Abstract
Coccidiosis represents a major driver in the economic performance of poultry operations, as coccidia control is expensive, and infections can result in increased feed conversion ratios, uneven growth rates, increased co-morbidities with pathogens such as Salmonella, and mortality within flocks. Shifts in broiler production to antibiotic-free strategies, increased attention on pre-harvest food safety, and growing incidence of anti-coccidial drug resistance has created a need for increased understanding of interventional efficacy and methods of coccidia control. Conventional methods to quantify coccidia oocysts in fecal samples involve manual microscopy processes that are time and labor intensive and subject to operator error, limiting their use as a diagnostic and monitoring tool in animal parasite control. To address the need for a high-throughput, robust, and reliable method to enumerate coccidia oocysts from poultry fecal samples, a novel diagnostic tool was developed. Utilizing the PIPER instrument and MagDrive technology, the diagnostic eliminates the requirement for extensive training and manual counting which currently limits the application of conventional microscopic methods of oocysts per gram (OPG) measurement. Automated microscopy to identify and count oocysts and report OPG simplifies analysis and removes potential sources of operator error. Morphometric analysis on identified oocysts allows for the oocyst counts to be separated into 3 size categories, which were shown to discriminate the 3 most common Eimeria species in commercial broilers, E. acervulina, E. tenella, and E. maxima. For 75% of the samples tested, the counts obtained by the PIPER and hemocytometer methods were within 2-fold of each other. Additionally, the PIPER method showed less variability than the hemocytometer counting method when OPG levels were below 100,000. By automated identification and counting of oocysts from 12 individual fecal samples in less than one hour, this tool could enable routine, noninvasive diagnostic monitoring of coccidia in poultry operations. This approach can generate large, uniform, and accurate data sets that create new opportunities for understanding the epidemiology and economics of coccidia infections and interventional efficacy.
Key words: coccidiosis monitoring, Eimeria enumeration, Eimeria oocyst, coccidia
INTRODUCTION
Coccidiosis represents a significant determinant in the economic performance of poultry operations (Williams, 1999; Chapman et al., 2013). Infections with protozoan parasites of the genus Eimeria cause coccidiosis in poultry, with E. acervulina, E. brunetti, E. hagani, E. maxima, E. mitis, E. mivati, E. necatrix, E. praecox, and E. tenella causing disease in chickens (McDougald and Fitz-Coy, 2013). Clinical and subclinical coccidiosis can cause increased feed conversion ratios, poor uniformity within a flock, co-morbidities with increased instances of necrotic enteritis (NE) or salmonellosis, and increased mortality (Baba et al., 1982; Williams, 2005; McDougald and Fitz-Coy, 2013). Recent estimates of the global economic costs of coccidiosis put the figure at greater than 10 billion US dollars annually (Williams, 1999; Dalloul and Lillehoj, 2006; Kadykalo et al., 2018; Blake et al., 2020).
Conventional disease control of coccidiosis has included chemical treatments, ionophores, antibiotics, and vaccines (Peek and Landman, 2011; Chapman, 2014; Noack et al., 2019). The emergence of resistant populations and the relatively uneven or poor performance of anticoccidial drugs has led to the frequent use of multiple products, such as combining 2 synthetic compounds or a synthetic compound and an ionophore (Peek and Landman, 2011; Chapman, 2014). Recently, there has been increasing pressure on food service companies to only buy poultry raised without any products classified as antibiotics, which includes ionophores in the United States (Chapman et al., 2010). This, together with the passage of the Veterinary Feed Directive (2015), emphases on animal welfare (Thaxton et al., 2016), and the increasing prevalence of anticoccidial-resistant populations, has prompted a need for increased understanding of interventional efficacy and alternate methods of coccidia control (Peek and Landman, 2011).
Monitoring the efficacy of interventions has economic implications for the poultry producers and health implications for the birds (Jenkins et al., 2017; Snyder et al., 2021). The World Association for the Advancement of Veterinary Parasitology (WAAVP) recommends measuring coccidia oocysts in feces or litter to understand field efficacy of interventional methods, using either the hemocytometer or McMaster method (Holdsworth et al., 2004), but oocyst counts have not been used historically as a diagnostic tool. On poultry farms fresh feces must be collected from the enclosure floor and then sent to a laboratory to measure oocysts per gram (OPG). Both the hemocytometer and McMaster methods are time and labor intensive, requiring personnel trained to identify, count, and report the number of oocysts, in addition to microscopes and other laboratory equipment (Ricciardi and Ndao, 2015; Intra et al., 2016).
In addition to quantification of infection, identification of Eimeria species is needed for evaluation and control of the disease, as each species may respond differently to management strategies (McDougald et al., 1986; Lee et al., 2010). Conventionally, identification of Eimeria species is based on morphological features of the sporulated oocyst, sporulation time, and location/scoring of pathological lesions in the intestine (evaluated on necropsy), but the procedures require specialized expertise and are subjective (Long and Joyner, 1984). Molecular methods are currently available to identify the Eimeria species (Jenkins et al., 2006; Morris and Gasser, 2006; Haug et al., 2007; Blake et al. 2008; Cantacessi et al., 2008; Kawahara et al. 2008; Vrba et al., 2010; Barkway et al., 2011; Lalonde and Gajadhar, 2011; Kumar et al., 2014; Blake et al. 2015; Nolan et al., 2015), but the lack of rapid, low-cost, quantitative tests prevents their broad implementation. Additionally, one study suggested that region-specific differences in DNA sequences of Eimeria species may affect the accuracy of molecular detection methods (Loo et al., 2019).
