Abstract
Despite the large toll of opioid use disorder (OUD), genome-wide association studies (GWAS) of OUD to date have yielded few susceptibility loci. We performed a large-scale GWAS of OUD in individuals of European (EUR) and African (AFR) ancestry, optimizing genetic informativeness by performing MTAG (Multi-trait analysis of GWAS) with genetically correlated substance use disorders (SUDs). Meta-analysis included seven cohorts: the Million Veteran Program, Psychiatric Genomics Consortium, iPSYCH, FinnGen, Partners Biobank, BioVU, and Yale-Penn 3, resulting in a total N = 639,063 (Ncases = 20,686;Neffective = 77,026) across ancestries. OUD cases were defined as having a lifetime OUD diagnosis, and controls as anyone not known to meet OUD criteria. We estimated SNP-heritability (h2SNP) and genetic correlations (rg). Based on genetic correlation, we performed MTAG on OUD, alcohol use disorder (AUD), and cannabis use disorder (CanUD). A leave-one-out polygenic risk score (PRS) analysis was performed to compare OUD and OUD-MTAG PRS as predictors of OUD case status in Yale-Penn 3. The EUR meta-analysis identified three genome-wide significant (GWS; p ≤ 5 × 10−8) lead SNPs—one at FURIN (rs11372849; p = 9.54 × 10−10) and two OPRM1 variants (rs1799971, p = 4.92 × 10−09; rs79704991, p = 1.11 × 10−08; r2 = 0.02). Rs1799971 (p = 4.91 × 10−08) and another OPRM1 variant (rs9478500; p = 1.95 × 10−08; r2 = 0.03) were identified in the cross-ancestry meta-analysis. Estimated h2SNP was 12.75%, with strong rg with CanUD (rg = 0.82; p = 1.14 × 10−47) and AUD (rg = 0.77; p = 6.36 × 10−78). The OUD-MTAG resulted in a GWAS Nequivalent = 128,748 and 18 independent GWS loci, some mapping to genes or gene regions that have previously been associated with psychiatric or addiction phenotypes. The OUD-MTAG PRS accounted for 3.81% of OUD variance (beta = 0.61;s.e. = 0.066; p = 2.00 × 10−16) compared to 2.41% (beta = 0.45; s.e. = 0.058; p = 2.90 × 10−13) explained by the OUD PRS. The current study identified OUD variant associations at OPRM1, single variant associations with FURIN, and 18 GWS associations in the OUD-MTAG. The genetic architecture of OUD is likely influenced by both OUD-specific loci and loci shared across SUDs.
Subject terms: Genetics, Addiction
Introduction
Opioid use disorder (OUD) has a serious negative impact on public health and is a leading cause of preventable death [1]. Although opioid misuse and progression to OUD [2] are influenced by heritable factors, discovery of OUD risk loci has been limited [3–7]. Difficulties in advancing OUD genetic discovery are largely due to lack of adequately powered cohorts of genetically informative samples [8, 9].
Genome-wide association studies (GWAS) examining single nucleotide polymorphism (SNP) effects on OUD have been underpowered [8, 9]. Nevertheless, recent progress in GWAS of OUD include the identification and confirmation of a genome-wide significant (GWS) functional variant (rs1799971) in OPRM1 [7]. Earlier OUD GWAS identified associations with variation in several genes including KCNG2, KCNC1, APBB2, CNIH3, RGMA, and OPRM1 [3–6], but the validity of those associations remains largely untested due to the lack of powerful independent OUD cohorts. OUD GWAS have also demonstrated genetic correlations (rg) with other substance use disorders (SUDs) (e.g. alcohol use disorder [AUD]; rg = 0.73) and psychiatric disorders (e.g. attention-deficit hyperactivity disorder; [rg = 0.36]) [7].
Large-scale GWAS meta-analyses have advanced discovery of novel loci for SUDs (e.g., AUD, problematic alcohol use (PAU), cannabis use disorder (CanUD) [10–12]. This study applies similar meta-analytic methods for OUD, combining GWAS effects across multiple studies and two ancestral groups.
Multi-trait methods (e.g., MTAG; Multi-trait analysis of GWAS) [13] have the potential to increase power. MTAG capitalizes on the rg between genetically-related traits (e.g., rg > 0.70) to increase the equivalent sample size. MTAG is an attractive option for boosting power for sets of similar traits like SUDs [11, 14], and holds particular promise for disorders such as OUD for which only limited cases are available for analysis. MTAG can generate estimates of trait-specific effects that leverage information from multiple GWAS summary statistics while accounting for both known and unknown sample overlap across the discovery samples [13]. MTAG can maximize the genetic information available for OUD by leveraging the statistical power of GWAS of non-opioid SUDs.
We conducted a large-scale GWAS meta-analysis of OUD in samples of African (AFR) and European (EUR) ancestry individuals. We maximized the informativeness of the available samples by performing a multi-trait analysis that incorporates SUDs that are highly genetically correlated with OUD.
Methods
Data and participants
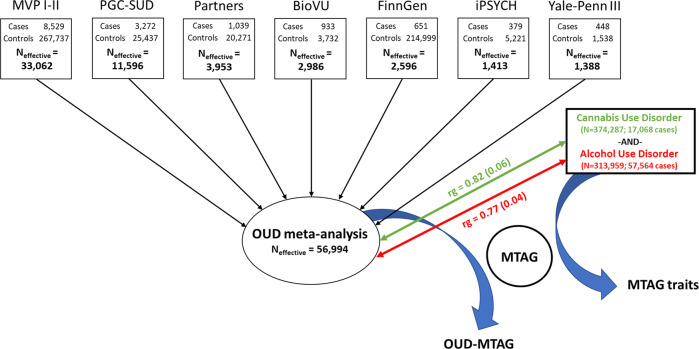
The meta-analysis includes summary statistics across seven cohorts examining OUD case vs. OUD control status in AFR and EUR ancestry individuals. We included both published and unpublished OUD GWAS. Previously published GWAS include data from Yale-Penn [3, 6, 7], PGC-SUD [6], and the Partners Biobank [15]. For MVP Releases 1 and 2 (the data releases used in the present analysis), a previous GWAS of OUD cases vs. opioid-exposed controls was reported [7]. MVP data included in the current meta-analysis use a different control definition (unscreened controls) to align better with the control definitions available in most other included samples. GWAS summary data for FinnGen [16] was accessed via a publicly available repository (https://r5.finngen.fi/). GWAS of OUD from iPSYCH [17], BioVU [18], and newly-available data from Yale-Penn subjects (Yale-Penn 3), previously unpublished, were performed by analysts at their respective study sites (Supplemental Materials). We had a total AFR N = 84,877 (Ncase = 5435 Neffective = 20,032), a total EUR N = 554,186 (Ncase = 15,251; Neffective = 56,994), and an overall N = 639,063 (Ncase =20,686; Neffective = 77,026). Other than Yale-Penn, this study involved de-identified data. The work was approved as appropriate by the Central Veterans Affairs (VA) institutional review board (IRB) and site-specific IRBs, including Yale University School of Medicine and VA Connecticut, and was conducted in accordance with all relevant ethical regulations. Cohort-specific summaries of AFR and EUR OUD subjects are presented in Table 1. Specific OUD diagnostic codes are provided in Supplementary Table 1. Additional phenotyping considerations are described in Supplemental Materials.
