Highlights
-
•
Microglia accumulate around the brain metastatic sites in mice and patients.
-
•
Exosomes of non-small cell lung cancer can change cellular functions of microglia.
-
•
miR1246 as a major microRNA mediates the effects exosomes on microglia.
-
•
miR-1246 is highly expressed in the plasmatic exosomes of lung cancer patients.
Keywords: Brain metastases, Non–small cell lung cancer, Microglia, Exosomes, miR1246
Abbreviations: NSCLC, non–small-cell lung cancer; TEM, tumor microenvironment; TAM, tumor-associated macrophages; BBB, blood-brain barrier; GBM, glioblastoma
Abstract
Non–small cell lung cancer (NSCLC) is the most common tumor that metastasizes to the brain. It is now accepted that the successful colonization and growth of tumor cells are determined by the interaction between tumor cells and the tumor microenvironment (TME). Microglia, brain innate immune cells, have been reported to play a vital role in the establishment of brain metastases. As essential mediators of intercellular communications, tumor-derived exosomes have an important role in the pathogenesis and progression of cancer by transferring their cargos to specific recipient cells. The crosstalk between microglia and tumor-derived exosomes has been extensively described. However, it is still unclear whether metastatic NSCLC cells secret exosomes to microglia and regulate the microglial functions. Here, our results showed that microglia aggregated in the brain metastatic sites. Meanwhile, microglia could take up the exosomes derived from NSCLC cells, leading to alterations of microglial morphology and increased proliferation, phagocytosis, and release of inflammatory cytokines including interleukin-6, interleukin-8, and CXCL1. Further investigation indicated that miR1246 was the most enriched microRNA in NSCLC-derived exosomes and mediated the partial effects of exosomes on microglia. Notably, miR1246 was also upregulated in the plasmatic exosomes of NSCLC patients. These results offer a new insight into the impact of NSCLC-derived exosomes on microglia and provide a new potential biomarker for diagnosing NSCLC.
Introduction
According to global cancer statistics, lung cancer remains the leading cause of cancer mortality, with 1.8 million (18%) deaths around the world in 2020, and non–small cell lung cancer (NSCLC) is the most common type, accounting for approximately 85% of lung cancer cases [1,2]. Tumor metastasis causes about 90% of cancer-related deaths [3]. In particular, the brain has a predisposition for colonization by disseminated lung cancer cells, with up to 50% of lung cancer patients developing brain metastases [4,5]. Historically, the brain has been regarded as a shelter for lung cancer cells due to its unique anatomical structures, immune environment, and metabolic constraints; therefore, brain metastases have been considered a terminal disease stage with a poor prognosis and a median overall survival period of approximately 3–8 months [6,7]. Although promising advances have been made to decipher the mechanism of brain metastases, the prognosis of lung cancer patients with brain metastasis remains poor [8], [9], [10]. Therefore, novel understandings of the process of brain metastases in lung cancer are greatly needed.
It is generally accepted that the successful colonization and growth of tumor cells in the brain is determined by the interaction between tumor cells and the brain microenvironment [11,12]. The brain microenvironment is isolated from peripheral circulation by the blood-brain barrier (BBB). The major cellular elements consist of neurons and glial cells, including astrocytes, microglia, pericytes, and oligodendrocytes. In recent years, significant progress has been made in studying the mechanism of tumor cells crossing the BBB and the interaction between tumor cells and astrocytes [13], [14], [15], [16], [17]. As the brain's innate immune cells, microglia have been reported to play a vital role in regulating the immunosuppressive microenvironment, the growth and mesenchymal-to-epithelial transition of tumor cells, extracellular matrix remodeling, and angiogenesis [18], [19], [20], [21], [22]. As essential mediators of cell-cell communications, tumor-derived exosomes containing proteins, DNA, and RNA have been increasingly recognized as contributors to the crosstalk between tumor and stromal cells [23]. In brain metastases of breast cancer, tumor cells that released more exosomal miR-503 triggered microglia polarization and upregulated immune-suppressive cytokines in microglia [18]. Similarly, microglia also took up exosomal microRNA (miRNA) of glioblastomas, leading to increased proliferation and shifting cytokine profiles toward immune suppression. Moreover, extracellular vesicles of metastatic melanoma cells instigated a pro-inflammatory gene signature in stromal cells in multiple distant organs, including brain metastases [24]. However, it is unclear whether metastatic NSCLC secret exosomes to the microglia and regulates the microglial functions. Hence, our study focused on the effects of NSCLC-derived exosomes on microglia.
In this study, we observed that microglia aggregated around the brain metastatic sites in mouse models and NSCLC patients’ specimens. Meanwhile, microglia took up the exosomes derived from NSCLC cells, leading to the alterations in microglial morphology and increased proliferation, phagocytosis, and release of inflammatory cytokines, including interleukin (IL)-6, IL-8, and CXCL1. Importantly, we found that exosomal miR1246 is a critical miRNA to mediate the effects of exosomes on the microglia. Moreover, we also discovered that the expression of serum exosomal miR1246 was significantly higher in NSCLC patients than in healthy controls. Ultimately, our study elucidated the underlying mechanism involving the effects of NSCLC-derived exosomes on the microglia.
Materials and methods
Cell culture
The HMC3 human microglia cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The A549 human NSCLC cell line was purchased from the China Center for Type Culture Collection (CCTCC, Wuhan, China). A549-BrM was a gift from Dr. Yuan (Department of Oncology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology). HMC3 cells were maintained in Minimum Essential Medium (MEM) medium (Gibco Laboratories, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS) in a 37 °C, 5% CO2 atmosphere. A549 cells were maintained in RPMI 1640 (Gibco Laboratories) with 10% FBS and 100 U/mL of penicillin-streptomycin at 37 °C in a humid atmosphere of 5% CO2.
Animal studies
All animal experiments were performed following protocols approved by the ethics committee of the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University Science and Technology University. Male BALB/c nude mice (6–8 weeks of age) were used and randomly allocated to each group. For the brain metastases assay, first mice were anesthetized with 1.5% sodium pentobarbital, then 2 × 105 luciferase-labeled A549-BrM cells suspended in 100 µL of PBS were injected into the left cardiac ventricle of mice using 1ml syringe with a 26G needle, while the control group received 100 µL of PBS. To confirm successful brain metastases, mice were intraperitoneally injected with D-luciferin (150 mg/kg) and the photo flux from whole body was observed and imaged at 4 weeks after injection tumor cells by using the Lago X (Spectral Instruments Imaging, Tucson, AZ, USA) coupled with the Living Image Acquisition and Analysis software program (PerkinElmer, Waltham, MA, USA).
Patient sample
Serum samples were collected from NSCLC patients (n = 39) diagnosed with lung adenocarcinoma at Tongji Hospital of Huazhong University of Science and Technology between 2019 and 2022 and healthy donors (n = 25). Eligible patients for study enrollment were those who did not receive any type of treatment before collecting the serum samples. The features of these participants are displayed in Supplementary Table S1. The serum was contained in a fresh anticoagulant tube and centrifugated at 3000 rpm for 10 min to remove cells. Next, serums were stored at −80 °C for further analysis. Five human lung cancer brain metastases specimens were obtained from the Department of Pathology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All tumor tissues were obtained by surgical resection and diagnosed as brain metastases of lung cancer by a pathologist. All procedures were approved by human research ethics committees at Tongji Hospital. Informed consent was obtained from all subjects involved in the study.
Exosomes isolation and identification
For exosome isolation, A549 cells were cultured in a standard medium until reaching 80%–90% confluence; after that, the medium was replaced with RPMI-1640 medium with 2% exosome-free FBS for 48 h. The supernatant was collected and centrifuged at 300 g for 10 min, 2000 g for 10 min, and 10,000 g for 20 min to remove residual cells and debris. After that, the supernatant was centrifugated at 120,000 g for 2 h at 4 °C, and pellets were resuspended in 6 mL of PBS. Then, the pellets in PBS were centrifugated again at 120,000 g for 70 min, and exosomes were suspended in 100–200 µL, then stored at −80 °C for further analysis. The analysis of exosome size was completed using nanoparticles tracking analysis (N30E; NanoFAM, Xiamen, China). Lastly, the morphology of exosomes was identified by transmission electron microscopy (HT-7700; Hitachi, Tokyo, Japan).
