Abstract
Lung adenocarcinoma (LUAD) is one of the most common cancer types and has various treatment options. Better biomarkers to predict therapeutic response are needed to guide choice of treatment modality and improve precision medicine. Here we utilized a consensus hierarchical clustering approach on 509 LUAD cases from The Cancer Genome Atlas (TCGA) to identify five robust LUAD expression subtypes. Genomic and proteomic data from patient samples and cell lines was then integrated to help define biomarkers of response to targeted therapies and immunotherapies. This approach defined subtypes with unique proteogenomic and dependency profiles. Subtype 4 (S4)-associated cell lines exhibited specific vulnerability to loss of CDK6 and CDK6-cyclin D3 complex gene (CCND3). S3 was characterized by dependency on CDK4, immune-related expression patterns, and altered MET signaling. Experimental validation showed that S3-associated cell lines responded to MET inhibitors, leading to increased expression of PD-L1. In an independent real-world patient dataset, patients with S3 tumors were enriched with responders to immune checkpoint blockade (ICB). Genomic features in S3 and S4 were further identified as biomarkers for enabling clinical diagnosis of these subtypes. Overall, our consensus hierarchical clustering approach identified robust tumor expression subtypes, and our subsequent integrative analysis of genomics, proteomics, and CRISPR screening data revealed subtype-specific biology and vulnerabilities. These lung adenocarcinoma expression subtypes and their biomarkers could help identify patients likely to respond to CDK4/6, MET, or PD-L1 inhibitors, potentially improving patient outcome.
Keywords: Lung adenocarcinoma, Multiomics, Molecular subtypes, Biomarkers, Precision medicine
Introduction
Lung cancer is the most prevalent cause of death from cancer worldwide (1). The two major histological classes of lung cancer are: (i) non-small-cell lung cancer (NSCLC) and (ii) small-cell lung cancer (SCLC). NSCLC is the most common histological type and is further divided into two major subtypes: lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LSCC, previously termed “LUSC”). Previous studies classified LUAD into molecular subtypes based on genomic (2, 3, 4) and proteogenomic (5) profiling of tumors and then associated these subtypes with clinical outcomes. Two of the largest published studies on LUAD subtypes are: (i) the original The Cancer Genome Atlas (TCGA) LUAD subtyping paper published in 2014 that used 230 patients (the largest number of patients available at the time) to identify three subtypes based on mRNA expression –– Proximal Inflammatory (PI), Proximal Proliferative (PP), and Terminal Respiratory Unit (TRU) (2); and (ii) the TCGA Pan-Lung study in 2017 that analyzed both the LSCC and LUAD cohort and identified 8 subtypes, 6 of which are enriched with LUAD tumors (4). To date, the TCGA LUAD cohort has increased to a total of 509 LUAD cases, offering increased power to identify higher resolution subtypes.
Robust LUAD subtyping can substantially aid in determining the most effective therapies that target subtype-specific vulnerabilities. Thus far, molecular therapies for LUAD have focused on targeting various genomic alterations, such as the RAS/RAF/RTK pathway. These include therapies targeting EGFR, ALK, and ROS1 alterations, as well as the more recently approved therapies such as those, targeting MET, RET, NTRK1/2, BRAF kinases and KRASG12C mutations (6, 7). Moreover, additional therapies are currently still under development, such as ERBB2 inhibitors (8). Recently, immune checkpoint blockades (ICB) have been approved to treat lung cancer, including inhibitors for PD-1 (pembrolizumab and nivolumab) and PD-L1 (atezolizumab and durvalumab). Previously reported biomarkers of response or resistance to immunotherapy in LUAD include PD-L1 expression (9, 10), tumor mutational burden (TMB) (11, 12, 13), mismatch repair deficiency/microsatellite instability (14), and STK11 mutation (15). However, most LUAD tumors continue to progress on therapy, underscoring the need for novel therapeutic approaches. Therefore, more precise and robust subtyping of LUAD tumors and their association with specific treatments can help improve patient prognosis and outcome.
In this study, we integrated multiple data sets: (i) the full 509 LUAD patient cohort in TCGA; (ii) vulnerability data in LUAD cell lines from the Cancer Cell Line Encyclopedia (CCLE) (16, 17) and the Dependency Map (DepMap) (18) repositories; (iii) an independent cohort from the Samsung medical center (SMC) including 164 patients with response to ICB therapy; and (iv) proteomic data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) cohort of 110 LUAD patients (5) to more precisely define therapeutically relevant LUAD subtypes (Figure 1A). We show that our analysis indeed yielded distinct subtypes compared with the previously published expression-based subtypes, with higher-resolution partitioning of previously defined subtypes. Moreover, our experimental work in vitro highlights potential subtype-specific therapeutic targets, and we identify a small number of biomarkers that could be used in the clinic to classify patients into our most clinically relevant subtypes, which could help guide clinical decision making.
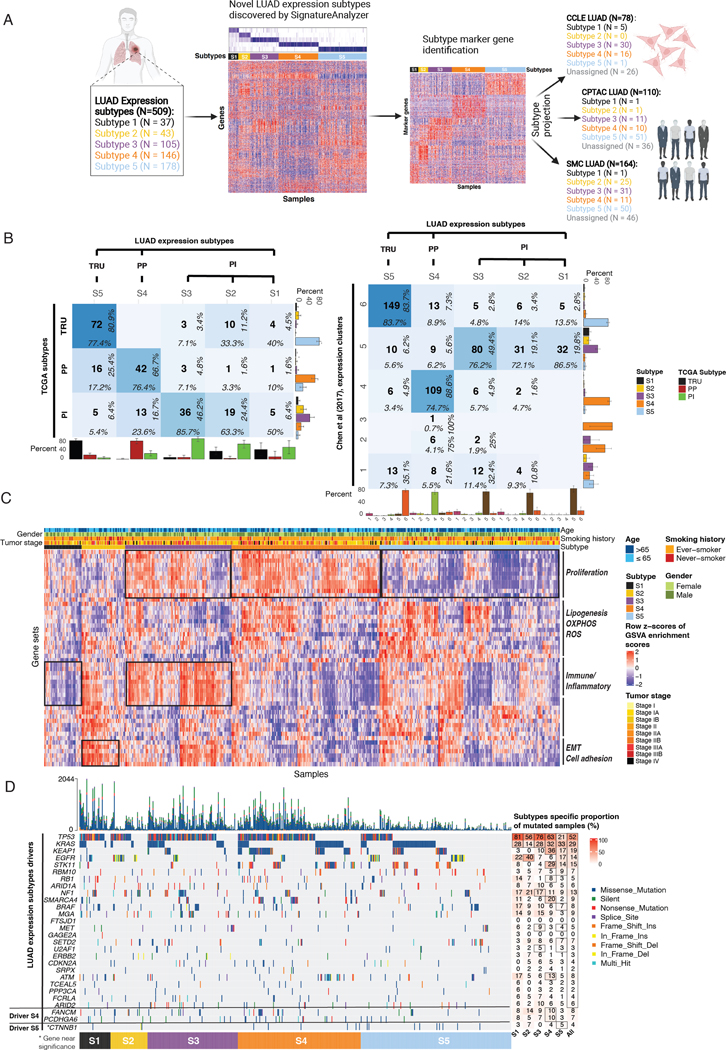
Figure 1. Study design and LUAD expression subtypes landscape.
(A) Study overview: 509 TCGA LUAD samples classified into 5 expression subtypes by SignatureAnalyzer. Normalized H matrix (left heatmap) mRNA expression of 100 subtype marker genes for each subtype (right heatmap). LUAD subtypes projection onto 3 datasets: CCLE LUAD (n=78), CPTAC LUAD (n=110), and SMC LUAD (n=164).
(B) Confusion matrix showing concordance between new and previously defined LUAD expression subtypes. Sample overlap (center number), column-wise proportion (bottom), row-wise proportion (right). Bottom bar plots show column-wise proportion, right bar plots show row-wise proportion.
(C) Heatmap represents GSVA pathway activation profiles in LUAD expression subtypes. Top tracks reflect age, gender, smoking history, and tumor stage.
(D) Co-mutation plot represents subtype-specific LUAD driver genes identified by MutSig2CV (point mutations, indels; Q value < 0.01) and their associated proportion. Black box denotes significantly mutated genes with Q < 0.01.
Methods
Data availability
For this study, we used publicly available data from multiple resources: (i) human tumor tissue data (from the (TCGA cohort, https://portal.gdc.cancer.gov/), (ii) proteomic data from CPTAC (https://cptac-data-portal.georgetown.edU/cptac/s/S056), and (iii) cell line data (via the DependencyMap [https://depmap.org/portal/]). In addition, we used a LUAD immunotherapy cohort data from Korean patients at Samsung Medical Center (SMC) for validation. All raw and processed sequencing data generated in this study have been submitted to the European Nucleotide Archive (EGA; https://ega-archive.org/) under accession number EGAS00001006461. Analysis scripts used in this study are available at https://github.com/getzlab/LUAD_subtypes.
Ethics approval and consent to participate
All datasets analyzed are publicly available. Ethics approval and consent were obtained in the original papers as required. The Korean LUAD immunotherapy cohort study was approved by the Samsung Medical Center Institutional Review Board (SMC 2018–03-130 and SMC 2013–10-112), and informed written consent was obtained from all patients enrolled in the study.
TCGA LUAD data
In this study, we used the full LUAD TCGA dataset including 509 samples. Batch-corrected upper quartile normalized RSEM (RNA-Seq by Expectation-Maximization) data for TCGA LUAD cohort (level 3) was obtained from the PanCanAtlas study (19). TCGA LUAD somatic mutation call data (level 2) was obtained from the MC3 (“Multi-Center Mutation Calling in Multiple Cancers”) data repository (https://gdc.cancer.gov/about-data/publications/mc3-2017). TCGA LUAD copy number segmentation file (level 3) (LUAD.snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.seg.txt) was obtained from Firehose (doi:10.7908/C11G0KM9). Survival data for TCGA LUAD samples was obtained from the integrated TCGA pan-cancer clinical data (20).