To address the limitations of existing OPG measurement methods (low throughput, labor intensive), a method was developed for the automated enumeration of coccidia oocysts in poultry feces. This was accomplished using PIPER (Ancera Inc., Branford, CT), an instrument which exploits ferrofluid-based cytometry to concentrate, manipulate, and count cells within a disposable microfluidic device. The assay enables a single technician to process up to 196 samples in a single work shift and eliminates the extensive training and subjectivity of manual microscopy methods by employing automated image analysis. This study introduces the PIPER coccidia enumeration assay and compares the accuracy and variability of the PIPER diagnostic to a conventional hemocytometer counting method. It also correlates the size-based separation of oocyst counts with known individual species of oocysts. We discuss how this diagnostic could be used to assess coccidia load at a populational level by surveying individual birds, providing more granularity in understanding the efficacy of coccidia interventions than previously possible.
MATERIALS AND METHODS
Samples Tested
Fecal samples were provided as blinded samples from commercial broiler producers throughout the United States. Additional purified oocyst samples were provided from Merck Animal Health (MAH, Madison, NJ) in the form of individual species samples of E. maxima, E. tenella, and E. acervulina, confirmed by oocyst morphology, lesion location in infected birds, and PCR testing. Southern Poultry Research, Inc. (Athens, GA) also provided purified and concentrated samples of E. acervulina, E. tenella, and E. maxima oocysts. All samples were shipped refrigerated (wet ice) and stored at 4°C.
Sample Preparation
Sample preparation as described by the Coccidia Detection and Quantification Kit (P/N ANC-EIM001, Ancera Inc.) is shown in Figure 1A. One gram of homogenized feces or sample to be analyzed (or 100 µL of cleaned, single species) was measured into a 7 oz Whirl-Pak filter bag (B01385WA, Nasco, Fort Atkinson, WI). Five mL of 1 M NaOH (SK-80044-64, Cole-Palmer, Vernon Hills, IL) was added to the bag. The sample and NaOH were massaged by hand for 30 s to create uniform slurry. The sample was allowed to sit for 15 min and then 5 mL of a saturated sugar solution (Sample Additive, P/N ANC-EIM001-02, Ancera Inc.) added to the bag to achieve neutral buoyancy, and the sample massaged a second time. At this point, portions of the sample were removed for the PIPER protocol or for counting via hemocytometer.
Figure 1.
Schematic of Assay Workflow. (A) Sample preparation. 1 g of a fecal sample is mixed with 5 mL of 1M NaOH in a filter bag (side A). The bag is massaged for 30 s to thoroughly mix the sample followed by incubation for 15 min at room temperature. Another 5 mL of a saturated sugar solution (Sample Additive) is subsequently added to prevent oocyst settling, and the sample is mixed again. A 280 µL aliquot of the slurry is then removed from the filtered side of the bag (side B) to avoid any solids that could clog the microfluidic device and transferred to a new tube. The sample is mixed with 20 µL of Ferrofluid and 3 µL of a nucleic acid intercalating dye (Detection Reagent), vortexed to mix, and loaded into a single well of a PIPER cartridge. (B) Separation of oocysts on PIPER. Sample is mixed with a biocompatible ferrofluid and loaded into a cartridge. Microvalves in the cartridge control a pumping layer in the cartridge which pulls the sample over the magnetic PCB, which pushes target cells up for imaging by a built-in epifluorescent microscope above the cartridge. Data can be transferred to a cloud-based system or to a connected laboratory information management system based on the needs of the end user.
Reference Method: Hemocytometer
A Neubauer hemocytometer was used to count oocysts as the reference method as described in Conway and McKenzie (2007). Ten microliters of the prepared sample slurry (above) were loaded into the counting chamber. The oocysts in the four corner and center squares of the etched slide (the quincunx) were counted to generate the hemocytometer count. The total number of oocysts counted was multiplied by the dilution and divided by the approximate volume of the area counted to report oocysts per gram of feces (OPG). By this preparation, one oocyst counted in a chamber represented 20,000 OPG. Additional hemocytometer chambers were loaded with the same initial slurry and counted to generate sample replicate counts. All hemocytometer counts were performed by the same trained technicians.
PIPER Instrument
Once the sample was prepared as above, 280 µL of the homogenized sample was added to a 2 mL microcentrifuge tube. Three µL of a nucleic acid intercalating dye (Detection Reagent, P/N ANC-EIM001-04, Ancera Inc.) and 20 µL of Ferrofluid (P/N ANC-EIM001-03, Ancera Inc.) were added to the tube containing 280 µL of the homogenized sample, which was then mixed via vortex to create the assay mixture. During the incubation step of the sample preparation described above, the PIPER instrument was initialized, and the disposable MagDrive coccidia cartridge (P/N 5.1.2.LT, Ancera Inc.) was loaded on the prepared machine. The assay mixture was loaded into one of the 12 lanes in the disposable cartridge. Each lane is an independent, simultaneous test such that the PIPER is capable of running 12 unique or replicate (or some combination thereof) coccidia samples at the same time. After loading the cartridge, the user started the assay run using the PIPER's user interface. PIPER uses pneumatically driven microvalves in the consumable cartridge to create a peristaltic pumping without sample exposure to the instrument. This eliminates the need to flush or otherwise clean the instrument between runs. PIPER generates magnetic fields using a printed circuit board (PCB) under the cartridge to push targets suspended within the ferrofluid mixture to the top of the cartridge (Figure 1B) while the sample is flowing within the cartridge channel. The flow is stopped once the assay mixture has sufficiently filled the channel and imaging window. While the flow is stopped, the targets are immobilized at the top of the cartridge purely by the action of ferrofluid and magnetic force (Kose and Koser, 2012), and a built-in fluorescent microscope with a digital camera above the cartridge obtains images of the immobilized samples.
