Abstract
The monkeypox virus (MPXV) outbreak confirmed in May 2022 in non-endemic countries is raising concern about the pandemic potential of novel orthopoxviruses. Little is known regarding MPXV immunity in the context of MPXV infection or vaccination with vaccinia-based vaccines (VACV). As with vaccinia, T cells are likely to provide an important contribution to overall immunity to MPXV. Here, we leveraged the epitope information available in the Immune Epitope Database (IEDB) on VACV to predict potential MPXV targets recognized by CD4+ and CD8+ T cell responses. We found a high degree of conservation between VACV epitopes and MPXV and defined T cell immunodominant targets. These analyses enabled the design of peptide pools able to experimentally detect VACV-specific T cell responses and MPXV cross-reactive T cells in a cohort of vaccinated individuals. Our findings will facilitate the monitoring of cellular immunity following MPXV infection and vaccination.
Keywords: MPXV, VACV, monkeypox, mpox, vaccinia virus, orthopoxvirus, T cell epitope, infectious disease, sequence conservation
Graphical abstract

Grifoni et al. developed orthopox- and monkeypox-specific epitope pools to measure monkeypox T cell responses in natural infection and vaccination. The pools were validated by detection of memory T cell responses in PBMCs from Dryvax vaccinees. A majority of the Dryvax-vaccinee CD4 responses were cytotoxic and produced granzyme B.
Introduction
On August 24th, 2022, the World Health Organization (WHO) reported 41,664 confirmed cases of monkeypox virus (MPXV) infection and five deaths in non-endemic regions.1 Although MPXV infections and outbreaks have been reported on the African continent in the past three decades, this current outbreak is unprecedented in size and scope; having spread globally to almost 100 countries, the vast majority of these countries have not historically reported MPXV cases, including European countries and the US.2
With the current outbreak, it is important to understand immunity against MPXV, but only a few studies have addressed immune responses to MPXV infections in humans.3 , 4 , 5 First, little information is available on the quality and duration of immune responses to MPXV infection in humans. Second, little data on efficacy in humans are available for the MPXV vaccines based on the vaccinia virus (VACV). Third, it is unknown to what extent human cellular immune responses induced by VACV vaccination are cross-reactive with MPXV. These knowledge gaps should be addressed in MPXV and also in the strain associated with the current outbreak.6
Information is available on immune responses and correlates of protection from VACV infection,7 , 8 , 9 the virus utilized as a vaccine to protect from smallpox disease caused by variola virus (VARV) infection.10 Several VACV studies demonstrate that antibody responses are crucial for disease prevention,8 , 9 whereas T cell responses are important to control and terminate pox-virus infections.11 , 12 Many studies describe T cell epitopes for Orthopoxviruses (OPXVs), and VACV in particular;12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 however, only two studies have investigated T cell responses against MPXV in humans.20 , 21
VACV was utilized under the brand name of Dryvax to eradicate smallpox in the 1980s,22 but Dryvax-vaccinated individuals with impaired cellular immunity were deficient in VACV control23 , 24 and risked severe reactions and safety issues.25 Dryvax and Acambis 2000 (the related attenuated vaccine) vaccination was largely discontinued after 2001 and replaced by the modified vaccinia Ankara (MVA) virus (under the brand name JYNNEOS), which has a superior safety profile26 and induces similar levels of antibody responses.27
Studies in non-human primate models using VACV vaccination to prevent MPXV infection show efficacy in preventing infection and/or attenuating disease severity.5 , 28 , 29 , 30 Data from human studies are limited to an observational study demonstrating 85% efficacy in preventing MPXV disease in subjects vaccinated with Dryvax.31 The JYNNEOS vaccine is approved for use to prevent MPXV infection/disease based on serological responses, but no data are available addressing clinical efficacy in humans. An additional knowledge gap is the degree of T cell epitope conservation elicited by VACV vaccination for MPXV infection. MPXV shares 90% overall sequence homology with VACV,32 suggesting that VACV-induced T cell responses might be largely cross-reactive with MPXV epitopes. VACV epitopes are largely conserved in VARV,33 suggesting a similar strategy can be applied to other OPXVs. Here, we used the Immune Epitope Database and Analysis Resource (IEDB)34 and Virus Pathogen Resource (ViPR)35 to compile known OPXV T cell epitopes to determine protein conservation and assess immunodominance.
We previously showed that large pools of peptides (megapools [MPs])36 can be used to measure CD4+ and CD8+ T cell responses against a number of allergens, as well as bacterial and viral targets.37 , 38 , 39 , 40 , 41 Here, we develop and validate pools of previously defined epitopes to assess T cell responses to VACV and MPXV.
Results
Orthopox T cell epitopes curated in IEDB and conservation of Orthopox epitopes within MPX
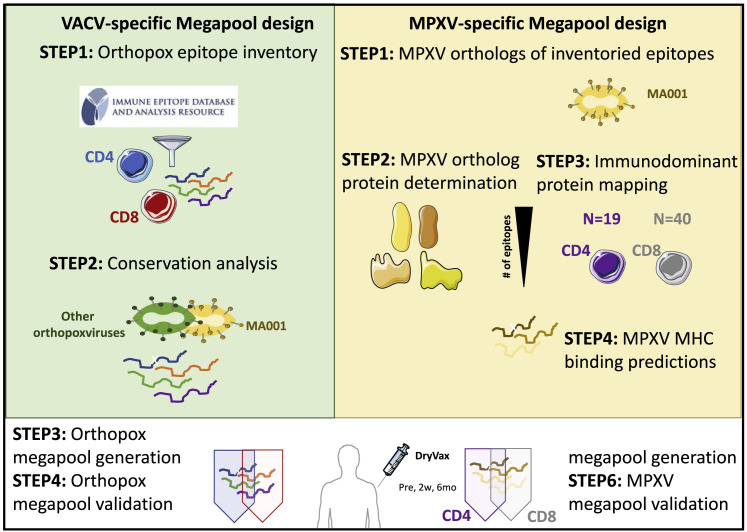
Pox viruses have relatively large genomes, encoding ∼200 different open reading frames (ORFs),32 with studies reporting broad immune responses to many ORFs.19 To define MPs to measure T cell responses in MPXV vaccination and infection, we inventoried experimentally defined epitopes described in the literature and curated by IEDB, as of May 2022 (step 1, Figure 1 A). The analysis identified 318 CD4+ and 659 CD8+ T cell epitopes derived from OPXVs (Table S1). The vast majority of epitopes (88%) have been described in the context of VACV, of which 78% of CD4+ epitopes and 36% of CD8+ epitopes were associated with responses in humans (Table S1).
Figure 1.
Schematic representation of peptide pools design
Based on these data, we developed two MPs: OPX-CD8-E and OPX-CD4-E. For OPX-CD8-E, we selected the 238 CD8+ epitopes recognized in humans, whereas for OPX-CD4-E, we included all 318 CD4+ epitopes recognized in any species, based on the high degree of overlap between binding repertoires of MHC class II.42 For the CD4+ epitopes, we performed a clustering analysis to create epitope regions of up to 22 residues that encompass nested or overlapping epitopes. Accordingly, a set of 300 CD4+ epitopes was generated and is listed in Table S1 along with the CD8+ epitopes.