CCLE LUAD cell line data
The ‘omics data and CRISPR knockout data for CCLE LUAD cell line samples were obtained from the Dependency Map (DepMap) portal (https://depmap.org/portal/; DepMap Public 21Q2 dataset) (18). Out of the 1379 cell lines available, we used 78 LUAD cell lines.
CPTAC LUAD data
Data used in this publication were generated by the National Cancer Institute CPTAC. Genomics and proteomics data for CPTAC LUAD samples were obtained from the previous study (5).
In this study, we included the full CPTAC LUAD cohort including 110 patients. We used the processed tables from the original study, specifically the two-component-normalized log2-transformed protein and phosphosite expression tables (5). From this previous study (5), 10,699 expressed proteins and 41,188 phosphorylation sites were detected and analyzed in the present study.
SMC LUAD data
Samples from histologically confirmed NSCLC adenocarcinoma patients who were treated with either PD-1 or PD-L1 inhibitors were collected. Patients with samples available for the whole transcriptome sequencing (WTS) were included in the analyses. The data from 164 patients with WTS results were used. Clinical information was collected from patients’ electronic medical records. Tumor response was assessed using the Response Evaluation Criteria in Solid Tumors version 1.1. Patients with complete/partial response (CR/PR) were considered “responsive” to therapy. Patients with progressive disease or stable disease (PD/SD) were considered “non-responsive” to therapy. PD-L1 immunohistochemistry (IHC) results were recorded based on tumor proportional score (TPS) using the 22C3 pharmDx antibody (Agilent, USA). This study was conducted with the approval of the SMC institutional review board (IRB number: 2018–03-130). RNA was purified from formalin-fixed paraffin-embedded (FFPE) or fresh tumor samples using the AllPrep DNA/RNA Mini Kit (Qiagen, USA). RNA concentration and purity were measured using the NanoDrop and Bioanalyzer (Agilent, USA). The library was prepared following the manufacturer’s instructions using either the TruSeq RNA Library Prep Kit v2 (Illumina, USA) or the TruSeq RNA Access Library Prep Kit (Illumina, USA). FASTQ files were mapped against hg19 using the 2-pass mode of STAR version 2.4.0. The first-pass-aligned reads to the hg19 genome reference were used to generate the sample-specific reference. The second pass was used to align the reads to the newly generated hg19 genome and the sample-specific reference. Raw read counts mapped to genes were analyzed for transcript abundance using RSEM version 1.2.18 and get TPM value. ComBat was used to merge gene expression generated with different platforms to eliminate batch effects between different kits.
Identification of TCGA LUAD expression subtypes using BayesNMF
For expression subtyping, Bayesian non-negative matrix factorization (BayesNMF) (21) with a consensus hierarchical clustering approach was applied to the log2(RSEM) TCGA LUAD gene expression data of n = 509 samples as described previously (22, 23, 24). In brief, for the preprocessing step, the genes with NA values in more than 10% across samples were removed, and the top 25% most varying genes with standard deviation of gene expression across samples were selected. The resulting matrix R (3619 by 509) was then transformed to the matrix R* of fold changes centered at the median expression. Using the distance matrix of 1 - C (Cij represents the Spearman correlation between sample i and j across 3,619 genes in R*), a consensus matrix, MK, was computed by iterating a standard hierarchical clustering K * 500 times with the average linkage method and 80% resampling in sample space (Mij represents the number of co-clustering of sample i and j. K represents the number of clusters). Next, the cumulative consensus matrix, M, was computed by summing up all MK with K increasing from 2 to 10. The normalized M* was obtained by normalizing M with the total number of iterations. For selection of optimal number of clusters, K*, that best explain the observed M*, BayesNMF with a half-normal prior was applied so that the best approximation of M* ~ HT H could be found, where hkj in H (K* by N) represents a clustering affinity or an association of the sample j to the cluster k. 23 out of 50 independent BayesNMF runs with different initial conditions converged to the solution of K* = 5. Therefore, K* = 5 was chosen as the optimal number of clusters. To provide additional internal validation for the robustness of the subtypes and test the effect of having fewer discovery samples, we performed re-clustering of downsampled datasets of sizes N=200,220,…,500 from our full discovery cohort (n=509). We performed 10 random downsamplings per value of N (“runs”); within each run, we performed 30 independent BayesNMF iterations (each initialized with a unique random seed) and selected the best number of clusters (denoted by K) following the BayesNMF criteria. K=5 or K=4 was the dominant solutions for most of the tested downsampling sizes, and K=5 was the soley dominant solution as the downsampled cohort size increased to N=480 and =500 (See results section Genomic characterization reveals five LUAD expression subtypes).
Subtype projection in independent cohorts (CCLE, CPTAC, SMC)
Expression subtype classifiers were derived as previously described (23). In brief, the subtype-specific marker genes were selected by performing an additional non-negative matrix factorization to the log2(RSEM) gene expression data X with the fixed K* and H* (a column-wise normalization of H) to determine the optimal W (20,502 by K*) in X ~ WH*, where wik represents an inferred association of the gene i to the cluster k. The genes with clustering association with any clusters higher than 0.5 were considered as subtype-specific marker genes. Next, the clustering membership of the gene i was determined by the maximum association criterion as k* = max_k [wik] (k = 1 through K*) (285 candidate subtype-specific marker genes for S1; 637 genes for S2; 397 genes for S3; 864 genes for S4; and 513 genes for S5). The subtype-specific marker genes were defined as the top 100 genes in descending order of wik, with dik >= 0.5, where dik is the mean difference of log2 fold change between samples in the cluster k and other samples. The association of samples from CCLE, CPTAC, or SMC RNA-seq samples to the TCGA LUAD expression subtypes (normalized hnew matrix) was determined by modeling the gene expression matrix of CCLE/CPTAC/SMC RNA-seq samples Xnew conditioned on W*TCGA to best approximate Xnew ~ W*TCGA hnew for the differentially over-expressed subtype markers (100 marker genes in each subtype) in TCGA LUAD expression subtypes. CCLE RNA-seq samples were assigned to one of the five identified TCGA LUAD expression subtypes if the normalized association (normalized hnew matrix) with one of the TCGA subtypes was larger than 0.6. Since there were not enough cell lines representing subtypes, a cutoff of 0.5 was used with the additional requirement that the difference of the normalized association values between the highest subtype and the second highest subtype was larger than 0.2. Using these thresholds, we confidently assigned 30 cell lines to S3, and 16 cell lines into S4. CPTAC/SMC RNA-seq samples were assigned to one of the five identified TCGA LUAD expression subtypes if the normalized association (normalized hnew matrix) with one of the TCGA subtypes was larger than 0.6 (the cutoff of 0.6 instead of 0.5 was used to be more conservative). The subtype projection code is available on GitHub (https://github.com/getzlab/LUAD_subtypes) so that any independent LUAD expression samples can be projected to our LUAD expression subtypes using our subtype projection code as a single sample classifier.
Mutation significance analysis
MutSig2CV (25, 26) was applied to identify significantly mutated genes (Q value ≤ 0.1) (impute_full_cov_when_promotes_significance = false, max_neighbors = 1000, num_neighbor_patients = 1, qual_min = 0.1, enforce_target_list = true), and GISTIC 2.0 (27) was applied to identify significant focal copy number alterations in a cohort of samples of interest (all TCGA LUAD samples, each of five TCGA LUAD expression subtypes) (Amplification Threshold = 0.1, Deletion Threshold = 0.1, Cap Values = 1.5, Broad Length Cutoff = 0.5, Remove X-Chromosome = 0, Confidence Level = 0.99, Join Segment Size = 4, Arm Level Peel Off = 1, Maximum Sample Segments = 2000). Gene amplification in TCGA LUAD was based on the entries having values of +2 (high-level threshold) or +1 (low-level threshold) in the ‘all_thresholded.by_genes.txt’ from GISTIC 2.0. Gene deletion in TCGA LUAD was based on the entries having values of −2 (high-level threshold) or −1 (low-level threshold) in the ‘all_thresholded.by_genes.txt’ from GISTIC 2.0. Gene amplification and deletion in CCLE LUAD was based on a log2 copy number ratio threshold of 0.3. Due to small sample size of CPTAC LUAD cohort (n=1 for S1, n=2 for S2, n=13 for S3, and n=13 for S4), MutSig2CV and GISTIC 2.0 could not be applied for CPTAC LUAD cohort. As an alternative, the proportion of samples with recurrent SCNAs in the TCGA LUAD cohort with those in the CPTAC LUAD cohort was compared.
Pathway analysis
Single-sample gene set variance analysis (GSVA) was performed on the log2(RSEM) TCGA LUAD gene expression data, CPTAC LUAD gene expression data (TPM), and SMC LUAD gene expression data (TPM) using the gsva function (method=“gsva”, mx.diff=TRUE) from the R package ‘GSVA’ (v.1.30.0). Using SMC data, we performed GSVA using the remove batch effect expression data. GSVA implements a non-parametric method of gene set enrichment to generate an enrichment score for each gene set within a sample. The Molecular Signatures Database (MSigDB) gene sets v.6.1 were used to represent broad biological processes. The pathways with significantly different activities across the subtypes were identified based on (i) Q < 0.05 and (ii) mean difference of GSVA enrichment scores between subtypes of interest vs. all other samples > 0.2 or < −0.2.