Table 1.
Overview of samples included in GWAS meta-analysis of OUD cases vs. OUD controls.
| Cohorts | EUR cases | EUR controls | EUR Neffective | AFR cases | AFR Controls | AFR Neffective | Case definition | Control definitiona |
|---|---|---|---|---|---|---|---|---|
| MVP 1-2 combined | 8529 | 267,737 | 33,062 | 4032 | 71,511 | 15,265 | ICD-9/ICD-10 | unscreened |
| MVP Release 1 | 6367 | 202,636 | 24,692 | 3151 | 54,178 | 11,911 | ||
| MVP Release 2 | 2162 | 65,101 | 8370 | 881 | 17,333 | 3354 | ||
| PGC-SUDb | 3272 | 25,437 | 11,596 | 1231 | 7063 | 4193 | DSM-IV | unexposed |
| Partners Biobank | 1039 | 20,271 | 3953 | - | - | - | ICD-9/ICD-10 | no SUD diagnosis |
| BioVU | 933 | 3,732 | 2,986 | - | - | - | ICD-9/ICD-10 | unscreened |
| FinnGen | 651 | 214,999 | 2596 | - | - | - | ICD-9/ICD-10 | unscreened |
| Yale-Penn 3 | 448 | 1538 | 1388 | 172 | 868 | 574 | DSM-IV | no OUD diagnosis |
| iPSYCH | 379 | 5221 | 1413 | - | - | - | ICD-9/ICD-10 | no OUD diagnosis |
| Ancestry-specific subtotals | 15251 | 538,935 | 56,994 | 5435 | 79,442 | 20,032 | ||
| Overall total cases | 20,686 | |||||||
| Overall total controls | 618,377 | |||||||
| Overall total N | 639,063 | Total Neffective | 77,026 | |||||
| EUR total N | 554,186 | EUR Neffective | 56,994 | |||||
| AFR total N | 84,877 | AFR Neffective | 20,032 |
aControl definition: “no OUD diagnosis”—controls assessed for OUD and not diagnosed. “Unscreened”—controls not assessed for OUD and thus may have unassessed opioid-related problems.bThe PGC-SUD OUD analysis included AFR and EUR participants from Yale-Penn 1 (N = 3922; Ncases = 1656) and Yale-Penn 2 (N = 2483; Ncases = 846)—data from Yale-Penn supplied 67.4% of AFR cases and 51.1% of EUR cases for the overall PGC-SUD meta-analysis. Yale-Penn 3 is included as a separate cohort in the current analysis. Effective sample size was calculated as: Neffective = 4/[(1/ncase) + (1/ncontrol)].
Ancestry-specific and cross-ancestry GWAS meta-analysis
GWAS samples were combined using an effective sample-size weighted meta-analysis in METAL[19]. Ancestry-specific and cross-ancestry meta-analyses were performed. Measures of cross-sample heterogeneity (Cochran’s Q, I2) and genomic inflation (λGC) were used to evaluate potential bias influenced by heterogeneity between cohorts or by population stratification. Included GWAS summary statistics were limited to variants present in at least 80% of the analysis-specific effective sample size (e.g., 80% of EUR Neffective). The 80% effective sample size inclusion threshold ensured that variant effects present only in smaller cohorts did not disproportionately influence the overall results. This effectively meant that a variant needed to be present in MVP, PGC-SUD, and at least one additional cohort for it to be included (Fig. 1).
Fig. 1. Summary of OUD GWAS, meta-analysis, and MTAG study design.
Overview of European-ancestry opioid use disorder (OUD) genome-wide association study and OUD multi-trait analysis.
Data from the 1000 Genome Project (1000 G) phase 3 [20] was used for LD reference. Variants were mapped to the nearest gene based upon physical position (<10 kb from assigned gene) and further characterized by gene-mapping approaches using expression quantitative trait locus (eQTL) associations and 3D chromatin interactions (Hi-C)(Supplemental Materials). Conditional analyses were conducted using GCTA-COJO [21] to examine the conditional independence of genome-wide significant (GWS; p = 5.00 × 10−08) OPRM1 variants in low LD (r2 < 0.1).
SNP-heritability and Linkage-Disequilibrium (LD) Score Regression
EUR OUD GWAS summary statistics were used to estimate SNP-heritability (h2SNP), and to characterize OUD genetic correlations (rg) using LD score regression (LDSC) [22]. LDSC analyses were restricted to HapMap3 variants [23]. Effective sample-size was used in all LDSC-based analytic steps. Genetic correlations were estimated with 54 other traits including SUDs, substance use, psychiatric traits, chronic pain, sociodemographic factors, and additional traits of interest (data sources are described in Supplemental Tables). Bonferroni-corrected significance was p = 9.26 × 10−04 (0.05/54). LDSC analyses were not performed in AFR and cross-ancestry meta-analyses because of the inability to use an LD reference panel for recently-admixed populations such as African-Americans or for analyses integrating datasets from diverse ancestry groups [22].
Multi-trait analysis of GWAS summary statistics (MTAG)
Based on LDSC estimates of genetic correlations with OUD, a joint-analysis that included the EUR OUD GWAS and GWAS summary statistics for AUD [11] and CanUD [12] was conducted using MTAG [13]. MTAG enhances statistical power by leveraging the genetic correlation between traits to generate trait-specific estimates for each SNP. The AUD GWAS summary statistics used in the present analysis are from a broader GWAS of problematic alcohol use [11]. MTAG used study-specific effective sample sizes for the respective GWAS. Study-specific effect sizes were transformed to Z-scores to be on a uniform scale across the three included GWAS. Included genetic variants were filtered using default MTAG parameters [13]. Briefly, variants were restricted to those common to all three of the GWAS, with a minor allele frequency (MAF) > 0.01, and present in at least two-thirds of the 90th percentile of the study-specific SNP sample sizes. These MTAG parameters guard against heterogeneity in the distribution of common vs. rare variant effects, ensuring that SNP effects generated from relatively small subsets of the contributing discovery GWAS do not bias the effect estimates across traits [13].
Phenome-wide Association Study (PheWAS)
To examine phenome-wide relationships for OUD and the OUD-MTAG analysis, and to compare their relationships with other clinically-relevant outcomes, we performed phenome-wide association studies (PheWAS) in BioVU [18], a cohort of >66,000 genotyped patients, with phenotypic data currently available for 1338 clinical outcomes from electronic health records [18]. Additional details on the BioVU cohort are provided in Supplemental Materials. Polygenic risk scores (PRS) for OUD and OUD-MTAG were computed using PRS-CS [24], excluding the subset of BioVU participants included in the meta-analysis. The respective PRS were then included in individual logistic regression models regressed on 1291 clinical outcomes with case counts ≥100, covarying for sex, age, and the first 10 genetic principal components. Statistical significance for the PheWAS was defined as p = 3.87 × 10−05 (0.05/1291).