The MPXV and VACV viruses have been reported32 to share a high degree of sequence homology and conservation, suggesting that Orthopox-specific T cell epitopes may be conserved in MPX.6 We ascertained whether each epitope was conserved in the MPXV isolate MA001 (step 2, Figure 1A).
The results indicate that both CD4+ and CD8+ epitopes are highly conserved with 94% and 82%, respectively, being 100% conserved in MPXV (Table S1), with high conservation (range 74%–96%) irrespective of the viral species in which the epitopes were originally defined. The results indicated that most previously defined Orthopox epitopes were highly conserved and could be used to generate MPs as a potential reagent (step 3, Figure 1A).
T cell immune responses after Dryvax vaccination are detected by Orthopox MPs
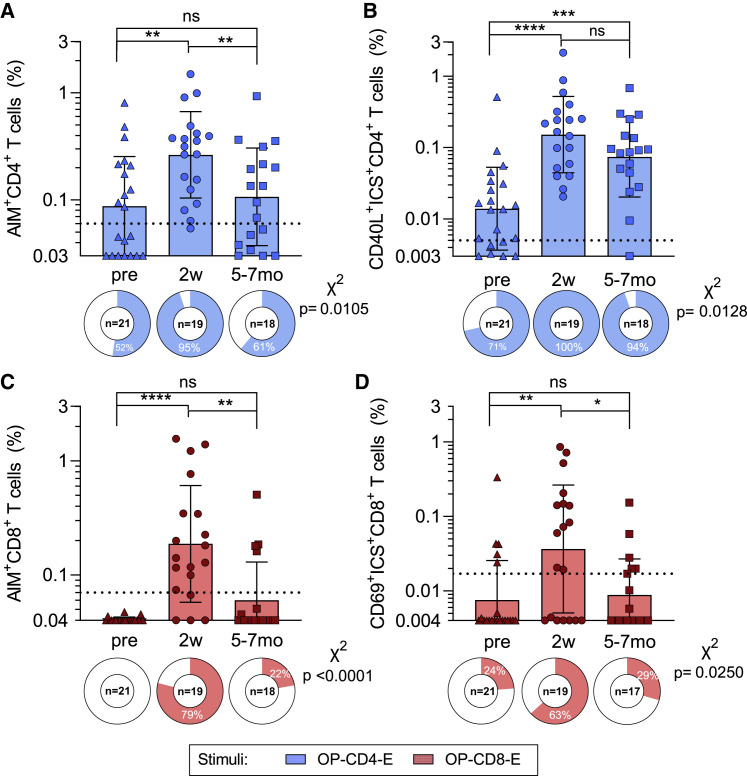
We evaluated T cell responses for their capacity to recognize the Orthopox MPs, using peripheral blood mononuclear cells (PBMCs) in cohorts of Dryvax-vaccinated and non-vaccinated subjects (step 4, Figure 1A). Cohort characteristics are provided in Table S2 and STAR Methods. To measure the T cell responses to OPX-CD8-E and OPX-CD4-E, we combined activation-induced marker (AIM) assays with cytokine intracellular staining (ICS) (Figure S1A). For CD4+ T cells, the highest responses were observed in recently vaccinated subjects. The magnitude of T cell reactivity was comparable pre-vaccination and 5–7 months post-vaccination (geometric mean [GM] ± geometric standard deviation [GSD]; pre = 0.09 ± 2.90, 2 weeks = 0.26 ± 2.53, and 5–7 months = 0.11 ± 2.86; p = 0.004 Kruskal-Wallis test; Figure 2 A). The frequency of CD4+ T cell responders increased from 52% to 95% 2 weeks post-vaccination with a decline to 61% 5–7 months post-vaccination (χ2 p = 0.01).
Figure 2.
T cell responses against Orthopox MPs after vaccinia vaccination; refers to Figures S1 and S2 and Table S1. List of orthopox IEDB epitopes, related to Figures 1 and 3, Table S4. List of predicted MPX CD4 and CD8 epitopes, related to Figures 1 and 4
PBMCs from each time point were tested in AIM/ICS assays with experimentally defined OPXV MPs, OP-CD4-E (blue), and OP-CD8-E (red).
(A and B) OP-CD4-E-specific CD4 + T cell reactivity is shown for (A) AIM and (B) ICS.
(C and D) OP-CD8-E specific CD8 + T cell reactivity is shown for (C) AIM and (D) ICS. The y axis of each bar graph starts at the limit of detection (LOD), and the limit of sensitivity (LOS) is indicated with a dotted line. Columns represent the geometric mean and error bars indicate the geometric standard deviation. Pie charts below each bar indicate the frequency of positive responders. Mann-Whitney t test was applied to each graph, and p values symbols are shown when significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. X2 test was applied to the frequencies of positivity, and p values are listed on the right of each graph.
Similar findings were observed by ICS, although CD4+ T cell responses showed a more modest and insignificant decline 5–7 months post-vaccination; responses post-vaccination were significantly higher in magnitude and frequency than in pre-vaccination (GM ± GSD; pre = 0.01 ± 3.80, 2 weeks = 0.15 ± 3.42, and 5–7 months = 0.07 ± 3.64; p < 0.0001 Kruskal-Wallis test, χ2 p = 0.0128; Figure 2B). Unexposed subjects did not yield appreciable responses (Figures S1B and S1C). The ICS assays demonstrated that CD40L+CD4+ T cell responses were Th1 or cytotoxic responses, encompassing mostly granzyme B (GZMB) and IFNγ, followed by TNF-α and IL-2 (Figure S2). Significant increases in TNF-α and IL-2 and a decrease in GZMB production were observed at 5–7 months post-vaccination, as compared with 2 weeks (TNF-α: 2 weeks = 18% and 5–7 months = 33%, p = 0.0088; IL-2: 2 weeks = 3% and 5–7 months = 13%, p = 0.0059; GZMB: 2 weeks = 53% and 5–7 months = 35%, p = 0.0292). A prevalence of CD40L+IFNγ+GZMB+ population followed by CD40L+GZMB+, CD40L+IFNγ+, and CD40L+TNF-α+ populations were observed (Figures S2A and S2E).
In CD8+ T cells in AIM assays, the highest responses were observed in recently vaccinated subjects, with the 5–7 months post-vaccination samples reverting to a magnitude similar to pre-vaccination (GM ± GSD; pre = 0.04 ± 1.04, 2 weeks = 0.19 ± 3.24, and 5–7 months = 0.06 ± 2.15; p < 0.0001 Kruskal-Wallis test; in Figure 2C). The frequency of positive CD8+ T cell responses increased to 79% 2 weeks post-vaccination and decreased to 22% at the 5–7 months’ time point (χ2 p < 0.0001). Similar results were observed in the ICS assay, where IFNγ, GZMB, TNF-α, and IL-2 secreting CD69+CD8+ T cells were quantified (GM ± GSD; pre = 0.01 ± 3.39, 2 weeks = 0.04 ± 7.22, and 5–7 months = 0.01 ± 3.06; p = 0.01 Kruskal-Wallis test, χ2 p = 0.025; Figure 2D). CD8+ T cell functionality was also assessed based on IFNγ, TNF-α, IL-2, and/or GZMB expression and showed a prevalence of IFNγ followed by IL-2 and GZMB (Figures S2C–S2F).
