Abstract
Climate change has a significant effect on the productivity of livestock including milk, meat, and reproduction. This could be attributed to the internal diversion of energy resources towards adaptive mechanisms. Among the climate change variables, thermal stress seems to be the major limiting factor in animal agriculture. A better understanding of the effects of climate change-influenced ecological factors on the genetic diversity of livestock species is warranted. Sheep is an ideal livestock species to be used in investigating environmental adaptation due to its wide range of agroecological habitats, genetic and phenotypic variability. There is a heavy reliance on sheep genetic diversity for future animal protein security, but the implications of climate change on their genetic diversity receive less attention.
Here, the potential environmental factors influencing natural selection in sheep populations are presented. We argue that prolonged exposure to these factors plays a major role in influencing the development of adaptation traits in indigenous sheep breeds, consequently leading to the alteration of genetic diversity at specific loci. The factors discussed include hot temperatures (heat stress), insufficient water, low quantity and quality of forage, and prevalence of parasites, pests, and diseases. In addition, genetic diversity, some signatures of selection for adaptation and economic angles of selection are also briefly discussed.
A better understanding of environmental factors influencing the genetic diversity of sheep populations will inform breeding and management programs and may offer an opportunity for greater production efficiency with low input costs.
Keywords: Genetic diversity, Sheep populations, Environmental factors, Economic factors, Climate change
1. Introduction
Animal breeding involves selecting and mating of parent animals with the desired superior phenotypes to produce the next generation that performs better than the average of the parental population. In the current animal farming systems, the selection of livestock breeds is done for improved efficiency and productivity. Therefore, livestock breeders prefer the performance of superior breeds over native breeds whose production potential is limited by their genetics. Little attention is given to adaptation traits in the development of breeding programs (Dudu et al., 2016). Thus, the authors have cautioned that the state of native farm animal species and breeds is increasingly becoming uncertain with the current breeding trend.
Genetic diversity is one of the biodiversity components alongside ecosystems and species. It is the foundation for species survival and provides the “raw material” for adaptation. Genetic diversity consists of variation in the sequence of four base pairs (ATGC) which are part of the nucleic acid and a component of the genetic code (Reed & Frankham, 2003). Genetic diversity is required for a population or species to evolve in the presence of changes in climatic or environmental conditions as well as to increase animals’ fitness in the existing environmental conditions (McNeely et al., 1990). Further genetic diversity promotes coexistence between fauna within the ecosystem (Clark, 2010).
Being one of the components of biodiversity, genetic diversity within species and among populations has attracted considerable attention including and not limited to being included in international agreements, for instance, sustainable development goal (SDG) 2.5, which addresses the concerns on genetic diversity of both domesticated plants and animal species and their relatives. Several other international agreements like European Union (EU) Biodiversity strategy and action plans, Global strategy for plant conservation, and USA and Canada endangered species legislation (Hoban et al., 2020) have also considered genetic diversity an essential factor for sustained food security.
A larger proportion of ovine species that are genetically diversified eco-regionally are local breeds (Kusza et al., 2009, Kusza et al., 2015). However, they are productively disadvantaged due to their genetic potential. They face stiff competition from their commercial counterparts since the latter are performance superior to the former. Breeding programs for exotic breeds are developed to target traits of economic importance neglecting non-additive traits like adaptation traits harbored by native breeds. Objective selection in native breeds is uncommon and when it occurs, it is usually on a small population chosen for conservation. Most of the local breeds kept by small-scale farmers and pastoralist communities breed anyhow leading to inbreeding.
To improve the breeding of native breeds, their value must be improved first. This can be done by developing value chains for breed-specific products (Belibasaki et al., 2012), or producing local breeds organically (Gavojdian et al., 2016) since the market for organic products is gaining popularity. These efforts will encourage in-situ native breed conservation by farmers. For example, Mediterranean native sheep breeds from the Apulian region (Altamurana and Apulian Merino) produce milk and cheese (pugliese cheese) of high nutritional and good sensory characteristics than non-local breeds. Similarly, local goats (Girgentana) breeds in the same region produce cheese (Ricotta cheese) with a distinguishable fatty acid profile with a high nutritional index and good sensory properties than their exotic counterparts (Di Trana et al., 2015). Hungarian grey, Angus, and Hungarian grey X Charolais produced meat with higher intramuscular fat content than Hungarian Simmental and Charolais breeds (Holló et al., 2012). Intramuscular fat beef contains high levels of Polyunsaturated fatty acids (PUFA) and monounsaturated fatty acids (MUFA) (Troy et al., 2016) which are beneficial to human health. Similar studies on pigs revealed that Mangalica a native pig breed produced under the free-range system had higher oleic acid and total monounsaturated acid, while a cross of Mangalica and Duroc had higher linoleic acid and total polyunsaturated fatty acid (Despotović et al., 2018). Generally, the quality of the native breed and their crosses’ products would be advisable to promote their commercial production as well as maintain genetic diversity. It is essential, however, that the economic aspects of farming local breeds should be also considered, thus the farmers would be interested in breeding native breeds. Developing marketing of local agricultural products is a rather complex effort including several inter-dependent activities (Karthick et al., 2020). One of its important factors is how to reach potential customers. Using the innovations of e-commerce can be efficient in this field (Salunke et al., 2018). Another possible way of promoting the in-situ conservation of native breeds is to promote crossbreeding programs in which exotic sires are used to breed native dams as terminal sires and all the crossbred progenies are slaughtered.
Selection pressure caused by both artificial and natural selection affects sheep’s genetic diversity. Climate change is a precursor of natural selection (Bemmels & Anderson, 2019). Furthermore, as climate change effects are predicted to continue, even worsen in the future (Meehl et al., 2007), sheep productivity and genetic diversity are also expected to be affected unless contingent measures are put in place. Even though farmers may put in contingent measures, individual animals and breeds also need to remain resilient to extreme environmental conditions. In this regard, they need to combine several adaptive mechanisms to remain productive in stressful environments (Bemmels & Anderson, 2019).
Climate change causes increased temperatures, reduced rainfall, increased concentration of carbon dioxide in the atmosphere, drought, flash floods, and formation of agroecological zones (Rosenblatt & Schmitz, 2014). Prolonged exposure to these factors exerts selection pressure causing selection sweeps and reduction of genomic diversity at certain loci. In the long run, the genetic architecture of the population will be altered, and thus, those individuals with the genetic background that fits the environment survive and reproduce. Several generations down the line are expected to have similar genetic architecture within the population, hence reduced diversity.
The present paper discusses the selected environmental factors that could impact the genetic diversity of indigenous sheep populations. There is a consensus that fitness and adaptation to the local environment are influenced by a myriad of factors and the process takes several generations for certain beneficial alleles or mutations to be fixed in the population. The environmental factors discussed in the succeeding section are attributed to the effects of climate change that could impact the genetics of the sheep populations.
2. Potential climate change variables impacting on sheep genetic diversity
A combination of climate change factors affects sheep production and their welfare directly or indirectly. Therefore, climate change causes several constraints that the sheep must cope with for survival. They include (I) extreme temperatures (heat and cold stress), (II) insufficient water, (III) low quality and (IV) quantity of forage, and (V) prevalence of parasites, pests, and diseases that vary with ecological zones. Such environments alter the pattern of allele frequency within the population leading to changes in population structure. In most cases, if the genetic pattern change is favorable, it is expected to rapidly increase in the population over a few generations. This would result in loss of genetic diversity, alteration of haplotype structure, and recombination patterns. Sheep populations living and performing well in such environments are most likely adapted to it. Individuals exposed to such environmental pressure undergo natural selection pressure. Certain genes underpinning fitness in such environments come to play in individuals with favorable genetic composition and over time, these genes would flow within the population to fixation level hence improving the adaptation of the population. Some of the environmental factors are discussed and their possible influence on the genetics of indigenous sheep populations.
2.1. Extreme temperatures (Heat and cold stress)
The thermal environment is the major limiting factor in animal production. A heat-stressed animal will exhibit several symptoms including increased body temperature, respiration rate, and reduced feed intake. Increased environmental temperatures lead to a drastic physiological response like redistribution of blood flow and endocrine changes that negatively affect the production, growth, and reproductive performance of an animal. The physiological changes are beneficial to the animal since they aid in maintaining normal body temperature and hypothermia prevention (Al-Haidary, 2004).