Polygenic risk score analysis
PRS are described in Supplemental Materials. Briefly, a leave-one-out PRS analysis was performed by excluding the EUR and AFR Yale-Penn 3 (YP3) cohorts from the respective OUD GWAS and OUD-MTAG analyses allowing for YP3 OUD cases and controls to be used as ancestry-specific PRS target samples.
Results
Ancestry-specific and cross-ancestry GWAS meta-analyses
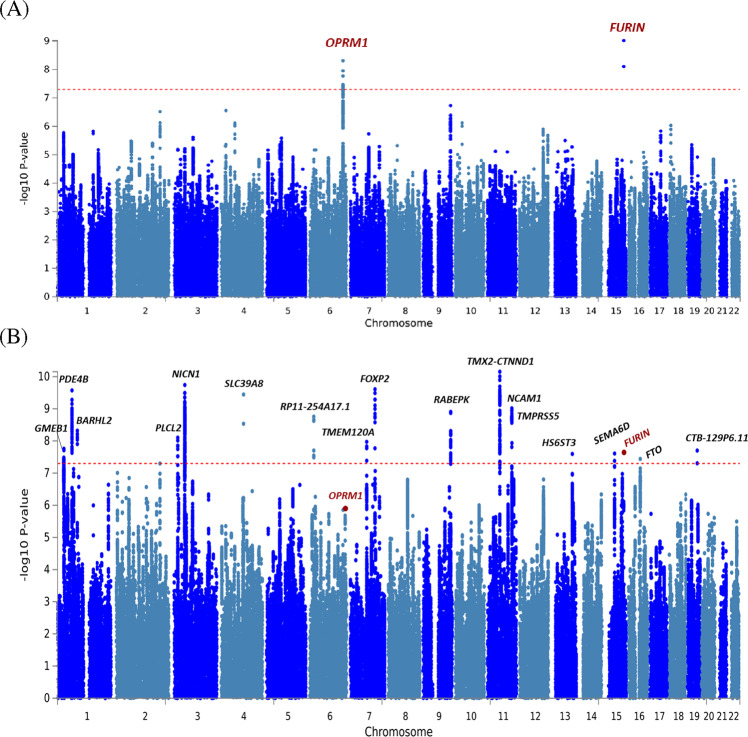
In the ancestry-specific analyses, there were three GWS variants (Fig. 2) in EUR (Table 2). The top association (rs11372849; p = 9.54 × 10−10) mapped to FURIN on chromosome 15, one of two GWS SNPs in the FURIN gene (rs17514846; r2 = 0.91). The second strongest association was with the OPRM1 functional variant (rs1799971; p = 4.92 × 10−09). An additional OPRM1 variant was also identified (rs79704991; p = 1.11 × 10−08; r2 = 0.02)(OPRM1 regional plots—Supplementary Fig. 1). GCTA-COJO [21] was used for conditional analysis of the two GWS OPRM1 variants demonstrating low LD (rs1799971 conditioned on rs79704991 and vice versa). In these analyses, each variant fell below GWS when conditioning on the effect of the other (conditioned rs1799971-pconditioned = 1.66 × 10−06; rs79704991-pconditioned = 3.71 × 10−06); although, there were no statistically significant differences in effect estimates for the respective OPRM1 variants conditioned vs. unconditioned effects. No GWS variants were identified in the AFR GWAS (Supplementary Fig. 2).
Fig. 2. OUD and OUD-MTAG manhattan plots.
Manhattan plots of (A) European-ancestry OUD GWAS results and (B) OUD-MTAG multi-trait GWAS results.
Table 2.
Genome-wide significant (p ≤ 5.00E-08) associations in (A) EUR OUD GWAS, and (B) cross-ancestry OUD GWAS.
| (A) EUR OUD analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Position | Marker | A1 | A2 | Gene | EUR MAF | EUR Z | EUR P-value | Direction | AFR MAF | AFR Z | AFR P-value | ||
| 15 | 91419432 | rs11372849 | T | TC | FURIN | 0.46 | 6.12 | 9.54E-10 | + + + ??? + ? | - | - | - | ||
| 6 | 154360797 | rs1799971 | A | G | OPRM1 | 0.13 | 5.85 | 4.92E-09 | + + + + + + + + | 0.03 | 0.83 | 0.407 | ||
| 15 | 91416550 | rs17514846 | A | C | FURIN | 0.46 | −5.77 | 7.87E-09 | ---- + --? | 0.18 | −0.94 | 0.347 | ||
| 6 | 154319449 | rs79704991 | T | G | OPRM1 | 0.13 | 5.71 | 1.11E-08 | + + + + + + + + | 0.08 | 0.59 | 0.552 | ||
| 6 | 154315310 | rs12200046 | T | C | OPRM1 | 0.13 | 5.64 | 1.70E-08 | + + + + + + + + | 0.08 | 0.67 | 0.505 | ||
| 6 | 154309808 | rs10499276 | T | C | OPRM1 | 0.13 | 5.52 | 3.38E-08 | + + + + + + + + | 0.08 | 0.49 | 0.621 | ||
| 6 | 154304242 | rs34069531 | T | C | OPRM1 | 0.13 | 5.51 | 3.67E-08 | + + + + + + + + | 0.08 | 0.61 | 0.542 | ||
| 6 | 154377925 | rs3778146 | T | C | OPRM1 | 0.17 | −5.5 | 3.84E-08 | --- + ---- | 0.08 | −1.54 | 0.125 | ||
| 6 | 154378223 | rs9478500 | T | C | OPRM1 | 0.17 | −5.49 | 4.12E-08 | --- + ---- | 0.08 | −1.76 | 0.078 | ||
| 6 | 154379152 | rs3823010 | A | G | OPRM1 | 0.17 | 5.48 | 4.25E-08 | + + + - + + + + | 0.08 | 1.59 | 0.111 | ||
| 6 | 154378739 | rs3778147 | A | G | OPRM1 | 0.17 | 5.46 | 4.83E-08 | + + + - + + + + | 0.08 | 1.47 | 0.141 | ||
| (B) Cross-ancestry OUD analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Position | Marker | A1 | A2 | Gene | Z | P-value | Direction | EUR MAF | EUR Z | EUR P-value | AFR MAF | AFR Z | AFR P-value |
| 6 | 154378223 | rs9478500 | T | C | OPRM1 | −5.62 | 1.95E-08 | --- + ----- + - + | 0.17 | −5.49 | 4.12E-08 | 0.18 | −1.76 | 0.078 |
| 6 | 154379934 | rs9285542 | T | C | OPRM1 | 5.56 | 2.73E-08 | + + + - + + + + + - + + | 0.17 | 5.43 | 5.61E-08 | 0.16 | 1.74 | 0.082 |
| 6 | 154379152 | rs3823010 | A | G | OPRM1 | 5.53 | 3.26E-08 | + + + - + + + + + + -- | 0.17 | 5.48 | 4.25E-08 | 0.11 | 1.59 | 0.111 |
| 6 | 154381012 | rs3778148 | T | G | OPRM1 | 5.52 | 3.34E-08 | + + + - + + + + + + - + | 0.17 | 5.43 | 5.73E-08 | 0.10 | 1.68 | 0.094 |
| 6 | 154355100 | rs6936615 | A | G | OPRM1 | −5.52 | 3.38E-08 | --- + --?--- + - | - | - | - | 0.10 | −1.72 | 0.085 |
| 6 | 154377925 | rs3778146 | T | C | OPRM1 | −5.51 | 3.54E-08 | --- + ------ + + | 0.17 | −5.5 | 3.84E-08 | 0.12 | −1.54 | 0.125 |
| 6 | 154383658 | rs3778150 | T | C | OPRM1 | −5.49 | 3.97E-08 | --- + ----- + - + | 0.17 | -5.38 | 7.31E-08 | 0.18 | −1.69 | 0.091 |
