Summary
Signaling cascades can act in series or in parallel. Here, we describe a convenient and robust protocol for dual, sequential knockdown of two proteins using RNA interference. We detail the steps for a quantitative mapping of signaling circuitry. We used this approach to study kinases in human ovarian cancer cells, but the protocol can be applied to many other posttranslational modifications.
For complete details on the use and execution of this protocol, please refer to Gocher et al. (2017).1
Subject areas: Cell Biology, Molecular Biology, Signal Transduction, Protein Biochemistry
Graphical abstract

Highlights
-
•
Knockdown of two proteins via a dual sequential RNA interference
-
•
Quantitative distinction of in-parallel versus in-series signaling cascades
-
•
Specific steps for kinases, but applicable to many posttranslational modifications
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Signaling cascades can act in series or in parallel. Here, we describe a convenient and robust protocol for dual, sequential knockdown of two proteins using RNA interference. We detail the steps for a quantitative mapping of signaling circuitry. We used this approach to study kinases in human ovarian cancer cells, but the protocol can be applied to many other posttranslational modifications.
Before you begin
The protocol below provides detailed instructions on a dual sequential transfection of adherent NIH:OVCAR-3 (OVCAR-3, ATCC) human ovarian cancer cells with targeted small interfering RNAs (siRNAs) towards phosphoinositide-dependent protein kinase-1 (PDPK1, gene encoding for PDK1) and calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2, gene encoding for CAMKK2) to inhibit the phosphorylation of AKT (protein kinase B/PKB). We have also successfully used this protocol in Caov-3 and SK-OV-3 (ATCC) human ovarian cancer cell lines. In our experience, dual sequential transfections have decreased cell toxicity in these cell lines when compared to a dual simultaneous transfection approach.
Design and/or purchase of individual siRNAs
Timing: 1 h (for step 1)
Determine your target genes of interest. Typically, one gene is a validated canonical upstream modulator, and the second gene is a putative upstream modulator. The following steps provide guidance for selecting specific siRNAs towards your target genes.
-
1.
Design and/or purchase siRNAs towards your protein of interest.
Note: Pre-designed, validated and non-validated siRNAs, for humans, mice, and rats can be purchased from ThermoFisher Scientific. Custom siRNAs are available. Validated and pre-designed SilencerR Select siRNAs towards human CAMKK2 and PDPK1 were used in this assay.
Note: A locked nucleic acid (LNA) was incorporated into the first-generation SilencerR siRNA, by ThermoFisher Scientific, to generate their more potent and stable SilencerR Select siRNA. Additional chemical modifications to first generation siRNAs such as the addition of a 2′-OMethyl, 2′-Deoxy-2-fluoro or Phosphorothioate have been made to improve stability, RNA binding efficiency, potency and improve pharmacokinetics in clinical settings.2
Note: It is recommended to purchase two siRNAs per target for thorough validation of results. It is necessary that the sequences of the replicate siRNAs are non-overlapping.
-
2.Purchase the relevant non-specific (NS) siRNAs for use as a control.
-
a.We used NS SilencerR Select siRNA.
-
a.
Alternatives: siRNA-resistant mutants of the targeted gene can be used as a control for proving targeting specificity and to rescue the gene function. A web tool to aid in designing siRNA-resistant gene constructs can be found at Ong et al.3
-
3.Prepare working stocks of the siRNAs.Note: The concentration will depend on the volume of siRNA used. Ideal volume to transfect cells is 2–20 μL.
-
a.We reconstituted each siRNA to a 50 μM stock.
-
i.5 nmol siRNA + 100 μL nuclease-free water = 50 μM stock.Note: The use of aseptic techniques is required when working with siRNAs, medium and cell lines to prevent contamination by microorganisms. The use of gloves and DNase-, and RNase-, free barrier/filter tips is strongly encouraged.
-
i.
-
a.
Prepare medium for growth and transfection of the cell line
Timing: 0.5–1 h (for step 4)
The following steps detail the various culture medium supplementations used throughout the transfection procedures.
-
4.Medium for growth of cells should be prepared.
-
a.Recommended complete formulation of medium for passaging of cells (complete medium).Note: This complete medium formulation is often unique to individual cell lines. Please refer to manufacturers’ guidelines for the recommended supplementation of complete medium.