The animal reduces or removes the impacts of extreme environmental temperature stress by using increased maintenance energy requirements, reducing the energy available for other functions like production, growth, and reproduction leading to a decline in production (Collier et al., 2017). According to the authors, animals’ responses to environmental stressors could be chronic or acute, both of which require an alteration of energy balance and metabolism to maintain balanced homeostasis (McManus et al., 2020). More concerns have been raised on the implications of heat than cold stress on animal production possibly because of the projected rise in global temperatures, however, cold stress is equally important in animal agriculture.
When animals are exposed to an uncomfortable environment, they develop environmental stressors coping mechanisms to minimize the impacts of these stressors on their biological system (Collier et al., 2019). Animals can either respond by acclimation, acclimatization, and adaptation. Authors defined acclimation as the coordinated phenotypic response developed by the animal to a specific stressor in the environment, acclimatization is a coordinated response to several simultaneous stressors while adaptation involves genetic changes as adverse environments persist over several generations of a species. Generally, environmental stressors are a combination of factors and therefore, the animal undergoes acclimatization than acclimation. However, it is important to note that acclimation and acclimatization are phenotypic responses aiming to improve tolerance of fitness to new environmental conditions (Collier et al., 2019). The adaptive response to environmental stressors is long-term, it occurs when the population is exposed to environmental stressors over a long period and through generations leading to a genetically fixed population (Collier et al., 2019). Some physical features aiding in the adaptation of sheep breeds to extreme environments include hair and wool, skin and coat color, and body size as reviewed by McManus et al. (2020). These adaptive physical features on their own may not be adequate to suppress extreme environmental stress. Therefore, a combination of mechanisms like behavioral and physiological (Dwyer & Lawrence, 2005) will be needed to supplement physical features.
The immediate heat stress acclimatization responses are reduced feed intake, increased respiration rate, enhanced sweating, and panting (McManus et al., 2020). Reduced feed intake has serious implications on the intrinsic energy budget of the animal. For example, heat-stressed animal reduces feed intake, exacerbated by increased energy requirement for maintenance, hence reducing the energy available for growth, production, and reproduction. Huber (2018) illustrated the energy budget and priorities of an animal, indicating that an animal partitions its energy expenditure to maintenance, ontogenic growth, production, and lastly reproduction correspondingly. Consequently, reproductive efficiency suffers most in environmentally stressful conditions. The reproductive aspects affected most are a disruption in spermatogenesis and oocyte development, oocyte maturation, early embryonic development, fetal and placental growth, and lactation (Hansen, 2009). The profitability of sheep farming enterprise depends on reproductive efficiency determined by age at first mating, age at first conception, inter-lambing interval, litter size, number of lambs weaned as well as service period (Rather et al., 2020).
The immunity of an animal is vital in the current state of changing climate. This is because climate change is strongly associated with the re-emergence of different health hazards. However, environmental stress especially heat stress compromises the immunity of the animal. Heat stress affects the synthesis of heat shock proteins (HSPs) at the cellular level. HSPs prevent the formation of non-specific protein aggregates and assist cellular proteins in the acquisition of native nature hence playing vital cellular homeostasis (Singh et al., 2017) enhancing inert immunity which acts as a first-level defense mechanism (Sevi & Caroprese, 2012). There aren't many studies on how terrestrial mammals adapt to cold temperatures, but it is known that these adaptations entail processes that help with thermal insulation. McManus et al. (2020) note that larger, heavier, shorter, and woolly/hairy/fleecy breeds are more adapted to colder climates than smaller, taller, and lighter breeds. Prolonged exposure to cold temperatures can also cause a variety of acclimation reactions including a rise in metabolism, the development of fleece, a reduction in growth (Webster et al., 1969), and possibly even reproductive rates.
It can be difficult to measure a livestock species' tolerance to heat stress, however some writers have suggested using indicators such as body temperature, heart rate, respiration rate, sweating rate and productivity on a scale of temperature to humidity index-THI (Carabaño et al., 2019). As a result, it is possible to test the genetics and heritability (h2) of traits associated with heat tolerance using progressive THI. For instance, Menéndez-Buxadera et al. (2014) found that the comfort (THI > 47) and heat stressed (THI > 48) zones differed in the heritability of the milk traits i.e., milk yield (MY), protein yield (FY), and dry matter yield (DMY). For MY, FY, and DMY, respectively, h2 values in the comfort zone were 0.161, 1.151, and 0.157 whereas those in the heat stress zones were 0.0081, 0.0094, and 0.105.
Several studies on sheep have been conducted to determine selection signatures under heat stress environments. Results infer that there are genes responsible for adaptation suggesting an impact of heat on the genomics architecture of the sheep population. For instance, Kim et al. (2016) studied signals of selection in Barki goats and sheep indigenous to the hot environment and identified suggestive genes believed to be underlying adaptation phenotypes and traits. They include thermotolerance (melanogenesis) (FGF2, GNAI3, PLCB1), body size and development (BMP2, BMP4, GJA3, GJB2), energy and digestive metabolism (MYH, TRHDE, ALDH1A3), and nervous and autoimmune response (GRIA1, IL2, IL7, IL21, IL1R1). Alike, HERC2 and CYFIP1 genes that are relevant to regulate innate and acquired immune responses, as well as cytokine signaling, GJB2, and GJA3 for body size and development, BMPR1B associated with prolificacy in sheep, TSHR gene for reproduction, NFI gene for litter size as well as BMP2 for prolificacy and fecundity, were under selection in Hetian, Karakul and Yabuyi Chinese indigenous sheep breeds (Abied et al., 2020).
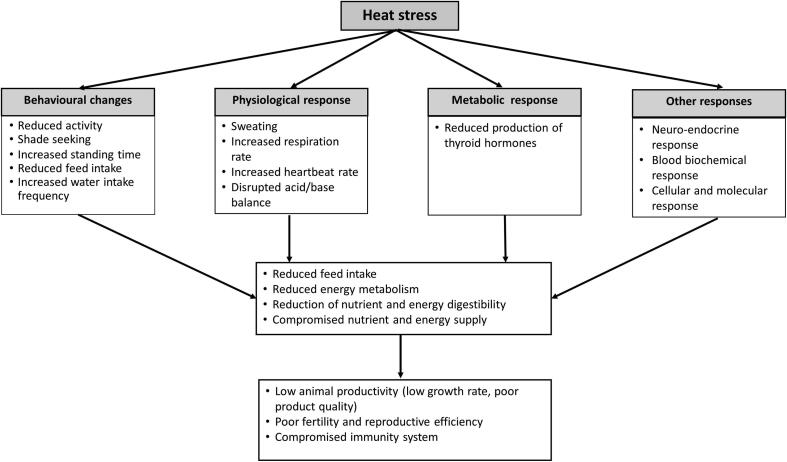
A species and/or population that is genetically endowed and has individuals that can efficiently combine adaptation mechanisms to adapt to the effects of climate change variables especially heat stress which is the main limiting factor (Fig. 1), stands a chance to survive for many generations.
Fig. 1.
A summary of a potential model of the mode of action of response to heat stress in sheep.
2.2. Insufficient water
The animal's live body weight is composed of 70 % water. This implies that water is an essential requirement in animal life and hence should be regarded as a major part of animal nutrition as well as the greatest constraint to performance. Despite its vitality, few studies have been conducted on the effects of water on animal production and other related factors. Water availability can be addressed to be either in terms of quantity or quality, both affect the performance of livestock (Umar et al., 2014).
The capacity of sheep to endure water stress depends on the breed hence the genetic background. Generally, indigenous sheep breeds mostly found in arid and semi-arid regions have a more efficient tolerance mechanism to water stress. The effects of water stress are more aggravated by heat stress (Chedid et al., 2014). When the animal is subjected to water stress for a prolonged period, it undergoes dehydration, which is a process of losing water.
Dehydration is positively correlated with heat stress, heat stress causes increased body temperatures which stimulates physiological responses like sweating and vasodilation to increase heat dissipation in the environment (Akerman et al., 2016), which in turn increases water consumption by an animal. Notwithstanding the impacts of water stress on animal production, some studies have suggested that ruminants have developed mechanisms of coping with the negative effects of both heat and water stress (Hussein et al., 2020).