| 6 | 154382139 | rs3778149 | C | G | OPRM1 | −5.48 | 4.20E-08 | --- + ------ + + | 0.17 | −5.44 | 5.42E-08 | 0.12 | −1.58 | 0.114 |
| 6 | 154382473 | rs7772959 | A | G | OPRM1 | 5.48 | 4.34E-08 | + + + - + + + + + + -- | 0.17 | 5.43 | 5.66E-08 | 0.12 | 1.58 | 0.114 |
| 6 | 154362254 | rs9322445 | A | G | OPRM1 | −5.47 | 4.54E-08 | --- + ------ + - | 0.17 | −5.44 | 5.49E-08 | 0.10 | −1.56 | 0.120 |
| 6 | 154382367 | rs7773995 | T | C | OPRM1 | 5.46 | 4.78E-08 | + + + - + + + + + + -- | 0.17 | 5.42 | 5.84E-08 | 0.12 | 1.56 | 0.120 |
| 6 | 154360797 | rs1799971 | A | G | OPRM1 | 5.46 | 4.91E-08 | + + + + + + + + + - + + | 0.13 | 5.85 | 4.92E-09 | 0.03 | 0.83 | 0.407 |
| Bold = Lead SNP | ||||||||||||||
The cross-ancestry OUD GWAS identified two GWS risk variants mapping to OPRM1 (Supplementary Fig. 3) (Table 2). The top association was with rs9478500 (p = 1.95 × 10−08), an intronic variant. Rs1799971 was also GWS in the cross-ancestry meta-analysis (p = 4.91 × 10−08), and is not in strong LD with rs9478500 (EUR r2 = 0.03; AFR r2 = 0.002; ALL r2 = 0.04). The top FURIN association in EUR (rs11372849) was uninformative in three of four AFR ancestry cohorts and did not meet the threshold (80% of Neffective) we set to be included in the analysis. The second top FURIN association (rs17514846) fell below GWS in the cross-ancestry GWAS (p = 6.00 × 10−08).
Gene-based analysis
Gene-based analyses are described in Supplemental Materials. Both FURIN (p = 3.09 × 10−07) and OPRM1 (p = 3.59 × 10−07) were significant in EUR gene-based analysis (Supplementary Fig 4). Additional results are reported in Supplemental Table 3–5 and Supplementary Fig 5.
SNP-heritability and Linkage-Disequilibrium (LD) Score Regression
The liability scale SNP-heritability (h2SNP) estimate was 12.75% (s.e. = 1.1%) in EUR using effective sample-size adjusted prevalence rates and a population prevalence of 0.021 [25]. Genome-wide inflation was mild with respect to sample size and favored OUD polygenicity as indicated by the LDSC inflation factor (λGC = 1.18), intercept = 1.01 (s.e. = 0.011), and attenuation ratio = 0.05 (s.e. = 0.049).
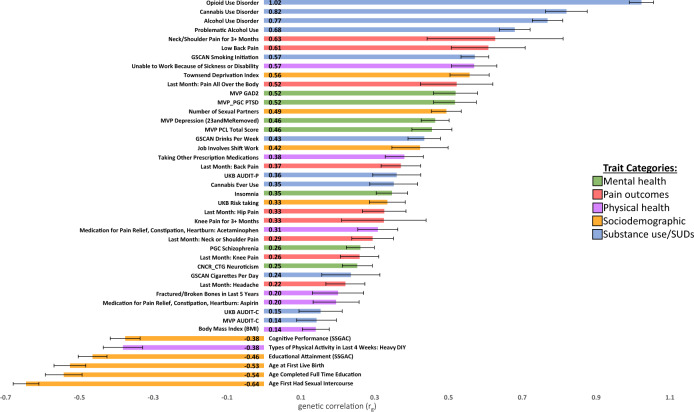
OUD showed statistically significant (p ≤ 9.26 x 10−04) genetic correlations with 40 traits including substance use, SUDs, psychiatric traits, pain outcomes, physical health, and sociodemographic characteristics (Fig. 3; Supplementary Table 6). The OUD trait in the current study was strongly genetically correlated with the largest published GWAS of OUD to date (rg = 1.02; p = 2.38 x 10−214) [7], suggesting that OUD is being captured consistently across the studies, as might be expected given the substantial overlap in OUD cases between the two studies, although the control definitions differed between analyses. OUD was also strongly genetically correlated with other SUDs, including CanUD (rg = 0.82; p = 1.14 x 10−47) [12] and AUD (rg = 0.77; p = 6.36 x 10−78) [11]. Modest genetic correlations were found for measures of substance use (e.g., the quantity/frequency alcohol use measure AUDIT-C) (rg = 0.14; p = 8.15 x 10−03) [10].
Fig. 3. OUD genetic correlation results.
EUR OUD GWAS genetic correlations (rg) with mental health, pain, physical health, sociodemographic, and substance use traits of interest.
OUD also demonstrated statistically significant genetic correlations with many mental health, pain, physical health, and sociodemographic traits. The strongest positive correlations across the respective domains were with Generalized Anxiety Disorder (rg = 0.52; p = 2.89 x 10−18) and PTSD (rg = 0.52; p = 3.87 x 10−19), lower back pain (rg = 0.61; p = 1.22 x 10−09), inability to work due to being sick or disabled (rg = 0.57; p = 1.31 x 10−20), and scores on the Townsend Deprivation Index (rg = 0.56; p = 1.13 x 10−25). OUD was negatively genetically correlated with measures of sexual activity (age of first sexual intercourse [rg = −0.64; p = 4.43 x 10−76]), indices of educational attainment (age of school completion [rg = −0.54; p = 9.41x10−28]) and cognitive performance (rg = −0.38; p = 1.54 x 10−20), and levels of past month “Heavy Do It Yourself” physical activity (rg = −0.38; p = 7.37 x 10−13), among others (Supplementary Table 6).