Image Recognition Algorithm
To automate the PIPER analysis, an image recognition algorithm was developed to identify and enumerate oocysts in the acquired images. The image recognition algorithm was developed using a two-step deep learning approach that first runs a U-NET and Stardist based segmentation model (Ronneberger et al., 2015; Schmidt et al., 2018), followed by a modified shallow Convolutional Neural Network (CNN) classification model (Lei et al., 2020). Metadata metrics such as major axis, minor axis, area, and intensity are available for every target identified. This additional data is used to categorize the oocysts into large, medium, and small sizes based on their major and minor axes’ lengths. For this analysis, large oocysts were characterized as having major axis length greater than 27 microns, medium oocysts were categorized as having major axis length less than 27 microns and minor axis length greater than 18 microns, and small oocysts were characterized as having major axis length less than 27 microns and minor axis length less than 18 microns. A portion of a processed PIPER image is shown in Figure 2 with identified oocysts automatically outlined by the algorithm and color coded by size categories. To assess the accuracy of the algorithm, trained analysts reviewed and manually counted oocysts in 67 PIPER images with oocyst counts ranging from <10 oocysts per image to greater than 1,000 oocysts per image using the ImageJ software (US National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij/) and compared those results to that of the algorithm (Figure 3).
Figure 2.
Example of PIPER image detecting oocysts. Image is magnified 100%. Detected oocysts are indicated by circles. Color discriminates oocysts based on size: large (yellow), medium (blue), or small (green).
Figure 3.
Accuracy of image recognition algorithm. Plot of counts obtained by manual review of 67 images (corresponding to replicates from 3 individual, fecal samples) to counts obtained for the same images by the image recognition algorithm. A plot of manual counts to algorithm counts for each of the images shows a linear regression line with a slope near 1 and coefficient of determination (R-squared) of 0.99.
Calibration to OPG
A total of 77 unique samples were assessed using both hemocytometer and PIPER to calibrate the PIPER counts to OPG. Each sample had 4 individual hemocytometer counts performed and most had 4 individual PIPER counts (6 samples had 12 PIPER measurements; 4 samples had 3 PIPER measurements). The samples had OPG levels ranging from 5,000 (1 oocyst seen in 4 replicate hemocytometer counts) to 5 million OPG by hemocytometer. The means and standard deviations of the replicates were calculated for each of the 77 samples for each method. Average PIPER counts for each sample were plotted against average hemocytometer OPG, and the calibration from PIPER count to OPG (as measured by hemocytometer) was determined by fitting a regression line through the origin. The conversion factor of PIPER counts to OPG was calculated from the slope of the regression line (425) and the coefficient of determination (R-squared) was 0.9835 (Figure 4).
Figure 4.
Correlation to Hemocytometer. Paired average hemocytometer and average PIPER counts for 77 independent samples were plotted against each other to determine the calibration of PIPER counts to oocysts per gram (OPG). The calibration from PIPER count to OPG (as measured by hemocytometer) was determined by fitting a regression line through the origin. The slope of the regression line was 425, generating the following calibration equation: PIPER total OPG = 425 × [PIPER total oocyst count]. The coefficient of determination (R-squared) for the regression line was 0.9835.
Linearity
Four samples of purified oocysts were prepared as described for cleaned, single species sample preparation above, with the modification that 3 preparations were made of each sample using different sampling volumes (100 µL, 30 µL, and 10 µL) in 5 mL of 1M NaOH. Each pseudo-dilution of sample was loaded on PIPER in quadruplicate (4 individual lanes) and used to assess the accuracy of counts below the limit of detection of the hemocytometer (generally accepted to be 2.5 × 10^5 (Cadena-Herrera et al., 2015)). Predicted counts were generated by multiplying the average 100 µL count for each sample by either 0.3 or 0.1, respectively, and compared to the actual counts as determined by PIPER.
Morphometrics and Classification
Individual metrics on each identified oocyst were collected by the PIPER instrument and used to classify the oocysts as small, medium, or large using the criteria defined for the algorithm above. Hundred microliters samples of individual species of oocysts (E. acervulina, E. maxima, E. tenella) were prepared as described above and assessed by the PIPER. The individual species samples utilized were cleaned oocyst samples prepared by MAH, with species identity confirmed by PCR.
Confirmatory Testing
Paired PIPER and hemocytometer counts were obtained for an additional 96 field fecal samples across three independent testing laboratories, one internal at Ancera (Site C) and 2 field laboratories (Sites A and B). Each of the 44 samples at Site A included between 1 and 6 PIPER replicates and between 1 and 3 hemocytometer chambers. The 20 samples at Site B included 2 or 3 PIPER replicates and 2 hemocytometer chambers, and the 32 samples at Site C included 2 PIPER replicates and 2 hemocytometer chambers.
Comparison of PIPER, McMaster, and Hemocytometer Counting Methods
Three separate aliquots of each of 138 poultry ingesta samples were taken at an independent testing laboratory for analysis by hemocytometer, McMaster, or PIPER, respectively. The sample preparation for the PIPER method and hemocytometer method was as described above. For the McMaster method, 100 µL of intestinal sample was diluted in 900 µL of a saturated (37.5%) sodium chloride solution. Six-hundred microliters of the prepared sample was then added to one side of the McMaster Chamber, filling the entire chamber. The slide was then allowed to sit for a minimum of 5 min for the oocysts to float to the surface. All oocysts inside of the grid, which is made up of 6 columns, were counted. If there were too many oocysts to count, the sample was diluted further before adding to the McMaster Chamber. OPG was determined by using the following formula: (counts per chamber/150 µL per chamber) × dilution factor × 1,000 µL.
Statistical Analysis
Graphics and data analyses were all performed using R (R Core Team, 2018). To determine the variability in measurements by the PIPER method or hemocytometer method, the coefficient of variation (CV), which is the standard deviation divided by the mean, was calculated for each sample. A LOESS (locally estimated scatterplot smoothing) local regression was used to generate a smoothed curve representing each CV (Figure 5). To determine the agreement in counts between 2 OPG measurement methods, the relative percent difference (RPD) was calculated. The relative percent difference between 2 non-negative numbers (assuming at least one is nonzero) is calculated by dividing the difference between the numbers by the average.
