Abstract
Objective
This study aimed to develop a deep learning-based approach to automatically segment the femoral articular cartilage (FAC) in 3D ultrasound (US) images of the knee to increase time efficiency and decrease rater variability.
Design
Our method involved deep learning predictions on 2DUS slices sampled in the transverse plane to view the cartilage of the femoral trochlea, followed by reconstruction into a 3D surface. A 2D U-Net was modified and trained using a dataset of 200 2DUS images resliced from 20 3DUS images. Segmentation accuracy was evaluated using a holdout dataset of 50 2DUS images resliced from 5 3DUS images. Absolute and signed error metrics were computed and FAC segmentation performance was compared between rater 1 and 2 manual segmentations.
Results
Our U-Net-based algorithm performed with mean 3D DSC, recall, precision, VPD, MSD, and HD of 73.1 ± 3.9%, 74.8 ± 6.1%, 72.0 ± 6.3%, 10.4 ± 6.0%, 0.3 ± 0.1 mm, and 1.6 ± 0.7 mm, respectively. Compared to the individual 2D predictions, our algorithm demonstrated a decrease in performance after 3D reconstruction, but these differences were not found to be statistically significant. The percent difference between the manually segmented volumes of the 2 raters was 3.4%, and rater 2 demonstrated the largest VPD with 14.2 ± 11.4 mm3 compared to 10.4 ± 6.0 mm3 for rater 1.
Conclusion
This study investigated the use of a modified U-Net algorithm to automatically segment the FAC in 3DUS knee images of healthy volunteers, demonstrating that this segmentation method would increase the efficiency of anterior femoral cartilage volume estimation and expedite the post-acquisition processing for 3D US images of the knee.
Keywords: Femoral articular cartilage segmentation, 3D ultrasound knee imaging, Knee osteoarthritis, Deep learning, Convolutional neural networks
1. Introduction
Knee osteoarthritis (KOA) is a progressive multifactorial joint disease that has a current global prevalence of 22.9% among individuals above the age of forty and is rising with increasing rates of obesity and population aging [1]. KOA affects all components of the joint and is characterized by cartilage degradation, synovitis, subchondral bone remodeling, and changes in muscular and ligamentous structures. Cartilage degradation is considered the defining characteristic of KOA and has been the focus of research efforts to characterize disease severity and progression [2,3]. Measures of femoral articular cartilage (FAC) thickness are integral to disease characterization, where a decrease in the quantity or quality is interpreted as an increase in disease severity. Semi-quantitative KOA grading scales focus on tibiofemoral joint space narrowing (JSN) as a surrogate marker for FAC loss. Currently, radiographic identification of KOA is based on the semi-quantitative Kellgren-Lawrence grading system, which relies on two radiographic features, joint space width and the presence of osteophytes [4]. This has traditionally been visualized using weight-bearing radiography, where a decrease in tibiofemoral JSN is equated to decrease in FAC volume. Although radiographic JSN is used to represent cartilage loss, it is critical to consider the fact that radiography lacks sensitivity to changes in soft tissue structures such as FAC quantity and quality, especially in the earlier stages of KOA. Furthermore, radiography captures two-dimensional (2D) images of inherently three-dimensional (3D) anatomical structures [5].
Magnetic resonance imaging (MRI) is a well-established imaging modality that overcomes many of the limitations associated with radiographic measurements of JSN in KOA. Conventional MRI allows for the assessment of anatomical changes in KOA, while compositional MRI techniques allow for the investigation of early biochemical compositional changes of the articular cartilages in KOA patients. While MRI is a promising technology, limitations including the general inaccessibility, long image acquisition times, and high manufacturing and operational costs limit its availability in cost-constrained healthcare systems and as a feasible point of care method of classifying KOA.
Currently, 2D musculoskeletal ultrasound (US) is used to assess KOA using semi-quantitative grading scales such as the Outcome Measures in Rheumatology (OMERACT). 2D US is a useful imaging tool for real-time point of care assessment of KOA as it provides high-resolution images rapidly and safely at the patient's bedside. However, 2D US is limited since the quality of the images is highly dependent on transducer placement and operator experience. 2D US also only provides 2D images of 3D anatomical structures and is at the mercy of a small fields-of-view for a large joint such as the knee. Thus, the limitations of radiography, MRI, and 2D US point to a clinical need for a new point of care imaging methods that can overcome some of these limitations.
3D US is an attractive bed-side alternative imaging technique that compensates for the limitations associated with MRI [6]. 3D US techniques have been used for applications in cardiology, gynecology, urology, neonatology, and most recently musculoskeletal (MSK)/rheumatology. Adding 3D US imaging to clinical assessments could provide clinicians with an easily accessible, time-efficient, and cost-effective method for assessing FAC status for longitudinal disease monitoring.
Segmentation of knee cartilage from US images is essential for various clinical tasks in the diagnosis and treatment planning of KOA [7]. Segmentation of the FAC is an important first step in acquiring measurements to quantify joint degeneration. In current clinical practice, tissue segmentation is typically performed manually by a highly trained rater. This process is time-consuming, as segmentations must be completed on each image slice [8]. Segmentations are typically performed after the clinical session, requiring approximately 30 min for the FAC. Efficiency and repeatability vary as inter- and intra-rater reliability is influenced by the user's level of expertise [9].