Multi-trait analysis of European GWAS summary statistics (MTAG)
MTAG was supported by strong genetic correlation for OUD with CanUD (rg=0.82; p = 1.14 x 10−47) and AUD (rg = 0.77; p = 6.36 x 10−78) in EUR. The OUD-MTAG analysis resulted in an increase in effective sample size from the original EUR OUD GWAS Neffective = 56,994 (GWAS mean x2 = 1.18) to an equivalent sample size of N = 128,748 (GWAS mean x2 = 1.40). The increase resulted in the identification of 18 independent GWS OUD-MTAG risk loci (Fig. 2; Table 3), some previously associated at either the variant level, or that reside in genes associated with, psychiatric and substance use outcomes in previous GWAS. Seven of the OUD-MTAG loci mapped to the nearest gene via brain eQTL data and Hi-C interactions; 8 loci were not mapped to the nearest gene via brain eQTL and Hi-C data but were implicated with additional genes in their respective genomic regions. Some loci fell in complex genomic regions with many mapped genes. These OUD-MTAG loci regions are summarized in Supplemental Fig. 6, 7 and Supplementary Tables 12–15 along with the OUD loci.
Table 3.
Genome-wide significant (p ≤ 5.00 × 10−08) Lead SNP associations in OUD-MTAG analysis (of 441 total GWS SNPs).
| Chr | Pos | MarkerName | Allele 1 | Allele 2 | Gene(s) | Z | P-value | p-value in AUD | p-value in CanUD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 57535966 | rs11229119 | T | C | TMX2-CTNND1 | −6.50 | 7.03E-11 | 2.60E-07 | 6.68E-05 | |
| 3 | 49469449 | rs77648866 | A | G | NICN1 | 6.33 | 1.82E-10 | 5.90E-07 | 5.89E-03 | |
| 7 | 114137940 | rs1989903 | A | G | FOXP2 | −6.20 | 2.47E-10 | 1.23E-03 | 3.52E-09 | |
| 1 | 66434743 | rs7519259 | A | G | PDE4B | 5.73 | 2.68E-10 | 9.94E-08 | 1.36E-05 | |
| 4 | 103198082 | rs13135092 | A | G | SLC39A8 | 6.51 | 3.60E-10 | 4.89E-18 | 0.88 | |
| 11 | 112869404 | rs1940701 | T | C | NCAM1 | -6.10 | 9.63E-10 | 4.91E-06 | 1.67E-03 | |
| 9 | 127968109 | rs864882 | T | C | RABEPK | −5.60 | 1.24E-09 | 2.45E-05 | 0.06 | |
| 6 | 19076417 | rs9350100 | T | C | RP11-254A17.1 | 5.62 | 1.76E-09 | 3.37E-05 | 2.89E-03 | |
| 1 | 91208451 | rs2166171 | T | C | BARHL2 | −5.88 | 4.80E-09 | 1.18E-06 | 2.76E-05 | |
| 3 | 16850764 | rs55855024 | A | C | PLCL2 | 5.73 | 7.89E-09 | 1.50E-06 | 0.20 | |
| 7 | 75622281 | rs6467958 | T | C | TMEM120A | −5.50 | 1.06E-08 | 2.24E-05 | 4.02E-03 | |
| 11 | 113477081 | rs11214677 | T | C | TMPRSS5 | −5.65 | 1.16E-08 | 1.23E-04 | 1.65E-05 | |
| 1 | 28989020 | rs6667501 | A | G | GMEB1 | 5.55 | 1.74E-08 | 3.66E-04 | 2.21E-03 | |
| 19 | 45453763 | rs10422888 | A | G | CTB-129P6.11 | 5.55 | 1.99E-08 | 5.83E-05 | 6.22E-03 | |
| 15 | 91416550 | rs17514846 | A | C | FURIN | −5.59 | 2.30E-08 | 2.85E-03 | 0.09 | |
| 15 | 47645174 | rs73403005 | A | G | SEMA6D | −5.69 | 2.46E-08 | 6.06E-09 | 2.38E-03 | |
| 13 | 96932868 | rs2389631 | A | C | HS6ST3 | −5.61 | 2.53E-08 | 8.72E-05 | 0.06 | |
| 16 | 53834607 | rs7188250 | T | C | FTO | 5.64 | 3.63E-08 | 4.41E-12 | 0.61 | |
| Bold = GWS in non-OUD MTAG sumstats | ||||||||||
The top OUD-MTAG association was with rs11229119 (p = 7.03 x 10−11) on chromosome 11 mapping to both TMX2 and CTNND1. The second strongest was with NICN1 (rs77648866; p = 1.82 x 10−10) on chromosome 3. Additional GWS associations included FOXP2 (rs1989903; p = 2.47 x 10−10), PDE4B (rs7519259; p = 2.68 x 10−10), SLC39A8 (rs13135092; p = 3.60 x 10−10), NCAM1 (rs1940701; p = 9.63 x 10−10), RABEPK (rs864882; p = 1.24 x 10−09), PLCL2 (rs55855024; p = 7.89 x 10−09), and FTO (rs7188250; p = 3.63 x 10−08). One of the FURIN variants identified in the EUR OUD GWAS was also GWS in the OUD-MTAG (rs17514846; p = 2.30 x 10−08). The top OPRM1 association was with rs1799971 (p = 1.39 x 10−06). Of the 18 GWS loci, three were GWS in the AUD GWAS and one was GWS in the CanUD GWAS used for MTAG (Table 3).
The OUD-MTAG gene-based analysis resulted in 66 Bonferroni significant (p ≤ 0.05/15,927 = 3.14 x 10−06) genes (Supplementary Table 7; Supplementary Fig 8).
The OUD-MTAG GWAS was significantly genetically correlated (Bonferroni p ≤ 9.26 x 10−04) with 46 traits including the largest previously published GWAS of OUD to date at rg = 0.98 (p = 1.22 x 10−77) [7]. All estimates of genetic correlation for the OUD-MTAG analysis can be found in Supplementary Table 8.
Phenome-wide Association Study (PheWAS)
The top PheWAS association for OUD was with substance addiction and disorders (OR = 1.53; p = 2.12 x 10−69). Additional top OUD associations included tobacco use disorder (OR = 1.26; p = 3.38 x 10−56), chronic pain (OR = 1.25; p = 2.32 x 10−28), alcohol-related disorders (OR = 1.35; p = 1.04 x 10−23), mood (OR = 1.13; p = 1.27 x 10−22) and anxiety (OR = 1.14; p = 1.00 × 10−21) disorders, viral hepatitis C (OR = 1.33; p = 3.04 x 10−20), and suicidal ideation or attempt (OR = 1.49; p = 2.17 x 10−19), amongst others (Supplementary Fig 9; Supplementary Table 9).