When symmetry is desired, sometimes the absolute value of the difference is used in the numerator. But since this can hide a bias in the measurement methods, if one of them is larger or smaller on average than the other, we did not take absolute values. Dividing by the average is useful when the differences scale with the size of the numbers. Since the oocyst per gram of sample (OPG) values in these experiments covers many orders of magnitude, this was appropriate. A relative percent difference of ±2/3 (66%) indicates a 2-fold difference between 2 positive numbers. For example, the relative percent difference between 20 and 10 is the difference divided by the average, which in this case is (20-10) / 15 = 10/15 = 2/3 (66%). The relative percent difference (RPD) is a useful measure of agreement between methods when what is being measured is away from zero. Since the hemocytometer lower limit of quantification with 2 quincunx measurements is 10,000 OPG, in Supplemental Figure 1 (where samples with OPG counts below 10,000 were observed), we calculated RPD only for samples with OPG greater than or equal to 10,000 by all counting methods.
Figure 5.
Comparison of PIPER to hemocytometer variability. Graph of the average log hemocytometer OPG versus the CV of hemocytometer (circles) or PIPER (plus symbols) counts. The line is a LOESS local regression used to generate a smoothed curve representing each CV (smooth line: hemocytometer; dashed line: PIPER). The shadows represent the standard error (95% confidence interval) of the mean CV of each measurement. Note that the mean PIPER count CV is more consistent and less than the hemocytometer count CV below 100,000 (log = 5). Above 100,000 OPG, there is not an appreciable difference in method mean CVs in this data set.
RESULTS
PIPER Counting Algorithm Performance
We have developed a coccidia detection and quantification assay on the PIPER platform that eliminates the subjective, labor intensive, manual microscopy steps that are associated with standard oocyst counting techniques (Figure 1). This is driven by the integrated deep learning oocyst detection algorithm that processes the scanned images (Figure 2). To assess performance of the image recognition algorithm, more than 60 images generated by the coccidia detection assay on PIPER were manually reviewed by technicians trained to identify oocysts and simultaneously processed through the image recognition algorithm. A plot of manual counts against algorithm counts shows a linear regression line with a slope near one and coefficient of determination (R-squared) of 0.99 (Figure 3), suggesting algorithmic performance consistent with human oocyst identification.
Calibration of PIPER Automated Oocyst Enumeration Assay With Hemocytometer Counting Method
The PIPER MagDrive technology enables concentration of oocysts in the imaging window as the sample flows through the cartridge (Kose and Koser, 2012). To calibrate the oocyst counts obtained by the PIPER assay to oocysts per gram as measured by the conventional hemocytometer method, paired hemocytometer and PIPER data was collected for 77 unique samples prepared by the PIPER sample preparation method. Average counts obtained by the two methods for each sample were plotted against each other (Figure 4), and the linear fit of a regression line through the origin was used to generate a calibration equation to convert PIPER counts to OPG. Based on this equation, one oocyst count on PIPER corresponds to an oocyst concentration in the sample of 425 OPG (Figure 4). Using this calibration, fewer measurements are needed to detect low OPG levels by the PIPER method, where one oocyst in the imaging window corresponds to 425 OPG, compared to the hemocytometer method, in which a single oocyst detected in 1 quincunx of a hemocytometer chamber is equivalent to 20,000 OPG.
Repeatability of PIPER Automated Oocyst Enumeration
The coefficient of variation (CV) was calculated for replicates of each unique sample (n = 77) from the calibration study, and the CVs of hemocytometer and PIPER counts are graphically represented in Figure 5. Variability between measurements for the same sample was on average between 20 and 30% across a 3-log range (5000 OPG to 5 million OPG) with the PIPER method (Figure 5). While the variability between measurements by hemocytometer was comparable to PIPER for samples with OPG above 100,000, PIPER CVs were consistently lower than hemocytometer CVs for samples under 100,000 OPG (Figure 5).
PIPER Assay Linearity and Performance at Low Concentrations
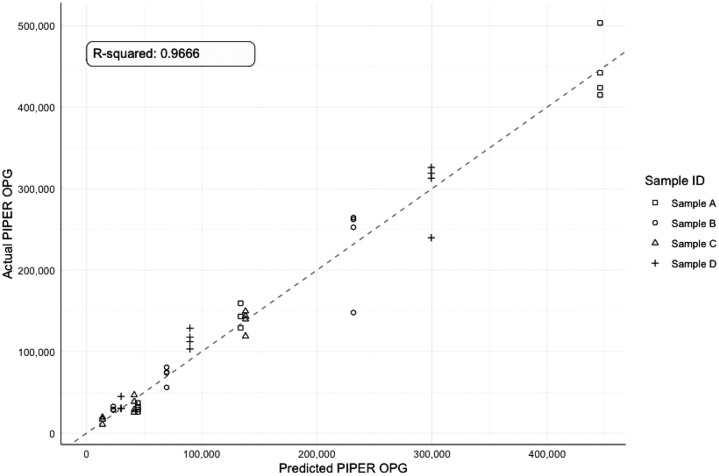
The comparison of CVs showed that hemocytometer OPG counts were increasingly variable at concentrations less than 100,000 OPG, even with the same technicians. This is consistent with the reported lower limit for accurate counting of cells via hemocytometer (Cadena-Herrera et al. 2015). To assess a predicted linear relationship of sample concentrations below the levels at which the hemocytometer could be considered reliable, we further investigated the linearity of the assay across 3 different volumes of each of 4, cleaned, oocyst samples. The average total counts on PIPER for four replicates of each sample at each volume were determined. The average count for the 1× volume of each sample was multiplied by 0.3 or 0.1 to calculate a predicted count for the smaller volumes of the same sample. These predicted counts were plotted against the actual counts for all samples and a linear regression performed to assess the linearity of the assay below the lower limit of accuracy or repeatability for the hemocytometer method. This analysis showed a linear relationship between the actual and predicted values with a linear regression line having a coefficient of determination (R-squared) of 0.97 (Figure 6), suggesting that the PIPER counts were reliable at lower OPG counts where the hemocytometer comparison could not be used to assess performance.