Deep learning using convolutional neural networks (CNNs) are well suited to solve image-based problems and have been used in an increasing number of musculoskeletal applications, including lesion detection, classification, segmentation, and non-interpretive tasks [10]. CNNs have already shown promising results for segmenting cartilage and bone using several imaging modalities. Schwartz et al. showed that a CNN can identify and classify KOA from radiographs as accurately as a fellowship-trained arthroplasty surgeon [11]. A study by Zhou et al. showed that a CNN trained using MR images was well-suited for performing rapid and accurate comprehensive tissue segmentation of knee joint structures [12]. They demonstrated that most musculoskeletal tissues had a mean value of average symmetric surface distance less than 1 mm. While most musculoskeletal deep learning applications focus on MR and radiographic applications, US segmentation has been explored in recent work. Kompella et al. report on segmentation of FAC from 2D knee US images using a mask R–CNN architecture, trained using 512 2D US slices taken from two 3D US volumes. The best results were achieved with Gaussian filter preprocessing and pretraining with the COCO 2016 image dataset, reporting an average DSC of 80% with a maximum of 88%. A key limitation is the lack of 3D segmentation, with reported results limited only to segmentation in 2D US images.
We hypothesize that the addition of deep learning to 3D US imaging will provide fast and accurate 3D segmentation of the FAC in 3D US images of the knee. We aim to demonstrate that this is possible with a 2D deep learning plus 3D reconstruction approach, the first of its kind for knee 3D US imaging to our knowledge. The application of deep learning-based automatic segmentation in 3D US-based KOA monitoring could greatly reduce segmentation time, potentially allowing for FAC segmentation to be completed during the imaging session ultimately reducing the time for examinations and easing physician burden.
2. Materials and methods
2.1. Clinical dataset
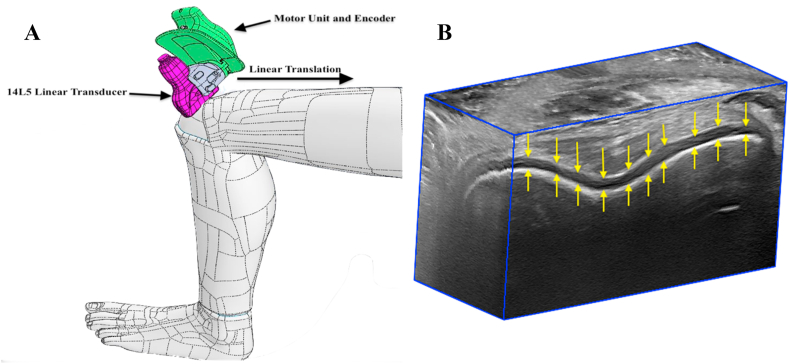
3D US images were acquired of 25 volunteers without prior history of knee joint pathology using a linear mechanical scanning approach (Fig. 1). Images were acquired using an Aplio i800 US machine (Canon Medical Systems Corporations, Otawara, Tochigi, Japan) equipped with a 14L5 linear transducer with an operating frequency of 10 MHz and a footprint length of 58 mm. This 2D transducer was attached to our 3D US scanner via a custom-designed 3D printed mount. The 3D US scanning device consisted of a motorized drive mechanism that linearly translated the transducer over 4.0 cm along the patient's skin, continuously acquiring 120 2D US images at regular spatial intervals (0.33 mm) and reconstructing them using custom software to generate a 3D US image with a size of 968 × 694 x 120 voxels and voxel sizes of 0.058 × 0.058 × 0.33 mm3 [6]. The average 3D US image acquisition time was 15 s per knee. This 3D US scanner was previously validated in a study by Papernick et al. [13], where the knees of 25 healthy volunteers were imaged at the trochlea of the femur, with the subject in a supine position and the knee positioned in maximum flexion. The previously described scanning position allows for visualization of the trochlear cartilage and is accessible with ultrasound. The total dataset consisted of 25 3D US images of the knee. 20 3D US were used for training, while 5 3D US were set aside as a hold-out testing dataset. Manual segmentations of the trochlear femoral articular cartilage (FAC) volumes were completed by two raters to assess inter-rater variability.
Fig. 1.
(A) A schematic of our handheld mechanical three-dimensional ultrasound (3D US) device acquiring a series of images of the suprapatellar region using a linear scanning approach. (B) An example of the resulting 3D US image with the femoral articular cartilage highlighted by arrows.
Additional 3D US images were acquired of four patients with diagnosed KOA using the same scanning approach and US system. These images were selected from a database of previously acquired three-dimensional ultrasound images from various KOA studies and a skilled musculoskeletal sonographer identified features of OA using the OMERACT grading scale. Demographic data for the healthy volunteers and patients is represented in Table 1. The five clinical 3D US images were included in the hold-out testing dataset, with the training dataset including only healthy volunteer images. Manual segmentations were once again completed by two raters.
Table 1.
Demographic information for four knee osteoarthritis patients and five healthy volunteers.
| Patients | Volunteers | |
|---|---|---|
| N | 4 | 5 |
| % Women | 25 | 20 |
| Age [Years] (mean ± SD) | 32.5 ± 17.3 | 33.6 ± 21.5 |
| Height [m] (mean ± SD) | 170.6 ± 9.7 | 174.4 ± 11.8 |
| Weight [kg] (mean ± SD) | 71.525 ± 12.2 | 75.12 ± 15.1 |
| BMI [kg/m^2] (mean ± SD) | 24.7 ± 4.24 | 24.5 ± 3.1 |
2.2. Deep learning training dataset
To utilize the 3D training dataset most efficiently, each image was resliced into 10 2D US planes extracted along the acquisition direction, maximizing image resolution. Only 10 transverse 2D images were resliced per 3D volume due to the similarity in FAC appearance between 2D slices. This resulted in a training dataset of 200 2D US images that was further split into 80% training (160 2D images) and 20% validation (40 2D images). All 2D images were resized to 256 × 256 pixels with no preprocessing. Manual segmentations completed by rater 1 (SP) were used as the gold standard for training the neural network.