Survival analysis
Disease-specific survival information of TCGA LUAD patients (‘OS’: overall survival event, ‘OS.time’: overall survival time) and other clinicopathologic variables were obtained from an integrated TCGA pan-cancer clinical data resource (20). Cox proportional hazard multivariate analysis was performed using the coxph function in the R package ‘survival’ (v.2.43–1).
Subtype-specific cancer vulnerability analysis
The CERES scores (gene dependency scores) obtained from the Cancer Dependency Map (DepMap) were used as the measure of cancer vulnerability in cell lines. CERES score is a computational method that estimates the dependency of each tested cell line to a given gene knockout. Conventionally, CERES scores are interpreted as follows: a score of 0 indicated that the gene is not essential in a given cell line, and a −1 score is highly dependent on the given gene (e.g. common essential genes), whereas scores < −0.5 are considered as the cutoff for dependency. For subtype-specific cancer vulnerability analysis, only the LUAD driver oncogenes (genes with recurrent point mutations, indels, and SCNAs) identified from this study (n=21) were tested. Top genes with subtype-specific cancer vulnerabilities were selected as the genes that meet the following two criteria: (i) First, we check for dependency, ensuring that each of the driver genes has a median CERES score lower than −0.5 in the subtype of interest, reflecting the fact that the cell lines of this subtype are dependent on the gene. (ii) Second, we check for specificity by requiring that the genes should have a median CERES score in the subtype of interest at least 0.2 lower than the other cell lines. The common essential genes (Achilles common essential genes) were filtered out from the top gene list. P values were calculated using the Wilcoxon rank sum test.
Biomarker analysis
Biomarker discovery was performed by applying lasso logistic regression on either gene expression data or reverse-phase protein array (RPPA) data (level 4 RPPA data were obtained from the Cancer Proteome Atlas Portal) from the TCGA LUAD cohort (randomly split into 80% training data and 20% test data) to predict subtypes of interest (S3 vs. others or S4 vs. others). For gene expression data, 100 subtype marker genes were used as the potential features to test. The best lambda value was chosen to minimize the prediction error rate using the cv.glmnet() function in the R package ‘glmnet’ (v.4.1–1). Threshold values from 0.1 to 1 in increments of 0.1 were tested for the best threshold selection that maximizes area under the curve (AUC) values. Accuracy of the model was based on the agreement of the predicted subtypes and the true subtype label in the test data. To reduce the number of features down to five for the 5-feature models, we forced the model to reduce the number of features down to five by increasing the lambda value that controls the amount of the coefficient shrinkage.
Antibodies and reagents
The following antibody was used for immunofluorescence staining: Recombinant Alexa Fluor® 488 Anti-PD-L1 antibody (ab209959). DAPI was used for nuclear staining (10236276001; Sigma-Aldrich). C-Met inhibitor tivantinib was purchased from Selleck Chemicals (Houston, TX, USA). CDK4/6 inhibitor Palbociclib (PD 0332991 isethionate) was purchased from Sigma-Aldrich. CDK4/6 Inhibitor IV (CAS 359886–84-3) was purchased from Calbiochem.
Cell cultures
Cell lines described in this study were received from Dr. Matthew Meyerson’s lab at the Broad Institute. The Meyerson lab received the ABC1 and NCIH1833 cell lines directly from the Cancer Cell Line Encyclopedia (CCLE) and are authenticated as part of their common processing. All other listed cell lines were profiled by SNP fingerprinting at the time of screening. SNP fingerprinting matches a panel of reference SNP genotypes for each known cell line. The reference set of SNP genotypes used was derived from the Affymetrix SNP6.0 array birdseed genotypes from the CCLE project. Tests for mycoplasma contamination were performed upon arrival at the Getz Lab (December 2020), and cell lines were screened for mycoplasma contamination at least quarterly.
Based on the subtype projection, we used 4 cell lines from the S3 subtype (HCC78, HCC827, NCIH1975, NCIH1838), 3 from the S4 subtype (NCIH1395, NCIH1833, NCIH1755), and two that were assigned to other subtypes (ABC1 – S1; CALU3 – S5). Cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. For each experiment, cell lines were thawed and undergone 2–3 passages before the indicated experiments.
Proliferation assay
Cells were seeded in duplicate (1 × 104 in 96 well plates) and treated with DMSO, 3 μM tivantinib (a non-ATP-competitive c-Met inhibitor that induces G2/M arrest and apoptosis; ), CDK4/6 inhibitor (Palbociclib-CDK4 concentration - 11nM; CDK4/6 concentration - 16 nM), or 1.5 μM CDK4/6 Inhibitor IV (CAS359886–84-3, a triaminopyrimidine compound that acts as a reversible and ATP-competitive inhibitor; abbreviated here as CINK4). Different cell lines have a varying growth rate; therefore, the media and drugs were replenished at different times for each cell line, either when the cell line confluency reached 60% (measured using IncuCyte or by the PAULA Cell Imager [Leica]) or after 3 days from the last media and drug replenishing. Continuous cell growth was monitored in 96-well plates every 3 hrs for 4 days using the IncuCyte Kinetic Imaging System. The relative confluency was analyzed using IncuCyte software. The reported response percentage for each cell line was calculated as the percent of confluency compared to a DMSO-treated control counterpart. Proliferation assays were repeated 4 times.
Immunofluorescence microscopy
Cells were seeded in duplicate (5 × 104 in 24 well plates), treated with DMSO or 3 μM of tivantinib, and grown for 2–3 days. Cells were then fixed in 4% paraformaldehyde for 10 min and washed twice in cold PBS. Fluor® 488 Anti-PD-L1 antibody was added for a 1 hr incubation in a light-protected environment at room temperature followed by staining the nuclei with DAPI. Fluorescence images were captured using Invitrogen™ EVOS™ FL Imaging System by Thermo Fisher Scientific. The increase in fluorescence was further quantified using ImageJ software.
Immune phenotype analysis based on H&E image
Immune phenotyping of tumor microenvironment was conducted by applying Lunit SCOPE IO, an artificial intelligence (AI)-powered spatial tumor infiltrating lymphocyte (TIL) analysis as published previously (28). Briefly, the AI model detected TIL and segmentized cancer area and cancer stroma. It then defined microscopic immune phenotype (IP) in 1 mm2-sized grid-level as follows: immune-inflamed was defined as TIL density in the cancer area above the threshold (106/mm2); immune-excluded was defined as TIL density in the cancer area below the threshold, and TIL density in the cancer stroma area above the threshold (357/mm2); immune-desert was defined as the TIL density in the cancer area and that in the cancer stroma area both below the thresholds. Inflamed, immune-excluded, and immune-desert scores of whole slide image (WSI) were defined by the number of grids annotated to a certain IP divided by the total number of grids analyzed in the WSI. We defined macroscopic IP in WSI-level, as follows: immune-inflamed IP as inflamed score ≥ 33.3%, otherwise immune-excluded IP as immune-excluded score ≥ 33.3%, otherwise immune-desert IP.
Statistical analysis
Statistical analysis was performed using R. Statistical tests included a two-sided Wilcoxon rank-sum test and Chi-squared test.
Results
Genomic characterization reveals five LUAD expression subtypes
Since previous studies showed that expression data are the most predictive genomic features of cancer dependencies (18), we sought to identify robust and novel expression subtypes which can be potentially associated with cancer vulnerabilities. Applying a consensus clustering approach using Bayesian Non-Negative Matrix Factorization (BayesNMF) to the expression data representing the 509 LUAD cases from TCGA, similar to the approach we used for bladder cancer (22), our analysis revealed five LUAD expression subtypes, designated Subtype 1 (S1) to S5 (Figure 1A, Table S1).
To further explore our expression subtypes, we compared them to the previously defined LUAD expression subtypes –– PI, PP, and TRU (Supplementary Note 1) (2). Among the 230 TCGA LUAD tumors, we found that S5 was most closely related to the TRU subtype (77.4% of S5 tumors were from the TRU subtype, and 80.9% of TRU subtype mapped to S5 [Fisher’s exact test P = 3.3×10−24]), and S4 was enriched with the PP subtype (76.4% of S4 were PP; P = 9.4×10−9). The S1, S2, and S3 subtypes mostly matched the PI subtype, suggesting that the PI subtype can be further split into three subgroups. Of these, S3 was the most enriched with the PI subtype (85.7% of S3 tumors matched to PI; P = 1.7×10−14) (Figure 1B). We also compared our expression subtypes to the 6 LUAD-enriched mRNA expression clusters from the more recent TCGA Pan-Lung study using cluster-of-clusters analysis (COCA) (4) and found a high concordance across the two TCGA studies and our analysis (Figure 1B, Figure S1A–C, Table S2, Supplementary Note 1).
To explore the biological differences among our five subtypes, we identified significantly differentiated pathways across the subtypes using single-sample gene set variance analysis (GSVA) on the Molecular Signatures Database (MSigDB) hallmark gene sets (Methods). S1 showed a low immune/inflammatory signature, and S2 showed high activity of gene sets associated with epithelial–mesenchymal transition (EMT) and cell-adhesion. Both S3 and S4 showed increased proliferation signatures (Q values < 2.5×10−10), but only S3 showed high immune/inflammatory signatures (Q values < 3.7×10−17). S5 distinctively showed low proliferation signatures (Figure 1C, Table S3). Additionally, association of clinical parameters with our subtypes (Figure 1C, Table S4–S5) showed that S5 tumors were more enriched in older (>65) patients (Q = 0.0022). Interestingly, Both S2 and S5 tumors were more enriched in never-smokers than S3 and S4 (Q < 0.012).
To further support partitioning the 60 tumors originally assigned to the PI subtype into our S1-S3 subgroups, we found a consistent set of differentially active pathways in each subgroup: low immune/inflammatory signature in S1; high EMT in S2; high E2F, MYC targets, G2M markers, and interferon alpha/gamma response in S3, etc. These findings demonstrate that the differences between the S1-S3 subgroups already existed in the original TCGA cohort but were likely not detected due to the small sample size (Figure S1D–F, Figure 1C, Table S3, Methods). Altogether, we revealed novel, biologically distinct subtypes that were previously grouped together within the single PI subtype.