Figure 6.
Assay linearity. Plot of PIPER counts obtained for 3 different amounts of each of four, cleaned oocyst samples versus the predicted counts for each sample portion. The average count for replicates of the 1× portion of each sample was multiplied by 0.3 or 0.1, respectively, to calculate a predicted count for the smaller portions. The r2 value for the line y = x was 0.9666.
Morphometrics and Classification
Metadata such as major axis, minor axis, area, and intensity are documented for every oocyst identified by the image recognition algorithm. These data are used by the algorithm to classify oocysts as small, medium, or large (Figure 2). We evaluated the difference in size classification by the algorithm for the three most prevalent species in broilers globally, E. acervulina, E. tenella, and E. maxima (Mesa et al., 2021; Moraes et al., 2015; Gyorke et al., 2013; Haug et al., 2008). Twelve lanes replicates each of PCR-confirmed, cleaned, individual species of E. acervulina, E. tenella, and E. maxima were analyzed on PIPER. In this study, 76% of E. acervulina oocysts were classified as small oocysts, 81% of E. tenella were classified as medium oocysts, and 96% of E. maxima were classified as large oocysts (Table 1). This data is consistent with previous morphometric analysis of these species (Haug et al., 2008) and suggests that the ability of the image recognition algorithm to bin oocyst counts into size categories can help provide additional information about the population of coccidia that is present in the samples tested.
Table 1.
Morphometric classification of oocysts. For each of 3 individual species of Eimeria: E. acervulina, E. tenella, and E. maxima, the automated image algorithm binned oocysts into categories based on size. The number and percentages of small, medium, and large oocysts are indicated for each individual species.
| Sample | Total oocyst counts | Small oocyst counts | Medium oocyst counts | Large oocyst counts | % Small oocyst counts | % Medium oocyst counts | % Large oocyst counts |
|---|---|---|---|---|---|---|---|
| E. acervulina | 13,141 | 9,993 | 3,148 | 0 | 76% | 24% | 0% |
| E. maxima | 3,791 | 40 | 117 | 3,634 | 1% | 3% | 96% |
| E. tenella | 8,503 | 1,511 | 6,862 | 130 | 18% | 81% | 1% |
Confirmatory Testing
To assess the accuracy of the PIPER calibration to OPG, an additional 96 field fecal samples with hemocytometer OPG counts in the range of 100,000 to 2 million were evaluated across 3 independent laboratories, including 2 field sites (Sites A & B) and Ancera's internal testing lab (Site C). All samples included paired PIPER and hemocytometer counts. Agreement between the 2 methods was assessed by plotting the relative percent difference between methods for each sample against the average OPG of the 2 methods for each sample. This metric was chosen because it directly measures the agreement between the 2 methods for each sample. This is an advantage over a metric such as an R-squared value, which is highly dependent on the range of OPG values of the samples used and can be artificially inflated by samples with unusually large OPG values (Montgomery et al., 2012; https://www.stat.cmu.edu/∼cshalizi/mreg/15/lectures/10/lecture-10.pdf). Based on this analysis, 75% of the samples fell within a 2-fold difference or less (Figure 7). In addition to evaluating the agreement between PIPER and hemocytometer, we further evaluated the agreement between measurements by PIPER and McMaster, another commonly used reference method for determining OPG. For this study, 138 poultry intestinal samples were collected, and three separate aliquots of each sample were prepared for analysis by the PIPER, hemocytometer, or McMaster method. Scatterplots comparing measurements by any of the two methods for each of the samples showed a similar spread relative to the line y = x (Supplemental Figure 1A), and the relative percent difference (RPD) between measurements by McMaster and PIPER was consistent with the RPD between measurements by hemocytometer and PIPER or between measurements by McMaster and hemocytometer (Supplemental Figure 1B). Taken together, these data suggest that the PIPER assay could provide comparable performance to conventional oocyst counting methods with improved throughput and ease of use, enabling monitoring of oocyst cycling in flocks to understand trends and efficacy of interventions that is not practical currently.
Figure. 7.
Relative percent difference between PIPER and hemocytometer. Ninety-six samples with hemocytometer OPG between 100,000 and 2 million were evaluated by paired hemocytometer and PIPER counts. 44 samples were evaluated at Site A (squares), 20 samples were evaluated at Site B (triangles), and 32 samples were evaluated at Site C (circles). Average counts for the replicates of each sample were calculated. The relative percent difference was computed using the formula RPD = hemocytometer OPG – average PIPER OPG//[(hemocytometer OPG + PIPER OPG)/2]. The dashed lines at ± 66.6% represent a two-fold difference.
DISCUSSION
The quantification of coccidia oocysts in feces (OPG) has been used to categorize flocks at risk of decreased performance levels (Haug et al., 2008). Recent data suggests that OPG measurements could also provide insight about the efficacy of intervention strategies, potentially linking performance metrics, such as average daily gain or feed conversion ratios (Chasser et al., 2020), and providing guidance for veterinarians and producers on the timing and appropriateness of adding different anticoccidial compounds, nutraceuticals, or vaccination protocols (Jenkins et al., 2017). Despite the need for quantitative tools that can rapidly and noninvasively determine the abundance of these parasites, conventional microscopy methods are time consuming, subjective, and low throughput (Joyner and Long, 1974; Long and Joyner, 1984). The PIPER assay described in this paper enables the automated identification and quantification of coccidia oocysts from fecal samples. Unlike traditional hemocytometer and McMaster methods which require manual counting of oocysts in each sample one at a time, a single PIPER cartridge can support the analysis of twelve fecal samples in parallel. While the hands-on time for sample preparation on PIPER is similar to that for hemocytometer or McMaster, the run time on the instrument for identifying and counting oocysts in 12 independent samples is less than an hour. Increased scalability can conceivably be achieved with incorporation of robotics and additional PIPER instruments. Since the PIPER assay can generate large amounts of data on oocyst populations quickly, this method will enable veterinarians and poultry professionals to monitor coccidia cycling at a populational level across multiple time points throughout the flock life cycle, providing more granularity than conventional diagnostics, like necropsy. Necropsy analysis of the intestinal tracts for macroscopic lesions typical of coccidiosis is generally only done on a few chickens in the flock (far below what would be needed for a representative sample; generally, 5 per house which may contain 20,000 birds) as opportunistic sampling for monitoring or when clinical coccidiosis is suspected (Johnson and Reid, 1970; Williams, 2005). Routine monitoring at a population level by analyzing OPG in feces samples throughout the flock life cycle could enable veterinarians to make changes to intervention strategies based on trends in coccidia cycling before an impact on performance is observed.