2.3. Deep learning architectures and 3D reconstruction
The widely prevalent U-Net architecture and its variants have demonstrated high performance in various medical image segmentation tasks [14,15]. We previously developed a modified U-Net approach applied to prostate segmentation in transrectal 3D US images [16]. This method was implemented using Keras with TensorFlow for FAC segmentation in US images of the knee and made use of the Adam optimizer, 1 × 10−4 learning rate, 100 epochs, and 600 steps per epoch [17,18]. For comparison, a state-of-the-art U-Net++ network was implemented with matched hyperparameters, as it has been shown to improve segmentation performance with small datasets [19]. Our U-Net++ implementation utilized a ResNet-50 network with batch normalization and a batch size of 10 as a backbone [20]. To expand the training dataset, data augmentation including random combinations of horizontal and vertical flips, shifts up to 20%, rotations up to 20%, and zooms up to 20% were applied. Neural network training and inferencing were completed on a personal computer with an i7-9700K central processing unit (CPU) at 3.60 GHz (Intel Corporation, Santa Clara, CA, USA), 64 GB of RAM, and a 24 GB NVIDIA TITAN RTX (NVIDIA Corporation, Santa Clara, CA, USA) graphics processing units (GPU). After inferencing the transverse 2D US images, the resulting 2D predictions were placed back in their position within the 3D volume where the boundary points between adjacent slices were connected. A smoothing filter was applied, and the ends of the volume were closed resulting in the complete 3D surface.
2.4. Testing dataset, evaluation metrics, and comparisons
A hold-out testing dataset, consisting of 5 3D US images of healthy knees and 5 3D US images of knees with diagnosed KOA, was used to evaluate the FAC segmentation performance of our method and was resliced as described for the training dataset. Using the ATS 538NH Beam Profile & Slice Thickness Phantom (CIRS Inc, Norfolk, VA, USA) the spatial elevational resolution of the 14L5 transducer was found to range from 2.35 mm at 2 cm depth to 4.04 mm at 4 cm depth. To account for the maximum elevational resolution at a depth of 4 cm, 2D transverse slices were extracted every 4 mm, corresponding to 10 slices per full 3D volume. This resulted in a total testing dataset of 50 transverse 2D US images of healthy volunteers and 50 2D US images of KOA patients. The characterization of our method's performance made use of both the 2D segmentations and the reconstructed 3D surfaces, including computation of standard pixel map metrics (DSC, recall, and precision). These metrics are calculated using the confusion matrix values defined as true positive (overlapping cartilage surface/volume between gold standard and algorithm), false positive (surface/volume identified as cartilage in the algorithm but not in the gold standard), true negative (surface/volume identified as not cartilage in both the gold standard and algorithm), and false negative (surface/volume identified as cartilage in the gold standard that was missed by the algorithm). In addition, absolute and signed variants of area/volume percent difference (A/VPD) and boundary distance metrics including mean surface distance (MSD), and Hausdorff distance (HD) were computed. All evaluation metrics aside from the 3D boundary distance metrics were computed using custom software in MATLAB R2019a (MathWorks, Natick, MA, United States). 3D MSD and HD were computed using the open-source program CloudCompare (v2.10.2, Girardeau-Montaut). 2D slice segmentation time, reconstruction time, and overall 3D segmentation time for our algorithm were recorded.
Automatically generated 2D FAC segmentations and corresponding 3D reconstructions were compared to the manual segmentations generated by rater 1 (SP). In addition, reconstructed 3D FAC surfaces generated by our algorithm were compared to manual segmentations generated by a different rater (RD). All comparisons utilized the evaluation metrics described above.
2.5. Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, Inc., San Diego, CA, USA). Distribution normality was assessed using the Shapiro-Wilk test. The significance level was chosen such that the probability of making a type I error was less than 5% (p < 0.05). Comparisons between 2D transverse slice segmentation and 3D reconstructed segmentation accuracy as well as between the U-Net and U-Net++ architectures were completed using two-tailed paired t-tests, or Wilcoxon matched-pairs signed-rank tests if normality assumptions were violated. FAC segmentation performance compared to rater 1 and rater 2 manual segmentations were subsequently compared using two-tailed paired t-tests or Wilcoxon matched-pairs signed-rank tests.
3. Results
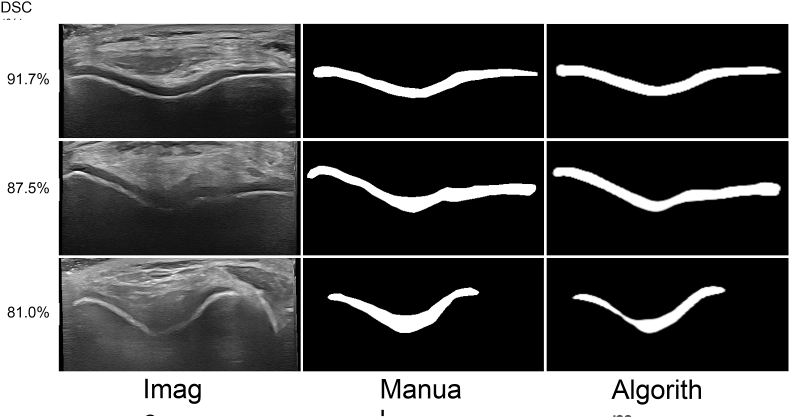
Fig. 2 depicts 2D US segmentation results at various performance levels. A comparison between manual segmentations and segmentations completed using our method are shown in Fig. 3.