Furthermore, environmental water stress affects forage quantity but has less effect on fodder quality depending on the type of fodder crop. For instance, Kuchenmeister et al., 2013, Allahdadi and Bahreininejad, 2019 found that water stress had less pronounced effects on nutrition (crude proteins (CP), neutral detergent fiber (NDF), and acid detergent fiber (ADF)). Affected forage quantity has implications on the animal’s total energy intake which subsequently impacts animal productivity and health. Some breeds are genetically enabled to minimize water loss and improve water reabsorption. For example, sheep breeds that are adapted to the arid and semi-arid regions exhibit different morphological adaptations like different fleece colors and coats, body height, and body size that aid in the tolerance to heat stress and water shortage as well (Chedid et al., 2014).
2.3. Inadequate feed availability, digestibility, and absorption
Feed availability in this context refers to both feed quantity and quality. Animal nutrition is positively correlated with growth, reproduction, and production. Poor and/or inadequate nutrition negatively affects the performance of the aforementioned. With the projected increase in atmospheric CO2 and air temperatures coupled with changed soil quality because of climate change, the quality and quantity of forage are expected to be affected. Further, intensive rainfall and longer dry periods are also projected leading to varied inter-annual precipitation variability (Grant et al., 2014). Persistent inter-annual rainfall variability leads to a continuous variation of soil moisture leading to crops subjected to moisture stress (Fay, 2009).
To understand the importance of feed available on the genetic architecture of the local sheep population, it is vital to understand the basic ingredients of forage first. In forage, protein, digestible energy, and the passage rate are the most important elements of ruminant nutrition determining forage quality. Crude proteins are converted from forage nitrogen by ruminal microbes, soluble carbon compounds, as well as cellulose and hemicellulose, are main sources of energy. Cellulose and hemicellulose are also digestible, but the rate of passage is limiting, digestibility can be hampered by the presence of lignin. The rate of passage in ruminants is more complex, but generally, “the greater the digestibility, the lower the lignin and the higher the rate of passage” (Milchunas et al., 2005) hence a higher absorption.
A combination of soil water stress and warming reduces forage quality by increasing lignin content, reducing starch and crude protein digestibility of C4 grass (Habermann et al., 2019). C4 grasses belong to the sub-families panicoide and Eragrostoidae a Gramineae family. They are mainly found in cool temperate to hot-wet tropics with a high concentration in semi-arid and arid regions which are characterized by high solar radiation, high daytime temperatures, daily and/or seasonal water stress (Doliner & Jolliffe, 1979).
Elevated carbon dioxide (CO2) concentration in the atmosphere combined with the rise in global temperatures, affects forage quality and quantity (Babinszky et al., 2011). Forage quality is determined by the digestibility and absorption of the ingested feed. Digestibility is one of the components of feed nutritive value, others are feed consumption and energetic efficiency (Van Soest, 2018). Digestibility can be apparent digestibility; the balance of feed less the feces or true digestibility; a balance between respective feed residues from the diet escaping digestion and arriving in the feces excluding metabolic products (Soest, 1963). It is also worth noting that plants, most importantly forage plants, respond differently to climate change effects depending on their photosynthesis types either C3, C4, or CAM (Babinszky et al., 2011). Therefore, as the dynamics of climate change events increase, the possibility of vegetation change also increases as more uncommon vegetation types emerge, not used to by existing livestock species, forcing them to either change their feeding habits, migrate, or starve to death at the worst scenario.
Also, a combination of climate change variables may alter the forage quality since the existing forage changes may also be forced to adjust their intrinsic mechanisms to adapt to the environmental conditions. Furthermore, the impact of climate change on quantity will also depend on photosynthesis pathway crops, however, Babinszky et al. (2011) noted that C3 crops will be more negatively affected, reducing their herbage quantity and quality in the event of increased temperatures and reduced rainfall. This could lead to reduced feed utilization by animals, compromising energy intake and in the long term, the species could face risks of extinction if it does not have an adequate adaptive mechanism. Extinction leads to the loss of vital domestic animal genetic resources.
Prolonged exposure to elevated CO2 causes a reduction in photosynthesis leading to reduced growth in a phenomenon referred to as photosynthetic downregulation (Sanz-Sáez et al., 2012). The authors also emphasized that some other environmental variables also interact with CO2 and surface temperature to affect plant growth. They include soil nitrogen, soil water content, atmospheric humidity, and solar radiation. Amongst them all, Nitrogen is the limiting factor and its presence is positively correlated with forage quality, in other words, elevated CO2 concentration with a deficiency in nitrogen supply causes reduced photosynthesis hence reduced yield (Sanz-Sáez et al., 2012). Further, nitrogen deficiency in the soil causes reduced shoot nitrogen content under elevated CO2 concentration, causing reduced crude protein and enhanced fiber concentration content hence lowering forage quality. The quality of neutral detergent fiber (NDF), acid detergent fiber (ADF), and acid detergent lignin (ADL) (Reid et al., 1988, Sanz-Sáez et al., 2012, Soest, 1963) determines digestibility, ingestion, and palatability of grass forages. NDF includes cellulose, hemicellulose, and lignin while ADF includes cellulose and lignin (Sanz-Sáez et al., 2012). Some studies have indicated that elevated concentration of CO2 significantly reduces nitrogen concentration in herbage while increasing lignin concentration and carbon: nitrogen and lignin: nitrogen ratios (Cha et al., 2017) reducing forage quality and negatively affecting digestibility.
Under heat stress, animal productivity is reduced since feed intake, digestibility (Maia et al., 2020), and absorption (Ríus, 2019) are decreased. Further, heat stress negatively affects rumination activity, a vital activity in the digestive physiology of ruminants (Maia et al., 2020, Reiter et al., 2018). Behavioral traits like chewing and rumination play an important role in feed intake and digestion efficiency (Beauchemin, 2018). A negative effect on these two activities negatively affects feed intake, thereby, affecting energy available for their output.
2.4. Prevalence of parasites, pests, and disease
Climate change is predicted to cause major ecological and biological changes that directly or indirectly impacts on sheep’s health within the ecosystem. It is, therefore, unreasonable not to consider the impacts of climate change on animal health and the possible intrinsic mechanisms animals use to tolerate the prevalence of health hazards. Majorly, these mechanisms are genetically driven hence the need to have an adequate genetic base that would provide opportunities to withstand threats to animal health. In this context, disease and parasite tolerance refer to an animal constantly being productive with normal reproductive efficiency despite the presence of parasites, pests, and disease-causing agents and to a further extent break the life cycle of pathogens by developing internal conditions unfavorable for pathogen, survivability, reproduction, and pathogenicity. It is challenging to select for parasite resistance using pedigree because several research publications suggest that the hereditability to parasite resistance is low and may range between h2 = 0.04 to h2 = 0.16 (Pacheco et al., 2021).
Climate change affects the distribution, prevalence, and seasonality of parasites, pests, and diseases. Pathogens and vectors habitats change with the climate, thus, with the projected increase in changing climate events, the distribution, prevalence, and seasonality of pests and diseases are also projected to change (Medlock & Leach, 2015), especially vector-borne diseases that have seasonal and spatial patterns influenced by environmental heterogeneity (de La Rocque et al., 2008). IPPC report of 2007 envisaged an increase in the spatial distribution of animal disease-causing vectors that are most sensitive to temperature changes (IPCC, 2007), inferring the role of climate change to the spread of infectious diseases by allowing disease-causing agents e.g. “Fungi, Viruses, bacteria and protozoa” to move to new ecosystems where they can cause harm to livestock, humans, and wildlife. Furthermore, the recent spread of vector-borne diseases such as trypanosomiasis, Lyme disease, tick-borne encephalitis, blue tongue, and dengue (de La Rocque et al., 2008) was linked to climate change. For instance, Higher temperatures have a greater impact on the lifecycle of disease-causing pathogens like aiding in shortening the incubation period of pathogens, increasing the incubation rates, and enhancing the chances of the vectors to live longer to transmit infections (Kutz et al., 2005). The authors averred that an increase in temperature by 1 °C shifted the protostronglid nematodes (U. pallikuukensis) larval development from 2 years to a 1-year cycle, expanded the period of transmission of the third stage of larvae development (L3) in slugs, and increased larvae infectious period. Some studies have shown a positive linkage between temperature and expansion of geographical ranges of arthropod vectors like bluetongue virus while others infer a contrasting trend like tsetse flies in sub-Saharan Africa. Moreover, there is a reported positive association between extreme events (cold and hot) and disease outbreaks, such as Rift valley fever outbreaks in East Africa (Bett et al., 2017, Kutz et al., 2005). Distribution, prevalence, and seasonality of pests, parasites, and diseases are not influenced by climate change alone, but other factors both biophysical (i.e. land cover, landscapes, and host abundance) and directly anthropogenic (i.e. public health, diet, socio-political, and other human behaviors) modulate contact between pathogens, vectors and reservoirs shifting disease behavior (de La Rocque et al., 2008).
