354. doi: 10.1007/s00018-018-2805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parker L., Lin X., Garnham A., et al. Glucocorticoid-induced insulin resistance in. J. Bone Miner. Res. 2019;34(1):49–58. doi: 10.1002/jbmr.3574. [DOI] [PubMed] [Google Scholar]
- 75.Barnabei M.S., Martindale J.M., Townsend D., Metzger J.M. Exercise and muscular Dystrophy : implications and analysis of effects on musculoskeletal and cardiovascular systems. 2011;1:1353–1363. doi: 10.1002/cphy.c100062. (July) [DOI] [PubMed] [Google Scholar]
- 76.Yu X., Li S. Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene. 2016;6:1–8. doi: 10.1038/onc.2016.410. (July) [DOI] [PubMed] [Google Scholar]
- 77.Nielsen S., Pedersen B.K. Skeletal muscle as an immunogenic organ. Curr. Opin. Pharmacol. 2008;8(3):346–351. doi: 10.1016/j.coph.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Petersen A.M.W., Pedersen B.K. The anti-inflammatory effect of exercise. J. Appl. Physiol. 2005;98(4):1154–1162. doi: 10.1152/japplphysiol.00164.2004. [DOI] [PubMed] [Google Scholar]
- 79.Polacek M., Bruun J.A., Johansen O., Martinez I. Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by SILAC technology. J. Orthop. Res. 2010;28(8):1040–1049. doi: 10.1002/jor.21067. [DOI] [PubMed] [Google Scholar]
- 80.Raschke S., Eckel J. Adipo-Myokines: two sides of the same coin - mediators of inflammation and mediators of exercise. Mediat. Inflamm. 2013;2013 doi: 10.1155/2013/320724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manabe Y., Ogino S., Ito M., et al. Evaluation of an in vitro muscle contraction model in mouse primary cultured myotubes. Anal. Biochem. 2016;497:36–38. doi: 10.1016/j.ab.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Sullivan J.F.O., Schinzel R.T., Lewis G.D., et al. Article b -aminoisobutyric acid induces browning of white fat and hepatic b -oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metabol. 2014;19(1):96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J.H., Jun H. Role of myokines in regulating skeletal muscle mass and function. Front Physiol. 2019;10:1–9. doi: 10.3389/fphys.2019.00042. (January) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seldin M.M., Peterson J.M., Byerly M.S., Wei Z., Wong G.W. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J. Biol. Chem. 2012;287(15):11968–11980. doi: 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao R.R., Long J.Z., White J.P., et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. 2015;157(6):1279–1291. doi: 10.1016/j.cell.2014.03.065. Meteorin-like. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebhardt H.A., Degen S., Tadini V., et al. Comprehensive proteome analysis of human skeletal muscle in cachexia and sarcopenia: a pilot study. J. Cachexia Sarcopenia Muscle. 2017;8(4):567–582. doi: 10.1002/jcsm.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. vol. 15. Taylor & Francis; 2018. (Proteomic Serum Biomarkers for Neuromuscular Diseases). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.