To associate each of our five expression subtypes with driver events (point mutations, indels, and copy-number alterations), we applied MutSig2CV (25) and GISTIC 2.0 (27) (Figure 1D, Figure S2A–B, Table S6–S7). Consistent with the original TCGA paper (2), S3 (mapping to PI-subgroup) was enriched with co-mutation of TP53 and NF1; S4 (mapping to PP) was enriched with activating KRAS mutations, inactivating STK11 mutations, and KEAP1 mutations; and S5 (mapping to TRU) was enriched with EGFR mutations/amplifications and (2). Interestingly, S2 (one of the subgroups of the PI subtype) was also enriched with EGFR mutations/amplifications, even with a higher frequency than S5 (40% vs 17%, P = 0.0023).
Beyond the previous associations of driver events, our new subtyping approach in a larger dataset enabled further identification of significantly recurrent SMARCA4, ATM, FANCM, and PCDHGA6 mutations as well as amplification of MET, FGFR1, and PIK3CA in S4; and BRAF, SETD2, and CTNNB1 recurrent mutations in S5 (Figure 1D, Figure S2A–B). Thus, our new subtypes show more enriched mutations compared to the previously identified subtypes. STK11 mutations in particular were enriched in KRAS-mutant S4 tumors (21 STK11-mutant tumors among 46 KRAS-mutant S4 tumors) (Fisher’s exact test P = 0.0029), suggesting that S4 tumors might be more resistant to PD-1 inhibitors (15, 29). Using a previously reported immune phenotype (IP) artificial intelligence (AI) assay that searches for TILs in H&E slides (Table S8) (28), S4 tumors also showed a trend of lower inflamed scores and higher immune-excluded scores in KRAS/STK11 co-mutated samples versus all other samples, whereas non-S4 co-mutated samples showed no difference (Supplementary Note 2).
Following this, we leveraged 16 previously calculated genomic features for TCGA samples (30) to further explore differences across the S1-S5 subtypes (Figure S3). S2 and S5 (both significantly enriched with EGFR mutations) have lower overall somatic tumor mutation burden (TMB) than the other subtypes, including significantly lower frequencies of both nonsilent mutations and indels, which influence the number of predicted neoantigens (Q < 0.0011) (Figure S3A–D). In features related to somatic copy-number alterations (SCNAs), we observed that S1 and S4 have significantly higher number of copy-number segments and fraction of genome altered by SCNAs; higher levels of homologous recombination defects and aneuploidy score (Q < 0.0011) (Figure S3E–H); and relatively lower stromal fraction and intratumor heterogeneity (Figure S3I–J). These genomic differences provide orthogonal evidence for splitting the prior PI subtype into S1 (higher SCNAs) and S2 (lower TMB) subtypes, with the remaining PI-like tumors falling into S3. Finally, we associated our subtypes with immune cell populations defined by Thorsson et al. (30) using CIBERSORT (31) for deconvolving expression data (Figure S4A). We found that S2 showed significantly higher TGF-beta levels and a higher fraction of M2 macrophages (Figure S4B), which may be explained by secretion of TGF-beta by M2 macrophages to promote immune suppression in S2 (32). Collectively, the genomic characterization of our five expression subtypes shows that each subtype has distinct biology.
Next, we tested whether our expression subtypes associated with clinical outcome. While the TRU expression subtype showed significantly longer overall survival compared to other subtypes (PP and PI combined) (P = 0.023; log rank test) in the TCGA LUAD study (2), this association was no longer significant when age, gender, and tumor stages were corrected for (Cox proportional hazard P = 0.45) (Table S9). On the other hand, S5 subtype enriched with TRU subtype (Figure 1B) showed significantly longer overall survival compared to other tumors (P value = 0.02) (Figure S4C), highlighting that S5 tumors have a stronger association with overall survival.
Subtype-specific cancer vulnerabilities
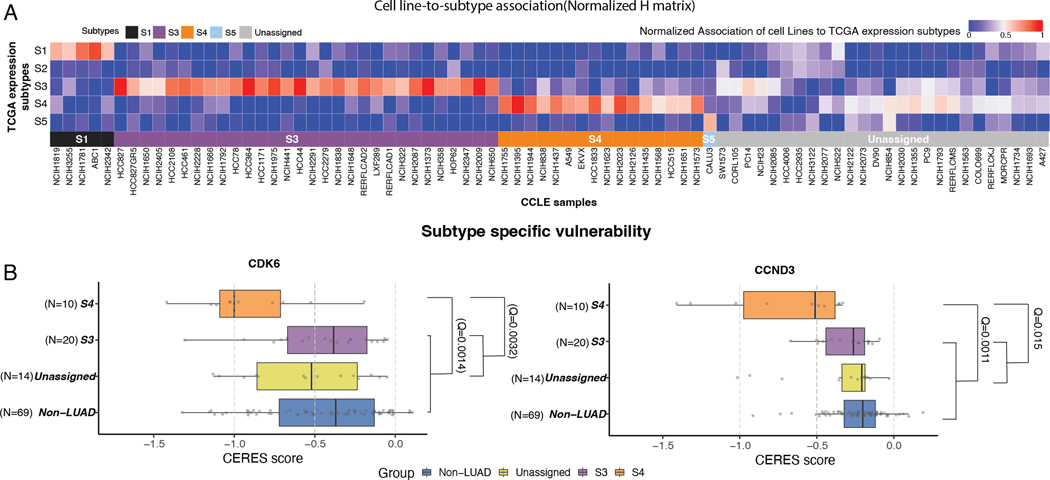
We next leveraged the CCLE (16, 17) and DepMap (18) resources, which collectively provide expression data as well as CRISPR and drug screening data for ~1,100 cell lines, to find subtype-specific cancer vulnerabilities. We first probabilistically classified the 78 CCLE LUAD cell lines into the LUAD expression subtypes using subtype-specific marker genes (Figure 1A, Figure 2A, Table S10–11, Methods). Since only S3 and S4 were assigned a sufficient number of cell lines (30 and 16, respectively), we focused our downstream analysis on these subtypes. To validate our subtype classification, we confirmed that the S3- and S4-associated cell lines harbored genetic events, somatic point mutations, and copy-number alterations (Figures S5A and S5B) that were consistent with patients associated with S3 and S4 tumors. Comparison of the cancer vulnerabilities of 21 LUAD driver oncogenes between S3/S4 and the other CCLE LUAD cell lines (Figure 2B) did not yield significant S3 vulnerabilities (Table S12) but did identify two significant vulnerabilities for S4: CDK6 and the CDK6-cyclin D3 complex gene, CCND3 (Table S12). This finding suggests that S4 tumors may be dependent on the CDK6 pathway and thus potentially vulnerable to CDK6 inhibition.
Figure 2. Subtype-specific cancer vulnerabilities.
(A) Heatmap shows cell line to subtype association based on marker genes.
(B) Boxplots show CERES scores of CDK6 (left panel) and CCND3 (right panel) in S4 versus other cell lines (Q values correspond to Wilcoxon rank sum test). * S1 not shown due to small sample size.
Although we did not find significant CRISPR vulnerabilities associated with S3, CDK4 was the nominally significant top potential S3 vulnerability (P = 0.014; Q = 0.14; median CERES scores in S3 = −0.68 and in the other cell lines −0.47) (Table S12), consistent with the recurrent genomic alterations in CDK4 (Figure S2B; Figure S5B) in S3. We therefore functionally tested the sensitivity of S3-associated cell lines to CDK4 specific inhibition using two CDK4 inhibitors: Palbociclib and CDK4/6 Inhibitor IV (abbreviated as CINK4). Both compounds are known CDK4/6 inhibitors at high concentrations; however, at low concentrations, they are potent CDK4-only inhibitors that induce G1 cell cycle arrest and senescence in retinoblastoma protein (Rb)-proficient cell lines (33). We evaluated proliferation in 9 cell lines treated with either palbociclib or CINK4 –– 4 from the S3 subtype, 3 from the S4 subtype and two that were assigned to other subtypes (see details in methods). As expected, the S3 cell lines showed significantly lower proliferation (higher response) compared to the S4 and unassigned cell lines (Palbociclib: P = 1.6×10−5 and P = 4.1.x10−6 respectively [Figure S5C, left panel, Table S13] and CINK4: P = 3.5×10−3 and P = 3.3×10−3 [Figure S5C, middle panel, Table S13]). These results show that the S3 subtype depends on CDK4, suggesting that therapy that includes a CDK4 inhibitor may benefit patients with S3 assigned tumors.
Since palbociclib inhibits both CDK4 and CDK6 at higher concentrations, we could not test CDK6-only inhibition in S4 cell lines, and higher doses of palbociclib inhibited proliferation in all cell lines (Figure S5C, right panel, Table S13). Taken all together, the CRISPR data and drug sensitivity experiments demonstrate specific vulnerabilities related to CDK4 in S3 and CDK6/CCND3 in S4 subtypes.
Proteogenomic analysis reveals distinct protein regulation between S3 and S4
To further characterize the expression subtypes at the proteomics level, we first classified the CPTAC LUAD samples (5) to S1-S5 based on the expression of subtype-specific marker genes (Figure 1A; Figure 3A; Table S10, Table S14). Since S3 (n=11), S4 (n=10), and S5 (n=51) were the major subtypes represented in the CPTAC LUAD cohort, we focused our downstream proteogenomic analysis on these subtypes. Consistent with the TCGA data analysis, both S3 and S4 showed increased proliferation signatures, and S3 also showed an increased immune/inflammatory signature (Figure 3A). Comparing our expression subtypes with the CPTAC multi-omics clusters (5), we found a good agreement: S3 was enriched with CPTAC multi-omics cluster C1 tumors (PI enriched; 11/11), S4 was enriched with C3 tumors (PP enriched; 9 /10) (Figure S6A), and S5 was enriched with C4 tumors (TRU enriched; 33/51). The multi-omic subtypes in CPTAC (5) also significantly overlapped with TCGA mRNA subtypes and provided additional evidence of a more refined partitioning of the PI subtype.