Similar patterns of association were found for the OUD-MTAG PheWAS. The top associations were with tobacco use disorder (OR = 1.30; p = 1.37 x 10−68), substance addiction and disorders (OR = 1.42; p = 1.15 x 10−46), and alcohol-related disorders (OR = 1.42; p = 7.36 × 10−31). OUD-MTAG also demonstrated significant associations with mood (OR = 1.12; p = 7.76 x 10−21) and anxiety (OR = 1.13; p = 1.45 x 10−20) disorders, chronic pain (OR = 1.20; p = 2.51 x 10−20), viral hepatitis C (OR = 1.32; p = 1.73 x 10−18), and suicidal ideation or attempt (OR = 1.47; p = 4.52 x 10−18) (Supplementary Fig 9; Supplementary Table 10).
OUD polygenic risk score (PRS) analysis
In Yale-Penn 3 (YP3) EUR (N = 1959, 440 OUD cases), the EUR OUD PRS was a significant predictor of OUD (beta = 0.45; s.e. = 0.058; p = 2.9 x 10−13) with the PRS specifically accounting for 2.41% of OUD variance (Supplementary Table 11). The OUD-MTAG PRS was a stronger predictor of OUD (beta = 0.61;s.e. = 0.066; p = 2.0 x 10−16) explaining 3.81% of OUD variance.
In YP3 AFR (N = 1017, 171 OUD cases), both AFR-derived and EUR-derived OUD PRS were generated. The EUR-derived PRS (beta = 0.19;s.e. = 0.0945; p = 0.042; R2 = 0.29%) explained a small proportion of OUD variance in YP3 AFR but the AFR-derived PRS (beta = 0.073;s.e. = 0.049;p = 0.136; R2 = 0.11%) was not a significant predictor.
Discussion
We present a large genetic study of OUD, with an overall sample size of 639,063 (EUR = 554,186; AFR = 84,877) individuals (Ncases = 20,686 [EUR = 15,251; AFR = 5435]). This study is the first to provide evidence of a GWS single-variant GWAS association between FURIN and OUD. We support findings from previous OUD GWAS implicating OPRM1 as a risk locus for OUD [7], including the coding variant rs1799971 and additional OPRM1 associations that remained statistically significant in a cross-ancestry analysis of AFR and EUR populations. We add evidence of gene-based associations with OUD and provide estimates of OUD SNP-heritability and genetic correlations with numerous traits. Further, we apply a multi-trait approach for OUD genetic discovery utilizing the high degree of genetic correlation across SUDs (OUD, AUD, CanUD) to increase power, yielding an equivalent sample size of 128,748 and 18 GWS OUD-MTAG risk loci. PheWAS of OUD and OUD-MTAG demonstrated similar patterns of associations across the phenome, and the OUD-MTAG PRS explained a larger amount of variance in OUD case status (3.81%) compared to the OUD PRS (2.41%), suggesting that the OUD-MTAG and OUD analyses are capturing similar phenomenon.
Compared to other complex psychiatric traits, there are comparatively small samples available for genetic analyses of SUDs, particularly those involving illegal substances (heroin, cocaine) [8, 9]. A strategy that increases statistical power by incorporating other sets of samples—for example, from GWAS of closely-related but non-identical traits such as other SUDs—could help advance our understanding of the genetic architecture of OUD. This study brought much more information to bear on the analysis of OUD risk variation, resulting in the identification of many more loci. These associations included two loci from the EUR OUD GWAS (OPRM1 and FURIN), and 18 loci identified in the OUD-MTAG analysis (also including FURIN). The OUD-MTAG loci did not include OPRM1. The absence from the MTAG analysis of any statistically significant association mapped to OPRM1, a locus that should be highly-specific to OUD, was unexpected.
FURIN was associated with OUD risk in both SNP-based and gene-based analyses. FURIN (Furin, Paired Basic Amino Acid Cleaving Enzyme) encodes the endoprotease furin enzyme that serves a primary role in regulating synaptic neuronal activity, including the synthesis of brain-derived neurotropic factor and regulation of neurotrophin levels in the brain [26]. Genetic variation in FURIN has been associated with multiple psychiatric outcomes including schizophrenia [27, 28] and studies examining genetic and phenotypic features shared between schizophrenia and bipolar disorder [29, 30]. The two top FURIN SNPs associated with OUD are in strong LD (r2 = 0.91); the second strongest FURIN association, rs17514846, has been significantly associated with multiple cardiovascular and hypertension outcomes [31, 32], and was also GWS in a GWAS of parents’ attained age (current age of parents or parental age at death) [33]. A statistically-significant FURIN gene-level association being driven by rs17514846 was reported between FURIN and opioid addiction [34]. In a targeted follow-up in the FURIN gene region, there was significant association between rs11372849 (also lead SNP in the current study) and opioid addiction. Accumulating evidence linking FURIN and opioid outcomes, including the FURIN GWAS associations reported herein along with evidence of gene-based associations with opioid addiction [34], reflect the high degree of co-morbidity between OUD and psychiatric and physical health traits.
Our findings support previous OUD GWAS implicating OPRM1 genetic variation in opioid addiction and OUD [7, 34] and extend GWS findings for OPRM1 as a risk factor in a cross-ancestry analysis. The top association in the EUR OUD GWAS was with the OPRM1 coding variant (rs1799971) identified in an earlier OUD GWAS, all cases of which are also included in the current study, plus an additional OPRM1 variant (rs79704991) in low-LD with rs1799971 (r2 = 0.02). Two OPRM1 variants were also GWS in the cross-ancestry OUD GWAS (rs1799971 and rs9478500; r2 = 0.02). Rs9478500 was previously GWS for opioid addiction in EUR [34]. There is evidence of OPRM1’s complex haplotype structure and the potential for multiple independent OPRM1 risk loci influencing risk for OUD and SUDs dating to 2006 [35]. Conditional analysis of the top OPRM1 variants (rs1799971 and rs79704991; r2 = 0.02) demonstrated that these variants are not independent as indicated by each variant falling below GWS when conditioned on the other; however, the variant effects remained nominally significant and there were no significant differences in the conditioned vs. unconditioned effect sizes. Future studies of larger cohorts with diverse ancestral backgrounds will be needed to distinguish the effects of OUD risk alleles across the OPRM1 region, including the known-functional rs1799971 variant which may or may not be the variant motivating previous findings.
We found an estimated SNP-heritability (h2SNP) of 12.75% (Z = 11.28) compared to the previous largest OUD GWAS (h2SNP = 11.30%; Z = 6.27) [7]. However, the comparison between these two studies is not direct: the largest previous GWAS [7] used opioid-exposed controls, while we used a broader control definition, including not only individuals who were opioid-exposed, but subjects with no OUD assessment. This was necessitated by the fact that many of the available datasets did not define exposed controls and would have been excluded had we used the exposed control definition. Findings from the current study do not establish whether the control definition impacted the detection of genetic loci or the genetic architecture of OUD.