-
i.For OVCAR-3 cells we used RPMI-1640 medium supplemented with the following components added per manufacturer’s recommendations (ATCC): 20% FBS, 10 mM HEPES, 1 mM sodium pyruvate, 1× penicillin/streptomycin, 2 mM L-glutamine, 2.4 mg/mL D-glucose, and 1% insulin.
-
i.
-
b.Plating medium is an antibiotic-free form of the complete medium.
-
i.Here we omitted 1× penicillin/streptomycin from our complete medium.Note: Cationic lipid reagents such as Lipofectamine 2000, increase cell permeability. Antibiotics were omitted during the transfection procedure since the increased cell permeability has been shown by the manufacturer (ThermoFisher Scientific) to increase the uptake of antibiotics, thus resulting in increased cell death.
-
i.
-
c.Transfection medium is an antibiotic-free and serum-free form of the complete medium.
-
i.Here we omitted 1× penicillin/streptomycin and FBS from our complete medium.Note: Some serums contain RNases which can breakdown the siRNA, therefore we omitted serum from the transfection media.Alternatives: Transfection medium can be replaced with Opti-MEMTM (ThermoFisher Scientific, #31985062) to increase cell survival during serum-free incubations.
-
i.
-
a.
Optimize the transfection of individual siRNAs
Transfection of cell lines with individual siRNAs must be conducted to determine the optimal concentration of the siRNAs used. Once the ideal concentrations are determined, these concentrations will be implemented in the dual sequential knockdown procedure.
-
5.
Determine the optimal concentration of siRNA by conducting a titration above and below the recommended concentration.
Note: The recommended concentration of SilencerR Select CAMKK2 siRNA is 5 nM however, we determined in-house that 40 nM is needed for efficient knockdown of CAMKK2 in OVCAR-3 cells.
Note: Include 2–3 replicates per experimental group.
-
6.Assess knockdown efficiency at 3-, 5- and 7-, days post transfection.
-
a.Determine the percent knockdown of targeting protein, i.e., knockdown efficiency, via Western blotting. Refer to “Harvesting cells for protein to assess the dual transfection knockdown efficiency and consequential signaling circuitry via Western blotting” for experimental detains.
-
i.Normalize the targeting protein, herein CAMKK2, to a housekeeping gene such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin or β-tubulin. Herein we used GAPDH.
-
ii.Determine the percent decrease in the normalized CAMKK2 levels achieved with the targeting siRNA to that of the NS siRNA.
-
i.
-
a.
Alternatives: Knockdown efficiency at the protein level can also be determined by flow cytometry if monoclonal antibodies are available for the target protein. Please refer to Holmes et al. for steps on the fluorescent labeling of antibodies and conduction of flow cytometry.4 If neither a polyclonal nor a monoclonal antibody are available, the knockdown efficiency at the transcript level can be determined via reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). Please refer to Bookout et el. for designing and conducting RT-qPCR of your target gene.5
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal Anti-CAMKK2 human (1:1000–2500) | Abnova | #H00010645-M01 |
| Anti-PDK1 (1:500) | BD Transduction Laboratories | #611070 |
| Anti-GAPDH (1:5000) | Santa Cruz | #sc-47724 |
| Anti-p-AKT Thr308 (1:200–5000) | Cell Signaling | #13038 |
| Rabbit polyclonal Anti-AKT (1:1000–10,000) | Cell Signaling | #9272 |
| Goat anti-rabbit (1:3000) | Cell Signaling | #7074 |
| Horse anti-mouse (1:3000) | Cell Signaling | #7076 |
| Chemicals, peptides, and recombinant proteins | ||
| LipofectamineTM 2000 Transfection Reagent | Thermo Fisher Scientific | #1668027 |
| Protease inhibitor cocktail | Sigma | #P8340 |
| Tris base | Sigma | #10708976001 |