Exosomes labeling and tracking
Purified exosomes isolated from the culture medium were labeled with PKH67 (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. Then, the labeled exosomes were added to HMC3 cells for exosome-uptake studies. After incubation for 12 h at 37 °C, these cells were stained with 4′,6-diamidino-2-phenylindole (Servicebio, Wuhan, China) and observed by fluorescence microscopy (FV1000; Olympus Corporation, Tokyo, Japan).
Western blotting
The cells and isolated exosomes were lysed on ice in RIPA buffer with a proteinase inhibitor (Beyotime Biotechnology, Shanghai, China). The lysates were collected and centrifugated at 4 °C and 12,000 rpm for 15 min. Protein concentrations were determined using a bicinchoninic acid assay (Servicebio, Wuhan, China). Equal amounts of protein lysates were loaded and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to NC membranes (Merck Millipore, Burlington, MA, USA). The membranes were blocked with 5% bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at room temperature and incubated overnight with primary antibodies, including ALIX, TSG-101, CD16, and CD80 (1:1000; Abcam, Cambridge, UK); Arg1 and MRC1 (1:1000; BOSTER, Wuhan, China); and β-actin (1:1000; Abclonal, Wuhan, China), followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000, Jackson ImmunoResearch, West Grove, PA, USA). The densitometry of protein bands was normalized with β-actin.
Immunofluorescent and immunohistochemical staining
For immunofluorescent staining, brain slices or HMC3 cells seeded on 24-well plates were washed 2 times with PBS, fixed with 4% formaldehyde for 15 min at room temperature, washed 3 times with PBS, permeabilized with 0.3% Triton X-100 for 15 min, blocked with 5% bovine serum albumin in PBS for 1 h, and incubated with phalloidin (1:40; Invitrogen, Carlsbad, CA, USA) for 30 min at room temperature or the primary antibody IBAI (1:400, WAKO Pure Chemical Industries, Osaka, Japan), Human Nuclear (1:500, Millipore, Billerica, MA, USA) overnight at 4 °C followed by Cy3 Affinity-purified Donkey Anti-Rabbit secondary antibody, Alexa Fluor 488 Donkey Anti-Mouse secondary antibody (1:200, Jackson ImmunoResearch, West Grove, PA, USA) for 1 h in the dark. The cells were stained with DAPI (Beyotime Biotechnology, Shanghai, China) and imaged using a confocal microscope with 20 × and 40 × objectives (FV1000; Olympus, Tokyo, Japan). For immunohistochemical staining, a secondary antibody (HRP anti-rabbit, 1:500; Jackson ImmunoResearch, West Grove, PA, USA) was incubated at room temperature for 50 min, then visualized with the peroxidase substrate DAB (Servicebio, Wuhan, China) for a maximum of 10 min. Finally, the slices were observed and imaged by microscopy (BX51; Olympus Corporation, Tokyo, Japan).
Phagocytosis assay
HMC3 cells were seeded into 24-cell plates at 2 × 104 cells per well for the phagocytosis assay. Following specific treatments, cells were added to 0.4 µL/mL of red fluorescent microspheres (Invitrogen, Carlsbad, CA, USA) and incubated at 37 °C for 2 h. Then, the cells were fixed with 4% paraformaldehyde (Servicebio, Wuhan, China) and incubated with phalloidin. Four randomly selected visual fields per coverslip were photographed by fluorescence microscopy (BX51; Olympus Corporation, Tokyo, Japan).
Proliferation assay
Cells were seeded into 96-well plates and transfected with miRNA mimic or incubated with a medium with 20 µg/mL of exosomes. Then cells were added to 10 µL of Cell Counting Kit (CCK)-8 solution (Dojindo, Kumamoto, Japan) per well and incubated for 2 h at 37 °C. The absorbance was measured by a microplate reader at 450 nm (Synergy H1; BioTek Instruments, Winooski, Vermont, USA).
Wound healing assay
The cells were seeded into 12-well plates until confluence, and a 10 µL pipette tip was used to scratch them. Then, the cells were washed 2 times with PBS to discard dead cells and treated with a control medium (PBS) or conditional medium (20 µg/mL of exosomes). Photographs were taken using an inverted fluorescence microscope (IX81; Olympus Corporation, Tokyo, Japan) at 0, 3, 12, and 24 h. The area of the scratches was calculated using the ImageJ software program (U.S. National Institutes of Health, Bethesda, MD, USA).
Cytokine assay
The Proteome Profiler human cytokine array (R&D Systems, Minneapolis, MN, USA) was used to detect the presence of 36 cytokines in conditioned media according to the manufacturer's instruction. The membranes were blocked for 1 h with array-blocking buffer, then incubated with 1.5 mL of conditioned media overnight on an orbital shaker at 4 °C, washed 3 times, and incubated with streptavidin–HRP for 30 min. Lastly, members were exposed by imaging (Bio-Rad Laboratories, Hercules, CA, USA), and spots were quantified using the ImageJ software program, with each spot normalized to the positive controls. Enzyme-linked immunosorbent assay (ELISA) kits were performed to detect the level of CXCL1 (R&D Systems) as well as those of IL-6 and IL-8 (Neobioscience, Shenzhen, China) according to the manufacturer's protocols.
RNA sequencing
A549 exosomes were isolated from A549 cell culture supernatants, and total RNA was extracted. The amount and quality of small RNA were tested by EpiBioteck (Guangzhou, China). The raw sequence data were evaluated by the Fast-QC analysis software program. The obtained small RNA sequences were compared to the human miRNA database (miRBase, http:\\www.mirbase.org\), and miRNA expression was obtained.
RNA extraction and real-time polymerase chain reaction (RT-PCR)
Total RNA in exosomes was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were measured using a multi-detection microplate reader (Synergy H1; BioTek Instruments, Winooski, Vermont, USA). Then, mir-X miRNA first-strand synthesis (TaKaRa, Tokyo, Japan) was used to convert miRNA to complementary DNA according to the manufacturer's instructions. Quantitative RT-PCR (qRT-PCR) was performed using SYBR Green PCR Master Mix (TOYOBO, Osaka, Japan). Expression data were normalized to the internal control U6, and the 2−ΔΔCt method was used to analyze the differences in relative expression levels between groups. The primer sequences are listed in Supplementary Table S2.
Transfection
The miR1246 mimic and miR1246 inhibitor, normal control, and inhibitor control were synthesized by Sangon Biotech (Sangon Biotech, Shanghai, China). These RNA oligonucleotides were then transfected into HMC3 cells by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) at a final concentration of 100 nM according to the manufacturer's instructions. Total RNA or culture supernatants were collected for further assay 24 h after transfection.
Statistical analysis
Graphs were created, and statistical analysis was performed using the GraphPad Prism version 8 software program (GraphPad Software, La Jolla, CA, USA). Each experiment was carried out ≥3 times, and data are shown as mean ± standard error of the mean values. A student's t-test was used to define differences between 2 groups, a 1-sample t-test was used when the control group was standardized, and a 1-way analysis of variance was used to analyze differences between 3–4 datasets. P < 0.05 was considered to be statistically significant.
Results
Microglia infiltrated into metastatic brain lesions in the mouse brain metastasis model and human specimens
Seeking the role of microglia in brain metastases of NSCLC, we inoculated luciferase-labeled A549-BrM cells into the left cardiac ventricle of mice to establish a metastatic brain model, then observed the formation of brain tumors by bioluminescence imaging (Fig. S1A). Essentially, 90% of mice developed brain metastases 4 weeks after injection, and there was no significant change in body weight compared to the control group (Fig. S1B). Immunohistochemical staining of brain slices from metastatic brain mice revealed the formation of tumor masses (Fig. S1C).