Individual differences in immune responses are observed widely and many studies have associated them with genetic differences between individuals. Therefore, understanding the gene and environment interaction and its role in generating immune response variation within and between populations is important for sustained sheep breeding and selection. It would be much more judicious, therefore, to consider the importance of having the animal genetic base that would be a backbone for sustainable production in the presence of these health risk challenges, thus sheep populations that are more genetically diverse provide opportunities for improved adaptation. Breeding for resistance will minimize drug use, ultimately enhancing food safety by reducing drug residues in sheep products (Aguerre et al., 2018). Recently, a consensus on the possibility of genetic selection for immunity response in sheep was reached and some studies reported successful experiments in this effect e.g. (Bouix et al., 1998, Gauly and Erhardt, 2001).
An example of a major disease of concern and whose immunity and genetic architecture are widely studied is mastitis. Mastitis is a disease-causing inflammation of mammary glands and causes major economic losses in the sheep industry (Giannakopoulos et al., 2019, Larsgard and Vaabenoe, 1993). The disease is environmentally influenced hence climate change catalyzed. Due to its economic importance and the risk, it poses to the affected population, genetic diversity within and among populations would enhance the survivability of the breed in the face of climate change events. Recently, studies on the genetics of disease resistance have been performed. For example, Larsgard & Vaabenoe (Larsgard & Vaabenoe, 1993) estimated heritability from ewe’s resistance to mastitis to be 0.13 (±0.16) by the LS-method and 0.49 by the nonlinear threshold method. Conington et al. (2008) have reviewed and discussed extensively breeding for mastitis resistance and its economic ramifications. Further use of marker-assisted selection for resistance has been embraced in the past few years with the aim of enhancing genetic improvement as well as maintaining genetic diversity, an opportunity for sustained production in the current uncertainties of climate change. Using genome-wide association studies, genes SOCS2, CTLA4, C6, C7, C9, PTGER4, DAB2, CARD6, OSMR, PLXNC1, IDH1, ICOS, FYB, and LYFR were identified based upon analysis of an ovine transcriptional atlas and transcriptome data derived from milk somatic cells to be under selection confirming the presence of animal genetic variability in mastitis resistance among 609 ewes of the Greek Chios breed (Banos et al., 2017). Other studies and evaluations of the role of sheep genetics on adaptation were conducted on nematodes. Different sheep breeds, like Rhön sheep, which a local sheep breeds in Germany (Gauly & Erhardt, 2001), and Polish longwool sheep from Poland (Bouix et al., 1998) have been evaluated on their resistance to nematodes facilitating selection for resistance.
3. Sheep genetic diversity and potential selection signatures for adaptation traits
In the recent past, scientists have developed interest in studying the genetic diversity of different animal species globally using different genetic markers. These studies aimed at elucidating either the origin, migratory paths, genetic diversity status within the breed and among populations resulting in classifying breeds according to extinction risk status and recommending conservation measures if need be. Little attention has been placed on the effects of genotype – environment (GxE) interaction on the loss of diversity in different species. Since domestication, sheep have been subjected to forces of natural and artificial selection leading to several mutations, allele realignment and frequencies in several generations in response to changing environmental conditions resulting from climate change (Gauly & Erhardt, 2001). These genetic changes were meant to align animals to meet the production needs of the environment as well as the farmer. However, due to the increased demand for animal products in the recent past, pressure for improved production has ensued, leading to enhanced selection intensity, especially in exotic sheep breeds. An intensive selection coupled with reduced population size leads to a loss of diversity (Eusebi et al., 2020). Previously, the emphasis was put on breeding strategies that ensured the maintenance of heterozygosity disregarding “allelic diversity”. Allelic diversity in this context refers to a full range of potentially adaptive alleles in the species (Notter, 1999), but this was less useful since heterozygosity did not effectively address genetic diversity corresponding with the changing environmental conditions. Intensive selection for additive traits of economic importance alongside inbreeding are the possible causes of declining allele diversity (Caballero et al., 2020, Simm et al., 1996). Another cause of declining genetic diversity is reduced effective population size (Zhao et al., 2014). The loss of genetic variability has also been greatly impacted by the transfer of breeds to new environments without taking GxE interaction into account. According to the fundamental theory of the GxE interaction, phenotypes of one breed may vary between two different environmental situations (Gavojdian et al., 2014). Hence, introducing breeds in new production environments may lead to decreased fitness traits, putting the breed at risk of extinction. Diversity loss reduces the breed and species' ability to withstand the extreme environments caused by climate change.
Identification of differences in allele frequencies and genetic diversity at a specific genomic region has enabled the possibility of associating the selection pressure to different ecological regions. Evidence of selection pressure for adaptation has been investigated recently and several genomic regions underlying adaptations have been detected. Analysis of allele frequencies between populations to infer selective pressure in one population and analysis of the reduction of genetic diversity at a certain genomic region are the main methods of detecting selection signatures. The latter method allows detection of possible “selective sweeps” arising from the regions where favorable alleles have risen in frequencies hence reducing levels of diversity in that genomic region or locus (Saravanan et al., 2020).
Among many genetic markers used in mapping out genomic variations and loss of diversity at a particular locus, single nucleotide polymorphisms (SNPs) are currently widely used. The bioinformatics analysis methods use the information from thousands of SNPs along the whole genome to estimate their effect to identify or select loci involved in the phenotypic variation. In other words, SNP markers are used to identify candidate genes whose nucleotides influence trait differences are located (Cardona Tobar et al., 2020). Selection signatures studies have been contacted in multiple domestic animal species for several adaptation traits e.g., cattle (Shen et al., 2020), goats (Guo et al., 2018), buffaloes (Mokhber et al., 2018), and sheep (Wiener et al., 2021).
Studies on the genetic diversity of worldwide indigenous sheep populations by Kijas (2009) revealed clustering based on geographic origin and known breed history while Ethiopian sheep breeds demonstrated clustering based on morphology (tail type) and geographic origin for example, short fat-tailed (very cool high altitude), long fat-tailed (mid to high-altitude) and fat-rumped (arid low-altitude) (Edea et al., 2017). Thus, suggesting possible structuring based on allele frequencies and realignments influenced by ecological conditions. Further, it was reported that similar populations had high levels of SNP diversity (Kijas et al., 2012), suggesting a wide range of genomic diversity between populations, possibly due to the natural selection pressure.
Several selection signatures (Table 1.) believed to be responsible for indigenous sheep adaptation have been identified in sheep breeds inferring the existence of genetic variation for adaptation traits among populations. This offers a good opportunity for breeding for adaptation traits that would aid in breed adapting to extreme environments resulting from climate change. It is worth noting that selection signatures differ between populations, even in populations exposed to similar ecological conditions.
Table 1.