Other technologies, such as flow cytometry, can be used to sort and enumerate cells based on fluorescent signals. However, flow cytometry is prone to clogging, requires instrument-specific calibration, and cannot be used with complex environmental samples like feces without extensive sample preparation. The PIPER MagDrive technology uses a uniquely patterned printed circuit board to generate magnetic force on a ferrofluid composed of superparamagnetic nanoparticles for flow-based manipulation of cells (Dhlakama et al., unpublished data). We have shown that oocysts can be efficiently detected and enumerated from complex field samples, including feces and intestinal contents, using the PIPER technology, and the results of confirmatory testing at field sites showed a strong correlation with the classical hemocytometer method. Furthermore, CVs for the PIPER assay were consistent across a 3-log range of OPG and were lower than hemocytometer CVs at concentrations below 100,000 OPG. Based on the calibration of PIPER counts to hemocytometer counts, a single oocyst count observed on PIPER translates to approximately 425 OPG, which is more sensitive than a single hemocytometer chamber (1 oocyst = 20,000 OPG), suggesting that this system requires fewer measurements than hemocytometer to detect low oocyst levels. The McMaster counting method has been reported to detect as few as 50 OPG (Alowanou et al., 2021), which is lower than the theoretical lower limit of the current PIPER assay (425 OPG). Further sensitivity could conceivably be achieved in the PIPER assay through a sample concentration step using centrifugation or filtration.
We showed that the PIPER assay can automatically count oocysts and separate those counts into size categories. The distribution of oocyst sizes differed for the three most common species in North American broiler production, E. acervulina, E. tenella, and E. maxima, with the majority of E. acervulina representing small oocysts, the majority of E. tenella representing medium oocysts, and the majority of E. maxima representing large oocysts. Future testing of the size categorizations and refinement of the algorithmic categories should be performed with a larger set of purified oocysts representing all Eimeria species. Although the PIPER algorithm cannot currently discriminate all nine species of Eimeria, even this greater granularity of classification of Eimeria into size categories could allow for better evaluation and control of the disease than looking at total oocyst counts alone, as each size category may respond differently to management strategies, and co-infection with multiple species can complicate diagnosis (Williams et al., 1996; Lee et al. 2010).
Given the advantages of the PIPER system for throughput, reliability, and ease of use, we believe that this novel assay will open new opportunities for understanding the epidemiology of coccidia infections and monitoring intervention efficacy.
ACKNOWLEDGMENTS
We acknowledge Merck Animal Health (MAH) for their partnership in helping fund development of this assay, providing access to samples, and performing confirmatory testing. We also acknowledge Dr. Lorraine Fuller and Southern Poultry Research, Inc. (Athens, GA) who provided purified and concentrated samples of E. acervulina, E. tenella, and E. maxima oocysts.
DISCLOSURES
All authors employed at Ancera, Inc. or Merck Animal Health has an indirect financial interest in the coccidia monitoring product described in this manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102252.
Appendix. Supplementary materials
REFERENCES
- Alowanou G.G., Adenile A., Akouedegni D., Bossou G.C.,, Zinsou A.C., Akakpo F.T., Kifouly G.-C.A., Rinaldi H.A., von Samson-Himmelstjerma L., Cringoli G., Hounzangbé-Adoté S. A comparison of Mini-FLOTAC and McMaster techniques in detecting gastrointestinal parasites in West Africa Dwarf sheep and goats and crossbreed rabbits. J. Appl. Anim. Res. 2021;49:30–38. [Google Scholar]
- Baba E., Fukata T., Arakawa A. Establishment and persistence of Salmonella typhimurium infection stimulated by Eimeria tenella in chickens. Res. Vet. Sci. 1982;33:95–98. [PubMed] [Google Scholar]
- Barkway C.P., Pocock R.L., Vrba V., Blake D.P. Loop-mediated isothermal amplification (LAMP) assays for the species-specific detection of Eimeria that infect chickens. BMC Vet. Res. 2011;7:67. doi: 10.1186/1746-6148-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D.P., Qin Z., Cai J., Smith A.L. Development and validation of real-time polymerase chain reaction assays specific to four species of Eimeria. Avian Pathol. 2008;37:89–94. doi: 10.1080/03079450701802248. [DOI] [PubMed] [Google Scholar]