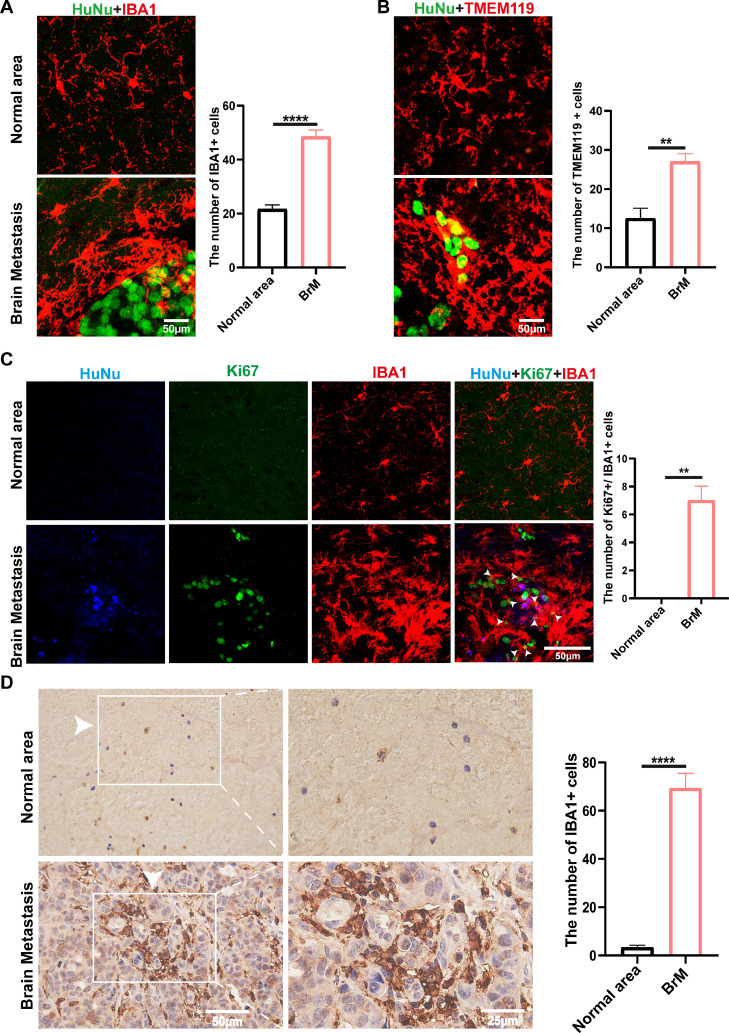
Next, we observed the distribution of microglia in metastatic brain lesions of mice. We found that abundant Iba1+ cells (a microglial marker) surrounded the tumor cells (HuNu+ cells) (Fig. 1A). The same observations were found in images using TMEM119 to label microglia (Fig. 1B). Moreover, the portion of these microglia showed Ki67 positivity, indicating increased proliferation (Fig. 1C). Furthermore, immunochemistry staining of metastatic brain specimens derived from NSCLC patients showed that numerous microglia had infiltrated into metastatic sites (Fig. 1D). Microglial morphology around the tumor cells became more extensive and rounder, with shorter ramifications, showing an apparent activation state (Fig. 1A, B). The results indicated that we successfully established a mouse brain metastasis model and could observe microglial aggregates in the brain metastatic sites in mice and NSCLC patients.
Fig. 1.
Microglia were abundantly infiltrated in metastatic brain lesions. A. Immunofluorescent staining of microglia (IBA1+, red) and tumor cells (HuNu+, green) in mouse brains with metastases. The numbers of IBA1+ cells were counted in the normal area or brain metastatic area (n = 6 / group). Scale bar, 50 µm. B. Representative images of HuNu+ (tumor cells) and TMEM119+(microglia) in the mice brain. Quantification of TMEM119+ cells numbers was shown (n = 4 / group). Scale bar, 50 µm. C. Representative immunofluorescence images of Ki67 staining (green) in mouse brain metastatic lesions were shown. HuNu (Blue) marks cancer cells, IBA1 (red) marks microglia (n = 6 / group). Scale bar, 50 µm. D. Representative images of immunohistochemical staining for IBA1 Positive cell numbers in normal brain lesions and metastatic brain lesions of NSCLC patients were counted (n = 5/ group). Scale bar, 50 µm (left) and 25 µm (right). (**P < 0.01, ****P < 0.0001).
Microglia took up exosomes derived from A549 cells
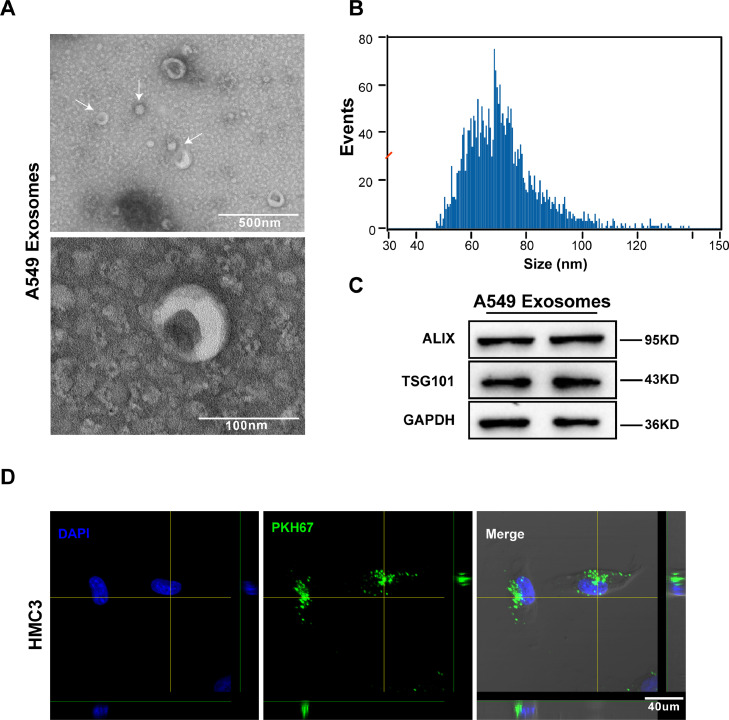
Previous studies have suggested that exosomes derived from cancer cells are involved in the formation of the TME [25]. To determine the effects of NSCLC-derived exosomes on the microglia, we firstly isolated exosomes from the culture supernatant of A549 cells via ultracentrifugation. Then, transmission electron microscopy and nanoparticle tracking analysis revealed that the exosomes were typical rounded particles 30–150 nm in diameter (Fig. 2A, B). Furthermore, western blot analysis of protein extracted from exosomes demonstrated that exosomes were positive for ALIX and TSG101 (Fig. 2C). Next, to prove that exosomes derived from A549 cells could enter microglia, the exosomes were labeled with PKH67 and incubated with HMC3 cells at 37 °C for 12 h. Confocal imaging confirmed the uptake of PKH67-labeled exosomes by microglia (Fig. 2D).
Fig. 2.
Microglia took up exosomes derived from A549 cells. A. Electron microscopy images of exosomes isolated from the medium of A549 cells. Scale bar, 500 nm (left) and 100 nm (right). B. Size distribution of exosomes was analyzed by nanoparticle tracking analysis. Most particles are in the range of 30–150 nm. C. Western blot analysis for exosomal markers ALIX and TSG101. GAPDH was used as a control for protein concentration. D. Representative images show the internalization of PKH67-labeled A549-derived exosomes (green) by microglia. Scale bar, 40 µm.