Fig. 2.
Example femoral articular cartilage segmentation results using our modified U-Net in healthy volunteer knee ultrasound images. The original two-dimensional ultrasound images, manual segmentations, and algorithm segmentations are shown in the columns from left to right. Each row shows a different performance level based on Dice similarity coefficient (DSC), including mean plus one standard deviation (top row), mean (middle row), and mean minus one standard deviation (bottom row).
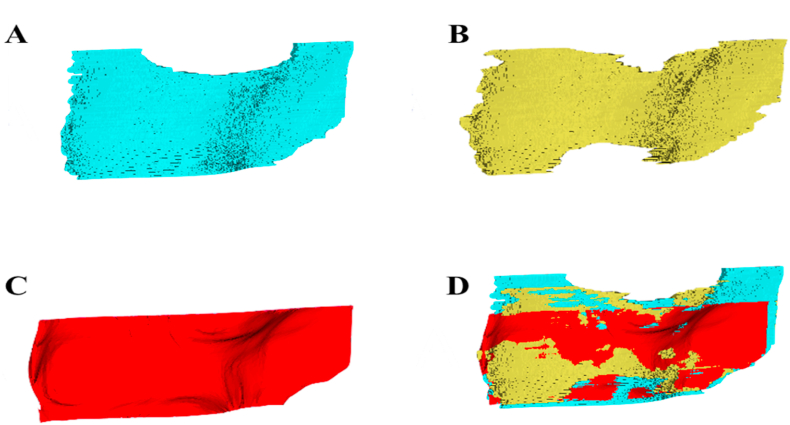
Fig. 3.
Manual segmentations by rater 1 (A) and rater 2 (B) for a healthy volunteer knee US image. Resultant three-dimensional (3D) reconstructed surface generated by our algorithm after two-dimensional predictions using the modified U-Net (C). The manual and algorithm segmentations registered and overlaid in 3D Slicer (D).
Table 2, Table 3 show results on healthy volunteer images for our 2D transverse slice segmentations and the 3D surface reconstructions for the absolute and signed metrics, respectively. For the U-Net, recall was the only metric that showed statistically significant differences when comparing 2D predictions to the corresponding 3D reconstructed segmentations. In contrast, the U-Net++ showed a significant reduction in performance for the precision, MSD, and a/VPD metrics between the 2D segmentations and the 3D reconstructed surface. Overall, the results indicated that there was a decrease in performance after 3D reconstruction, with decreases in recall, precision, and DSC. Interestingly, A/VPD (%) and sHD (mm) were lower for the 3D reconstructions than the 2D segmentations when using the U-Net but were not found to be statistically significant. The U-Net outperformed the U-Net ++ on all the tested metrics, with the exception of Hausdorff distance. When comparing the resultant volume measurements to the manually segmented volumes, the U-Net showed a larger percent difference for the 2D predictions than observed with the U-Net++. The opposite was true for the 3D reconstructed results, where the U-Net showed a smaller percent difference than the U-Net++. The Computation time was 0.029 s (U-Net) and 0.088 s (U-Net++) for each 2D segmentation, with 10 2D US slices per 3D US volume. Reconstruction time was 0.27 s for a total mean 3D segmentation time of 0.56 s (U-Net) and 1.15 s (U-Net++).
Table 2.
Mean ± standard deviation results comparing two-dimensional (2D) U-Net and U-Net ++ healthy participant segmentations on transverse ultrasound images to subsequent three-dimensional (3D) reconstructed segmentations. The p-value corresponds to a comparison between 2D and 3D segmentation performance.
| Method | Segmentation | Precision (%) | Recall (%) | DSC (%) | MSD (mm) | HD (mm) | A/VPD (%) |
|---|---|---|---|---|---|---|---|
| U-Net | 2D Transverse | 83.1 ± 4.6 | 91.5 ± 5.4 | 87.1 ± 4.6 | 0.22 ± 0.10 | 1.06 ± 0.46 | 10.1 ± 4.7 |
| 3D Reconstruction | 74.8 ± 6.1 | 72.0 ± 6.3 | 73.1 ± 3.9 | 0.30 ± 0.06 | 1.55 ± 0.67 | 1.55 ± 0.67 | |
| P-value | 0.062 | 0.015 | 0.063 | 0.256 | 0.256 | 0.946 | |
| U-Net++ | 2D Transverse | 82.4 ± 4.9 | 85.6 ± 3.6 | 83.8 ± 2.9 | 0.33 ± 0.10 | 1.04 ± 0.16 | 6.73 ± 3.9 |
| 3D Reconstruction | 75.0 ± 4.8 | 74.5 ± 3.7 | 74.3 ± 1.7 | 0.31 ± 0.06 | 1.76 ± 0.99 | 11.3 ± 4.8 | |
| P-value | 0.014 | 0.458 | 0.064 | 0.032 | 0.159 | 0.012 |
Table 3.