OUD was positively genetically correlated with other SUDs (CanUD, AUD) and psychiatric conditions (PTSD, depression, schizophrenia), with lower correlations for measures of substance use (as opposed to dependence; e.g., AUDIT-C), suggesting that OUD is more akin to measures of substance dependence than use per se. OUD was genetically correlated with multiple forms of chronic pain (e.g., lower back pain) and indicators of impairment (inability to work, decreased physical activity) and significantly genetically correlated with socioeconomic hardship (Townsend Deprivation Index) and lower levels of educational attainment. These patterns of genetic correlation are consistent with and may reflect high rates of co-occurrence of OUD with SUDs and psychiatric disorders in epidemiologic studies [25, 36]. Beyond epidemiologic estimates, SUDs and psychiatric disorders have also been demonstrated to be risk markers for severe opioid-related outcomes, such as opioid overdose [37]. Lower educational attainment and greater economic hardship have also been associated with higher rates of opioid overdose and opioid overdose-related deaths [38]. These patterns of genetic correlation are consistent with the complex clinical presentation of OUD.
We examined the utility of using MTAG to increase the information available from the limited number of genotyped OUD subjects. The OUD multi-trait analysis was feasible given the high genetic correlations with CanUD (rg = 0.82) and AUD (rg = 0.77) and increased by an order-of-magnitude the number of GWS risk loci detected. While this provides proof-of-concept for this approach given that many of the loci identified via OUD-MTAG have previously been implicated with psychiatric and substance use outcomes, OPRM1 was not GWS in the OUD-MTAG analysis, so increased detection may have come at the cost of specificity for OUD. However, only 4 of the 18 OUD-MTAG GWS associations were GWS in the respective AUD and CanUD GWAS used as MTAG instruments, so the MTAG results did not simply reflect the findings from AUD and CanUD GWAS. The OUD-MTAG PRS also outperformed the OUD PRS in predicting OUD case status in the Yale-Penn holdout analysis (OUD-MTAG PRS: 3.81%, OUD PRS: 2.41%) indicating that the OUD-MTAG is capturing OUD risk, and that the OUD-MTAG PRS is more powerful than the OUD PRS alone as also reflected by the comparative number of GWS loci identified in the respective GWAS.
A PheWAS across 1291 clinical outcomes also demonstrated convergent patterns of association between OUD and OUD-MTAG with common comorbidities (including SUDs, psychiatric traits, chronic pain, viral hepatitis C), supporting that these two analyses capture genetic factors that underlie similar clinical presentations and related impairment. Additionally, summary data from the OUD MTAG analysis including multiple SUDs was highly genetically correlated (rg = 0.98) with OUD [7], so it appears that the OUD-MTAG did capture genetic information relevant to OUD risk, though measuring the risk for OUD through a genetic liability for SUDs more broadly. That is, genetic risk for OUD may be a combination of a broader addiction liability (OUD-MTAG loci) combined with the opioid-specific genetic effects (e.g., OPRM1) that were found in the OUD single-trait analysis that are also influencing risk.
The distinction between substance-specific genetic effects and general SUD liability is of interest. Quantitative genetic studies have demonstrated both substance-specific influences, as well as heritable factors that contribute to SUDs more broadly [39, 40]. Up to 38% of variation in opioid dependence was reportedly accounted for by opioid-specific factors that were not shared with other SUDs [41]. Molecular genetic studies have begun to disentangle common vs. substance-specific genetic influences, reporting evidence to suggest the presence of a common unitary addiction factor that can account for risk across SUDs, in addition to substance-specific influences [42, 43]. Larger-scale OUD studies will be needed to parse genomic influences specific to OUD from those underlying risk for SUDs more broadly, but this will require many more genotyped OUD cases, because it cannot be accomplished via statistical methods alone.
The present study has limitations. Despite including all genotyped OUD subjects available, the OUD-only component of the present study is smaller than GWAS for other substance use behaviors [14] because OUD cases are underrepresented in available datasets. MTAG yielded a much larger sample, but at the apparent cost of a reduction in specificity marked by the non-significance of OPRM1 in the OUD-MTAG. To maximize sample size while maintaining OUD diagnosis to define case status in extant datasets, we used an unscreened control group, which although not optimal, allowed for the inclusion of additional subject cohorts. Important consideration must be given to OUD control definitions [6, 34]. Additionally, inadequate subject numbers limited our ability to identify risk variants in non-EUR populations. This must be addressed by purposeful recruitment of AFR and other non-EUR OUD subjects.
We report novel findings from a large-scale GWAS meta-analysis of OUD and employed multi-trait approaches that advanced discovery. These identified genomic risk factors for the development of OUD and its underlying biology highlight the need to assemble large OUD datasets that include individuals from diverse ancestral backgrounds. To advance our scientific understanding of OUD risk will require study of a range of opioid-related traits (e.g., clinically diagnosed OUD, non-dependent opioid use, and prescription painkiller use) [44].
Supplementary information
Acknowledgements
This work was supported by NIH grants R01DA054869 (JG, HE, AA), R01 DA051906 (JG), R01 AA026364 (JG), T32 AA028259 (JDD), K01 AA028292 (RLK), U24 AA020794 (ACJ), U01 AA020790 (ACJ), U24 AA022001(ACJ), U10 AA013566 (ACJ), K02 DA032573 (AA), DP1 DA054394 (SSR), R33 DA047527 (RP), and F32 MH122058 (FRW); the Brain & Behavior Research Foundation (HZ); the California Tobacco-Related Disease Research Program (T29KT0526; SSR); The iPSYCH team was supported by grants from the Lundbeck Foundation (R102-A9118, R155-2014-1724, and R248-2017-2003), U01 MH109514-01 (ADB) and the Universities and University Hospitals of Aarhus and Copenhagen. The Danish National Biobank resource was supported by the Novo Nordisk Foundation. High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to ADB); and by funding from the Department of Veterans Affairs Office of Research and Development, USVA, grants I01CX001849, I01BX003342, I01BX004820, and I01 CX001734, and the VA Cooperative Studies Program study, no. 575B. This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award #CSP575B, MVP004, and MVP025. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. A previous version of this article was published medRxiv: https://www.medrxiv.org/content/10.1101/2021.12.04.21267094v2.
Author contributions
JDD and JG conceived the initial study design. JDD, HZ, FRW, ASH, ECJ, RLK, HJE, AA, RP, HRJ, and JG provided ongoing feedback and refinement of the analytic plan. JDD, HZ, MG, DFL, FRW, SS, ECJ, YZN, DD, ADB, VMR, MVJ, RLK, ACJ, HJE, AA, RP, HRK, and JG provided contributions to the individual GWAS included in the meta-analysis. JDD performed the GWAS meta-analysis, genetic correlation analysis, multi-trait analysis of GWAS, gene-based analysis, functional characterization analysis, and PRS analysis. SS and MVJ performed the PheWAS analysis. DFL performed the conditional analysis. JDD, HZ, DFL, FRW, SS, ASH, ECJ, RLK, HJE, AA, RP, HRK, and JG contributed to data interpretation. JDD, SS, MVJ, and JG contributed to data visualization. JDD drafted, and JDD and JG refined the manuscript. All authors critically evaluated and revised the manuscript.