Exosomes derived from A549 cells induced morphological changes in microglia and increased microglial proliferation
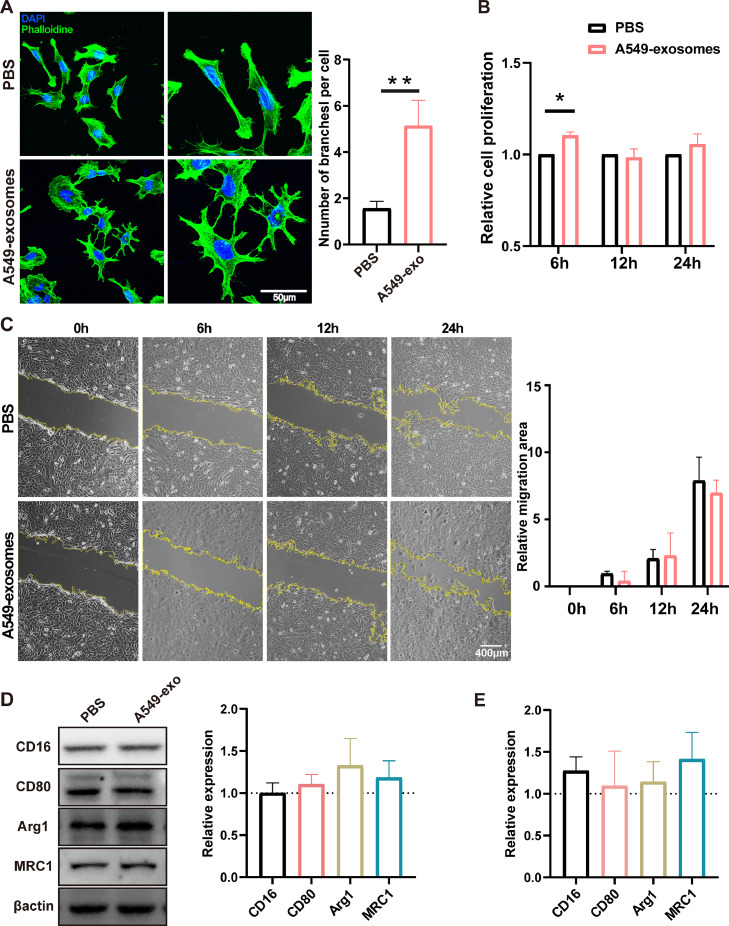
To define the exact role of exosomes derived from NSCLC cells on microglia, HMC3 cells were incubated with a medium containing A549-derived exosomes. First, we stained the microglia with phalloidin (staining cytoskeletal protein) to observe the alteration in microglia morphology, which became rounder and larger and had more branches (Fig. 3A). Furthermore, our results demonstrated that A549-derived exosomes significantly increased microglial proliferation at 6 h but not at 12 h or beyond (Fig. 3B). However, migration assay showed that the exosomes did not affect the microglial migration (Fig. 3C). Next, to elucidate the effect of A549-derived exosomes on the activation of microglia, western blot analysis, and qRT-PCR were used to quantify the conventional markers of microglial activation, including pro-inflammatory markers (CD16, CD80) and anti-inflammatory markers (Arg1, MRC1). Still, there were no significant changes in the expression of these inflammatory markers (Fig. 3D, E).
Fig. 3.
Exosomes derived from A549 cells induced morphological changes in microglia and increased microglial proliferation. A. Representative immunofluorescence images showed the morphological changes incubated with exosomes for 24 h. Microglia cytoskeletons are labeled with phalloidin (green), and nuclei are labeled with DAPI (blue). Quantitative results for the numbers of branches per cell were showed (n = 7). Scale bar, 50 µm. B. The proliferation of microglia cells incubated with tumor-derived exosomes (20 ng/mL) was detected by CCK-8 assay at 6, 12, and 24 h, and statistical analysis was performed (n = 4). C. Wound healing of HMC3 was used to examine microglial mobility incubated with exosomes. Images were taken 0, 6, 12, and 24 h after wound scratching. The migration area was quantified (n = 4). Scale bar, 400 µm. D, E. The effect of A549-derived exosomes on the activation of microglia was analyzed by western blot analysis (D) and qPCR. Histogram shows relative protein expressions of CD16, CD80, Arg1, and Mrc1 normalized to the PBS-treated group (n = 3) (*P < 0.05, **P < 0.01).
A549-derived exosomes increased microglial phagocytic activity and cytokine expression
Because recent studies have demonstrated that microglia alter their phagocytosis and increase the release of some inflammatory factors, such as IL-10, insulin-like growth factor (IGF)-1, CCL20, and transforming growth factor (TGF)-β, to contribute to the formation of the TME [7,26], we next investigated the effect of exosomes on microglia phagocytosis and the secretion of inflammatory cytokines. The results indicated that uptake of A549 exosomes led to an increase in the phagocytic ability of HMC3 cells relative to the untreated control group. We observed a significant increase at 48 h for HMC3 cells incubated with the exosomes (Fig. 4A, B). Then, we collected the supernatants of microglia to detect the expression levels of 36 human cytokines using human cytokine array kits and found that HMC3 cells incubated with A549 exosomes showed enhanced release of IL-6, IL-8, and CXCL1 (Fig. 4C, D). Individual cytokine concentration testing of IL-6, IL-8, and CXCL1 using ELISA kits confirmed these results (Fig. 4E). Collectively, these observations demonstrated that A549 exosomes could increase microglial phagocytosis and secretions of IL-6, IL-8, and CXCL1.
Fig. 4.
A549-derived exosomes increased microglial phagocytic activity and cytokine expression. A, B. Phagocytosis of red fluorescent microspheres in HMC3 cells treated with PBS or exosomes for 24 and 48 h was observed. The phagocytic ability of microglia in the exosome-treated group was increased at 48 h. Scale bar, 100 µm (n = 6). C. Microglia were incubated with exosomes (20 ng/mL) or PBS for 48 h. The expressions of 36 cytokines in the supernatants were assessed by a human cytokine array. D. The cytokines with increased expression in the exosome-treated group were further detected by a separate ELISA kit (IL-6, IL-8, CXCL1) (n = 5) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
MiR-1246 was highly expressed in exosomes derived from NSCLC cells
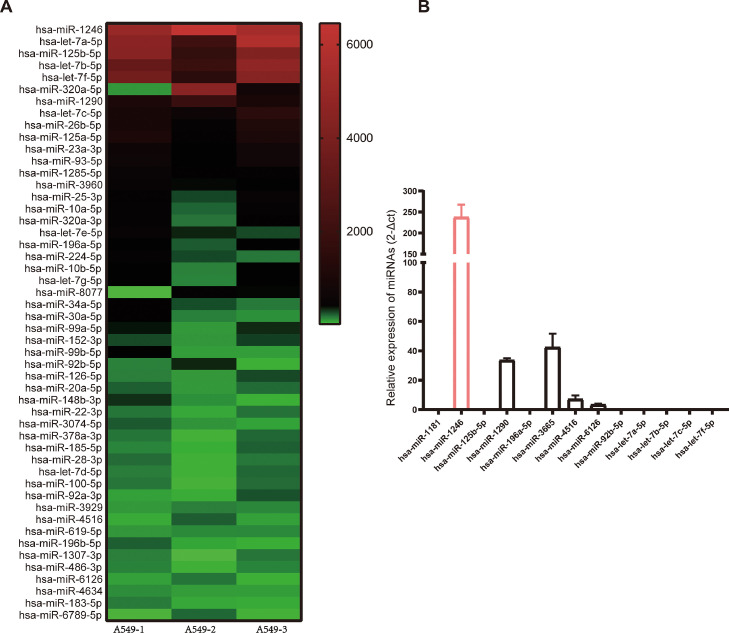
Recent studies have demonstrated that exosomes contain a variety of biologically active molecules, including proteins, RNA, DNA, and miRNAs—the most abundant type of RNA content in exosomes—have an essential role in intercellular communications [27,28]. Therefore, we conducted miRNA sequencing analysis on A549 exosomes and identified 875 miRNAs encapsulated in the exosomes. The heatmap revealed the top 50 miRNAs, among which miR1246 was the most abundant exosomal miRNA (Fig. 5A). Next, we confirmed that the expression level of miR1246 was indeed much higher than those of the other 12 miRNAs with high and stable expression levels in exosomes by using qRT-PCR (Fig. 5B). Consequently, we hypothesized that miR1246 plays a crucial role in mediating the functions of exosomes on the microglia.