Signed mean ± standard deviation results for healthy knee participants comparing U-Net and U-Net++ two-dimensional (2D) deep-learning segmentations on transverse ultrasound images to subsequent three-dimensional (3D) reconstructed segmentations. The p-value corresponds to a comparison between 2D and 3D segmentation performance.
| Method | Segmentation | sA/VPD (%) | sMSD (mm) | sHD (mm) |
|---|---|---|---|---|
| U-Net | 2D Transverse | 10.1 ± 4.71 | 0.10 ± 0.04 | 1.06 ± 0.46 |
| 3D Reconstruction | −3.01 ± 13.1 | 0.19 ± 0.07 | 0.59 ± 1.73 | |
| P-value | 0.8371 | 0.0566 | 0.5829 | |
| U-Net++ | 2D Transverse | 3.92 ± 7.79 | 0.08 ± 0.13 | 0.56 ± 1.18 |
| 3D Reconstruction | 28.2 ± 8.09 | 0.27 ± 0.07 | 1.56 ± 0.43 | |
| P-value | 0.021 | 0.002 | 0.007 |
Table 4, Table 5 show results on knee osteoarthritis patient images for our 2D transverse slice segmentations and the 3D surface reconstructions for the absolute and signed metrics, respectively. For the U-Net++, performance on all metrics decreased for 3D reconstructed surfaces compared to 2D segmentations, while for the U-Net, all metrics apart from recall and HD decreased in performance, similar to results seen in healthy volunteers.
Table 4.
Mean ± standard deviation results comparing two-dimensional (2D) U-Net and U-Net++ segmentations on knee osteoarthritis patient 2D transverse ultrasound images and subsequent three-dimensional (3D) reconstructed segmentations to the corresponding manual gold standard segmentations. The p-value corresponds to a comparison between 2D and 3D segmentation performance.
| Method | Segmentation | Precision (%) | Recall (%) | DSC (%) | MSD (mm) | HD (mm) | A/VPD (%) |
|---|---|---|---|---|---|---|---|
| U-Net | 2D Transverse | 83.9 ± 2.6 | 94.7 ± 1.7 | 88.9 ± 1.6 | 0.23 ± 0.03 | 1.28 ± 0.35 | 13.4 ± 9.4 |
| 3D Reconstruction | 73.2 ± 8.6 | 94.3 ± 1.3 | 82.3 ± 6.1 | 0.44 ± 0.13 | 1.91 ± 0.57 | 30.1 ± 15.6 | |
| P-value | 0.001 | 0.598 | 0.007 | <0.001 | 0.099 | 0.014 | |
| U-Net++ | 2D Transverse | 84.5 ± 5.8 | 93.0 ± 1.0 | 88.4 ± 3.5 | 0.24 ± 0.03 | 1.18 ± 0.43 | 10.4 ± 7.6 |
| 3D Reconstruction | 72.7 ± 8.0 | 95.0 ± 1.1 | 82.2 ± 5.7 | 0.43 ± 0.11 | 2.02 ± 0.64 | 31.4 ± 13.9 | |
| P-value | 0.002 | 0.005 | 0.011 | 0.037 | 0.038 | 0.007 |
Table 5.
Signed mean ± standard deviation results for knee osteoarthritis patients comparing U-Net and U-Net++ two-dimensional (2D) deep-learning segmentations on transverse ultrasound images to subsequent three-dimensional (3D) reconstructed segmentations. The p-value corresponds to a comparison between 2D and 3D segmentation performance.
| Method | Segmentation | sA/VPD (%) | sMSD (mm) | sHD (mm) |
|---|---|---|---|---|
| U-Net | 2D Transverse | 13.4 ± 9.41 | 0.14 ± 0.05 | 1.14 ± 0.45 |
| 3D Reconstruction | 30.1 ± 15.6 | 0.33 ± 0.09 | 1.91 ± 0.57 | |
| P-value | 0.014 | 0.025 | 0.038 | |
| U-Net++ | 2D Transverse | 10.4 ± 7.58 | 0.11 ± 0.04 | 0.69 ± 1.01 |
| 3D Reconstruction | 31.5 ± 13.9 | 0.34 ± 0.07 | 2.02 ± 0.64 | |
| P-value | 0.025 | 0.007 | 0.049 |
Table 6 shows the results of our 3D reconstructions compared to manual segmentation for two different raters for healthy volunteer images. The overall mean volume was found to be 2103.1 ± 368.2 mm3 for the manual segmentations by rater 1 and 2033.4 ± 487.4 mm3 for the algorithmic 3D reconstructions. Rater 1 demonstrated a 10.4 ± 6.0 mm3 and 9.54 ± 10.4 mm3 mean volume percent difference between the manual 3D segmentations and the algorithmic 3D reconstructions for the U-Net and U-Net++, respectively, and rater 2 demonstrated a volume percent difference of 14.2 ± 11.4 mm3 and 11.3 ± 4.8 mm3, respectively. Paired sample t-tests indicated that none of the differences observed were statistically significant.
Table 6.