| Sodium dodecyl sulfate (SDS) | Sigma | #L3771 |
| Ethylene glycol-bis (2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) Molecular Biology Grade | Sigma | #324626 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma | #E5134 |
| Sodium fluoride (NaF) 0.5 M solution | Sigma | #67414 |
| Sodium orthovanadate (Na3VO4) | Sigma | #6508 |
| Hydrochloric acid concentrate 10 N | Fisher Scientific | #SA49 |
| Enhanced chemiluminescence (ECL): Enhanced Western Lightning Plus Chemiluminescence Substrate | PerkinElmer | #NEL105001EA |
| Experimental models: Cell lines | ||
| OVCAR-3 | ATCC | #HTB-161 |
| Oligonucleotides | ||
| CAMKK2 SilencerR Select siRNA | Thermo Fisher Scientific | #4390826, s20926 |
| PDPK1 SilencerR Select siRNA | Thermo Fisher Scientific | #4392422, s10274 |
| NS SilencerR Select siRNA | Thermo Fisher Scientific | #4390844 |
| Software and algorithms | ||
| Quantity One 1-D software | Bio-Rad | https://www.bio-rad.com/en-us/product/quantity-one-1-d-analysis-software?ID=1de9eb3a-1eb5-4edb-82d2-68b91bf360fb |
| ImageJ software | Schneider et al. (2012)6 | https://imagej.nih.gov/ij/download.html |
| Other | ||
| Tissue culture incubator (37°C and 5% CO2) | ||
| ChemiDoc Imaging system | Bio-Rad | |
| Microcentrifuge | ||
| Water bath/Bead bath (37°C) | ||
| Sonicator | ||
Materials and equipment
Harvest buffer (pH 8.0) for western blotting
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris base | 50 mM | 605.7 mg |
| SDS | 2% w/v | 2 g |
| EGTA | 5 mM | 190.2 mg |
| EDTA | 5 mM | 86.1 mg |
| NaF | 25 mM | 5 mL |
| Na3VO4 | 1 mM | 18.4 mg |
| Water | N/A | 95 mL |
| 10 N HCl | N/A | Until pH reaches 8.0 |
| Total | N/A | 100 mL |
Storage at 20°C–22°C for up to 6 months.
Note: EGTA and EDTA will not dissolve until the solution is at a pH of 8.0. When ready to use for harvesting cell, freshly add 1% w/v of Protease inhibitor cocktail to Harvest buffer.
CRITICAL: SDS is corrosive, an irritant and is flammable. EDTA is toxic and is an irritant. Na3VO4 is an irritant and is toxic. HCl is corrosive and an irritant. Protease inhibitor cocktail is flammable and is an irritant. Use a chemical fume hood when handling these reagents.
Step-by-step method details
Plating cells—Day 1
Timing: 0.5–1 h
The following steps provide guidance in drafting the experimental layout for the dual sequential knockdowns of your genes of interest. The experimental design and number of experimental groups must be determined prior to plating the cell line.
Note: Include 2–3 replicate plates per group. Experimental groups are as follows:
| Group | siRNA (Day 2) | siRNA (Day 4) |
|---|---|---|
| NS (negative control) | NS | NS |
| PDPK1 knockdown | PDPK1 | NS |
| CAMKK2 knockdown | NS | CAMKK2 |
| PDPK1 and CAMKK2 knockdown | PDPK1 | CAMKK2 |
Note: In this sequential double knockdown design, each plate of cells will be transfected with an NS or a targeting siRNA on Day 2 and Day 4. This is a staggered protocol; we have experienced toxicity when co-transfecting siRNAs to PDPK1 and CAMKK2 at the same time.
-
1.
Warm up Plating medium to 37°C prior to use.
-
2.Prepare 60 mm or 100 mm tissue culture dishes for each condition.
-
a.Pipet 5 mL per 60 mm dish or 10 mL per 100 mm dish of Plating medium.
-
a.
-
3.Plate cells at 10%–20% density in a tissue culture dish.
-
a.For OVCAR-3 cells, SK-OV-3 and Caov-3 cells we found that 300,000 cells per 60 mm dish or 1,000,000 cells per 100 mm dish is ideal.
-
a.
Note: A 10%–20% seeding density will allow the cells to grow for 5 days without reaching maximal confluency. Cell seeding density is based on the population doubling time which is unique for each cell line. OVCAR-3, SK-OV-3 and Caov-3 cells population doubling time is around 50–80 h.7 If the assay requires >5 days in culture, decrease the seeding density to >10%.