Selection signatures responsible for indigenous sheep adaptation.
| Breeds | Country | Gene | Function | Reference |
|---|---|---|---|---|
| Prairie Tibetan, Valley Tibetan, Oula, Ayinbuluke, Cele Black, Hu, Tan, Small Tail Han, Wuzhumuqin and Australian Merino sheep | China | RXFP2 | Horn phenotype | Pan et al., 2018 |
| MITF, MSRB3, SLC26A4 | Hearing | |||
| SMDC1, SOX6 | Muscle development | |||
| PRD-SPRRII | Rumen development | |||
| Kefis, Adane, Arabo, Gafera-Washera, Molalu-Menz, Bonga, Gesses, Kido, Doyogena, ShubiGemo and Loya | Ethiopia | ALX4, HOXB13, BMP4 | Limp development and Tail formation | Ahbara et al., 2019 |
| NPR2, HINT2, SPAG8, INSR | Growth traits | |||
| TRPM8 | Regulation of body temperature | |||
| DNAJC18 | Response to heat stress | |||
| Barbarine, Noire de Thibar and Queue fine de l'Ouest | Tunisia | CDS2, PROKR1, BMP2 | Lipid storage /Tail fatness | Baazaoui et al., 2021 |
| Changthangi, Deccani,and Garole | India | TRPM8 | Cold adaptation | Saravanan et al., 2021 |
| Garole | RAD1 | Immunity | ||
| Changthangi and Deccani | IL2, JADE2, FGF2, SPP2, RMI1 and TSHR | Growth and body growth | ||
| Bariga Negra, Morada Nova, Rabo Largo, Santa Ines and Somalis. | Brazil | GNG2; PTGDR; PLEKHO2; USP3; APH1B; RAB8B | Immune response | Paim et al., 2022 |
| PTGDR; PLEKHO2; ZNF609, CAPSL; IL7R; | Inflammation response | |||
| CA12 | Dehydration and Kidney function, water balance | |||
| PTGDR | Body Temperature; Thermogenesis | |||
| Adane, Arabo, Bonga, Doyogena, Kefis, Segentu, Gesses, Kido, Menz, Shubi Gemo and Washera | Ethiopia | ITPR2, FAM162A, GATA6, GNGT1 and HIF3A. | Hypoxia | Wiener et al., 2021 |
RXFP2: Relaxin Family Peptide Receptor 2, MITF: Melanocyte inducing transcription factor, MSRB3: Methionine Sulfoxide Reductase B3, SLC26A4: solute carrier family 26 member 4, SMDC1: SAYSVFN motif domain containing 1, SOX6: SRY-Box Transcription Factor 6, PRD-SPRRII: Small proline-rich protein type II, ALX4: Aristaless-Like Homeobox 4, HOXB13: Homeobox Protein Hox-B13, BMP4: Bone Morphogenetic Protein 4, TRPM8: Transient Receptor Potential Cation Channel Subfamily M, member 8, NPR2: natriuretic peptide receptor 2, HINT2: Histidine Triad Nucleotide Binding Protein 2, SPAG8: Sperm Associated Antigen 8, INSR: Insulin Receptor, DNAJC18: DnaJ Heat Shock Protein Family (Hsp40) Member C18, CDS2: CDP-Diacylglycerol Synthase 2, PROKR1: Prokineticin Receptor 1, BMP2: Bone Morphogenetic Protein 2, RAD1 Checkpoint DNA Exonuclease, IL2: interleukin 2, JADE2: E3 Ubiquitin-Protein Ligase Jade-2, FGF2: Fibroblast Growth Factor 2, SPP2: Secreted Phosphoprotein 2, RMI1: RecQ Mediated Genome Instability 1: TSHR: thyroid stimulating hormone receptor, GNG2: G Protein Subunit Gamma 2, PTGDR: Prostaglandin D2 Receptor, PLEKHO2: Pleckstrin Homology Domain Containing O2, USP3: Ubiquitin Specific Peptidase 3; APH1B: Aph-1 Homolog B, Gamma-Secretase Subunit, RAB8B: Ras-Related Protein Rab-8B, PTGDR: Prostaglandin D2 Receptor; PLEKHO2: Pleckstrin Homology Domain Containing O2, ZNF609: Zinc Finger Protein 609, CAPSL: Calcyphosine Like, IL7R: Interleukin 7 Receptor, CA12: Carbonic Anhydrase 12, ITPR2: Inositol 1,4,5-Trisphosphate Receptor Type 2, FAM162A: Family With Sequence Similarity 162 Member A, GATA6: GATA Binding Protein 6, GNGT1: G Protein Subunit Gamma Transducin 1 and HIF3A: Hypoxia Inducible Factor 3 Subunit Alpha.
4. Conclusion
Climate change tremendously effects animal agriculture, most importantly sheep production. Furthermore, climate change leads to the formation of agroecological regions that experience different levels of climatic conditions. Among the climate change variables affecting sheep production and genetic diversity are droughts affecting water availability and increased surface temperatures, elevated levels of carbon dioxide affecting the quality of forage, and most importantly, heat stress that triggers all physiological and behavioral responses in animals to minimize and tolerate effects of extreme weather stress.
Genetic diversity is the foundation for the evolution and is the primary tool for the adaptability and survival of species in several generations. Moreover, since climate change is an essential player in natural selection, which leaves genomic signatures associated with selection sweeps at a specific locus, it is worthy to maintain genetic diversity for sustained production. It is believed that native sheep breeds, most of which are reared in Africa and Asia by pastoral communities are important genetic resources because they are adapted to extreme local conditions. These communities depend on native sheep breeds for meat, milk, and income thus, losing the breeds to forces of climate change will be detrimental to them.
Studies on selection signatures for adaptation offer an opportunity to improve sheep breeding programs to include selection for adaptation traits. Production of adapted breeds improves animal welfare and ensures environmental sustainability in the long term. Although commercial sheep breeders target genetic improvement for additive economic traits, they should include non-additive genes in their breeding program for sustained food security in the face of climate change.
To ensure the goals mentioned in the previous paragraph, it is indispensable to make farmers interested in breeding adaptive (local) breeds. Therefore, local breeds products and value chains should be advertised among consumers, and marketing efforts of these products may be supported by National Governments. These steps could lead to developing more sustainable agricultural management as well.
Multi-institutional collaboration is required for the success of the selection of eco-friendly traits in sheep. For instance, population geneticists should collaborate with quantitative geneticists, farmers, meteorologists, the natural resource management departments, and other stakeholders to ensure sustained food security in the wake of climate change.
CRediT authorship contribution statement
George Wanjala: Conceptualization, Writing – original draft, Writing – review & editing. Putri Kusuma Astuti: Writing – review & editing. Zoltán Bagi: Writing – review & editing. Nelly Kichamu: . Péter Strausz: Writing – review & editing. Szilvia Kusza: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
George Wanjala, Email: geog.wanjala@agr.unideb.hu.
Putri Kusuma Astuti, Email: astuti@agr.unideb.hu.
Zoltán Bagi, Email: bagiz@agr.unideb.hu.
Nelly Kichamu, Email: n.kichamu@uqconnect.edu.au.
Péter Strausz, Email: peter.strausz@uni-corvinus.hu.
Szilvia Kusza, Email: kusza@agr.unideb.hu.
References
- Abied A., Bagadi A., Bordbar F., Pu Y., Augustino S.M.A., Xue X., Xing F., Gebreselassie G., Han J.-L., Mwacharo J.M., Ma Y., Zhao Q. Genomic diversity, population structure, and signature of selection in five chinese native sheep breeds adapted to extreme environments. Genes. 2020;11(5):494. doi: 10.3390/genes11050494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguerre S., Jacquiet P., Brodier H., Bournazel J.P., Grisez C., Prévot F., Michot L., Fidelle F., Astruc J.M., Moreno C.R. Resistance to gastrointestinal nematodes in dairy sheep: genetic variability and relevance of artificial infection of nucleus rams to select for resistant ewes on farms. Vet. Parasitol. 2018;256:16–23. doi: 10.1016/j.vetpar.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Ahbara A., Bahbahani H., Almathen F., Al Abri M., Agoub M.O., Abeba A., Kebede A., Musa H.H., Mastrangelo S., Pilla F., Ciani E., Hanotte O., Mwacharo J.M. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet. 2019;Vol. 9 doi: 10.3389/fgene.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman, A. P., Tipton, M., Minson, C. T., & Cotter, J. D., 2016. Heat stress and dehydration in adapting for performance: Good, bad, both, or neither? Temperature: Multidisciplinary Biomedical Journal, 3(3), 412–436. https://doi.org/10.1080/23328940.2016.1216255. [DOI] [PMC free article] [PubMed]
- Al-Haidary A.A. Physiological responses of Naimey sheep to heat stress challenge under semi-arid environments. Int. J. Agric. Biol. 2004;2:307–309. [Google Scholar]
- Allahdadi, M., & Bahreininejad, B., 2019. Effects of water stress on growth parameters and forage quality of globe artichoke (Cynara cardunculus var. scolymus L.). 10. http://iar.shirazu.ac.ir/article_5596_0d82535bbf1a58118eac41634fd5e25c.pdf.