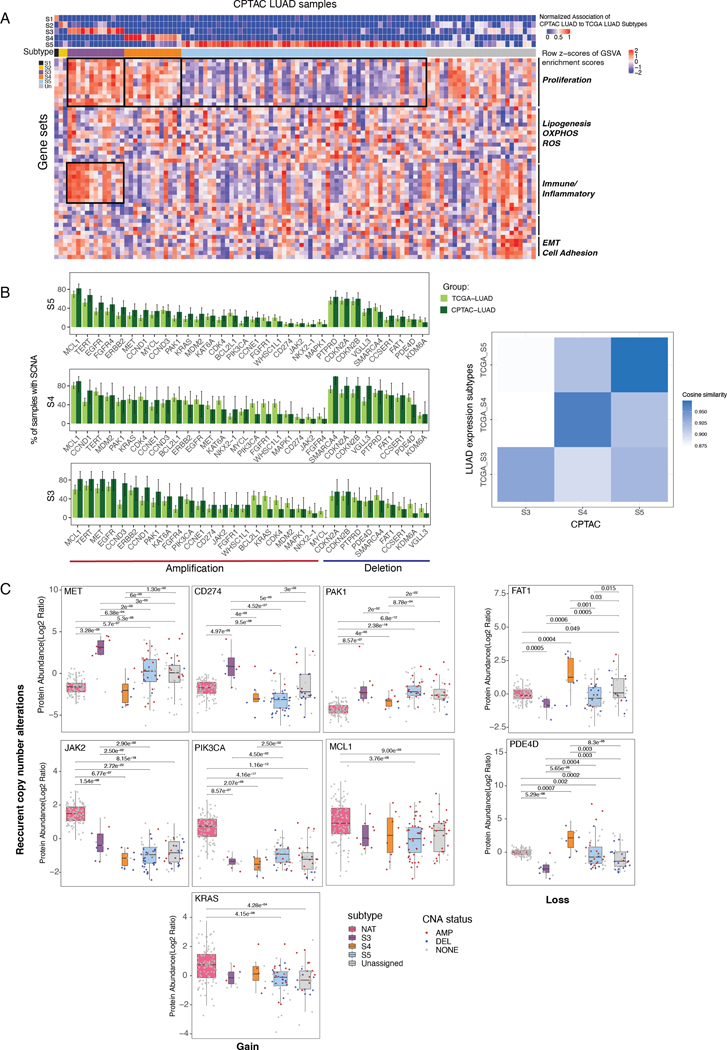
Figure 3. Proteogenomic analysis of genes with subtype-specific recurrent SCNAs.
(A) Top heatmap shows CPTAC LUAD sample association to the expression subtypes. Center heatmap shows the GSVA pathway activation profiles. Boxed pathways are consistent with TCGA LUAD.
(B) Barplots show the proportion of TCGA/CPTAC LUAD samples in S3-S5 with gene amplification (red) or deletion (blue) for genes with recurrent SCNAs. Right heatmap shows the cosine similarity among S3-S5 tumors in TCGA and CPTAC data.
(C) Boxplots show protein abundance of recurrent SCNAs genes across CPTAC LUAD expression subtypes and their normal adjacent tumors (NAT). Copy number states are denoted (red: amplification; blue: deletion; gray: no SCNAs).
Next, we focused on the genomic and proteomic features of each of our subtypes. The overall frequency profiles of amplification or deletion of significantly copy-number altered genes was similar between the TCGA and CPTAC cohorts for S3, S4, and S5 (cosine similarities > 0.93; Figure 3B). Since CPTAC and TCGA had different ethnicity distributions (Figure S6B), we tested whether differences are attributed to ethnicity. Among CPTAC samples, only S5 had sufficient numbers of caucasian samples (n=18) for statistical analysis. For S5, the cosine similarity to be very similar between all S5 tumors (0.974) and caucasian-only S5 tumors (0.970), suggesting that our subtype classification was robust with respect to ethnicity.
We next explored the effect of recurrent SCNAs on protein expression. Among the genes with recurrent SCNAs (in S3-S5, Table S15). JAK2 and CD274 (PD-L1) showed both recurrent gene amplification and significantly higher protein expression in S3 (Wilcoxon rank sum test, Q < 2.9×10−2, 4×10−3, respectively) (Figure 3C; Figure S6C). Interestingly, MET showed recurrent gene amplification in both S3 and S4, but its protein expression was significantly up-regulated only in S3 (Q < 2×10−3, Figure 3C), also exceeding the expression in control normal adjacent tissues (NATs; Q < 5.5×10−4) (Figure S6D). Moreover, S3 tumor samples with MET amplification showed much higher MET protein expression than S3 tumors with no MET amplification (Q < 5×10−3), whereas other subtypes showed weaker (or no) correlation between MET amplification and MET protein expression. Of all the genes that showed recurrent gene deletion in both S3 and S4, only FAT1 and PDE4D also exhibited significant changes to their proteomic expression. Moreover, only S3 exhibited significantly downregulated protein expression for both FAT1 and PDE4D that was associated with their respective gene loss when compared to both NAT and the other subtypes (Q < 6.3×10−4, 3.6×10−4, respectively). We observed a similar trend for mRNA expression in the TCGA LUAD cohort. These findings highlight the need to take into account not only copy-number alterations but also mRNA and protein expression for understanding downstream effects of genetic alterations (Figure S7A) (34).
MET is a core regulator of proliferation and PD-L1 expression in S3
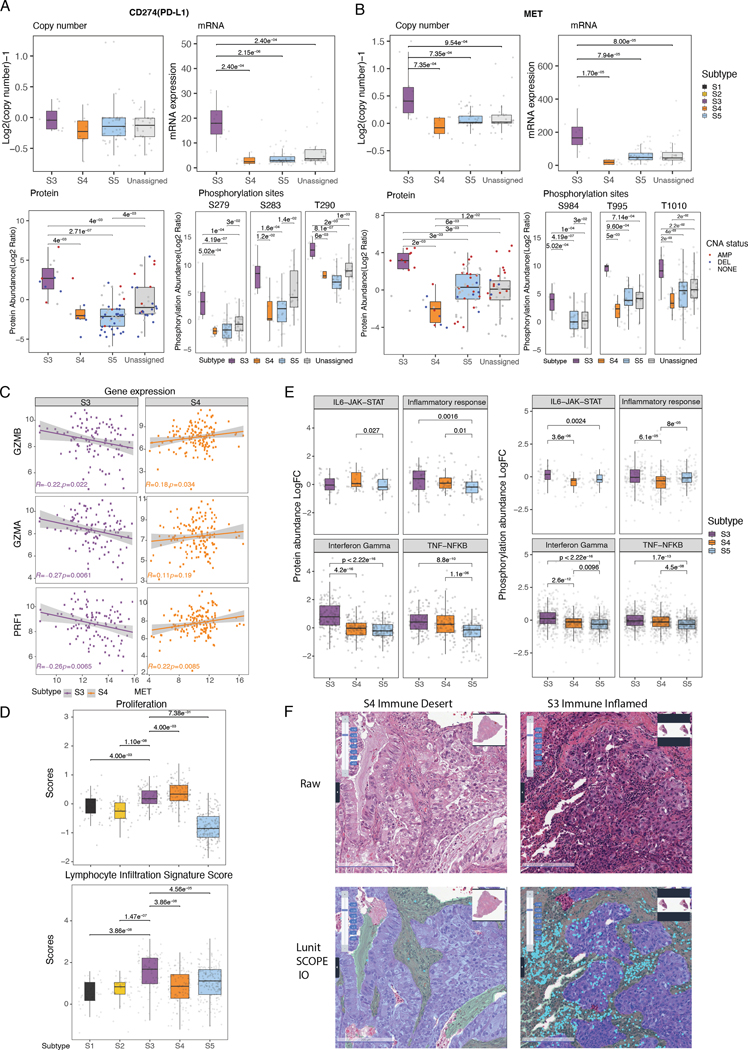
Our initial pathway activation analysis found that both S3 and S4 upregulate proliferation-associated genes, whereas only S3 highly expresses immune-related genes (Figure 1C). To gain additional insight into the underlying biological differences between S3 and S4, we performed a deeper proteogenomic characterization of our subtypes. We noted that CD274 (PD-L1) was among the two top markers that are both recurrently amplified in S3 and strongly associated with S3 (marker-subtype association values = 2.544 for SBSN gene to S3, 2.43 for CD274 gene to S3) (Table S7, Table S10). We further observed that PD-L1 copy number, mRNA expression, protein expression, and phosphorylation levels were significantly higher in S3 versus S4 (Figure 3B–C, Figure 4A, Figure S7B). Since both S3 and S4 showed high proliferation signatures and recurrent MET amplification (Figure 1C, Figure S2B, Figure S7C), we assessed MET copy number and protein expression across subtypes and found that MET copy number was significantly higher in S3 versus S4, and that its expression of mRNA, protein, and phosphorylation levels was also higher in S3 versus S4 (Figure 3B–C, Figure 4B), echoing the expression pattern of PD-L1. The mRNA and protein expression of MET were also significantly higher in S3 versus S4, even when restricting the analysis only to MET-amplified tumors (Q=1.1×10−8 for mRNA, Q=2.7×10−2) (Figure S7D). Additionally, we identified higher MET pathway activation in S3 versus S4 as evidenced by increased phosphorylation levels of GAB1 in S3, a known downstream substrate of MET (Figure S7E). Based on a previous study showing a negative correlation between MET expression and the expression of T cell effector molecules (granzyme A, granzyme B, and perforin) in the TCGA dataset (35), we asked whether the same pattern is present in both subtypes. Interestingly, the expression of MET negatively correlated with T cell effector molecules in S3, but not in S4 (Figure 4C), suggesting potential immune evasion of S3 tumors associated with MET overexpression.