Mean ± standard deviation results comparing algorithmic three-dimensional (3D) reconstructed femoral articular cartilage surfaces to manual 3D segmentations generated by rater 1 and rater 2 for healthy volunteer knee images.
| Segmentation | Recall (%) | Precision (%) | DSC (%) | MSD (mm) | HD (mm) | A/VPD (%) | |
|---|---|---|---|---|---|---|---|
| Rater 1 | U-Net | 72.0 ± 6.3 | 74.8 ± 6.1 | 73.1 ± 3.9 | 0.29 ± 0.05 | 1.55 ± 0.67 | 10.4 ± 6.0 |
| U-Net++ | 70.5 ± 3.1 | 74.5 ± 2.7 | 72.3 ± 1.6 | 0.30 ± 0.03 | 1.29 ± 0.27 | 9.54 ± 10.4 | |
| Rater 2 | U-Net | 71.4 ± 5.7 | 74.3 ± 8.1 | 72.3 ± 2.8 | 0.32 ± 0.15 | 2.69 ± 2.92 | 14.2 ± 11.0 |
| U-Net++ | 74.5 ± 3.7 | 75.0 ± 4.8 | 74.4 ± 1.7 | 0.31 ± 0.06 | 1.76 ± 0.99 | 11.3 ± 4.8 |
4. Discussion
While 3D US provides a cost-effective and accurate method for assessing FAC status, time-consuming and difficult manual segmentation of the FAC is required for each image. To ease the physician's burden and reduce segmentation time, we have developed a 2D deep learning algorithm with a 3D reconstruction approach allowing for fast and accurate 3D segmentation of the FAC in 3D US images of the knee.
Outputs shown in Fig. 3 highlight the impact of image artifacts such as shadowing on segmentation performance. In the top row, a high-performance case, the FAC boundary is clearly defined, with no shadowing present. As performance decreases, as shown in the middle and bottom row, shadowing artifacts are more prevalent, which obstruct FAC boundary visibility. Our trained U-Net is robust to some amount of shadowing, as shown in the middle row where the algorithm accurately segmented the FAC even in the presence of shadowing artifacts. In the bottom row, a lower-performance case, the extensive shadowing resulted in a thin algorithmic segmentation that does not agree as closely with the manual gold standard. It is important to note that even in this poor-quality image, the algorithm does not confuse nearby anatomy with the cartilage, demonstrated by the lack of characteristic “islands” in the segmentation.
We first compared the segmentation accuracy in 2D slices to the reconstructed 3D surfaces. As shown in Table 2, the mean segmentation performance on the 2D US slices was higher when compared to the 3D reconstructed segmentations for both networks. While the agreement between the manual 3D surface and our algorithmic 3D reconstruction is excellent in most areas of the FAC, these results also highlight a limitation of our 3D reconstruction approach. As seen in Fig. 3, the reconstruction method produces a rectangular surface, cutting off three regions that can be seen in the manual segmentations (Fig. 3). In addition, when combining 2D predictions during the reconstruction process, a smoothing filter was applied which may result in the loss of fine details from each 2D prediction. These two considerations resulted in reduced performance for metrics like recall, precision, and DSC. This smoothing process proved a benefit for the signed and absolute boundary distance metrics (MSD and HD) and A/VPD, as outliers were removed, resulting in similar evaluation metric values between the 2D and 3D segmentations. The mean recall metric showed the largest difference at almost 20% for the U-Net results. This greatly reduced mean recall for the 3D reconstructed surfaces is the result of increased false negatives or underprediction. Performance for the U-Net and U-Net++ in both the 2D segmentations and 3D reconstructed segmentations were very similar, demonstrating no preferred network when segmenting healthy knee images.
Assessing the performance of our segmentation algorithm on KOA patient images (Table 4, Table 5) demonstrated higher mean performance compared to healthy knee images in key metrics such as DSC, precision, and recall. Performance between the U-Net and U-Net++ was approximately the same, aligning with the results seen for healthy images. In contrast to the healthy knee images, the difference between the 2D and 3D results were also smaller for the KOA patient images. These observed differences may be due to several reasons. First, we have a small sample size of four KOA patient 3D US images, so high image quality or cartilage boundary visibility in these images compared to the healthy knee images may have lead to higher segmentation performance. In addition, the same US scanner and operator were used to acquire the patient images, so the algorithm was trained with the same image and voxel size. These results demonstrate the ability of our algorithm to accurately segment the cartilage in clinical US images of patients with KOA, providing the potential for efficient monitoring of KOA.
Table 6 shows a comparison of segmentation performance when two different raters provided gold-standard manual segmentations. Rater 1 provided manual segmentations for the training dataset, while the network had not seen any manual segmentation from rater 2 during training. Manual segmentations were similar between raters with a percent difference in mean volume of only 3.4%. Automated segmentation performance was also very similar regardless of rater, with a paired t-test showing no statistically significant difference. Difference in mean VPD comparing the algorithmic segmentation to raters 1 and 2 was 3.8% for the U-Net and 1.8% for the U-Net++, nearly matching the percent difference in mean volume between raters (3.4%). This highlights the robustness of our algorithm to the manual rater. The largest difference in mean was observed for the HD metric, likely due to the presence of outliers based on slight differences in segmentation technique between raters. As reported in Papernick et al. [13], manual segmentation of the FAC surface in 3D US images takes approximately 20–30 min per knee. With a total segmentation time of only 0.56 s for the U-Net or 1.15 s for the U-Net++, our deep learning algorithm offers a vast improvement, potentially allowing for FAC segmentation during the initial imaging exam which could speed up diagnosis time.