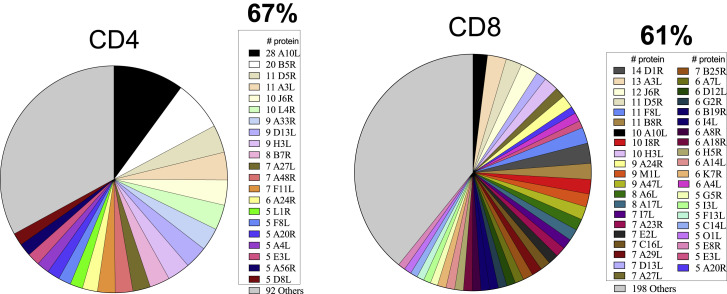
Definitionof immunodominant ORFs and prediction of MPX T cell epitopes
We next wanted to define MPs based on MPXV sequences that would be more suited to characterize responses in MPXV infection. We started with the same list of Orthopox epitopes (Table S1) and identified corresponding conserved sequences in MPXV (step 1, Figure 1B) with sequence identities of ≥70% for class II and ≥80% for class I epitopes. By mapping those epitopes to the corresponding MPXV ortholog proteins (step 2, Figure 1B), we derived a set of MPXV proteins that is potentially immunodominant (see Table S3). Accordingly, we identified 19 antigens for CD4+ and 40 for CD8+ (step 3, Figure 1B). The 19 MPXV CD4+ ORFs accounted for 67% of the homologous IEDB Orthopox CD4+ epitopes, and the 40 CD8+ ORFs accounted for 61% of the corresponding IEDB CD8+ epitopes (Figure 3 ). Overall, 48 proteins were immunodominant, corresponding to 25% of the MPXV proteome. T cell immunodominant targets identified were mostly early transcribed proteins.43 Several targets, such as A10L, A3L, D5R, A26L/A30L, H3L, J6R, A27L, D13L, A24R, A4L, and F8L, are of particular interest, since they were dominant for both CD4+ and CD8+ T cells. The study of immunodominant ORFs is also of particular relevance, since JYNNEOS vaccine is based on MVA, an attenuated VACV that lost 14% of the original genome,44 retaining only 157 ORFs (GenBank: U94848.1).45 Importantly, all the ORFs considered as dominant for CD4+, CD8+, or both were conserved in the MVA/JYNNEOS sequences. This confirms the large breadth of immunogenic ORFs in OPXVs.15 We predicted potential T cell epitopes from the MPXV orthologs of the 19 CD4+ and 40 CD8+ immunodominant antigens, using IEDB tools46 (step 4, Figure 1B). For CD4+, we predicted 276 promiscuous HLA class II binders (Table S3).47 In parallel, we predicted 1,647 potential CD8+ epitopes binding to a panel of common HLA class I alleles, as previously utilized in other viral systems39 , 48 (step 5, Figure 1B; Document S1. Figures S1 and S2 and Tables S2 and S3, Table S4. List of predicted MPX CD4 and CD8 epitopes, related to Figures 1 and 4).
Figure 3.
Orthopox protein immunodominance; refers to Table S1
The IEDB was mined for experimentally defined epitopes derived from Orthopoxviruses. A total of 47 different antigens were identified as the protein source for defined epitopes, corresponding to 19 and 39 antigens associated with CD4 + or CD8+ epitopes, respectively. The pie charts represent the total proteins recognized by CD4+ and CD8+ as defined by the IEDB and listed in Table S1. The most immunodominant antigens are listed in the figure legend, and the remaining antigens (“others”) are colored in gray.
Assessment of CD4+ and CD8+ T cell cross-reactive responses able to recognize MPXV in smallpox-vaccinated individuals
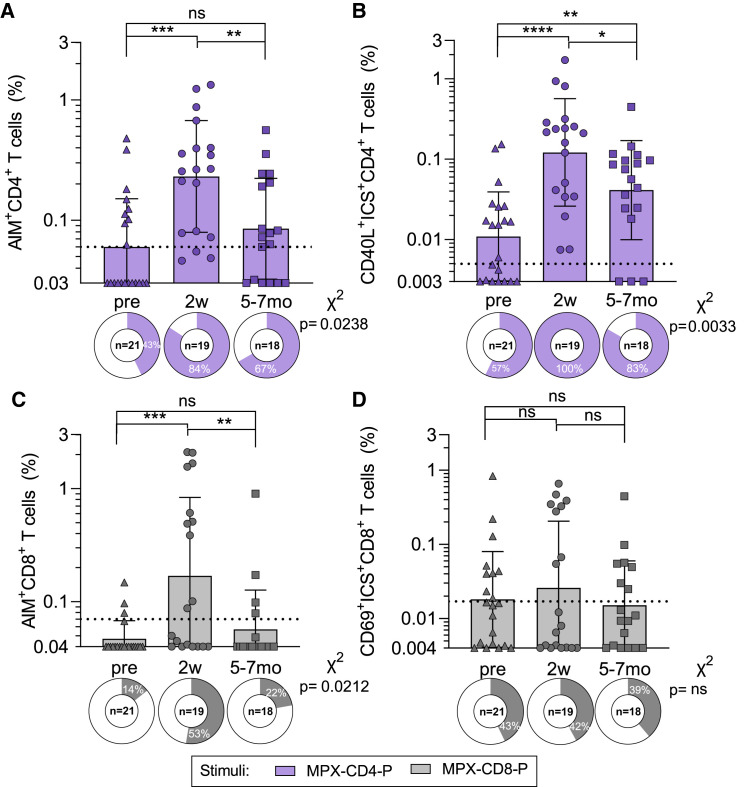
We then evaluated whether VACV-induced T cell responses could cross-recognize the MPXV-derived, predicted epitope pools (Figures 4 and S2). By the AIM assay, the magnitude of CD4+ T cell cross-reactivity peaked at 2 weeks post-vaccination and declined at 6–7 months, reaching comparable reactivity to pre-vaccination (GM ± GSD; pre = 0.06 ± 2.51, 2 weeks = 0.23 ± 2.91, and 5–7 months = 0.08 ± 2.63; p = 0.0005 Kruskal-Wallis test; Figure 4A). The frequency of CD4+ T cell responders significantly increased from 43% to 84% 2 weeks post-vaccination, with a decline to 67% 5–7 months post-vaccination (χ2 p = 0.024). Similar results were observed by ICS, although the post-vaccination decline was less pronounced and responses were still significantly higher than those observed pre-vaccination (GM ± GSD; pre = 0.011 ± 3.58, 2 weeks = 0.12 ± 4.67, and 5–7 months = 0.04 ± 4.13; p < 0.0001 Kruskal-Wallis test; Figure 4B). Finally, the functionality of CD4+ T cells was comparable to what was observed for the OPXV-specific T cell responses (Figures S2B and S2E).
Figure 4.
T cell responses able to cross-recognize MPXV MPs after vaccinia vaccination; refers to Figures S1 and S2 and Table S1. List of orthopox IEDB epitopes, related to Figures 1 and 3, Table S4. List of predicted MPX CD4 and CD8 epitopes, related to Figures 1 and 4
PBMCs from each time point were tested in AIM/ICS assays with predicted MPXV MPs, MPXV-CD4-P (purple), and 5 pools of MPXV-CD8-P1-P5 summed to get the overall MPXV-CD8-P reactivity (gray).
(A and B) MPXV-CD4-P-specific CD4 + T cell reactivity is shown for (A) AIM and (B) ICS.