- Blake D.P., Clark E.L., MacDonald S.E., Thenmozhi V., Kundu K., Garg R., Jatau I.D., Ayoade S., Kawahara F., Moftah A., Reid A.J. Population, genetic, and antigenic diversity of the apicomplexan Eimeria tenella and their relevance to vaccine development. Proc. Natl. Acad. Sci. 2015;112:E5343–E5350. doi: 10.1073/pnas.1506468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D.P., Knox J., Dehaeck B., Huntington B., Rathinam T., Ravipati V., Ayoade S., Gilbert W., Adebambo A.O., Jatau I.D., Raman M., Parker D., Rushton J., Tomley F.M. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020;51:115. doi: 10.1186/s13567-020-00837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena-Herrera D.J.E.Esparza-De Lara, Ramiréz-Ibañez N.D., López-Morales C.A., Pérez N.O., Florez-Ortiz L.F., Medina-Rivero E. Validation of three viable-cell counting methods: manual, semi-automated, and automated. Biotechnol. Rep. 2015;7:9–16. doi: 10.1016/j.btre.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C., Riddell S., Morris G.M., Doran T., Woods W.G., Otranto D., Gassera R.B. Genetic characterization of three unique operational taxonomic units of Eimeria from chickens in Australia based on nuclear spacer ribosomal DNA. Vet. Parasitol. 2008;154:226–234. doi: 10.1016/j.vetpar.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Chapman H.D., Jeffers T.K., Williams R.B. Forty years of monensin for the control for the control of coccidiosis in poultry. Poult. Sci. 2010;89:1788–1801. doi: 10.3382/ps.2010-00931. [DOI] [PubMed] [Google Scholar]
- Chapman H.D., Barta J.R., Blake D., Gruber A., Jenkins M., Smith N.C., Suo X., Tomley F.M. A selective review of advances in coccidiosis research. Adv. Parasitol. 2013;83:93–171. doi: 10.1016/B978-0-12-407705-8.00002-1. [DOI] [PubMed] [Google Scholar]
- Chapman H.D. Milestones in avian coccidiosis research: a review. Poult. Sci. 2014;93:501–511. doi: 10.3382/ps.2013-03634. [DOI] [PubMed] [Google Scholar]
- Chasser K.M., Duff A.F., Wilson K.M., Briggs W.N., Latorre J.D., Barta J.R., Bielke L.R. Evaluating fecal shedding of oocysts in relation to body weight gain and lesion scores during Eimeria infection. Poult. Sci. 2020;99:886–892. doi: 10.1016/j.psj.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway D.P., McKenzie M.E. 3rd ed. Blackwell Publishing Professional; Oxford, UK: 2007. Poultry Coccidiosis: Diagnostic and Testing Procedures. [Google Scholar]
- Dalloul R.A., Lillehoj H.S. Poultry coccidiosis: recent advancements in control measures and vaccine development. Expert Rev. Vaccines. 2006;5:143–163. doi: 10.1586/14760584.5.1.143. [DOI] [PubMed] [Google Scholar]
- Györke A., Pop L., Cozma V. Prevalence and distribution of Eimeria species in broiler chicken farms of different capacities. Parasite. 2013;20:50. doi: 10.1051/parasite/2013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug A., Thebo P., Mattsson J.G. A simplified protocol for molecular identification of Eimeria species in field samples. Vet. Parasitol. 2007;146:35–45. doi: 10.1016/j.vetpar.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Haug A., Gjevre A.-G., Thebo P., Mattsson J.G., Kaldhusdal M. Coccidial infections in commercial broilers: epidemiological aspects and comparison of Eimeria species identification by morphometric and polymerase chain reaction techniques. Avian Pathol. 2008;37:161–170. doi: 10.1080/03079450801915130. [DOI] [PubMed] [Google Scholar]
- Holdsworth P.A., Conway D.P., McKenzie M.E., Dayton A.D., Chapman H.D., Mathis G.F., Skinner J.T., Mundt H.C., Williams R.B., World Association for the Advancement of Veterinary Pathology World Association for the Advancement of Veterinary Parasitology (WAAVP) guidelines for evaluating the efficacy of anticoccidial drugs in chickens and turkeys. Vet. Parasitol. 2004;121:189–212. doi: 10.1016/j.vetpar.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Intra J., Taverna E., Sala M.R., Falbo R., Cappellini F., Brambilla P. Detection of intestinal parasites by use of the cuvette-based automated microscopy analyzer sediMAX. Clin. Microbiol. Infect. 2016;22:279–284. doi: 10.1016/j.cmi.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Jenkins M.C., Miska K., Klopp S. Improved polymerase chain reaction technique for determining the species composition of Eimeria in poultry litter. Avian Dis. 2006;50:632–635. doi: 10.1637/7615-042106R.1. [DOI] [PubMed] [Google Scholar]
- Jenkins M.C., Parker C., Ritter D. Eimeria oocyst concentrations and species composition in litter from commercial broiler farms during anticoccidial drug or live Eimeria oocyst vaccine control programs. Avian Dis. 2017;61:214–220. doi: 10.1637/11578-010317-Reg.1. [DOI] [PubMed] [Google Scholar]
- Johnson J., Reid W.M. Anticoccidial drugs: lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 1970;28:30–36. doi: 10.1016/0014-4894(70)90063-9. [DOI] [PubMed] [Google Scholar]
- Joyner L.P., Long P.I. The specific characters of the Eimeria, with special reference to the coccidia of the fowl. Avian Pathol. 1974;3:145–157. doi: 10.1080/03079457409353827. [DOI] [PubMed] [Google Scholar]
- Kadykalo S., Roberts T., Thompson M., Wilson J., Lang M., Espeisse O. The value of anticoccidials for sustainable global poultry production. Int. J. Antimicrob. Agents. 2018;51:304–410. doi: 10.1016/j.ijantimicag.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Kawahara F., Taira K., Nagai S., Onaga H., Onuma M., Nunova T. Detection of five avian Eimeria species by species-specific real-time polymerase chain reaction assay. Avian Dis. 2008;52:652–656. doi: 10.1637/8351-050908-Reg.1. [DOI] [PubMed] [Google Scholar]
- Kose A.R., Koser H. Ferrofluid mediated nanocytometry. Lab. Chip. 2012;12:190–196. doi: 10.1039/c1lc20864k. [DOI] [PubMed] [Google Scholar]