Fig. 5.
MiR-1246 is highly expressed in exosomes derived from NSCLC cells. A. Results of RNA sequencing analysis of exosomal miRNAs from A549 cells are presented in the heatmap. B. The expression levels of highly expressed miRNAs in exosomes were examined by qRT-PCR (n = 6). MiR1246 is the most enriched miRNA in A549-derived exosomes.
miR1246 can be transferred to human microglia and promote microglia release IL-6, IL-8, and CXCL1
To test the effect of exosomal miR1246 on the microglia, we transfected microglia with miR1246 mimic or inhibitor. qRT-PCR results showed that the miR1246 mimic increased the expression level of miR1246. In contrast, the expression of miR1246 was significantly lower in the miR1246 inhibitor group (Fig. 6A). The proliferation assay demonstrated that the mimic or inhibitor had no significant effect on microglial proliferation (Fig. 6B). The ELISA results showed that miR1246 mimic promoted the secretions of IL-6, IL-8, and CXCL1, whereas miR1246 inhibitor had the opposite effect on the release of IL-8 (Fig. 6C). Consequently, these results suggest that miR1246 mediated the effects of exosomes on the microglial secretory functions.
Fig. 6.
MiR1246 can be transferred to human microglia and promote microglia release of IL-6, IL-8, and CXCL1. A. Microglia were transfected with miR1246, NC, miR1246 inhibitor, and inhibitor NC. Levels of miR1246 in microglia were measured using qRT-PCR. The expression of miR1246 was upregulated by miR1246 mimic and downregulated by miR1246 inhibitor (n = 4). B. Microglia were transfected with miRNA1246, NC, miR1246 inhibitor, or inhibitor NC (100 ng/mL) for 24 h, and no significant differences were observed in cell viability measured by CCK-8 (n = 4). C. Microglia treatment was the same as described before, and the supernatant of 24 h cultures were used to measure the secretions of CXCL1, IL-6, and IL-8 by ELISA kits. MiR1246 induced an increase in the microglial secretion of IL-6, IL-8, and CXCL1 (n = 3) (*P < 0.05, **P < 0.01).
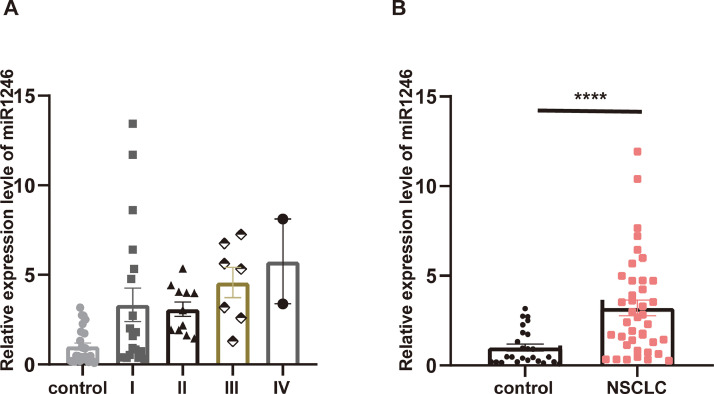
miR1246 was highly expressed in the plasmatic exosomes of NSCLC patients
The expression levels of miR1246 were also measured in the plasmatic exosomes of NSCLC patients. We firstly isolated serum exosomes from 56 NSCL patients and healthy controls using an exosome isolation reagent kit. Although the quantitative PCR results showed no significant difference between the clinical stages (Fig. 7A), the expression of exosomal miR1246 was significantly higher in NSCLC patients compared to healthy controls, which indicated that miR1246 has potential relevance in diagnosing NSCLC (Fig. 7B).
Fig. 7.
MiR1246 is highly expressed in the plasmatic exosomes of NSCLC patients. A. The differences in expression levels of miR1246 in different NSCLC clinical stages were detected, and there were no significant differences among the different stages. B. The expression levels of miR1246 were increased in the plasmatic exosomes of NSCLC patients (n = 39) compared to in the control group (n = 25) (****P < 0.0001).
Discussion
Brain metastases of peripheral tumors have been regarded as a terminal disease stage with a very poor prognosis and few therapeutic options [5,6]. Lung cancer is the most common tumor that metastasizes to the brain [29]. Therefore, exploring novel mechanisms of brain metastases in lung cancer is a vital and complex challenge. In recent years, exosomes derived from tumor cells were demonstrated to mediate the crosstalk between tumor cells and non-tumor cells in the tumor microenvironment (TME) [23]. In the central nervous system, microglia act as innate immune cells and have been reported to be enriched in brain tumors and promote tumor progression [[18], [19], [20], [21],30,31]. In this study, we focused on the effects of NSCLC-derived exosomes on microglia. Our results showed that microglia aggregated around the brain metastatic sites in mouse models and non-small cell lung cancer (NSCLC) patients’ specimens. Microglia around the tumor cells became larger and rounder with shorter ramifications, displaying an apparent activation state. Meanwhile, microglia took up the exosomes derived from NSCLC cells, leading to alterations in microglial morphology and increased proliferation, phagocytosis, and release of inflammatory cytokines, including IL-6, IL-8, and CXCL1. Additionally, we identified exosomal miR1246 as a critical miRNA able to mediate the effects of exosomes on microglia.
The fundamental cellular components in the brain tumor environment include neurons, astrocytes, microglia, endothelial cells, pericytes, and peripheral immune cells such as macrophages and lymphocytes [32,33]. Microglia and macrophages (together called tumor-associated macrophages [TAMs]) are the most abundant non-cancerous cell types composing up to 30% of the total tumor mass [34]. The historical view is that TAMs with a pro-inflammatory phenotype (M1 phenotype) are anti-tumorigenic; in contrast, anti-inflammatory (M2 phenotype) TAMs have shown tumor-supporting activity [35,36]. Many researchers suggested that tumor cells triggered M1-to-M2 conversion of TAMs to support tumor progression [19,26,[37], [38], [39]]. However, this view of dual-polarization is now regarded as an oversimplification. Polarization is a dynamic process, and many studies have instead focused on observing the functional diversity of TAMs [8,40,41]. In this study, we observed that microglia aggregated around the brain metastases in mouse models and NSCLC patients. Moreover, the microglial morphology and proliferation were altered by NSCLC-derived exosomes in vitro. Interestingly, the conventional inflammatory markers of M1–M2 polarization showed no significant changes in microglia incubated with NSCLC-derived exosomes. This result further confirmed that we should focus on observing the functional diversity of microglia.
In brain metastases, when macro-metastases are established, microglia and macrophages would lose phagocytosis and increase the production of specific inflammatory cytokines to play a tumor-supportive role [7]. Some studies showed that the functional profiles of microglia and macrophages were different. The microglial functional profile is rich in chemokines, cytokines, and complement components secretions, while macrophage signatures focus on wound healing, antigen presentation, and immune suppression [42,43]. Here, we found that NSCLC-derived exosomes did not affect microglia migration but did increase microglial phagocytic activity and secretions of IL-6, IL-8, and CXCL1. These results indicate that the effects of NSCLC-derived exosomes on microglial functions mainly focus on the phagocytosis and production of inflammatory cytokines. Enigmatically, the phagocytic activity of microglia was upregulated by NSCLC-derived exosomes. This result reminds us that microglia might exert an anti-tumor effect, especially before establishing macro-metastases. Future studies should be designed to dynamically observe microglial functions at different metastatic stages.