(C and D) MPXV-CD8-P-specific CD8 + T cell reactivity is shown for (C) AIM and (D) ICS. The y axis of each bar graph starts at the LOD, and the LOS is indicated with a dotted line. Columns represent the geometric mean and error bars indicate the geometric standard deviation. Pie charts below each bar indicate the frequency of positive responders. Mann-Whitney t test was applied to each graph, and p values symbols are shown when significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. X2 test was applied to the frequencies of positivity, and p values are listed on the right of each graph.
CD8+ T cell responses were also able to cross-recognize the MPXV pools. Reactivity peaked at 2 weeks and further declined 5–7 months post-vaccination, with comparable reactivity with the pre-vaccination by AIM (GM ± GSD; pre = 0.05 ± 1.44, 2 weeks = 0.17 ± 4.93, and 5–7 months = 0.06 ± 2.23; p = 0.0014 Kruskal-Wallis test, χ2 p = 0.0212; Figure 4C) and a less pronounced decline by ICS (GM ± GSD; pre = 0.02 ± 4.42, 2 weeks = 0.03 ± 7.97, and 5–7 months = 0.02 ± 3.96; p = 0.8653 Kruskal-Wallis test, χ2 p = 0.9961; Figure 4D). The quality of CD8+ T cells was comparable to and driven by IFNγ production, although an increase in IL-2 was also observed (Figures S2D and S2F). Little reactivity was again observed in non-vaccinated subjects for both CD4+ and CD8+ (Figures S1D and S1E).
Discussion
There is an urgent need to understand adaptive immune responses to MPXV in both natural immunity and vaccination. This study is focused on CD4+ and CD8+ T cell responses and peptide MPs to detect these responses. In the context of SARS-CoV-2, we have used three different approaches to design MPs. The first one uses experimentally defined epitopes of highly homologous viral species and typically generates pools with higher activity, but it is limited to described epitopes and may underrepresent less frequent HLAs. A second approach utilizes T cell epitope predictions targeting common HLA alleles.49 It is limited by the accuracy of bioinformatic predictions, but it can be valuable when little information is available on the actual epitopes recognized in a population and/or when the target pathogen is particularly large so use of complete peptide sets is unfeasible. The third approach utilizes overlapping peptides spanning the entire antigen sequence37 , 50 and is the most comprehensive agnostic approach. This approach requires synthesis and testing of many peptides and becomes unfeasible with larger genomes, such as the Orthopoxviridae, which contain ≥200 ORFs. Accordingly, we defined dominant cross-reactive ORFs to generate peptide MPs to measure T cell responses, which we plan to make available to the research community. These reagents were validated by assessing T cell responses induced by Dryvax vaccination in PBMC samples.
Because all current vaccinations against MPXV are based on VACV/MVA, it was important to determine the sequence conservation frequency of previously defined VACV T cell epitopes curated in the IEDB with the current outbreak MA001 strain reference. Overall, the median degree of amino acid sequence conservation was 61%–67%, implying that responses induced by VACV vaccination should recognize ortholog protein sequences in the MPXV genome. McKay and collaborators reported a similar sequence conservation frequency of 70% between MPXV-2022 and the MPXV-CB (Congo Basin) strains.6 VACV-reactive T cells recognizing MPXV-derived epitopes are not surprising because they cross-react to ectromelia virus epitopes as shown on a mousepox infection model.51 This observation enabled the definition of Orthopox-specific MP reagents.
The IEDB epitope data were utilized to define dominant ORFs recognized by Orthopox-specific T cell responses. This analysis revealed the remarkable breadth of CD4+ and CD8+ T cell immune responses, consistent with earlier reports.18 , 52 Nineteen ORFs were required to cover 67% of CD4+ T cell responses, and 40 ORFs were required to cover 61% of CD8+ T cell responses. This significant finding suggests that focusing on 10%–25% of the 200+ ORFs typically encoded in pox genomes will still capture the majority of T cell responses. This observation enabled the definition of MPXV-specific MP reagents. The definition of dominant ORFs provided insights into the mechanisms underlying the development of these responses, showing that dominant antigens are predominantly early ORFs, confirming earlier studies.52 In vaccination, we note that all dominant ORFs were conserved in MVA/JYNNEOS and had orthologs in MPXV. This suggest that the response directed to MPXV induced by MVA should mirror the response induced by previous VACV vaccines, with known clinical efficacy against MPXV in humans.
We developed peptide pools based on experimentally defined Orthopox T cell epitopes or predicted T cell epitopes derived from the most immunodominant ortholog proteins of MPXV, similar to the approach we used for SARS-CoV-2.37 , 48 The use of epitope pools has the important potential advantage of obviating the use of infected cells to quantify T cell responses, which is prone to interference by pox-virus expression of immune antagonizing genes53 and is associated with biosafety concerns.
These Orthopox MPs were validated using PBMC from subjects who received the Dryvax vaccine. Early time points (2 weeks from vaccination) were associated with positive responses in 100% and 63% of the subjects for CD4+ and CD8+ T cells, respectively, and decreased 5–7 months after vaccination. CD4+ and CD8+ T cell responses to the pools of predicted MPXV epitopes were similarly detected in 84% and 52% of vaccinees 2 weeks post-vaccination and also decreased 5–7 months after vaccination. Our results are consistent with the report of Amara et al. that shows a better persistence of CD4+ T cells, with a 2-fold contraction between effector and memory phase, in contrast with the CD8+ T cell response that shows a 7-fold contraction.54 Hammarlund and Slifka reported that the humoral and cellular T cell responses are long-lasting, with a half-life of 8–15 years.55 This apparent discrepancy can be reconciled considering that in the Hammarlund-Slifka report, the immune response is measured 1+ years after vaccination. Here, we compare pre-vaccination and 6-month post-vaccination samples. Considering cytokine+ CD4+ T cell responses, which mirror the readout of Hammarlund and Slifka, positive responses observed are 94% 6 months post-vaccination and 71% for pre-vaccinated with a previous history of vaccination in the 5–20 years range. Thus, the % of positivity is comparable. In the context of CD8+, we observed a frequency of response of 30%, whereas Hammarlund, noting several negative donors, reports an overall higher % of responses. Additionally, we observed that human CD4+ T cell responses are associated with a large fraction of GZMB-secreting, antigen-specific cells. It was previously reported that the role of CD4+ T cells in protection from VACV and MPXV infections outweighs the contribution provided by CD8+ T cells in macaques.29 , 56 The current data suggest that in addition to their role supporting the development of antibody responses, their longevity and a cytotoxic component could also contribute to a sustained antiviral function of CD4+ T cells.
In conclusion, the use of available information related to VACV epitopes in conjunction with bioinformatic predictions points to specific regions that are conserved across several OPXV species, including MPXV, making them suitable for vaccine evaluation. To the best of our knowledge, this is the first demonstration of the use of epitope MPs suitable to characterize vaccine-specific responses and also likely to detect immune responses in the context of MPXV infection and disease.