Recent work by Kompella et al. [7] reported on segmentation of FAC from 2D knee US images using a mask R–CNN architecture. A 2D mask-R-CNN was trained using 512 2D US slices taken from two 3D US volumes, with a testing dataset of 55 2D US images, similar in size to our training and testing dataset. Best results were achieved with Gaussian filter preprocessing and pretraining with the COCO 2016 image dataset, reporting a mean DSC of 80% with a maximum of 88%. Comparatively, our modified U-Net approach with no preprocessing and no pretraining demonstrated mean 2D DSC scores of 87.1% with a maximum of 93.5%, approximately 7% higher. In their study three-dimensional ultrasound images were acquired using a 4D ultrasound probe (13-5 MHz frequency) whereas our 3D images were acquired using a 14L5 linear transducer and then interpolated to create 3D reconstructions. Differences in image resolution may be a contributing factor to the variation observed in the DSC scores. Our patients were also scanned at 90° of flexion, in comparison to Kompella et al. [7] who scanned participants at 30° of knee flexion. Increased flexion of the knee opens the joint further and allows for better access to the cartilage. Critically, our 3D reconstruction method allowed for complete 3D segmentation of the FAC in 3D US images where Kompella et al. [7] were limited to 2D segmentations. This improved segmentation performance with a similar dataset size and ability to complete 3D segmentations demonstrates the advantage of our method.
While we have demonstrated promising performance with our automatic FAC segmentation method, we address several limitations. Although our testing dataset included manual segmentations from two different raters, the training dataset used gold standard manual segmentations from only one rater. While we could assess the robustness of our method to different raters, we could not assess the impact of inter- or intra-rater variability during training. Future work will explore the inclusion of multiple raters in the training dataset, which may improve algorithm robustness. In addition, the training dataset of 20 3D US images, resliced into 200 2D US images, is small in the context of deep learning applications. More importantly, all images were generated using the same US probe and system, with matching image and voxel size. This lack of diversity may limit the generalizability of our approach. To address this limitation, future work will utilize a larger training dataset, including data from multiple US machines, scanning protocols, and centers. This will improve algorithm robustness, ultimately improving the clinical translation potential. Our dataset of clinical images was small, so future work will look to expand this dataset, including augmenting the training dataset with clinical images. Results acquired from an expanded KOA patient study would enable us to determine how the pathological changes in cartilage tissue impact the accuracy and precision of our approach as well as improving the robustness and generalizability of our algorithm. Another limitation is our 3D reconstructions were all completed using 10 transverse 2D US slices. While this number was chosen due to constraints from the transducer elevational resolution and is suitable for the largely homogenous images in our healthy volunteer population, future work could explore the utilization of more 2D planes. This may be particularly important for KOA patient images, as pathology and tissue irregularities may be smaller than our 4 mm spacing, necessitating additional 2D slices with smaller spacing. Due to our low per-slice computation time, this change would result in minimal addition to the overall 3D segmentation time. Additionally, our current imaging technique did not provide a method to allow for imaging the same region of the knee in a follow-up procedure in a longitudinal study and did not provide a method for standardizing the region of the knee to be imaged for a cross-sectional study. To address these limitations, our future work will include a modification of the system, which will allow us to replicate exact knee flexion and position for subsequent follow-up examinations. In addition, we will also develop an image registration method to overlay the 3D US images of the anterior femoral cartilage to ensure that the same region of cartilage is quantified.
The greatest advantage of integrating deep learning technology into our 3D US system's image processing workflow is the ability to reconstruct patient cartilage volumes rapidly, accurately, and precisely. The addition of our proposed algorithm allows the volume reconstruction process to be conducted at a patient's bedside without requiring additional post-image processing or extensive additional training for clinicians. Using our algorithm decreases the segmentation processing time substantially from 20 to 30 min for manual segmentation, to approximately 0.56 s per knee cartilage [13]. This not only gives clinicians a safe, cost-effective, and non-invasive method for assessing FAC status but also provides a time-efficient method for evaluating and comparing volumes longitudinally. The importance of this new addition to our system is the potential impact it may have on the workflow of orthopedic, arthritis, and primary care clinics. It would allow clinicians to obtain 3D US images without the added complexity of having to manually process image volumes and surfaces or adding additional discomfort to patients. Its integration would relieve many of the limitations associated with current clinical methods for assessing cartilage health such as qualitative grading scales used in 2D US. The Kellgren-Lawrence grading scale is used to assess the progression of KOA via 2D US; however, it is insensitive to change and relies on indirect characteristics of cartilage thinning [21]. MRI-based volume measurement methods are far superior to radiography, but they are severely limited by cost, accessibility, and patient-related factors such as movement. This prevents the generalized use of MRI for quantitative assessment and monitoring of KOA.
5. Conclusions
This study investigated the development and validation of an automatic FAC segmentation approach for 3D US images of the knee in both healthy volunteers and KOA patients, utilizing a 2D deep learning architecture with 3D reconstruction. A fast and accurate FAC segmentation method would drastically reduce segmentation time, expediting clinical diagnosis times and potentially allowing for efficient longitudinal monitoring of KOA with 3D US.
Studies involving humans or animals
Clinical trials or other experimentation on humans must be in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Randomized controlled trials should follow the Consolidated Standards of Reporting Trials (CONSORT) guidelines and be registered in a public trials registry.
Funding
The authors are grateful for funding support from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (NSERC). C. Du Toit was supported in part by a Transdisciplinary Training Award from the Bone and Joint Institute at Western University, Canada. N. Orlando was supported in part by the Queen Elizabeth II Graduate Scholarship in Science and Technology.