Figure 4. MET as a core regulator of proliferation and PD-L1 expression in S3.
(A-B) Boxplots show copy number, mRNA expression, protein abundance, and phosphorylation of CD274 (PD-L1, Left) and MET (Right) genes across CPTAC LUAD expression subtypes.
(C) Scatter plots show correlation between MET expression and cytolytic marker expression (GZMB, GZMA, and PRF1) in S3 versus S4.
(D) Boxplots show proliferation and lymphocyte infiltration scores (Methods) across LUAD expression subtypes.
(E) Boxplots show protein and phosphorylation levels of genes in immune-related pathways among CPTAC LUAD S3-S5.
(F) Representative images (x20) of immune-desert S4 (left) and immune-inflamed S3 (right). H&E original image (top) and Lunit SCOPE IO–inferenced segmentation (bottom). Cancer area – purple; cancer stroma – green, tumor infiltrating lymphocytes – cyan.
To next characterize the proteogenomic differences by an alternative approach, we evaluated proliferation and immune signatures using previously developed scores for proliferation and lymphocyte-infiltration (30). Again, consistent with our results (Figure 1C), we found high proliferation scores in both S3 and S4, and a higher immune score only in S3 (Figure 4D). Additionally, this alternative characterization method showed that S3 had a significantly higher fraction of anti-tumoral M1 macrophages (Q = 3.77×10−9; Wilcoxon rank sum test of S3 vs. others), suggesting a favorable tumor immune microenvironment for therapy, whereas S4 showed a significantly higher fraction of pro-tumoral Th2 cells (Q = 8.75×10−23; Wilcoxon rank sum test of S4 vs. others) (Figure S4). We also observed increased IFNɣ pathway activity in S3 compared to S4 and S5 based on protein expression and phosphorylation data (Q<2.6×10−12, Figure 4E, Table S16–S17). This finding was further supported by the increased expression of proteins involved in antigen presentation and interferon signaling in S3 (Figure S8A). Based on a previous report of immune phenotype (IP) by AI-powered spatial TIL analysis (Table S8) (28), we categorized the samples in our subtypes into 3 defined immune phenotypes: inflamed, immune-excluded, and immune-desert (Methods). S3 showed a higher proportion of the inflamed IP than the immune-excluded IP (53.8% versus 34.4%, Q = 0.047, Figure 4F and S8B, Table S18). Taken together, these proteogenomic findings support increased immune/inflammatory activity in S3.
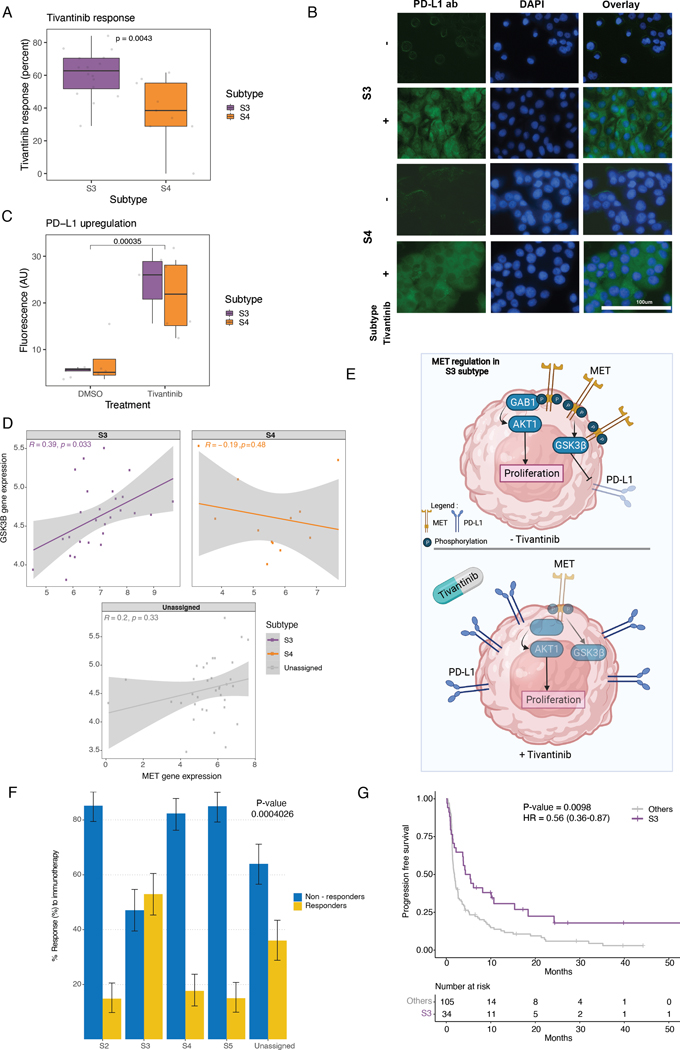
To further support and validate our proliferation findings in cell lines, we explored the response of our subtype-specific cell lines (described above) to the MET inhibitor, tivantinib. After 4 days post-treatment, S3 cell lines showed a significantly increased proliferation (P value > 0.001) to tivantinib treatment compared to the other assigned groups (data not shown), and in particular, compared to S4 (Figure 5A, Table S19). Previous reports in NSCLC cells suggested a direct relationship between PD-L1 and MET expression by showing enhanced PD-L1 expression in response to c-MET inhibition (36). To test whether we could also observe this relationship in our subtypes, we assessed PD-L1 levels by immunofluorescence. A significant increase in PD-L1 levels was detected in all subtypes in response to tivantinib (Wilcoxon test P value > 0.0001) (Figure 5B–C, Table S20). Since c-MET inhibition drives PD-L1 expression by suppressing glycogen synthase kinase 3 beta (GSK3β) (36), we next tested the correlation of mRNA expression between MET and GSK3β in LUAD cell line data and found a significant positive correlation only in the S3 subtype (Pearson correlation coefficient=0.46, P value=0.016) (Figure 5D).
Figure 5. c-MET inhibition drives PD-L1 expression in cell lines.
(A) Boxplots show response to tivantinib measured by the delta change in confluency between treated and untreated (DMSO only) cell lines.
(B) Immunofluorescence staining under tivantinib treatment (x40). Anti-PD-L1 antibody (Green; right); DAPI nuclear staining (Blue; middle), and overlay (left). Bar scale was adapted from another region on the same slide and superimposed on the figure.
(C) Fluorescence quantification shown in boxplots using ImageJ software after background correction.
(D) Scatter plots show correlation between MET expression and GSK3β expression in CCLE data across different subtypes.
(E) Schematic diagram shows MET as a proliferation core regulator and PD-L1 expression regulation by GSK3β in S3 tumors, both without (top) and with (bottom) tivantinib treatment.
(F) Proportion of responders and non-responders across subtypes. S1 excluded due to small sample size. (Chi-squared test P value shown).
(G) Kaplan-Meier curves for the disease-specific survival (DSS) between S3 tumors and others. (Log-rank test P value shown)
Collectively, these data suggest a model for S3 in which MET plays a key role in driving proliferation through GAB1/AKT1, and MET can also upregulate PD-L1 expression through the GSK3β axis, potentially for immune escape. Additional synergistic players, found to have higher protein expression in S3 vs S4, such as BCL2L1, PAK1, and RB1, also likely further contribute to the proliferation of cells in S3 (Figure 5E; Figure S8C). Hence, S3 tumors may respond to a combined therapeutic regimen of MET and PD-L1 inhibitors.
S3 is a robust biomarker for immune checkpoint blockade (ICB) response
Lastly, we tested our hypothesis that S3 tumors are more responsive to immune checkpoint blockade (ICB) due to high PD-L1 protein expression and immune-inflamed phenotype. Wwe first classified the independent Samsung Medical Center (SMC) LUAD cohort patients (n=164) treated with ICB to S1-S5 based on the expression of the subtype-specific marker genes (Figure 1A, Table S10, S21–S25). Consistent with the TCGA data analysis, S2 showed high activity of epithelial–mesenchymal transition (EMT) and cell-adhesion signatures (Figure S8D). Both S3 and S4 showed increased proliferation signatures, and S3 showed an increased immune/inflammatory signature. PD-L1 protein expression by IHC was also significantly higher in S3 compared to other subtypes (P value = 1.05 × 10−6) (Figure S8E). After confirming our subtypes in the SMC LUAD cohort, we studied whether responders to ICB are enriched in any of the subtypes. Interestingly, responders were enriched only in S3 (chi-square test P value = 0.0004) (Figure 5F). Even after correcting for age, gender, smoking status, and PD-L1 expression, S3 was still significantly associated with ICB response (P value = 0.00045) (Table S25). S3 was also significantly associated with longer progression-free survival (PFS) (P value = 0.0098, HR = 0.61) (Figure 5G), and multivariate analysis showed that S3 (P value = 0.012, HR = 0.64) was more strongly associated with PFS than PD-L1 expression (P value = 0.59, HR = 1.09) (Figure S8F). These results suggest that S3 subtype can potentially be used as a robust biomarker for predicting response to ICB in the clinic.