Limitations and future directions
The cohort selected has the following limitations: cross-sectional sample collection decreases the accuracy of kinetic analysis of responses, a lack of information on childhood vaccination status, and inclusion of healthcare workers previously exposed to occupational vaccination before the Dryvax vaccination administered in relation to this specific study. Antigen selection for MPXV was based on studies performed considering other OPXVs (mainly VACV). Future studies might use overlapping peptide pools spanning various ORFs to define in more detail the ORFs specifically recognized following MPX vaccination or infection. Most previously defined epitopes are focused on more common HLAs, and this might impact detecting responses from individuals with dominant responses restricted by rare HLAs, although this is partially addressed by the prediction of a set of alleles with over 95% coverage in the general population worldwide. The use of peptide pools representing immunodominant viral protein targets may not fully reflect physiological targets. The current analysis did not address antibody responses, which are the dominant correlate of vaccine-induced protection. A strong limitation of the current study is the lack of validation of the MPXV pools in MPXV-infected individuals. This is important because while OPXVs are highly related, their pathogenesis—including processing and presentation of T cell epitopes and being able to evade innate and adaptive immunity—within different hosts may vary. Future directions will include the following: (1) using the MPs to characterize immune responses in acute and convalescent MPXV natural infection, (2) addressing the Th and memory phenotypes of responding T cells, (3) comparing responses induced by different vaccines, (4) widely disseminating the MPs to the scientific community, and (5) further optimizing these reagents.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD8 BUV496 (clone RPA-T8) | BD Biosciences | Cat# 612942; RRID:AB_2870223 |
| Mouse anti-human CD3 BUV805 (clone UCHT1) | BD Biosciences | Cat# 612895; RRID:AB_2870183 |
| Mouse anti-human TNF alpha eFluor450 (clone MAb11) | Life Tech | Cat# 48-7349-42; RRID:AB_2043889 |
| Mouse anti-human CD14 V500 (clone M5E2) | BD Biosciences | Cat# 561391; RRID:AB_10611856 |
| Mouse anti-human CD19 V500 (clone HIB19) | BD Biosciences | Cat# 561121; RRID:AB_10562391 |
| Mouse anti-human CD4 BV605 (clone RPA-T4) | BD Biosciences | Cat# 562658; RRID:AB_2744420 |
| Mouse anti-human IFN gamma FITC (clone 4S.B3) | Invitrogen (Thermo Fisher Scientific) | Cat# 11-7319-82; RRID:AB_465415 |
| Rat anti-human IL-2 BB700 (clone MQ1-17H12) | BD Biosciences | Cat# 566405; RRID:AB_2744488 |
| Mouse anti-human CD69 PE (clone FN50) | BD Biosciences | Cat# 555531; RRID:AB_395916 |
| Mouse anti-human CD134 (OX40) PE-Cy7 (clone Ber-ACT35) | BioLegend | Cat# 350012; RRID:AB_10901161 |
| Mouse anti-human CD137 APC (clone 4B4-1) | BioLegend | Cat# 309810; RRID:AB_830672 |
| Mouse anti-human Granzyme B AF700 (clone GB11) | BD Biosciences | Cat# 560213; RRID:AB_1645453 |
| Mouse anti-human CD154 (CD40 Ligand) APC-ef780 (clone 24-31) | eBioscience (Thermo Fisher Scientific) | Cat# 47-1548-42; RRID:AB_1603203 |
| Biological samples | ||
| Pre-vaccinated donor PBMCs samples | LJI Clinical Core | N/A |
| Two weeks post vaccinated donor PBMCs samples | LJI Clinical Core | N/A |
| 5-7 months post vaccinated donor PBMCs samples | LJI Clinical Core | N/A |
| Unexposed donors PBMCs samples | LJI Clinical Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Brilliant Staining Buffer Plus | BD Biosciences | Cat# 566385; RRID:AB_2869761 |
| Live/Dead Viability Dye eFluor506 | Invitrogen (Thermo Fisher Scientific) | Cat# 65-0866-14 |
| Synthetic peptides | TC Peptide Lab | https://www.tcpeptide.com |
| Deposited data | ||
| OPXV and MPXV peptides | This study | Table S1 |
| OPXV and MPXV megapools (MP) | This study | Table S4 |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| FlowJo 10.8.1 | FlowJo | https://www.flowjo.com/; RRID:SCR_008520 |
| IEDB | Immune Epitope DataBase | https://www.iedb.org; RRID:SCR_006604 |
| ViPR | Virus Pathogen Resource | http://www.viprbrc.org; RRID:SCR_010685 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alessandro Sette (alex@lji.org).
Materials availability
Aliquots of synthesized sets of peptides identified in this study are available from the lead contact. There are restrictions to the availability of the peptide reagents due to cost and limited quantity.
Experimental model and subject details
Cohort of VACV vaccinees to assess T cell responses
The characteristics of the study population that donated the banked PBMC utilized for the present study was described previously.15 Healthy male and female donors between 7 and 62 years of age that had received a vaccinia virus (Dryvax) vaccination as a prophylactic measure, either because of their potential exposure to vaccinia in a laboratory or hospital setting, or because of their enrollment into military and health worker vaccination programs, within one year of providing the blood donation. PBMCs were collected at 2 weeks (n=19) or 5-7 months after vaccination (n=18). In addition, we also utilized two additional control cohorts. For the first control cohort, Pre-vaccination (“pre”) PBMC samples (n=21) were collected from a similar cohort of healthcare donors prior to vaccination. Since some of the pre-vaccinated donors had received childhood Smallpox vaccination, a second control cohort of truly unexposed donors (n=15) was enrolled; this cohort consisted of individuals born after 1980, the official year of worldwide eradication of Smallpox and eight years after the United States stopped childhood Smallpox vaccinations. Further, this unexposed cohort had no history of occupational vaccination with Smallpox. Characteristics of the donor cohorts are also summarized in Table S2. Institutional Review Board approval and appropriate consent were obtained for this study.
PBMCs
For all donors in this study, PBMCs were isolated from heparinized blood by density gradient centrifugation with a Histopaque-1077 and cryopreserved in 10% DMSO in FBS prior to long term storage in liquid nitrogen freezers. The PBMC isolation of these specific donors has been described in greater detail by Oseroff et al.57
Method details
IEDB analysis of Orthopoxvirus-derived T cell epitopes
Known OPXV-derived T cell epitopes reported in the published literature, or through direct database submission, were identified by searching the IEDB at the end of May, 2022. Queries were performed broadly for the Orthopox genus, using NCBI taxonomy ID 10242, and specifying positive T cell assays. This retrieved 1076 records, from which 31 were removed because responses had not been defined in the context of either MHC class I or class II. For epitopes with responses in the context of class II, the set was further filtered to select epitopes of 12-25 residues, comporting with the canonical size of class II ligands associated with CD4+ T cell responses. Epitopes with class I responses were filtered to select those of 8-11 residues, canonical for class I ligands associated with CD8+ T cell responses. As a result, a final set of 977 epitopes, including 318 associated with class II responses, and 659 with class I responses, was identified for subsequent analyses. About 70% of the epitope data in this set is derived from the peer-reviewed literature.
Identification of Orthopox T cell epitope homologs in MPXV
The MPXV_USA_2022_MA001 (MA001) isolate (GenBank accession ON563414) was selected as the representative strain of the 2022 MPXV outbreak because MA001 was the first sequence from the 2022 outbreak deposited in GenBank58 and had been annotated by BV-BRC (www.bv-brc.org). The protein sequences of MA001 were retrieved from the BV-BRC website (=https://www.bv-brc.org/view/Genome/10244.322#view_tab=overview) on May 25, 2022. To identify the MPXV homologs of VACV antigens, a BLAST based homology search was used. All the 26 VACV antigens related to IEDB T cell epitopes except for VP8 and A47 had high identity (>83%) and good length coverage in MA001. With regard to the VP8 antigen we removed 1 nt insertion which caused a frameshift and re-annotated with GATU tool in ViPR (www.viprbrc.org). For the A47 antigen, neither MA001 nor the RefSeq Zaire strain had a good match and it is unlikely to have an MPXV homolog.