Competing interests
HRK is a member of scientific advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals, and Enthion Pharmaceuticals; a consultant to Sophrosyne Pharmaceuticals; and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which during the past three years was supported by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi, and Otsuka. HRK and JG are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018 and issued on January 26, 2021 as U.S. Patent No. 10,900,082. JG and RP were paid for editorial work for the journal “Complex Psychiatry.” All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-022-01709-1.
References
- 1.Hedegaard, M, Miniño, A, & Warner, M NCHS data brief: January 2020: Drug overdose deaths in the United States, 1999–2018. 2020; Retrieved from https://stacks.cdc.gov/view/cdc/84647-h.pdf. [PubMed]
- 2.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: Diagnostic and statistical manual of mental disorders, fifth edition. 2013; Arlington, VA: American Psychiatric Association.
- 3.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-Wide Association Study of Opioid Dependence: Multiple Associations Mapped to Calcium and Potassium Pathways. Biol Psychiatr. 2014;76:66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson EC, Agrawal A, Heath AC, Bogdan R, Sherva R, Zhang B, et al. Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry. 2016;21:608–14. doi: 10.1038/mp.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Z, Zhou H, Sherva R, Farrer LA, Kranzler HR, Gelernter J. Genome-wide Association Study Identifies a Regulatory Variant of RGMA Associated With Opioid Dependence in European Americans. Biol Psychiatr. 2018;84:762–70. doi: 10.1016/j.biopsych.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polimanti R, Walters RK, Johnson EC, Mcclintick JN, Adkins AE, Adkins DE, et al. Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol Psychiatr. 2020;25:1673–87. doi: 10.1038/s41380-020-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, et al. Association of OPRM1 Functional Coding Variant With Opioid Use Disorder. JAMA Psychiatr. 2020;77:1072. doi: 10.1001/jamapsychiatry.2020.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deak JD, Johnson EC. Genetics of substance use disorders: a review. Psychological Medicine 2021;51:2189–220. [DOI] [PMC free article] [PubMed]
- 9.Gelernter J, Polimanti R. Genetics of substance use disorders in the era of big data. Nat Rev Genet. 2021;22:712–29. doi: 10.1038/s41576-021-00377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat.Commun. 2019; 10. 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed]
- 11.Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–18. doi: 10.1038/s41593-020-0643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7:1032–45. doi: 10.1016/S2215-0366(20)30339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–37. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song W, Kossowsky J, Torous J, Chen C-Y, Huang H, Mukamal KJ, et al. Genome-wide association analysis of opioid use disorder: A novel approach using clinical data. Drug Alcohol Depend. 2020;217:108276. doi: 10.1016/j.drugalcdep.2020.108276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finngen, Documentation of R5 Release. https://finngen.gitbook.io/documentation/. 2021; 2021-05-11.
- 17.Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, Grove J, Agerbo E, Bækvad-Hansen M, et al. The iPSYCH2012 case–cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roden D, Pulley J, Basford M, Bernard G, Clayton E, Balser J, et al. Development of a Large-Scale De-Identified DNA Biobank to Enable Personalized Medicine. Clin Pharmacol Therapeutics. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW Polygenic prediction via Bayesian regression and continuous shrinkage priors. NatCommun. 2019; 10. 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed]
- 25.Grant BF, Saha TD, Ruan WJ, Goldstein RB, Chou SP, Jung J, et al. Epidemiology of DSM-5 Drug Use Disorder. JAMA Psychiatry. 2016;73:39. doi: 10.1001/jamapsychiatry.2015.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–66. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruderfer DM, Fanous AH, Ripke S, Mcquillin A, Amdur RL, Gejman PV, et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatr. 2014;19:1017–24. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruderfer DM, Ripke S, Mcquillin A, Boocock J, Stahl EA, Pavlides JMW, et al. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018;173:1705–.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–91. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 32.Zhao G, Yang W, Wu J, Chen B, Yang X, Chen J, et al. Influence of a Coronary Artery Disease–Associated Genetic Variant on FURIN Expression and Effect of Furin on Macrophage Behavior. Arteriosclerosis, Thrombosis, Vasc Biol. 2018;38:1837–44. doi: 10.1161/ATVBAHA.118.311030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilling LC, Kuo C-L, Sicinski K, Tamosauskaite J, Kuchel GA, Harries LW, et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging. 2017;9:2504–20. doi: 10.18632/aging.101334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaddis N, Mathur R, Marks J, Zhou L, Quach B, Waldrop A, et al. Multi-trait genome-wide association study of opioid addiction: OPRM1 and Beyond. medRxiv 2021; 10.1101/2021.09.13.21263503. [DOI] [PMC free article] [PubMed]
- 35.Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang B-Z, Krupitsky E, et al. Association between two µ-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–19. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kessler RC. The epidemiology of dual diagnosis. Biol Psychiatr. 2004;56:730–7. doi: 10.1016/j.biopsych.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 37.Brady JE, Giglio R, Keyes KM, Dimaggio C, Li G Risk markers for fatal and non-fatal prescription drug overdose: a meta-analysis. Injury Epidemiol. 2017; 4. 10.1186/s40621-017-0118-7. [DOI] [PMC free article] [PubMed]
- 38.Dahlman D, Ohlsson H, Edwards AC, Sundquist J, Håkansson A, Sundquist K. Socioeconomic correlates of incident and fatal opioid overdose among Swedish people with opioid use disorder. Substance Abuse Treat Prev and Policy 2021; 16. 10.1186/s13011-021-00409-3. [DOI] [PMC free article] [PubMed]
- 39.Kendler KS, Myers J, Prescott CA. Specificity of Genetic and Environmental Risk Factors for Symptoms of Cannabis, Cocaine, Alcohol, Caffeine, and Nicotine Dependence. Arch Gen Psychiatr. 2007;64:1313. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- 40.Kendler KS, Prescott CA, Myers J, Neale MC. The Structure of Genetic and Environmental Risk Factors for Common Psychiatric and Substance Use Disorders in Men and Women. Arch Gen Psychiatr. 2003;60:929. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 41.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J. Co-occurrence of Abuse of Different Drugs in Men. Arch Gen Psychiatr. 1998;55:967. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 42.Hatoum AS, Johnson EC, Colbert SMC, Polimanti R, Zhou H, Walters RK, et al. The addiction risk factor: A unitary genetic vulnerability characterizes substance use disorders and their associations with common correlates. Neuropsychopharmacol. 2021; 10.1038/s41386-021-01209-w. [DOI] [PMC free article] [PubMed]
- 43.Hatoum AS, Colbert SMC, Johnson EC, Huggett SB, Deak JD, Pathak GA Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. medRxiv 2022; 10.1101/2022.01.06.22268753. [DOI] [PMC free article] [PubMed]
- 44.Sanchez-Roige S, Fontanillas P, Jennings MV, Bianchi SB, Huang Y, Hatoum AS Genome-wide association study of problematic opioid prescription use in 132,113 23andMe research participants of European ancestry. Mol Psychiatr. 2021. 10.1038/s41380-021-01335-3. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.