Author contribution
All authors have made substantial contributions to all of the following: (1) the conception and design of the study, or acquisition of data, or analysis and interpretation of data, (2) drafting the article or revising it critically for important intellectual content, (3) final approval of the version to be submitted.
Declaration of competing interest
The authors have no relevant conflicts of interest to disclose.
Acknowledgments
The authors would like to thank all the volunteers who made this study possible. In addition, the authors would like to acknowledge Dr. David Tessier for volunteer recruitment and coordination and Dr. Tom Appleton for providing clinical guidance and insight.
Contributor Information
Carla du Toit, Email: cdutoit@uwo.ca.
Nathan Orlando, Email: norlando2@uwo.ca.
References
- 1.Cui A., Li H., Wang D., Zhong J., Chen Y., Lu H. Global, regional prevalence, incidence and risk factors of knee osteoarthritis in population-based studies. Clin. Med. 2020;29–30:100587. doi: 10.1016/J.ECLINM.2020.100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poole A.R. Osteoarthritis as a whole joint disease. HSS J. 2012;8(1):4–6. doi: 10.1007/s11420-011-9248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felson D.T. Osteoarthritis of the knee. N. Engl. J. Med. 2006;354(8):841–848. doi: 10.1056/NEJMCP051726. [DOI] [PubMed] [Google Scholar]
- 4.Emrani P.S., Katz J.N., Kessler C.L., et al. Joint space narrowing and Kellgren–Lawrence progression in knee osteoarthritis: an analytic literature synthesis. Osteoarthr. Cartil. 2008;16(8):873–882. doi: 10.1016/J.JOCA.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guermazi A., Roemer F.W., Burstein D., Hayashi D. Why radiography should no longer be considered a surrogate outcome measure for longitudinal assessment of cartilage in knee osteoarthritis. Arthritis Res. Ther. 2011;13(6):1–11. doi: 10.1186/AR3488/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenster A., Downey D.B., Cardinal H.N. Three-dimensional ultrasound imaging. Phys. Med. Biol. 2001;46(5) doi: 10.1088/0031-9155/46/5/201. [DOI] [PubMed] [Google Scholar]
- 7.Kompella G., Antico M., Sasazawa F., et al. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBS. 2019. Segmentation of femoral cartilage from knee ultrasound images using mask R-CNN; pp. 966–969. [DOI] [PubMed] [Google Scholar]
- 8.Pham D.L., Xu C., Prince J.L. Current methods in medical image Segmentation1. Annu. Rev. Biomed. Eng. 2003;2(2000):315–337. doi: 10.1146/ANNUREV.BIOENG.2.1.315. [DOI] [PubMed] [Google Scholar]
- 9.McWalter E.J., Wirth W., Siebert M., et al. Use of novel interactive input devices for segmentation of articular cartilage from magnetic resonance images. Osteoarthr. Cartil. 2005;13(1):48–53. doi: 10.1016/J.JOCA.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Chea P., Mandell J.C. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol. 2020;49(2):183–197. doi: 10.1007/S00256-019-03284-Z/TABLES/5. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz A.J., Clarke H.D., Spangehl M.J., Bingham J.S., Etzioni D.A., Neville M.R. Can a convolutional neural network classify knee osteoarthritis on plain radiographs as accurately as fellowship-trained knee arthroplasty surgeons? J. Arthroplasty. 2020;35(9):2423–2428. doi: 10.1016/J.ARTH.2020.04.059. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z., Zhao G., Kijowski R., Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn. Reson. Med. 2018;80(6):2759–2770. doi: 10.1002/MRM.27229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papernick S., Dima R., Gillies D.J., Appleton C.T., Fenster A. Reliability and concurrent validity of three-dimensional ultrasound for quantifying knee cartilage volume. Osteoarthr. Cartil. Open. 2020;2(4) doi: 10.1016/J.OCARTO.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronneberger O., Fischer P., Brox T., U-Net Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformat.) 2015;9351:234–241. doi: 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 15.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. 3D U-net: learning dense volumetric segmentation from sparse annotation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformat.) 2016;9901:424–432. doi: 10.48550/arxiv.1606.06650. [DOI] [Google Scholar]
- 16.Orlando N., Gillies D.J., Gyacskov I., Romagnoli C., D'Souza D., Fenster A. Automatic prostate segmentation using deep learning on clinically diverse 3D transrectal ultrasound images. Med. Phys. 2020;47(6):2413–2426. doi: 10.1002/MP.14134. [DOI] [PubMed] [Google Scholar]
- 17.Chollet F., Keras 2015. https://github.com/fchollet/keras
- 18.Abadi M., Agarwal A., Barham P., et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv. 2016 doi: 10.48550/arxiv.1603.04467. [DOI] [Google Scholar]
- 19.Zhou Z., Rahman Siddiquee M.M., Tajbakhsh N., Liang J. UNet++: a nested U-net architecture for medical image segmentation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformat.) 2018;11045:3–11. doi: 10.48550/arxiv.1807.10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn. 2015:770–778. doi: 10.48550/arxiv.1512.03385. [DOI] [Google Scholar]
- 21.Kohn M.D., Sassoon A.A., Fernando N.D. Classifications in brief: kellgren-lawrence classification of osteoarthriti. Clin. Orthop. Relat. Res. 2016;474(8):1886–1893. doi: 10.1007/S11999-016-4732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]