To assess the sequence conservation of the OPX-CD8-E and OPX-CD4-E epitope pools in MPXV, a k-mismatch string search program was developed to find all matched sequences of an input epitope. The matched sequences meet the criteria of having the same length as the input epitope, and having at most k mismatched residues in comparison to the input epitope. In case of multiple matches for the same input epitope, the program also picks the optimal match. In order to identify the MPXV protein region homologous to the OPXV T cell epitopes, a k-mismatch string search method was used. Conceptually, the k-mismatch string search program searches through a protein sequence file using a fixed-size sliding window and identifies all windows with a maximum of k-mismatches compared to the input epitope sequence. In identifying OPXV epitope homologs in MPXV, the search pool used included all MA001 protein sequences, while the maximum number of k mismatches was set to be the larger of 20% of the input epitope length and 1, i.e., k = max (epitope length ∗ 0.2, 1). We additionally set up a maximum of 2 and 3 mismatches for class I and class II epitopes, respectively.
Besides finding all epitope homologs in proteins, the epitope search program also picks the best match if multiple matches were found. The best match was defined as the one with the smallest number of mismatches, and in case of ties, the one(s) with the least shift in the start coordinate compared with the input epitope.
In validating the epitope search result, two metrics were used: (1) whether the epitope hit resides in a protein homologous to the input epitope's parent protein identified from the pairwise analysis, and (2) whether the start coordinate of the epitope hit was near the start coordinate of the input epitope. In case of a start coordinate shift of 10 or more residues, the sequences of the parent proteins were aligned and then manually examined to see if the match was a false positive. For all epitopes evaluated using these criteria, only one was found to be a false positive following manual curation and was excluded from the downstream analysis.
T cell epitope predictions
Epitope prediction was carried out using the various dominant MPXV ORFs described above (Table S3). For CD4+ T cell epitope prediction, we applied a previously described algorithm that was developed to predict dominant HLA class II epitopes, using a median consensus percentile of prediction cutoff ≤ 20 percentile as recommended.47 For CD8+ T cell epitope prediction, we selected the 12 most frequent HLA class I alleles in the worldwide population,59 , 60 using a phenotypic frequency cutoff ≥ 6%. The specific alleles included were: HLA-A∗01:01, HLA-A∗02:01, HLA-A∗03:01, HLA-A∗11:01, HLA-A∗23:01, HLA-A∗24:02, HLA-B∗07:02, HLA-B∗08:01, HLA-B∗35:01, HLA-B∗40:01, HLA-B∗44:02, HLA-B∗44:03. HLA class I binding predictions were performed using the IEDB recommended class I prediction algorithm (as recommended in June, 2022) and selecting for each allele the top 1 percentile of peptides based on the total amino acid sequences of the 40 MPXV antigens selected. This initial list of epitopes was then filtered to eliminate redundancies and nested peptides by clustering61 to a single occurrence, and nested peptides were included within longer sequences, up to 12 residues in length, before assigning the multiple corresponding HLA restrictions for each region.
Peptide synthesis and Megapool preparation
OPXV and MPXV peptides were synthesized as crude material (TC Peptide Lab, San Diego, CA), and then individually resuspended in dimethyl sulfoxide (DMSO) at a concentration of 10–20 mg/mL. Aliquots of all peptides were pooled into megapools (MP) designated as OP-CD4-E, OP-CD8-E, MPX-CD4-P, MPX-CD8-P1, MPX-CD8-P2, MPX-CD8-P3, MPX-CD8-P4, and MPX-CD8-P5. These MPs underwent a sequential lyophilization. The resulting lyocake was resuspended in DMSO to yield a stock solution in which each individual peptide was present at a concentration of 1 mg/mL, as previously described,37 resulting in a final test concentration of 1ug/mL in the assay after dilution. All peptides and MPs are listed in Tables S1 and S4.
AIM/ICS assay
We performed the combined AIM/ICS assay as previously described.62 In brief, after thawing, 1–2x106 PBMCs per well were cultured with the OPXV- or MXPV- specific peptide MPs (1 μg/mL of each individual peptide contained in the peptide pool). An equimolar amount of DMSO was added to the cells in triplicate wells as a negative control. Phytohemagglutinin (PHA, Roche, 1 μg/mL) was used to stimulate cells as a positive control. Treated cells were incubated at 37°C in 5% CO2 for 22 hours before the addition of Golgi-Plug containing brefeldin A, Golgi-Stop containing monensin (BD Biosciences, San Diego, CA), and the CD137 APC antibody (2:100, Biolegend, Cat# 309810) for an additional 4-hour incubation. Then the cells underwent membrane surface staining for 30 minutes at 4°C protected from light with Fixable Viability Dye eFluor506 (1:1000, eBiosience, Cat# 65-0866-14) and the following antibodies: CD3 BUV805 (1:50, BD, Cat# 612895), CD8 BUV496 (1:50, BD, Cat# 612942), CD4 BV605 (1:100, BD, Cat# 562658), CD14 V500 (1:50, BD, Cat# 561391), CD19 V500 (1:50, BD, Cat# 561121), CD69 PE (1:10, BD, Cat# 555531), CD137 APC (1:50, Biolegend, Cat# 309810), and OX40 PE-Cy7 (1:50, Biolegend, Cat# 350012). After staining, the cells were fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO), permeabilized with saponin buffer (0.5% saponin [Sigma-Aldrich, St. Louis, MO], 1% bovine serum albumin, and 0.1% sodium azide), and blocked for 15 minutes with 10% human serum (Gemini Bio-Products, Sacramento, CA) in saponin buffer. After blocking, the cells were stained intracellularly for 30 minutes at room temperature with the following antibodies: TNFα ef450(3:100, Life Tech, Cat# 48-7349-42), IFNγ FTIC (1:100, Invitrogen, Cat# 11-7319-82), IL-2 BB700 (1:25, BD, 566405), IFNγ (1:100, Invitrogen, Cat# 11-7319-82), Granzyme B Alexa700 (1:100, BD, 560213), and CD40L APC-eFluor780 (3:100, eBioscience, Cat# 47-1548-42). All samples were acquired on a ZE5 5-laser cell analyzer (Bio-Rad laboratories) and were analyzed with FlowJo software (Tree Star Inc.).
The data was analyzed to establish the Limit of Detection (LOD) and Limit of Sensitivity (LOS) based on all the DMSO-only conditions for AIM and ICS. For ICS these calculations were done on the IFNγ data. The LOD was calculated as twice the upper 95% confidence interval of the geometric mean and the LOS was calculated as two times the standard deviation from the median. Only responses with Stimulation Index (SI) > 2 were considered significant for AIM (CD4: LOS = 0.06%, SI > 2; CD8: LOS = 0.07%, SI > 2). For ICS, responses with SI>2 were considered significant for CD4+ (LOS = 0.006%) and responses with SI>3 for CD8+ (LOS = 0.017%). For AIM, the CD8+ T cell response to MPXV-CD8-P was calculated by summing the background subtracted, SI>2 and >LOS AIM data. For ICS, the same calculation was performed for the ICS data but considering an SI>3.