Cytokines and chemokines are pivotal regulators in tumor metastases. They can determine the distribution of immune cells, affect stroma composition, drive angiogenesis, and directly regulate the growth and invasiveness of cancer cells in tumor progression. In brain tumors, nicotine skewed the polarity of microglia, thereby increasing the secretion of IGF-1 and CCL20, which promoted tumor progression and stemness [26]. Breast cancer cells trigger microglia polarization to upregulate the expression of immune-suppressive cytokines that could suppress T-cell proliferation [44]. Glioma-derived exosomes were shown to deliver miRNA to microglia/macrophages, inducing the secretions of cytokines such as IL-6, IL-10, and TGF-β1 to form the immunosuppressive environment [45]. Our results demonstrated that NSCLC-derived exosomes promoted microglia to produce more IL-6, IL-8, and CXCL1. A recent study suggests that targeting IL-6/JAK2/STAT3 signaling in microglia can reduce brain metastases of NSCLC [19]. Indeed, the IL-6 signaling pathway affects several stromal cells, including endothelial cells and immune cells, influencing the BBB permeability, angiogenesis, and immune-suppressive response to promote tumor proliferation and metastases [46,47]. Besides, IL-6 also plays an essential role in the epithelial-mesenchymal transition, invasion, and metastatic colonization of cancer cells [48,49]. Similarly, IL-8 has been described to have dual pro-tumorigenic roles, including regulating non-cancerous cells in the TME and directly influencing tumor cells [50]. IL-8 increases angiogenesis and preferentially recruits pro-tumorigenic immune cells, such as myeloid-derived suppressive cells (MDSCs), to suppress the anti-tumor efficacy of cytotoxic T-cells [51]. Moreover, some studies reported the polarization of TAMs further secreted IL-8, enhancing tumor motility and promoting tumor epithelial-mesenchymal transition [52,53]. As a member of the CXC class of chemokines, CXCL1 secreted by TAMs can recruit CXCR2-positive (CXCL1 receptor) MDSCs to establish a pre-metastatic niche and also play a role in increasing angiogenesis to promote tumor metastases [54]. Therefore, the NSCLC-derived exosomes induce microglia to release more IL-6, IL-8, and CXCL1 in favor of creating an adequate blood supply and immune-suppressive environment. Studies targeting cytokine and chemokine signaling pathways may play a crucial role in developing future clinical tumor therapies.
Exosomes (small vesicles containing nucleic acids and proteins) are emerging as essential components in TME formation that act by transferring their cargos to specific recipient cells [25,55]. Various studies indicated that exosomal miRNAs contribute to tumor proliferation, metastases, and chemoresistance in tumors [56]. In this study, we analyzed the profiles of miRNAs in A549 exosomes and found the most enriched miRNA was miR1246. Notably, miR1246 has been reported to be rich in various tumors, including pancreatic cancer, colorectal cancer, ovarian cancer, and glioblastoma (GBM). Moreover, some studies suggested that miR1246 could cause activation of STAT3 and increase the expressions of inflammatory cytokines in tumor cells and noncancerous cells [45,[57], [58], [59]]. For example, miR1246 in tumor extracellular vesicles caused IL-6 induction by Androgen receptor (AR) downregulation in tumor endothelial cells and activated STAT3 [58]. Hypoxic GBM-derived exosomal miR1246 can induce TAM polarization, facilitating the formation of the immunosuppressive microenvironment by targeting TERF2IP to regulate nuclear factor κB and STAT3 signaling and increase the expression of IL-10, IL-1RA, TGFB1, and CCL2 [45]. Our results showed that miR1246 enriched in NSCLC-derived exosomes promoted microglia to secrete IL-6, IL-8, and CXCL1. According to the previous studies, we speculate the possible mechanism is that miR1246 targets certain genes such as AR and TERF2IP to reprogram microglia and STAT3 activation is involved. The possible mechanism is that miR1246 targets certain genes such as AR and TERF2IP and STAT3 activation is involved. Furthermore, we found that the expression level of miR1246 was upregulated in the serum exosomes of NSCLC patients. All these results imply that miR1246 may be an essential player in the exosome-mediated functional changes of TAMs and could be a potential plasmatic marker for NSCLC diagnosis.
Nevertheless, our study has some limitations. Although the expression level of exosomal miR1246 was higher in NSCLC patients, there was no significant difference among different clinical stages. Future studies should include more patients at different clinical stages to confirm our results. Furthermore, our subsequent studies will investigate the exact functional changes in microglia in vivo and in vitro. Of course, the functions of other NSCLC-derived exosomal miRNAs on microglia should also be explored.
Conclusions
In summary, we demonstrated that NSCLC-derived exosomes could alter microglial morphology, increasing proliferation, phagocytosis, and the release of inflammatory cytokines, including IL-6, IL-8, and CXCL1. MiR1246 is a critical miRNA and plays a vital role in mediating the effects of exosomes on microglia. More importantly, our findings show that miR1246 in serum exosomes might be a potential plasmatic marker of NSCLC prognosis. The present results offer new insights into the effects of NSCLC-derived exosomes on microglia and provide a new potential biomarker for diagnosing NSCLC.
Funding
This study was supported by the National Nature Science Foundation of China (grant numbers 82171385), Ministry of Science and Technology China Brain Initiative Grant (grant numbers 2022ZD0204704), Key Research and Development Program of Hubei Province (grant numbers 2020BCA070), the Flagship Program of Tongji Hospital (grant numbers 2019CR106).
Ethical statement
The study was conducted in accordance with the guidelines of the Declaration of Helsinki and the Guide for the Care and Use of Laboratory Animals, and approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Permit number: TJ-C20121219 and TJ-IRB20210222). Informed consent was obtained from all subjects involved in the study.
Availability of data and materials
Data analyzed and generated during the study are available from the corresponding authors on reasonable request.
CRediT authorship contribution statement
Peng Chen: Methodology, Investigation, Formal analysis, Data curation, Visualization, Writing – original draft, Writing – review & editing. Ying Li: Investigation, Data curation. Rui Liu: Investigation. Yi Xie: Visualization. Yu Jin: Investigation. Minghuan Wang: Formal analysis, Data curation. Zhiyuan Yu: Visualization, Writing – review & editing. Wei Wang: Conceptualization, Funding acquisition. Xiang Luo: Conceptualization, Funding acquisition.
Declaration of Competing Interest
None.