Biomarkers for identifying patients with S3 or S4 tumors
The above findings suggest that BayesNMF projection can be used as a robust classifier for subtyping analysis, but this type of analysis requires the evaluation of 500 subtype marker genes. In the interest of developing a more clinically useful approach, we tested whether a fewer set of key genes could be used as biomarkers to reliably identify tumors belonging to S3 and S4. Using the subtype marker genes defined above from gene-expression data (Table S7) as the potential features to test, the best prediction model for S3 (23 genes) had an accuracy of 95%, and the best prediction model for S4 (27 genes) had an accuracy of 85% (Table 1, Table S26). To identify IHC markers, we also considered TCGA reverse-phase protein array (RPPA) data as potential proteomic features. The best prediction models for S3 and S4 contained 20 and 24 protein features, respectively (both 91% accurate). Additionally, for better clinical utility, we forced the model to reduce the number of features down to five (Methods). Interestingly, the five-feature model for S3 based on RPPA data (PD-L1, JAK2, MIG6, P70S6K1, GATA6) still showed a high model accuracy of 91%, whereas the five-feature model for S4 (BIM, CAVEOLIN1, FOXM1, PKCPANBETAII_pS660, NRF2) had a reduced accuracy of 74%. Overall, these results show that we can reach high prediction accuracies for S3 and S4 using both gene expression and RPPA data.
Table 1. Biomarker discovery for LUAD expression subtypes.
Biomarkers for LUAD expression subtypes S3 and S4 based on gene expression and reverse-phase protein array (RPPA) data. Best model and 5-feature model are shown. Blue – shared features included in both models.
| Gene Expression | Protein Expression (RPPA) | |||||
|---|---|---|---|---|---|---|
| Accuracy | Best 95% | 5-features 83% | Best 91% | 5-features 91% | ||
| S3 | CD274 | TBX21 | CHK1_pS345 | DJ1 | MIG6 | |
| TGM4 | CD70 | CD274 | ERALPHA | GATA3 | P70S6K1 | |
| ARNTL2 | GZMB | DCBLD2 | LCK | MIG6 | GATA6 | |
| CD8A | NKG7 | FBX032 | PI3KP110α | PEA15 | JAK2 | |
| DCBLD2 | CSF2 | MYBL | GATA6 | P63 | PDL1 | |
| AFAP1L2 | GPR84 | AIM2 | TIGAR | BRD4 | ||
| FBX032 | MYBL1 | JAK2 | PDL1 | |||
| CDA | BATF3 | PDCD1 | CD20 | |||
| C15orf48 | MET | TTF1 | P63 | |||
| TMEM156 | CATSPE R1 | ANNEXINVII | ||||
| S100A2 | KCNK12 | SYNAPTOPHYSIN | ||||
| PKCDELTA_pS664 | ||||||
| Accuracy | 85% | 67% | 91% | 74% | ||
| S4 | KCNU1 | ZMAT4 | AMPKALPHA | BIM | ||
| SLC38A8 | HOXD13 | CAVE0LIN1 | CYCLINB1 | |||
| PCSK1 | UGT3A1 | JNK2 | MIG6 | |||
| KLK14 | HEPACAM2 | TIGAR | TFRC | |||
| CPS1 | CALB1 | P38MAPK | PEA15 | |||
| AKR1C4 | F2 | UALUA | NRF2 | TTF1 | BIM | |
| MLLT11 | INSL4 | HOXD13 | YAP_pS127 | P90RSK | CAVEOLIN1 | |
| HOXD11 | AKR1C2 | AKR1C4 | ANNEXIN1 | MSH6 | FOXM1 | |
| WDR72 | F7 | MLLT11 | NCADHERIN | VEGFR2 | NRF2 | |
| UCHL1 | POPDC3 | PAH | MTOR_pS2448 | NAPSINA | PKCPANBETAII pS660 | |
| CSAG2 | C20orf70 | ACETYLATUBULINLYS40 | ||||
| GNG4 | C12orf39 | PKCALPHA pS657 | ||||
| IGF2BP1 | C12orf56 | SYNAPTOPHYSIN | ||||
| LOC100190940 | CASPASE7CLEAVEDD198 | |||||
Discussion
Over the past several years, multiple lung cancer subtype studies have revealed important biological insights and clinical outcomes (2, 3, 4, 5). While these studies were mostly consistent with the PI, PP, and TRU subtypes defined in the original TCGA study (2), we were sufficiently powered in the current study to further partition the PI subtype into 3 subgroups: S1, S2, and S3. By integrating genomic and proteomic data, we further identified distinct biology in each of our subtypes as well as subtype-specific recurrent mutations (point mutations, indels, and SCNAs). Notably, the biology of these subtypes was quite distinct from the biology of subtypes identified by the CPTAC LUAD study (5) because the CPTAC LUAD subtypes were based on multi-omics data from a smaller sample size than ours (< 5-fold, with only one S1 and no S2 tumors found in the CPTAC LUAD cohort). Furthermore, the unique features of the newly identified subtypes might also serve as biomarkers of response to targeted therapies (e.g., response to EGFR inhibitors and TGF-beta inhibitors for S2; response to PD-L1, MET, and CDK4 inhibitors for S3; and resistance to PD-1 inhibitors for S4 due to STK11 mutations (15) (Table 2)). An integrative analysis of TCGA and DepMap data also demonstrated the proof-of-concept idea that leveraging the genome-wide CRISPR screening data and expression subtypes of cell lines can identify novel therapeutic vulnerabilities for specific expression subtypes.
Table 2. Subtype-specific feature summary.
Subtype-specific features identified in this study and potential therapeutic targets. Highlighted features are noted by their respective subtype color.
| Point mutations/indels | CNAs | Activated pathways | TMB | Signaling | immune cell subsets | Cancer vulnerability in cell lines | Protein levels | Potential therapeutic targets | |
|---|---|---|---|---|---|---|---|---|---|
| S1 | High | ||||||||
| S2 | EGFR mutations | EGFR amplification | EMT, Cell adhesion | TGF-beta | M2 macrophage (pro-tumoral) | EGFR inhibitors. TGF-beta inhibitors | |||
| S3 | MET, PD-L1 amplification | Proliferation, Immune/ Inflammatory | High | IFN-gamma response | M1 macrophage (anti-tumoral) | CDK4 | High MET, High PD-L1 | CDK4/MET + PD-L1 inhibitors | |
| S4 | STK11 mutations | MET, FGFR1, PIK3CA amplification | Proliferation | High | Th2 cells | CDK6, CCND3 | CDK6/CCND3 inhibitors | ||
| S5 | CTNNB1 mutations | Lipogenesis, OXPHOS, ROS |
Our observation that MET amplification had a profound impact on its protein expression in S3 but not in the other subtypes suggests that the mRNA and protein expression of these genes may, in some cases, be affected by negative feedback loops or other types of regulation that reduces the effect of the increased DNA copy-number. These results highlight the importance of integrating the analysis between genomic and proteomic data to reveal underlying subtype-specific biology (37, 38, 39, 40).
Leveraging proteogenomic data to explore the subtype-specific effect of MET amplification, we show that MET amplification in S3 tumors may lead to cell proliferation through the GAB1/AKT1 axis. In addition, the observed S3 tumor–specific positive correlation between MET and PD-L1 expression suggests that the MET gene may regulate PD-L1 expression in S3 tumors through GSK3β, consistent with findings in lung cancer cell lines (35). A recent study demonstrated that MET amplification attenuates immunotherapy response by inhibiting STING in lung cancer and that targeted MET inhibition could increase the efficacy of immunotherapy (41). In our data, the MET–STING axis was attenuated only in S4, but not in S3, suggesting that the MET–GSK3β–PD-L1 axis may play a more important role in S3 than the MET–STING axis. Thus, in S3, MET might be a core regulator of two important cancer-related functions: (i) immune escape by upregulating PD-L1 expression, and (ii) proliferation through a synergistic effect with increased expression of BCL2L1 and MCM-family members (42, 43). Hence, it is possible that PD-L1 overexpression in response to MET inhibition causes a suppression of anti-tumor immunity that may help to explain the poor performance of the c-MET inhibitor tivantinib in a clinical trial (44); moreover, this trial being performed on mixed tumor subtypes rather than on a selected patient population makes it difficult to interpret whether tivantinib would show effectiveness specifically in LUAD patients belonging to a subset such as S3. Based on our experimental results and the higher response to immune checkpoint blockade in the S3 subtype (based on the SMC LUAD cohort), combination therapy targeting MET and PD-L1 could be synergistic for S3 tumors, benefiting from both inhibiting MET and avoiding the immune-suppression due to PD-L1 overexpression. Moreover, since S3 tumors also have relatively high TMB, IFNɣ gene expression signature, and a higher proportion of inflamed vs. immune-excluded phenotype, this S3 subtype that accounts for approximately 20% of all LUAD patients (105/509 TCGA LUAD tumors) further support the potential combination of MET inhibitors and PD-L1 blockade therapy (45).
Since S3 cell lines also had CDK4 cancer vulnerability and showed high response to CDK4 inhibitors (Figure S5C), S3 tumors might also respond well to combined CDK4 inhibitors and PD-L1 blockade, consistent with the findings from multiple mouse model studies on combined CDK4/6 inhibitors and ICBs (46, 47). Therefore, dedicated preclinical studies should be performed in tumor models representing the different tumor subtypes. These findings raise a potential clinical therapeutic hypothesis that membership in the S3 subtype can serve as a biomarker of response to combination immunotherapy targeting CDK4 or MET together with PD-L1 inhibitors. Since S4 tumors showed recurrent CCND3 amplification, and S4-associated cell lines showed a significantly stronger dependency on CDK6 (Figure 2B), future studies specifically targeting CDK6 in S4 cell lines and patients would also be of interest. S3 cell lines also showed a higher stem cell signature compared to other cell lines, suggesting that S3 might show more lineage plasticity than other subtypes (Supplementary Note 3). However, this observation needs to be experimentally validated in future studies.