Quantification and statistical analysis
All the statistical analyses are described separately in each section of the STAR Methods, results and figure legends.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Department of Health and Human Services under contract nos. 75N93019C00001 and 75N9301900065 to A.S. and HHS75N93019C00076 to R.H.S. A.T. was supported by a PhD student fellowship through the Clinical and Experimental Immunology Course at the University of Genoa, Italy.
Author contributions
Conceptualization, A.G., Y.Z., R.H.S., and A.S.; data curation and bioinformatic analysis, A.G., J.S., and Y.Z.; formal analysis, Y.Z., A.T., and A.G.; funding acquisition, R.H.S. and A.S.; investigation, A.T., M.R.-C., and A.G.; resources, J.M.D. and P.R.; supervision, G.F., A.G., R.H.S., and A.S.; writing, A.G., Y.Z., R.H.S., and A.S.
Declaration of interests
L.J.I. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in out reference list.
Published: December 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.11.003.
Supplemental information
Data and code availability
All data presented and analyzed in the present study was retrieved from the IEDB (www.IEDB.org) and ViPR (www.viprbrc.org), as described below. The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental information. This study uses publicly available algorithms and does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.WHO Multi-country outbreak of monkeypox, External Situation Report #10–16 November 2022. 2022. https://www.who.int/emergencies/situation-reports
- 2.Bunge E.M., Hoet B., Chen L., Lienert F., Weidenthaler H., Baer L.R., Steffen R. The changing epidemiology of human monkeypox-A potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022;16 doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammarlund E., Dasgupta A., Pinilla C., Norori P., Früh K., Slifka M.K. Monkeypox virus evades antiviral CD4+ and CD8+ T cell responses by suppressing cognate T cell activation. Proc. Natl. Acad. Sci. USA. 2008;105:14567–14572. doi: 10.1073/pnas.0800589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammarlund E., Lewis M.W., Carter S.V., Amanna I., Hansen S.G., Strelow L.I., Wong S.W., Yoshihara P., Hanifin J.M., Slifka M.K. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 2005;11:1005–1011. doi: 10.1038/nm1273. [DOI] [PubMed] [Google Scholar]
- 5.Karem K.L., Reynolds M., Hughes C., Braden Z., Nigam P., Crotty S., Glidewell J., Ahmed R., Amara R., Damon I.K. Monkeypox-induced immunity and failure of childhood smallpox vaccination to provide complete protection. Clin. Vaccine Immunol. 2007;14:1318–1327. doi: 10.1128/CVI.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed S.F., Sohail M.S., Quadeer A.A., McKay M.R. Vaccinia virus vaccination is expected to elicit highly cross-reactive immunity to the 2022 monkeypox virus. Preprint at bioRxiv. 2022 doi: 10.1101/2022.06.23.497143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy R.B., Ovsyannikova I.G., Jacobson R.M., Poland G.A. The immunology of smallpox vaccines. Curr. Opin. Immunol. 2009;21:314–320. doi: 10.1016/j.coi.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu R., Johnson A.J., Liggitt D., Bevan M.J. Cellular and humoral immunity against vaccinia virus infection of mice. J. Immunol. 2004;172:6265–6271. doi: 10.4049/jimmunol.172.10.6265. [DOI] [PubMed] [Google Scholar]
- 9.Amanna I.J., Slifka M.K., Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 2006;211:320–337. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 10.Carroll M.W., Moss B. Poxviruses as expression vectors. Curr. Opin. Biotechnol. 1997;8:573–577. doi: 10.1016/s0958-1669(97)80031-6. [DOI] [PubMed] [Google Scholar]
- 11.Pauli G., Blümel J., Burger R., Drosten C., Gröner A., Gürtler L., Heiden M., Hildebrandt M., Jansen B., Montag-Lessing T., et al. Orthopox viruses: infections in humans. Transfus. Med. Hemother. 2010;37:351–364. doi: 10.1159/000322101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing L., Chong T.M., McClurkan C.L., Huang J., Story B.T., Koelle D.M. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J. Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terajima M., Cruz J., Raines G., Kilpatrick E.D., Kennedy J.S., Rothman A.L., Ennis F.A. Quantitation of CD8+ T cell responses to newly identified HLA-a∗0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J. Exp. Med. 2003;197:927–932. doi: 10.1084/jem.20022222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler I., Staib C., Kastenmuller W., Stevanović S., Schmidt B., Lemonnier F.A., Rammensee H.G., Busch D.H., Bernhard H., Erfle V., Sutter G. Identification of vaccinia virus epitope-specific HLA-A∗0201-restricted T cells and comparative analysis of smallpox vaccines. Proc. Natl. Acad. Sci. USA. 2003;100:217–222. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oseroff C., Kos F., Bui H.H., Peters B., Pasquetto V., Glenn J., Palmore T., Sidney J., Tscharke D.C., Bennink J.R., et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc. Natl. Acad. Sci. USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone J.D., Demkowicz W.E., Jr., Stern L.J. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc. Natl. Acad. Sci. USA. 2005;102:3744–3749. doi: 10.1073/pnas.0407019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terajima M., Orphin L., Leporati A.M., Pazoles P., Cruz J., Rothman A.L., Ennis F.A. Vaccinia virus-specific CD8(+) T-cell responses target a group of epitopes without a strong immunodominance hierarchy in humans. Hum. Immunol. 2008;69:815–825. doi: 10.1016/j.humimm.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing L., Davies D.H., Chong T.M., Chun S., McClurkan C.L., Huang J., Story B.T., Molina D.M., Hirst S., Felgner P.L., Koelle D.M. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.H., Grey H., Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 20.Alzhanova D., Hammarlund E., Reed J., Meermeier E., Rawlings S., Ray C.A., Edwards D.M., Bimber B., Legasse A., Planer S., et al. T cell inactivation by poxviral B22 family proteins increases viral virulence. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song H., Sidney J., Wiseman R.W., Josleyn N., Cohen M., Blaney J.E., Jahrling P.B., Sette A. Characterizing monkeypox virus specific CD8+ T cell epitopes in rhesus macaques. Virology. 2013;447:181–186. doi: 10.1016/j.virol.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson E.M., Yang H., Menconi F., Wang R., Osman R., Skrabanek L., Li C.W., Fadlalla M., Gandhi A., Chaturvedi V., et al. Employing a recombinant HLA-DR3 expression system to dissect major histocompatibility complex II-thyroglobulin peptide dynamism: a genetic, biochemical, and reverse immunological perspective. J. Biol. Chem. 2009;284:34231–34243. doi: 10.1074/jbc.M109.041574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell M.D., Gallo R.L., Boguniewicz M., Jones J.F., Wong C., Streib J.E., Leung D.Y.M. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Redfield R.R., Wright D.C., James W.D., Jones T.S., Brown C., Burke D.S. Disseminated vaccinia in a military recruit with human immunodeficiency virus (HIV) disease. N. Engl. J. Med. 1987;316:673–676. doi: 10.1056/NEJM198703123161106. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs B.L., Langland J.O., Kibler K.V., Denzler K.L., White S.D., Holechek S.A., Wong S., Huynh T., Baskin C.R. Vaccinia virus vaccines: past, present and future. Antiviral Res. 2009;84:1–13. doi: 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Y., White A. Monkeypox virus emerges from the shadow of its more infamous cousin: family biology matters. Emerg. Microbes Infect. 2022;11:1768–1777. doi: 10.1080/22221751.2022.2095309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies D.H., Wyatt L.S., Newman F.K., Earl P.L., Chun S., Hernandez J.E., Molina D.M., Hirst S., Moss B., Frey S.E., Felgner P.L. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J. Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earl P.L., Americo J.L., Wyatt L.S., Eller L.A., Whitbeck J.C., Cohen G.H., Eisenberg R.J., Hartmann C.J., Jackson D.L., Kulesh D.A., et al. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428:182–185. doi: 10.1038/nature02331. [DOI] [PubMed] [Google Scholar]