- Baazaoui I., Bedhiaf-Romdhani S., Mastrangelo S., Ciani E. Genome-wide analyses reveal population structure and identify candidate genes associated with tail fatness in local sheep from a semi-arid area. Animal. 2021;15(4) doi: 10.1016/j.animal.2021.100193. [DOI] [PubMed] [Google Scholar]
- Babinszky, L., Halas, V., & W.A., M., 2011. Impacts of Climate Change on Animal Production and Quality of Animal Food Products. In H. Kheradmand (Ed.), Climate Change - Socioeconomic Effects. InTech. http://www.intechopen.com/books/climate-change-socioeconomic-effects/impacts-of-climate-change-on-animal-production-and-quality-of-animal-food-products.
- Banos G., Bramis G., Bush S.J., Clark E.L., McCulloch M.E.B., Smith J., Schulze G., Arsenos G., Hume D.A., Psifidi A. The genomic architecture of mastitis resistance in dairy sheep. BMC Genomics. 2017;18(1):624. doi: 10.1186/s12864-017-3982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin K.A. Invited review: Current perspectives on eating and rumination activity in dairy cows. J. Dairy Sci. 2018;101(6):4762–4784. doi: 10.3168/jds.2017-13706. [DOI] [PubMed] [Google Scholar]
- Belibasaki S., Sossidou E., Gavojdian D. Local breeds: can they be a competitive solution for regional development in the world of “globalization”? the cases of greek and romanian local breeds. Animal Sci. Biotechnol. 2012;45(1):243–254. [Google Scholar]
- Bemmels J.B., Anderson J.T. Climate change shifts natural selection and the adaptive potential of the perennial forb Boechera stricta in the Rocky Mountains. Evolution. 2019;73(11):2247–2262. doi: 10.1111/evo.13854. [DOI] [PubMed] [Google Scholar]
- Bett B., Kiunga P., Gachohi J., Sindato C., Mbotha D., Robinson T., Lindahl J., Grace D. Effects of climate change on the occurrence and distribution of livestock diseases. Prev. Vet. Med. 2017;137(Pt B):119–129. doi: 10.1016/j.prevetmed.2016.11.019. [DOI] [PubMed] [Google Scholar]
- Bouix J., Krupinski J., Rzepecki R., Nowosad B., Skrzyzala I., Roborzynski M., Fudalewicz-Niemczyk W., Skalska M., Malczewski A., Gruner L. Genetic resistance to gastrointestinal nematode parasites in Polish long-wool sheep. Int. J. Parasitol. 1998;28(11):1797–1804. doi: 10.1016/S0020-7519(98)00147-7. [DOI] [PubMed] [Google Scholar]
- Caballero, A., Villanueva, B., & Druet, T., 2020. On the estimation of inbreeding depression using different measures of inbreeding from molecular markers. Evolutionary Applications, n/a(n/a). https://doi.org/https://doi.org/10.1111/eva.13126. [DOI] [PMC free article] [PubMed]
- Carabaño María, J, Ramón Manuel, Menéndez-Buxadera Alberto, Molina Antonio, Díaz Clara. Selecting for heat tolerance. Animal Frontiers. 2019;9(1):62–68. doi: 10.1093/af/vfy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona Tobar K.M., Álvarez D.C.L., Franco L.Á.Á. Review; Rev Mex Cienc Pecu: 2020. Genome-wide association studies in sheep from Latin America; p. 25. [Google Scholar]
- Cha S., Chae H.-M., Lee S.-H., Shim J.-K. Effect of elevated atmospheric CO2 concentration on growth and leaf litter decomposition of Quercus acutissima and Fraxinus rhynchophylla. PLoS One. 2017;12(2):e0171197. doi: 10.1371/journal.pone.0171197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedid, M., S, J. L., Sylvie, G.-R., Christine, D.-P., & K, H., 2014. Review: Water stress in sheep raised under arid conditions. Canadian Journal of Animal Science. https://doi.org/10.4141/cjas2013-188.
- Clark J.S. Individuals and the variation needed for high species diversity in forest trees. Science. 2010;327(5969):1129–1132. doi: 10.1126/science.1183506. [DOI] [PubMed] [Google Scholar]
- Collier R.J., Renquist B.J., Xiao Y. A 100-Year Review: Stress physiology including heat stress. J. Dairy Sci. 2017;100(12):10367–10380. doi: 10.3168/jds.2017-13676. [DOI] [PubMed] [Google Scholar]
- Collier R.J., Baumgard L.H., Zimbelman R.B., Xiao Y. Heat stress: physiology of acclimation and adaptation. Anim. Front. 2019;9(1):12–19. doi: 10.1093/af/vfy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conington J., Cao G., Stott A., Bünger L. Breeding for resistance to mastitis in United Kingdom sheep, a review and economic appraisal. Vet. Rec. 2008;162(12):369–376. doi: 10.1136/vr.162.12.369. [DOI] [PubMed] [Google Scholar]
- de La Rocque, S., Rioux, J. A., & Slingenbergh, J., 2008. Climate change: effects on animal disease systems and implications for surveillance and control. Revue Scientifique Et Technique (International Office of Epizootics), 27(2), 339–354. http://www.ncbi.nlm.nih.gov/pubmed/18819664. [PubMed]
- Despotović A., Tomović V., Šević R., Jokanović M., Stanišić N., Škaljac S., Šojić B., Hromiš N., Stajić S., Petrović J. Meat quality traits of M. longissimus lumborum from White Mangalica and (Duroc× White Mangalica)× White Mangalica pigs reared under intensive conditions and slaughtered at about 180-kg live weight. Ital. J. Anim. Sci. 2018;17(4):859–866. [Google Scholar]
- Di Trana A., Sepe L., Di Gregorio P., Di Napoli M.A., Giorgio D., Caputo A.R., Claps S. The role of local sheep and goat breeds and their products as a tool for sustainability and safeguard of the Mediterranean environment. Sustain. Agro-Food Nat. Resour. Syst. Mediterranean Basin. 2015:77–112. [Google Scholar]
- Doliner, L. H., & Jolliffe, P. A., 1979. Ecological Evidence concerning the Adaptive Significance of the C₄ Dicarboxylic Acid Pathway of Photosynthesis. Oecologia, 38(1), 23–34. https://www.jstor.org/stable/4215762. [DOI] [PubMed]
- Dudu A., Ghiţă E., Costache M., Georgescu S.E. Origin and genetic diversity of Romanian Racka sheep using mitochondrial markers. Small Rumin. Res. 2016;144:276–282. doi: 10.1016/j.smallrumres.2016.10.016. [DOI] [Google Scholar]
- Dwyer C.M., Lawrence A.B. A review of the behavioural and physiological adaptations of hill and lowland breeds of sheep that favour lamb survival. Appl. Anim. Behav. Sci. 2005;92(3):235–260. [Google Scholar]
- Edea Z., Dessie T., Dadi H., Do K.-T., Kim K.-S. Genetic Diversity and population structure of ethiopian sheep populations revealed by high-density SNP markers. Front. Genet. 2017;8:218. doi: 10.3389/fgene.2017.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebi P.G., Martinez A., Cortes O. Genomic tools for effective conservation of livestock breed diversity. Diversity. 2020;12(1):8. doi: 10.3390/d12010008. [DOI] [Google Scholar]
- Fay, P. A., 2009. Precipitation Variability and Primary Productivity in Water-Limited Ecosystems: How Plants “Leverage” Precipitation to “Finance” Growth. The New Phytologist, 181(1), 5–8. https://www.jstor.org/stable/30225813. [DOI] [PubMed]
- Gauly M., Erhardt G. Genetic resistance to gastrointestinal nematode parasites in Rhön sheep following natural infection. Vet. Parasitol. 2001;102(3):253–259. doi: 10.1016/S0304-4017(01)00530-1. [DOI] [PubMed] [Google Scholar]
- Gavojdian D., Kusza S., Jávor A. Implications of genotype by environment interactions in dairy sheep welfare. Scient. Pap.: Anim. Sci. Biotechnol./Lucrari Stiintifice: Zootehnie si Biotehnol. 2014;47(1) [Google Scholar]