Overall, our study demonstrates that a BayesNMF approach can identify novel tumor expression subtypes, and that integrative analysis of multi-modal data can identify subtype-specific biology and vulnerabilities. Consistent subtype biology could be also observed in the heterogeneous (Korean never-smokers) real-world patient data (SMC LUAD), which shows the robustness of classifying patients into the subtypes and their biology. Generation of mouse models representative of our LUAD expression subtypes would allow in vivo experimental validation of drug response associated with each subtype (46, 47, 48). Since expression subtypes can represent both the tumor cells and their microenvironment –– both of which can contribute to treatment response or resistance –– the expression subtypes can potentially inform more subtype-specific clinical intervention. Future studies, at the single-cell level, could decouple the contribution of different cell types and potentially reveal new subtype-specific biology as well as cell types and states associated with clinical outcomes (47, 49, 50).
Supplementary Material
Significance.
Integrative analysis of multi-omic and drug dependency data uncovers robust lung adenocarcinoma expression subtypes with unique therapeutic vulnerabilities and subtype-specific biomarkers of response.
Acknowledgements:
We thank The Cancer Genome Atlas Analysis Network (TCGA) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) for generating the data and making it publicly available. We also thank Dr. Matthew Meyerson’s lab at the Broad institute for providing us all needed cell lines for this work. This work was supported by funding from the NIH U24 Genomic Data Analysis Network grant to G. Getz (U24CA210999) and NIH U24 CPTAC grant to G. Getz (U24CA210979), as well as U24CA210978 and U24CA264029 to A.D. Cherniack. G. Getz is also partially supported by the Paul C. Zamecnik Chair in Oncology at the Massachusetts General Hospital Cancer Center.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1A2C3006535) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HR20C0025). Schematic figures for this manuscript were created using BioRender.com.
Footnotes
Authors’ Conflict(s) of Interest
W. Roh Senior Computational Biologist at Pfizer Inc. Y. Geffen Consultant for Oriel Research Therapeutics. J. Kim Clinical Bioinformatics Director at GSK Inc. J.F.Gainor compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo/Lilly, Blueprint, Oncorus, Regeneron, Gilead, Moderna, AstraZeneca, Pfizer, Novartis, Merck, and GlydeBio; research support from Novartis, Genentech/Roche, and Ariad/Takeda; institutional research support from Bristol-Myers Squibb, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis, and Alexo; and has an immediate family member who is an employee with equity at Ironwood Pharmaceuticals. P. Laird Consultant and member of the scientific advisory board at AnchorDX. A.D. Cherniack receives research funding from Bayer. C.Y. Ock employee with equity at Lunit. G. Getz receives research funds from IBM & Pharmacyclics, and is a founder, consultant, and has privately held equity in Scorpion Therapeutics; G.Getz is also an inventor on patent applications filed by The Broad Institute related to MSMuTect, MSMutSig, POLYSOLVER, SignatureAnalyzer-GPU, and MSIDetect. M. Miller, S. Anand, and D. Heiman declare no potential conflicts of interest. W. Roh, Y. Geffen, and G. Getz are co-inventors on a patent application related to this work (U.S. Provisional Patent Application No.: 63/293,349).
Consortia
The participants in the National Cancer Institute (NCI) Center for Cancer Genomics (CCG) Tumor Molecular Pathology (TMP) Analysis Working Group are: Jean C. Zenklusen, Anab Kemal, Ina Felau, John A. Demchok, Liming Yang, Martin L. Ferguson, Roy Tarnuzzer, Samantha J. Caesar-Johnson, Zhining Today Wang, Rehan Akbani, Andre Schultz, Zhenlin Ju, Bradley M. Broom, Alexander J. Lazar, A. Gordon Robertson, Mauro A. A. Castro, Ioannis Tsamardinos, Vincenzo Lagani, Paulos Charonyktakis, Joshua M. Stuart, Christopher K. Wong, Verena Friedl, Toshinori Hinoue, Vladislav Uzunangelov, Peter W. Laird, Andrew D. Cherniack, Lindsay Westlake, Whijae Roh, Gad Getz, Stephanie H. Hoyt, Theo A Knijnenburg, Christina Yau, Jordan A. Lee, Lewis R. Roberts, Kyle Ellrott, Jasleen K. Grewal, Steven J.M. Jones, Chen Wang, Brian J Karlberg, Akinyemi I. Ojesina, Christopher C. Benz, Kami E Chiotti, Katherine A. Hoadley, Ilya Shmulevich, Bahar Tercan, Galen F. Gao, Ilya Shmulevich, Taek-Kyun Kim, Esther Drill, Ronglai Shen, Daniele Ramazzotti, Vinicius S. Chagas, Victor H. A. dos Santos, Paul T. Spellman, Adam Struck, Eve Lowenstein, D. Neil Hayes.
The members of this Consortia are listed in the Acknowledgements section of this article.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Network TCGAR, The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma [Internet]. Nature. 2014. page 543–50. Available from: 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, et al. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene. 2017;36:1384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillette MA, Satpathy S, Cao S, Dhanasekaran SM, Vasaikar SV, Krug K, et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell. 2020;182:200–25.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gainor JF, Curigliano G, Kim D-W, Lee DH, Besse B, Baik CS, et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol. 2021;22:959–69. [DOI] [PubMed] [Google Scholar]
- 7.Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li BT, Smit EF, Goto Y, Nakagawa K, Udagawa H, Mazières J, et al. Trastuzumab Deruxtecan in -Mutant Non-Small-Cell Lung Cancer. N Engl J Med. 2022;386:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 10.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018;378:2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–52.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. Mutations and PD-1 Inhibitor Resistance in -Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–16.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan VYF, Févotte C. Automatic relevance determination in nonnegative matrix factorization with the β-divergence. IEEE Trans Pattern Anal Mach Intell. 2013;35:1592–605. [DOI] [PubMed] [Google Scholar]
- 22.Taylor-Weiner A, Aguet F, Haradhvala NJ, Gosai S, Anand S, Kim J, et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Kwiatkowski D, McConkey DJ, Meeks JJ, Freeman SS, Bellmunt J, et al. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur Urol. 2019;75:961–4. [DOI] [PubMed] [Google Scholar]
- 24.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171:540–56.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S, Ock C-Y, Kim H, Pereira S, Park S, Ma M, et al. Artificial Intelligence-Powered Spatial Analysis of Tumor-Infiltrating Lymphocytes as Complementary Biomarker for Immune Checkpoint Inhibition in Non-Small-Cell Lung Cancer. J Clin Oncol. 2022;40:1916–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricciuti B, Arbour KC, Lin JJ, Vajdi A, Vokes N, Hong L, et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11- and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. J Thorac Oncol [Internet]. 2021; Available from: 10.1016/j.jtho.2021.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The Immune Landscape of Cancer. Immunity. 2019;51:411–2. [DOI] [PubMed] [Google Scholar]
- 31.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Kuang W, Zhou Q, Zhang Y. TGF-β1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int J Mol Med. 2018;42:3395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger T, Cox J, Mann M. Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet. 2010;6:e1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn HK, Kim S, Kwon D, Koh J, Kim YA, Kim K, et al. MET Receptor Tyrosine Kinase Regulates the Expression of Co-Stimulatory and Co-Inhibitory Molecules in Tumor Cells and Contributes to PD-L1-Mediated Suppression of Immune Cell Function. Int J Mol Sci [Internet]. 2019;20. Available from: 10.3390/ijms20174287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X, Li C-W, Wang W-J, Chen M-K, Li H, Lai Y-J, et al. Inhibition of c-MET upregulates PD-L1 expression in lung adenocarcinoma. Am J Cancer Res. 2020;10:564–71. [PMC free article] [PubMed] [Google Scholar]
- 37.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez H, Zenklusen JC, Staudt LM, Doroshow JH, Lowy DR. The next horizon in precision oncology: Proteogenomics to inform cancer diagnosis and treatment. Cell. 2021;184:1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satpathy S, Krug K, Jean Beltran PM, Savage SR, Petralia F, Kumar-Sinha C, et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell. 2021;184:4348–71.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Yang Q, Zeng X, Wang M, Dong S, Yang B, et al. MET Amplification Attenuates Lung Tumor Response to Immunotherapy by Inhibiting STING. Cancer Discov. 2021;11:2726–37. [DOI] [PubMed] [Google Scholar]
- 42.Warren CFA, Wong-Brown MW, Bowden NA. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019;10:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y-Z, Wang B-S, Jiang Y-Y, Cao J, Hao J-J, Zhang Y, et al. MCMs expression in lung cancer: implication of prognostic significance. J Cancer. 2017;8:3641–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scagliotti G, von Pawel J, Novello S, Ramlau R, Favaretto A, Barlesi F, et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol. 2015;33:2667–74. [DOI] [PubMed] [Google Scholar]
- 45.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science [Internet]. 2018;362. Available from: 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, et al. CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov. 2018;8:216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell. 2018;175:984–97.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2019;176:775–89.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For this study, we used publicly available data from multiple resources: (i) human tumor tissue data (from the (TCGA cohort, https://portal.gdc.cancer.gov/), (ii) proteomic data from CPTAC (https://cptac-data-portal.georgetown.edU/cptac/s/S056), and (iii) cell line data (via the DependencyMap [https://depmap.org/portal/]). In addition, we used a LUAD immunotherapy cohort data from Korean patients at Samsung Medical Center (SMC) for validation. All raw and processed sequencing data generated in this study have been submitted to the European Nucleotide Archive (EGA; https://ega-archive.org/) under accession number EGAS00001006461. Analysis scripts used in this study are available at https://github.com/getzlab/LUAD_subtypes.