- 29.Edghill-Smith Y., Golding H., Manischewitz J., King L.R., Scott D., Bray M., Nalca A., Hooper J.W., Whitehouse C.A., Schmitz J.E., et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 30.Nigam P., Earl P.L., Americo J.L., Sharma S., Wyatt L.S., Edghill-Spano Y., Chennareddi L.S., Silvera P., Moss B., Robinson H.L., Amara R.R. DNA/MVA HIV-1/AIDS vaccine elicits long-lived vaccinia virus-specific immunity and confers protection against a lethal monkeypox challenge. Virology. 2007;366:73–83. doi: 10.1016/j.virol.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jezek Z., Marennikova S.S., Mutumbo M., Nakano J.H., Paluku K.M., Szczeniowski M. Human monkeypox: a study of 2, 510 contacts of 214 patients. J. Infect. Dis. 1986;154:551–555. doi: 10.1093/infdis/154.4.551. [DOI] [PubMed] [Google Scholar]
- 32.Babkin I.V., Babkina I.N., Tikunova N.V. An update of Orthopoxvirus molecular evolution. Viruses. 2022;14:388. doi: 10.3390/v14020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sette A., Grey H., Oseroff C., Peters B., Moutaftsi M., Crotty S., Assarsson E., Greenbaum J., Kim Y., Kolla R., et al. Definition of epitopes and antigens recognized by vaccinia specific immune responses: their conservation in variola virus sequences, and use as a model system to study complex pathogens. Vaccine. 2009;27:G21–G26. doi: 10.1016/j.vaccine.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrasco Pro S., Sidney J., Paul S., Lindestam Arlehamn C., Weiskopf D., Peters B., Sette A. Automatic generation of validated specific epitope sets. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grifoni A., da Silva Antunes R., Westernberg L., Pham J., Birrueta G., Peters B., Sette A., Schulten V. Characterization and epitope identification of the T cell response in non-allergic individuals exposed to mouse allergen. World Allergy Organ. J. 2019;12 doi: 10.1016/j.waojou.2019.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grifoni A., Voic H., Dhanda S.K., Kidd C.K., Brien J.D., Buus S., Stryhn A., Durbin A.P., Whitehead S., Diehl S.A., et al. T cell responses induced by attenuated Flavivirus vaccination are specific and show limited cross-reactivity with other Flavivirus species. J. Virol. 2020;94 doi: 10.1128/JVI.00089-20. e00089–e00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.da Silva Antunes R., Paul S., Sidney J., Weiskopf D., Dan J.M., Phillips E., Mallal S., Crotty S., Sette A., Lindestam Arlehamn C.S. Definition of human epitopes recognized in tetanus toxoid and development of an assay strategy to detect ex vivo tetanus CD4+ T cell responses. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arlehamn C.S.L., Sidney J., Henderson R., Greenbaum J.A., James E.A., Moutaftsi M., Coler R., McKinney D.M., Park D., Taplitz R., et al. Dissecting mechanisms of immunodominance to the common tuberculosis antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ) J. Immunol. 2012;188:5020–5031. doi: 10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters B., Nielsen M., Sette A. T cell epitope predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B. Smallpox vaccines: targets of protective immunity. Immunol. Rev. 2011;239:8–26. doi: 10.1111/j.1600-065X.2010.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volz A., Sutter G. Modified vaccinia virus Ankara: history, value in basic research, and current perspectives for vaccine development. Adv. Virus Res. 2017;97:187–243. doi: 10.1016/bs.aivir.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antoine G., Scheiflinger F., Dorner F., Falkner F.G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 46.Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul S., Lindestam Arlehamn C.S., Scriba T.J., Dillon M.B.C., Oseroff C., Hinz D., McKinney D.M., Carrasco Pro S., Sidney J., Peters B., Sette A. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell reports. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar A., Suryadevara N.C., Wolf K.J., Wilson J.T., Di Paolo R.J., Brien J.D., Joyce S. Heterotypic immunity against vaccinia virus in an HLA-B∗07:02 transgenic mousepox infection model. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69897-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moutaftsi M., Bui H.H., Peters B., Sidney J., Salek-Ardakani S., Oseroff C., Pasquetto V., Crotty S., Croft M., Lefkowitz E.J., et al. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 53.Seet B.T., Johnston J.B., Brunetti C.R., Barrett J.W., Everett H., Cameron C., Sypula J., Nazarian S.H., Lucas A., McFadden G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 54.Amara R.R., Nigam P., Sharma S., Liu J., Bostik V. Long-lived poxvirus immunity, robust CD4 help, and better persistence of CD4 than CD8 T cells. J. Virol. 2004;78:3811–3816. doi: 10.1128/jvi.78.8.3811-3816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 56.Edghill-Smith Y., Bray M., Whitehouse C.A., Miller D., Mucker E., Manischewitz J., King L.R., Robert-Guroff M., Hryniewicz A., Venzon D., et al. Smallpox vaccine does not protect macaques with AIDS from a lethal monkeypox virus challenge. J. Infect. Dis. 2005;191:372–381. doi: 10.1086/427265. [DOI] [PubMed] [Google Scholar]
- 57.Oseroff C., Peters B., Pasquetto V., Moutaftsi M., Sidney J., Panchanathan V., Tscharke D.C., Maillere B., Grey H., Sette A. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve. J. Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gigante C.M., Korber B., Seabolt M.H., Wilkins K., Davidson W., Rao A.K., Zhao H., Hughes C.M., Minhaj F., Waltenburg M.A., et al. Multiple lineages of Monkeypox virus detected in the United States, 2021–2022. Preprint at bioRxiv. 2022 doi: 10.1101/2022.06.10.495526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul S., Kolla R.V., Sidney J., Weiskopf D., Fleri W., Kim Y., Peters B., Sette A. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clin. Dev. Immunol. 2013;2013 doi: 10.1155/2013/467852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Middleton D., Menchaca L., Rood H., Komerofsky R. New allele frequency database. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net [DOI] [PubMed] [Google Scholar]
- 61.Dhanda S.K., Vaughan K., Schulten V., Grifoni A., Weiskopf D., Sidney J., Peters B., Sette A. Development of a novel clustering tool for linear peptide sequences. Immunology. 2018;155:331–345. doi: 10.1111/imm.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented and analyzed in the present study was retrieved from the IEDB (www.IEDB.org) and ViPR (www.viprbrc.org), as described below. The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental information. This study uses publicly available algorithms and does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.