Acknowledgments
We thank Dr. Yuan (Department of Oncology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology) and Dr. Dai (Department of pathology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology) for their generous help and technical assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101594.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Thandra K.C., Barsouk A., Saginala K., Aluru J.S., Barsouk A. Epidemiology of lung cancer. Wspolczesna Onkol. Contemp. Oncol. 2021;25(1):45–52. doi: 10.5114/wo.2021.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehlen P., Puisieux A. Metastasis: a question of life or death. Nat. Rev. Cancer. 2006;6(6):449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 4.Fabi A., Vidiri A. Defining the endpoints: how to measure the efficacy of drugs that are active against central nervous system metastases. Transl. Lung Cancer Res. 2016;5(6):637–646. doi: 10.21037/tlcr.2016.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernani V., Stinchcombe T.E. Management of brain metastases in non-small-cell lung cancer. J. Oncol. Pract. 2019;15(11):563. doi: 10.1200/JOP.19.00357. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wasilewski D., Radke J., Xu R., Raspe M., Trelinska-Finger A., Rosenstock T., Poeser P., Schumann E., Lindner J., Heppner F., Kaul D., Suttorp N., Vajkoczy P., Frost N., Onken J. Effectiveness of immune checkpoint inhibition vs chemotherapy in combination with radiation therapy among patients with non-small cell lung cancer and brain metastasis undergoing neurosurgical resection. JAMA Netw. Open. 2022;5(4) doi: 10.1001/jamanetworkopen.2022.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boire A., Brastianos P.K., Garzia L., Valiente M. Brain metastasis. Nat. Rev. Cancer. 2020;20(1):4–11. doi: 10.1038/s41568-019-0220-y. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez H., Mei W., Robles I., Hagerling C., Allen B.M., Hauge Okholm T.L., Nanjaraj A., Verbeek T., Kalavacherla S., van Gogh M., Georgiou S., Daras M., Phillips J.J., Spitzer M.H., Roose J.P., Werb Z. Cellular architecture of human brain metastases. Cell. 2022;185(4):729–745. doi: 10.1016/j.cell.2021.12.043. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirschenbaum D., Amit I. Brain metastases: not all tumors are created equal. Neuron. 2022;110(7):1097–1099. doi: 10.1016/j.neuron.2022.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Parida P.K., Marquez-Palencia M., Nair V., Kaushik A.K., Kim K., Sudderth J., Quesada-Diaz E., Cajigas A., Vemireddy V., Gonzalez-Ericsson P.I., Sanders M.E., Mobley B.C., Huffman K., Sahoo S., Alluri P., Lewis C., Peng Y., Bachoo R.M., Arteaga C.L., Hanker A.B., DeBerardinis R.J., Malladi S. Metabolic diversity within breast cancer brain-tropic cells determines metastatic fitness. Cell Metab. 2022;34(1):90–105. doi: 10.1016/j.cmet.2021.12.001. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Custodio-Santos T., Videira M., Brito M.A. Brain metastasization of breast cancer. Biochim. Biophys. Acta Rev. Cancer. 2017;1868(1):132–147. doi: 10.1016/j.bbcan.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan E.S., Tan A.C., Anders C.K., Pendergast A.M., Sipkins D.A., Ashley D.M., Fecci P.E., Khasraw M. Salting the soil: targeting the microenvironment of brain metastases. Mol. Cancer Ther. 2021;20(3):455–466. doi: 10.1158/1535-7163.MCT-20-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Y., Watters A., Cheng N., Perry C.E., Xu K., Alicea G.M., Parris J.L.D., Baraban E., Ray P., Nayak A., Xu X., Herlyn M., Murphy M.E., Weeraratna A.T., Schug Z.T., Chen Q. Polyunsaturated fatty acids from astrocytes activate ppargamma signaling in cancer cells to promote brain metastasis. Cancer Discov. 2019;9(12):1720–1735. doi: 10.1158/2159-8290.CD-19-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng S.M., Tu S.H., Bai Z.W., Geng Y.X. Exosomal lncRNA LINC01356 derived from brain metastatic nonsmall-cell lung cancer cells remodels the blood-brain barrier. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.825899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Q., Michael I.P., Zhang P., Saghafinia S., Knott G., Jiao W., McCabe B.D., Galvan J.A., Robinson H.P.C., Zlobec I., Ciriello G., Hanahan D. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573(7775):526–531. doi: 10.1038/s41586-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morad G., Carman C.V., Hagedorn E.J., Perlin J.R., Zon L.I., Mustafaoglu N., Park T.E., Ingber D.E., Daisy C.C., Moses M.A. Tumor-derived extracellular vesicles breach the intact blood-brain barrier via transcytosis. ACS Nano. 2019;13(12):13853–13865. doi: 10.1021/acsnano.9b04397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues G., Hoshino A., Kenific C.M., Matei I.R., Steiner L., Freitas D., Kim H.S., Oxley P.R., Scandariato I., Casanova-Salas I., Dai J., Badwe C.R., Gril B., Tesic Mark M., Dill B.D., Molina H., Zhang H., Benito-Martin A., Bojmar L., Ararso Y., Offer K., LaPlant Q., Buehring W., Wang H., Jiang X., Lu T.M., Liu Y., Sabari J.K., Shin S.J., Narula N., Ginter P.S., Rajasekhar V.K., Healey J.H., Meylan E., Costa-Silva B., Wang S.E., Rafii S., Altorki N.K., Rudin C.M., Jones D.R., Steeg P.S., Peinado H., Ghajar C.M., Bromberg J., de Sousa M., Pisapia D., Lyden D. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019;21(11):1403–1412. doi: 10.1038/s41556-019-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing F., Liu Y., Wu S.Y., Wu K., Sharma S., Mo Y.Y., Feng J., Sanders S., Jin G., Singh R., Vidi P.A., Tyagi A., Chan M.D., Ruiz J., Debinski W., Pasche B.C., Lo H.W., Metheny-Barlow L.J., D'Agostino R.B., Jr., Watabe K. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal mirna to promote brain metastasis. Cancer Res. 2018;78(15):4316–4330. doi: 10.1158/0008-5472.CAN-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y., Kang Y.L., Wang M.H., Wu B.L., Su B.B., Yin H., Tang Y., Li Q.X., Wei W.J., Mei Q., Hu G.Y., Lukacs-Kornek V., Li J., Wu K.M., Yuan X.L., Wang W. Targeting polarized phenotype of microglia via IL6/JAK2/STAT3 signaling to reduce NSCLC brain metastasis. Signal Transduct. Target. Ther. 2022;7(1) doi: 10.1038/s41392-022-00872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maas S.L.N., Abels E.R., Van De Haar L.L., Zhang X., Morsett L., Sil S., Guedes J., Sen P., Prabhakar S., Hickman S.E., Lai C.P., Ting D.T., Breakefield X.O., Broekman M.L.D., El Khoury J. Glioblastoma hijacks microglial gene expression to support tumor growth. J. Neuroinflamm. 2020;17(1):120. doi: 10.1186/s12974-020-01797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izraely S., Ben-Menachem S., Sagi-Assif O., Telerman A., Zubrilov I., Ashkenazi O., Meshel T., Maman S., Orozco J.I.J., Salomon M.P., Marzese D.M., Pasmanik-Chor M., Pikarski E., Ehrlich M., Hoon D.S.B., Witz I.P. The metastatic microenvironment: Melanoma-microglia cross-talk promotes the malignant phenotype of melanoma cells. Int. J. Cancer. 2019;144(4):802–817. doi: 10.1002/ijc.31745. [DOI] [PubMed] [Google Scholar]
- 22.Pukrop T., Dehghani F., Chuang H.N., Lohaus R., Bayanga K., Heermann S., Regen T., Van Rossum D., Klemm F., Schulz M., Siam L., Hoffmann A., Trumper L., Stadelmann C., Bechmann I., Hanisch U.K., Binder C. Microglia promote colonization of brain tissue by breast cancer cells in a Wnt-dependent way. Glia. 2010;58(12):1477–1489. doi: 10.1002/glia.21022. [DOI] [PubMed] [Google Scholar]
- 23.Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell. 2019;49(3):347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Gener Lahav T., Adler O., Zait Y., Shani O., Amer M., Doron H., Abramovitz L., Yofe I., Cohen N., Erez N. Melanoma-derived extracellular vesicles instigate proinflammatory signaling in the metastatic microenvironment. Int. J. Cancer. 2019;145(9):2521–2534. doi: 10.1002/ijc.32521. [DOI] [PubMed] [Google Scholar]