- Gavojdian D., Padeanu I., Sauer M., Dragomir N., Ilisiu E., Kusza S.z., Rahmann G. Effects of using indigenous heritage sheep breeds in organic and low-input production systems on production efficiency and animal welfare in Romania. J. Appl. Agric. Forestry Res. - Landbauforschung Volkenrode. 2016;66:290–297. doi: 10.3220/LBF1483607712000. [DOI] [Google Scholar]
- Giannakopoulos A., Vasileiou N.G.C., Gougoulis D.A., Cripps P.J., Ioannidi K.S., Chatzopoulos D.C., Billinis C., Mavrogianni V.S., Petinaki E., Fthenakis G.C. Use of geographical information system and ecological niche modelling for predicting potential space distribution of subclinical mastitis in ewes. Vet. Microbiol. 2019;228:119–128. doi: 10.1016/j.vetmic.2018.11.021. [DOI] [PubMed] [Google Scholar]
- Grant K., Kreyling J., Dienstbach L.F.H., Beierkuhnlein C., Jentsch A. Water stress due to increased intra-annual precipitation variability reduced forage yield but raised forage quality of a temperate grassland. Agr. Ecosyst. Environ. 2014;186:11–22. doi: 10.1016/j.agee.2014.01.013. [DOI] [Google Scholar]
- Guo J., Tao H., Li P., Li L., Zhong T., Wang L., Ma J., Chen X., Song T., Zhang H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018;8(1):10405. doi: 10.1038/s41598-018-28719-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann, E., Oliveira, E. A. D. de, Contin, D. R., Delvecchio, G., Viciedo, D. O., Moraes, M. A. de, Prado, R. de M., Costa, K. A. de P., Braga, M. R., & Martinez, C. A., 2019. Warming and water deficit impact leaf photosynthesis and decrease forage quality and digestibility of a C4 tropical grass. Physiologia Plantarum, 165(2), 383–402. https://doi.org/https://doi.org/10.1111/ppl.12891. [DOI] [PubMed]
- Hansen P.J. Effects of heat stress on mammalian reproduction. Philos. Trans. R. Soc., B. 2009;364(1534):3341–3350. doi: 10.1098/rstb.2009.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S., Bruford M., Jackson J.D., Lopes-Fernandes M., Heuertz M., Hohenlohe P.A., Paz-Vinas I., Sjögren-Gulve P., Segelbacher G., Vernesi C. Genetic diversity targets and indicators in the CBD post-2020 global biodiversity framework must be improved. Biol. Conserv. 2020;248 [Google Scholar]
- Holló G., Nuernberg K., Somogyi T., Anton I., Holló I. Comparison of fattening performance and slaughter value of local Hungarian cattle breeds to international breeds. Arch. Animal Breeding. 2012;55(1):1–12. [Google Scholar]
- Huber K. Invited review: resource allocation mismatch as pathway to disproportionate growth in farm animals–prerequisite for a disturbed health. Animal. 2018;12(3):528–536. doi: 10.1017/S1751731117002051. [DOI] [PubMed] [Google Scholar]
- Hussein A.H., Puchala R., Gipson T.A., Tadesse D., Wilson B.K., Goetsch A.L. Effects of water restriction on feed intake, digestion, and energy utilization by mature female St. Croix sheep. Veter. Animal Sci. 2020;10 doi: 10.1016/j.vas.2020.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC, L. U.-I. P. on C. C., 2007. Climate change 2007: Synthesis report. Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Core Writing Team (R. R. Pachauri A (Ed.)). https://heronet.epa.gov/heronet/index.cfm/reference/download/reference_id/2242845.
- Karthick, S., Saminathan, R., Gopinath, R. Agricultural Marketing – An Overview. International Journal of Management (IJM)Volume 11, Issue 11, November 2020, 3007-3013. https://doi.org/10.34218/IJM.11.11.2020.285
- Kijas, J. W., Lenstra, J. A., Hayes, B., Boitard, S., Porto Neto, L. R., San Cristobal, M., Servin, B., McCulloch, R., Whan, V., Gietzen, K., Paiva, S., Barendse, W., Ciani, E., Raadsma, H., McEwan, J., Dalrymple, B., & Consortium, other members of the I. S. G., 2012. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biology, 10(2), e1001258. https://doi.org/10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed]
- Kijas, J. W., Townley, D., Dalrymple, B. P., Heaton, M. P., Maddox, J. F., McGrath, A., Wilson, P., Ingersoll, R. G., McCulloch, R., McWilliam, S., Tang, D., McEwan, J., Cockett, N., Oddy, V. H., Nicholas, F. W., Raadsma, H., & Consortium, for the I. S. G., 2009. A Genome Wide Survey of SNP Variation Reveals the Genetic Structure of Sheep Breeds. PLoS ONE, 4(3), e4668. https://doi.org/10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed]
- Kim E.-S., Elbeltagy A.R., Aboul-Naga A.M., Rischkowsky B., Sayre B., Mwacharo J.M., Rothschild M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity. 2016;116(3):255–264. doi: 10.1038/hdy.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenmeister, K., Kuchenmeister, F., Kayser, M., WRAGE, M. N., & Isselstein, J., 2013. Influence of drought stress on nutritive value of perennial forage legumes.
- Kusza S., Gyarmathy E., Dubravska J., Nagy I., Jávor A., Kukovics S. Study of genetic differences among Slovak Tsigai populations using microsatellite markers. Czech J. Anim. Sci. 2009;54(10):468–474. [Google Scholar]
- Kusza S., Zakar E., Budai C., Cziszter L.-T., Padeanu I., Gavojdian D. Mitochondrial DNA variability in Gyimesi Racka and Turcana sheep breeds. Acta Biochim. Pol. 2015;62(2) doi: 10.18388/abp.2015_978. [DOI] [PubMed] [Google Scholar]
- Kutz S.J., Hoberg E.P., Polley L., Jenkins E.J. Global warming is changing the dynamics of Arctic host–parasite systems. Proc. R. Soc. B Biol. Sci. 2005;272(1581):2571–2576. doi: 10.1098/rspb.2005.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsgard A.G., Vaabenoe A. Genetic and environmental causes of variation in mastitis in sheep. Small Rumin. Res. 1993;12(3):339–347. doi: 10.1016/0921-4488(93)90069-T. [DOI] [Google Scholar]
- Maia, G. G., Siqueira, L. G. B., Vasconcelos, C. O. de P., Tomich, T. R., Camargo, L. S. de A., Rodrigues, J. P. P., de Menezes, R. A., Gonçalves, L. C., Teixeira, B. F., Grando, R. de O., Nogueira, L. A. G., & Pereira, L. G. R., 2020. Effects of heat stress on rumination activity in Holstein-Gyr dry cows. Livestock Science, 239, 104092. https://doi.org/10.1016/j.livsci.2020.104092.
- McManus C.M., Faria D.A., Lucci C.M., Louvandini H., Pereira S.A., Paiva S.R. Heat stress effects on sheep: Are hair sheep more heat resistant? Theriogenology. 2020;155:157–167. doi: 10.1016/j.theriogenology.2020.05.047. [DOI] [PubMed] [Google Scholar]
- McNeely, J. A., Miller, K. R., Reid, W. V, Mettermeier, R. A., & Werner, T. B., 1990. Conserving the world’s biological diversity. UICN, Morges (Suiza) WRI, Washington DC (EUA) CI, Washington DC (EUA) WWF ….
- Medlock J.M., Leach S.A. Effect of climate change on vector-borne disease risk in the UK. Lancet Infect. Dis. 2015;15(6):721–730. doi: 10.1016/S1473-3099(15)70091-5. [DOI] [PubMed] [Google Scholar]
- Meehl, G. A., Stocker, T. F., Collins, W. D., Friedlingstein, P., Gaye, A. T., Gregory, J. M., Kitoh, A., Knutti, R., Murphy, J. M., Noda, A., Raper, S. C. B., Watterson, I. G., Weaver, A. J., Zhao, Z.-C., Alley, R. B., Annan, J., Arblaster, J., Bitz, C., Brockmann, P., … Pant, G. B., 2007. Global Climate Projections. The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change [Solomon, S., D. Qin, M. Manning, Z. Chen, M. Marquis, K.B. Averyt, M. Tignor and H.L. Miller (Eds.)]. Cambridge U, 100. https://www.ipcc.ch/site/assets/uploads/2018/02/ar4-wg1-chapter10-1.pdf.