- Kumar S.R.Garg, Moftah A., Clark E.L., Macdonald S.E., Chaudhry A.S., Sparagano O., Banerjee P.S., Kundu K., Tomley F., Blake D.P. An optimised protocol for molecular identification of Eimeria from chickens. Vet. Parasitol. 2014;199:24–31. doi: 10.1016/j.vetpar.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde L., Gajadhar A.A. Detection and differentiation of coccidian oocysts by real-time PCR and melting curve analysis. J. Parasitol. 2011;97:725–730. doi: 10.1645/GE-2706.1. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Kim W.H., Jeong J., Yoo J., Kwon Y.K., Jung B.Y., Kwon J.H., Lillehoj H.S., Min W. Prevalence and cross-immunity of Eimeria species on Korean chicken farms. J. Vet. Med. Sci. 2010;72:985–989. doi: 10.1292/jvms.09-0517. [DOI] [PubMed] [Google Scholar]
- Lei F., Liu X., Dai Q., Ling B.W.-K. Shallow convolutional neural network for image classification. SN Appl. Sci. 2020;2:97. [Google Scholar]
- Long P.I., Joyner L.P. Problems in the identification of species of Eimeria. J. Protozool. 1984;31:535–541. doi: 10.1111/j.1550-7408.1984.tb05498.x. [DOI] [PubMed] [Google Scholar]
- Loo S.-S., Lim L.-S., Efendi N.A., Blake D.P., Kawazu S.-I., Wan K.-L. Comparison of molecular methods for the detection of Eimeria in domestic chickens in Malaysia. Sains Malaysiana. 2019;48:1425–1432. [Google Scholar]
- Mesa C., Gómez-Osorio L.M., López-Osorio S., Williams S.M., Chaparro-Gutiérrez J.J. Survey of coccidia on commercial broiler farms in Colombia: frequency of Eimeria species, anticoccidial sensitivity, and histopathology. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D.C., Peck E.A., Vining G.G. 5th ed. John Wiley and Sons, Inc.; Hoboken, NJ: 2012. Pages 35–36 in Introduction to Linear Regression Analysis. [Google Scholar]
- Moraes J.C., França M., Sartor A.A., Bellato V., de Moura A.B., de Lourdes M., Magalhães B., de Souza A.P., Miletti L.C. Prevalence of Eimeria spp. in broilers by multiplex PCR in the southern region of Brazil on two hundred and fifty farms. Avian Dis. 2015;59:277–281. doi: 10.1637/10989-112014-Reg. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Gasser R.B. Biotechnological advances in the diagnosis of avian coccidiosis and the analysis of genetic variation in Eimeria. Biotechnol.Adv. 2006;24:590–603. doi: 10.1016/j.biotechadv.2006.06.001. [DOI] [PubMed] [Google Scholar]
- McDougald L.R., Fuller L., Solis J. Drug-sensitivity of 99 isolates of coccidia from broiler farms. Avian Dis. 1986;30:690–694. [PubMed] [Google Scholar]
- McDougald L.R., Fitz-Coy S.H. In: Pages 1148–1166 in Diseases of Poultry. Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L., Nair V.L., editors. Wiley-Blackwell; 2013. “Coccidiosis.”. [Google Scholar]
- Noack S., Chapman H.D., Selzer P.M. Anticoccidial drugs of the livestock industry. Parasitol. Res. 2019;118:2009–2026. doi: 10.1007/s00436-019-06343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan M.J., Tomley F.M., Kaiser P., Blake D.P. Quantitative real-time PCR (qPCR) for Eimeria tenella replication- Implications for experimental refinement and animal welfare. Parasitol. Int. 2015;64:464–470. doi: 10.1016/j.parint.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek H.W., Landman W.J.M. Coccidiosis in poultry: anticoccidial products, vaccines and other prevention strategies. Vet. Q. 2011;31:143–161. doi: 10.1080/01652176.2011.605247. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- Ricciardi A., Ndao M. Diagnosis of parasitic infections: what's going on? J. Biomol. Screen. 2015;20:6–21. doi: 10.1177/1087057114548065. [DOI] [PubMed] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. MICCAI. 2015;9351:234–241. [Google Scholar]
- Schmidt U., Weigert M., Broaddus C., Myers G. In: Medical Image Computing and Computer Assisted Intervention. Frangi A.F., editor. Springer International Publishing; Cham: 2018. Cell detection with star-convex polygons; pp. 265–273. MICCAI 2018: 21st International Conference, Granada, Spain, Sept. 16-20, Proceedings, Part II. [Google Scholar]
- Snyder R.P., Guerin M.T., Hargis B.M., Page G., Barta J.R. Monitoring coccidia in commercial broiler chicken flocks in Ontario: comparing oocyst cycling patterns in flocks using anticoccidial medications or live vaccination. Poult. Sci. 2021;100:110–118. doi: 10.1016/j.psj.2020.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaxton Y.V., Christensen K.D., Mench J.A., Rumley E.R., Daugherty C., Feinberg B., Parker M., Siegel P., Scanes C.G. Symposium: Animal welfare challenges for today and tomorrow. Poult. Sci. 2016;95:2198–2207. doi: 10.3382/ps/pew099. [DOI] [PubMed] [Google Scholar]
- Vrba V., Blake D.P., Poplstein M. Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Vet. Parasitol. 2010;174:183–190. doi: 10.1016/j.vetpar.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Williams R.B., Bushell A.C., Reperant J.M., Doy T.G., Morgan J.H., Shirley M.W., Yvore P., Carr M.M., Fremont Y. A survey of Eimeria species in commercially-reared chickens in France during 1994. Avian Pathol. 1996;25:113–130. doi: 10.1080/03079459608419125. [DOI] [PubMed] [Google Scholar]
- Williams R.B. A compartmentalized model for the estimation of the cost of coccidiosis to the world's chicken production industry. Int. J. Parasitol. 1999;29:1209–1229. doi: 10.1016/s0020-7519(99)00086-7. [DOI] [PubMed] [Google Scholar]
- Williams R.B. Intercurrent coccidiosis and necrotic enteritis of chickens: rational, integrated disease management by maintenance of gut integrity. Avian Pathol. 2005;34:159–180. doi: 10.1080/03079450500112195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.