- 25.Kahlert C., Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. JMM. 2013;91(4):431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu S.Y., Xing F., Sharma S., Wu K., Tyagi A., Liu Y., Zhao D., Deshpande R.P., Shiozawa Y., Ahmed T., Zhang W., Chan M., Ruiz J., Lycan T.W., Dothard A., Watabe K. Nicotine promotes brain metastasis by polarizing microglia and suppressing innate immune function. J. Exp. Med. 2020;217(8) doi: 10.1084/jem.20191131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita Y., Yoshioka Y., Ochiya T. Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 2016;107(4):385–390. doi: 10.1111/cas.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G., Drescher K.M., Chen X.M. Exosomal miRNAs: biological properties and therapeutic potential. Front. Genet. 2012;3:56. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Rassy E., Botticella A., Kattan J., Le Pechoux C., Besse B., Hendriks L. Non-small cell lung cancer brain metastases and the immune system: From brain metastases development to treatment. Cancer Treat. Rev. 2018;68:69–79. doi: 10.1016/j.ctrv.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Kabba J.A., Xu Y., Christian H., Ruan W., Chenai K., Xiang Y., Zhang L., Saavedra J.M., Pang T. Microglia: housekeeper of the central nervous system. Cell. Mol. Neurobiol. 2018;38(1):53–71. doi: 10.1007/s10571-017-0504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matias D., Predes D., Niemeyer Filho P., Lopes M.C., Abreu J.G., Lima F.R.S., Moura Neto V. Microglia-glioblastoma interactions: new role for Wnt signaling. Biochim. Biophys. Acta Rev. Cancer. 2017;1868(1):333–340. doi: 10.1016/j.bbcan.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Kienast Y., von Baumgarten L., Fuhrmann M., Klinkert W.E., Goldbrunner R., Herms J., Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 2010;16(1):116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 33.Valiente M., Obenauf A.C., Jin X., Chen Q., Zhang X.H.F., Lee D.J., Chaft J.E., Kris M.G., Huse J.T., Brogi E., Massague J. Serpins promote cancer cell survival and vascular Co-option in brain metastasis. Cell. 2014;156(5):1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevenich L., Bowman R.L., Mason S.D., Quail D.E., Rapaport F., Elie B.T., Brogi E., Brastianos P.K., Hahn W.C., Holsinger L.J., Massague J., Leslie C.S., Joyce J.A. Analysis of tumour-and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014;16(9):876–888. doi: 10.1038/ncb3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills C.D., Kincaid K., Alt J.M., Heilman M.J., Hill A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164(12):6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L., Qiu Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78(16):4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 38.Wolf-Dennen K., Gordon N., Kleinerman E.S. Exosomal communication by metastatic osteosarcoma cells modulates alveolar macrophages to an M2 tumor-promoting phenotype and inhibits tumoricidal functions. Oncoimmunology. 2020;9(1) doi: 10.1080/2162402X.2020.1747677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D., Wang X., Si M., Yang J., Sun S., Wu H., Cui S., Qu X., Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Sailor K.A., Agoranos G., Lopez-Manzaneda S., Tada S., Gillet-Legrand B., Guerinot C., Masson J.B., Vestergaard C.L., Bonner M., Gagnidze K., Veres G., Lledo P.M., Cartier N. Hematopoietic stem cell transplantation chemotherapy causes microglia senescence and peripheral macrophage engraftment in the brain. Nat. Med. 2022;28(3):517–527. doi: 10.1038/s41591-022-01691-9. [DOI] [PubMed] [Google Scholar]
- 41.Borriello L., Coste A., Traub B., Sharma V.P., Karagiannis G.S., Lin Y., Wang Y., Ye X., Duran C.L., Chen X., Friedman M., Sosa M.S., Sun D., Dalla E., Singh D.K., Oktay M.H., Aguirre-Ghiso J.A., Condeelis J.S., Entenberg D. Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat. Commun. 2022;13(1):626. doi: 10.1038/s41467-022-28076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman R.L., Klemm F., Akkari L., Pyonteck S.M., Sevenich L., Quail D.F., Dhara S., Simpson K., Gardner E.E., Iacobuzio-Donahue C.A., Brennan C.W., Tabar V., Gutin P.H., Joyce J.A. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17(9):2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z.H., Feng X., Herting C.J., Garcia V.A., Nie K., Pong W.W., Rasmussen R., Dwivedi B., Seby S., Wolf S.A., Gutmann D.H., Hambardzumyan D. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77(9):2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing F., Liu Y., Wu S.Y., Wu K.R., Sharma S., Mo Y.Y., Feng J.M., Sanders S., Jin G.X., Singh R., Vidi P.A., Tyagi A., Chan M.D., Ruiz J., Debinski W., Pasche B.C., Lo H.W., Metheny-Barlow L.J., D'Agostino R.B., Watabe K. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018;78(15):4316–4330. doi: 10.1158/0008-5472.CAN-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qian M., Wang S., Guo X., Wang J., Zhang Z., Qiu W., Gao X., Chen Z., Xu J., Zhao R., Xue H., Li G. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-kappaB pathways. Oncogene. 2020;39(2):428–442. doi: 10.1038/s41388-019-0996-y. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J.Y., Sadowska G.B., Chen X.D., Park S.Y., Kim J.E., Bodge C.A., Cummings E., Lim Y.P., Makeyev O., Besio W.G., Gaitanis J., Banks W.A., Stonestreet B.S. Anti-IL-6 neutralizing antibody modulates blood-brain barrier function in the ovine fetus. FASEB J. 2015;29(5):1739–1753. doi: 10.1096/fj.14-258822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018;18(12):773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 48.Rokavec M., Oner M.G., Li H.H., Jackstadt R., Jiang L.C., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., Slotta-Huspenina J., Bader F.G., Greten F.R., Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 2014;124(4):1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R., Wen A., Lin J. Pro-inflammatory cytokines in the formation of the pre-metastatic niche. Cancers. 2020;12(12) doi: 10.3390/cancers12123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fousek K., Horn L.A., Palena C. Interleukin-8: a chemokine at the intersection of cancer plasticity, angiogenesis, and immune suppression. Pharmacol. Ther. 2021;219 doi: 10.1016/j.pharmthera.2020.107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostrand-Rosenberg S., Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J. Immunol. 2018;200(2):422–431. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S.J., Lian G.D., Li J.J., Zhang Q.B., Zeng L.J., Yang K.G., Huang C.M., Li Y.Q., Chen Y.T., Huang K.H. Tumor-driven like macrophages induced by conditioned media from pancreatic ductal adenocarcinoma promote tumor metastasis via secreting IL-8. Cancer Med. 2018;7(11):5679–5690. doi: 10.1002/cam4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao P., Long X.X., Zhang L.J., Ye Y.N., Guo J.C., Liu P.P., Zhang R., Ning J.Y., Yu W.W., Wei F., Yu J.P. Neurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of tumor-associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncoimmunology. 2018;7(7) doi: 10.1080/2162402X.2018.1440166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D., Sun H., Wei J., Cen B., DuBois R.N. CXCL1 is critical for premetastatic niche formation and metastasis in colorectal cancer. Cancer Res. 2017;77(13):3655–3665. doi: 10.1158/0008-5472.CAN-16-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X.Y., Zhang Y., Zhang Y.G., Zhang S., Qiu L., Zhuang Z.X., Wei M.T., Deng X.B., Wang Z.Q., Han J.H. The key role of exosomes on the pre-metastatic niche formation in tumors. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.703640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Z.Q., Shi K., Yang S.X., Liu J.B., Zhou Q.B., Wang G.X., Song J.M., Li Z., Zhang Z.Y., Yuan W.T. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018;17 doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L.J., Guo Z., Zhou Y.M., Ni J., Zhu J.H., Fan X., Chen X.X., Liu Y.L., Li Z.P., Zhou H. microRNA-1246-containing extracellular vesicles from acute myeloid leukemia cells promote the survival of leukemia stem cells via the LRIG1-meditated STAT3 pathway. Aging. 2021;13(10):13644–13662. doi: 10.18632/aging.202893. Us. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torii C., Maishi N., Kawamoto T., Morimoto M., Akiyama K., Yoshioka Y., Minami T., Tsumita T., Alam M.T., Ochiya T., Hida Y., Hida K. miRNA-1246 in extracellular vesicles secreted from metastatic tumor induces drug resistance in tumor endothelial cells. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-92879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooks T., Pateras I.S., Jenkins L.M., Patel K.M., Robles A.I., Morris J., Forshew T., Appella E., Gorgoulis V.G., Harris C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018;9(1):771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data analyzed and generated during the study are available from the corresponding authors on reasonable request.