- Menéndez-Buxadera A., Serradilla J.M., Molina A. Genetic variability for heat stress sensitivity in Merino de Grazalema sheep. Small Rumin. Res. 2014;121(2–3):207–214. [Google Scholar]
- Milchunas D.G., Mosier A.R., Morgan J.A., LeCain D.R., King J.Y., Nelson J.A. Elevated CO2 and defoliation effects on a shortgrass steppe: forage quality versus quantity for ruminants. Agr. Ecosyst. Environ. 2005;111(1):166–184. doi: 10.1016/j.agee.2005.06.014. [DOI] [Google Scholar]
- Mokhber M., Moradi-Shahrbabak M., Sadeghi M., Moradi-Shahrbabak H., Stella A., Nicolzzi E., Rahmaninia J., Williams J.L. A genome-wide scan for signatures of selection in Azeri and Khuzestani buffalo breeds. BMC Genomics. 2018;19(1):449. doi: 10.1186/s12864-018-4759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notter D.R. The importance of genetic diversity in livestock populations of the future. J. Anim. Sci. 1999;77(1):61–69. doi: 10.2527/1999.77161x. [DOI] [PubMed] [Google Scholar]
- Pacheco A., McNeilly T.N., Banos G., Conington J. Genetic parameters of animal traits associated with coccidian and nematode parasite load and growth in Scottish Blackface sheep. Animal. 2021;15(4) doi: 10.1016/j.animal.2021.100185. [DOI] [PubMed] [Google Scholar]
- Paim, T. do P., Alves dos Santos, C., Faria, D. A. de, Paiva, S. R., & McManus, C., 2022. Genomic selection signatures in Brazilian sheep breeds reared in a tropical environment. Livestock Science, 258,
- Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., Miao B., Hu W., Wang X., Hu X., Xu Z., Wei D., He X., Yuan L., Guo X., Liang B., Wang R., Li X., Cao X., Li Y. Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization. GigaScience. 2018;7(4):giy019. doi: 10.1093/gigascience/giy019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather M.A., Hamadani A., Shanaz S., Alam S., Shabir M., Bukhari S., Khan N.N. Genetics of some reproduction traits in some sheep breeds from India: a review. J. Entomol. Zool. Studies. 2020;5 [Google Scholar]
- Reed D.H., Frankham R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003;17(1):230–237. doi: 10.1046/j.1523-1739.2003.01236.x. [DOI] [Google Scholar]
- Reid R.L., Jung G.A., Thayne W.V. Relationships between nutritive quality and fiber components of cool season and warm season forages: a retrospective study. J. Anim. Sci. 1988;66(5):1275–1291. doi: 10.2527/jas1988.6651275x. [DOI] [PubMed] [Google Scholar]
- Reiter S., Sattlecker G., Lidauer L., Kickinger F., Öhlschuster M., Auer W., Schweinzer V., Klein-Jöbstl D., Drillich M., Iwersen M. Evaluation of an ear-tag-based accelerometer for monitoring rumination in dairy cows. J. Dairy Sci. 2018;101(4):3398–3411. doi: 10.3168/jds.2017-12686. [DOI] [PubMed] [Google Scholar]
- Ríus A.G. Invited Review: Adaptations of protein and amino acid metabolism to heat stress in dairy cows and other livestock species. Appl. Anim. Sci. 2019;35(1):39–48. doi: 10.15232/aas.2018-01805. [DOI] [Google Scholar]
- Rosenblatt A.E., Schmitz O.J. Interactive effects of multiple climate change variables on trophic interactions: a meta-analysis. Climate Change Responses. 2014;1(1):1–10. [Google Scholar]
- Salunke, A., Shinde, S., Tone, A., Satpute, A., Deshmukh, J. 2018. Promote Local Agricultural Products Using E-Community Supported System. International Journal for Research in Applied Science & Engineering Technology (IJRASET), Volume 6 Issue I, January 2018, 2828–2831. https://www.ijraset.com/fileserve.php?FID=12193.
- Sanz-Sáez Á., Erice G., Aguirreolea J., Muñoz F., Sánchez-Díaz M., Irigoyen J.J. Alfalfa forage digestibility, quality and yield under future climate change scenarios vary with Sinorhizobium meliloti strain. J. Plant Physiol. 2012;169(8):782–788. doi: 10.1016/j.jplph.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Saravanan K.A., Panigrahi M., Kumar H., Bhushan B., Dutt T., Mishra B.P. Selection signatures in livestock genome: a review of concepts, approaches and applications. Livest. Sci. 2020;241 doi: 10.1016/j.livsci.2020.104257. [DOI] [Google Scholar]
- Saravanan K.A., Panigrahi M., Kumar H., Bhushan B., Dutt T., Mishra B.P. Genome-wide analysis of genetic diversity and selection signatures in three Indian sheep breeds. Livest. Sci. 2021;243:104367. doi: 10.1016/j.livsci.2020.104367. [DOI] [Google Scholar]
- Sevi A., Caroprese M. Impact of heat stress on milk production, immunity and udder health in sheep: a critical review. Small Rumin. Res. 2012;107(1):1–7. doi: 10.1016/j.smallrumres.2012.07.012. [DOI] [Google Scholar]
- Shen, J., Hanif, Q., Cao, Y., Yu, Y., Lei, C., Zhang, G., & Zhao, Y., 2020. Whole Genome Scan and Selection Signatures for Climate Adaption in Yanbian Cattle . In Frontiers in Genetics (Vol. 11). https://www.frontiersin.org/article/10.3389/fgene.2020.00094. [DOI] [PMC free article] [PubMed]
- Simm G., Conington J., Bishop S.C., Dwyer C.M., Pattinson S. Genetic selection for extensive conditions. Appl. Anim. Behav. Sci. 1996;49(1):47–59. doi: 10.1016/0168-1591(95)00667-2. [DOI] [Google Scholar]
- Singh K.M., Singh S., Ganguly I., Nachiappan R.K., Ganguly A., Venkataramanan R., Chopra A., Narula H.K. Association of heat stress protein 90 and 70 gene polymorphism with adaptability traits in Indian sheep (Ovis aries) Cell Stress Chaperones. 2017;22(5):675–684. doi: 10.1007/s12192-017-0770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soest P.J.V. Use of detergents in the analysis of fibrous feeds. II. a rapid method for the determination of fiber and lignin. J. Assoc. Off. Agric. Chem. 1963;46(5):829–835. doi: 10.1093/jaoac/46.5.829. [DOI] [Google Scholar]
- Troy D.J., Tiwari B.K., Joo S.-T. Health implications of beef intramuscular fat consumption. Korean J. Food Sci. Anim. Resour. 2016;36(5):577. doi: 10.5851/kosfa.2016.36.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar, S., Munir, M. T., Azeem, T., Ali, S., Umar, W., Rehman, A., & Shah, Y., 2014. Effects of Water Quality on Productivity and Performance of Livestock: A Mini Review. Veterinaria, 2, 11–15. https://www.researchgate.net/profile/Muahammad_Tanveer_Munir/publication/284550968_Effects_of_Water_Quality_on_Productivity_and_Performance_of_Livestock_A_Mini_Review/links/5654c53d08aeafc2aabc0d90.pdf.
- Van Soest P.J. Nutritional Ecology of the Ruminant. Cornell University Press; 2018. 10. Fiber and Physicochemical Properties of Feeds; pp. 140–155. [Google Scholar]
- Webster A.J.F., Hicks A.M., Hays F.L. Cold climate and cold temperature induced changes in the heat production and thermal insulation of sheep. Can. J. Physiol. Pharmacol. 1969;47(6):553–562. doi: 10.1139/y69-097. [DOI] [PubMed] [Google Scholar]
- Wiener P., Robert C., Ahbara A., Salavati M., Abebe A., Kebede A., Wragg D., Friedrich J., Vasoya D., Hume D.A. Whole-genome sequence data suggest environmental adaptation of ethiopian sheep populations. Genome Biol. Evol. 2021;13(3):evab014. doi: 10.1093/gbe/evab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Wang G., Zeng T., Wei C., Zhang L., Wang H., Zhang S., Liu R., Liu Z., Du L. Estimations of genomic linkage disequilibrium and effective population sizes in three sheep populations. Livest. Sci. 2014;170:22–29. doi: 10.1016/j.livsci.2014.10.015. [DOI] [Google Scholar]