Note: Determine the size of the tissue culture plate needed for subsequent applications. Scaling to 60 mm dishes is adequate for downstream applications such as Western blotting. Immunoprecipitation experiments may require more cellular material, therefore a 100 mm format can be used.
Alternatives: Although Lipofectamine™ 2000 was used here, the newly formulated Lipofectamine 3000 (ThermoFisher Scientific, #L30000001) has been reported to have increased transfection efficiency at lower doses. Please see manufacturer's protocol for required volumes of Lipofectamine 3000 to use.
Transfection of cells with first siRNA (PDPK1)—Day 2
Timing: 7 h
The following steps provide guidance for the knockdown of the canonical upstream modulator of Akt, PDPK1. This protocol is written for a 60 mm format, with the PDPK1 siRNA to be used at 40 nM in a final culture volume of 5 mL.
Note: Volume of reagents for 60 mm and 100 mm tissue culture dishes using a final concentration of 40 nM siRNA:
| Reagent | 60 mm dish | 100 mm dish |
|---|---|---|
| Final tissue culture volume (mL) | 5 | 10 |
| Lipofectamine™ 2000 (μL) | 10 | 13.3 |
| 50 μM siRNA (μL) | 4 | 8 |
-
4.
Warm up Transfection medium to 37°C prior to use.
-
5.Obtain two sets of Eppendorf tubes for each dish to be transfected.
-
a.Label tubes accordingly to decipher which set will be for the Lipofectamine + medium, and which set will be for the siRNA + medium.
-
a.
Note: Volumes below are designed to have a final volume of 500 μL in each set of Eppendorf tubes.
-
6.Pipet 490 μL of transfection medium and 10 μL of Lipofectamine™ 2000 into one set of tubes.
-
a.Vortex to mix and quickly spin down tubes in a microcentrifuge.
-
a.
Note: This quick spin is only used to pull-down any medium that may reside on the inside of the cap during the vortexing step.
-
7.
Incubate for 5 min.
-
8.Pipet 496 μL of transfection medium and 4 μL of siRNA into another set of tubes.
-
a.Vortex to mix and quickly spin down tubes in a microcentrifuge.
-
a.
-
9.Pipet medium + Lipofectamine™ 2000 from step 6 into the tube with medium + siRNA from step 8.
-
a.Vortex to mix and quickly spin down tubes in a microcentrifuge.
-
a.
-
10.
Incubate for 20 min. The final volume will be 1 mL.
Note: This incubation time can be reduced to 5 min when using Opti-MEMTM medium.
-
11.
Remove medium from plated cells and replace with 4 mL of Transfection medium.
-
12.
After 20 min (step 10), pipet 1 mL of medium + Lipofectamine™ + siRNA to each dish of cells slowly, to avoid breaking Lipofectamine/siRNA complex.
-
13.
Incubate for 6 h at 37oC in a tissue culture incubator.
-
14.
After 6 h, remove medium from dishes and pipet 4 mL of Plating medium into the plates.
-
15.
Incubate at 37oC in a tissue culture incubator for 2 days.
Transfection of cells with second siRNA (CAMKK2)—Day 4
Timing: 7 h (for step 16)
Timing: 2 days (for step 17)
This procedure is the same as the prior, except that this second siRNA is targeted to the putative upstream modulator (herein CAMKK2).
-
16.
Repeat steps 4–15 with second siRNA and incubate cells for 3 days.
Note: OVCAR-3 cells grow best if medium is changed every 2 days. On day 6, medium was removed from all plates and replaced with fresh and warmed Plating medium.
Harvesting cells for protein to assess the dual transfection knockdown efficiency and consequential signaling circuitry via Western blotting – Day 7.
The following protocol assess the knockdown efficiencies of the genes of interest (herein PDPK1 and CAMKK2) via the quantification of the protein expression in the single and double transfected cells with Western blotting. In this experiment, AKT was considered a putative target of PDK1 and CAMKK2. Therefore, in this experiment, the phosphorylation of AKT at the primary activation site, Thr308, was measured and normalized to total AKT protein levels.
-
17.
Remove media from cells.
-
18.Pipet 100–300 μL of Harvest buffer per dish and scrape cells from the plate.
-
a.Collect lysate into an Eppendorf tube and store on ice.
-
a.
Pause point: Cell lysates can be stored at −80°C.
-
19.
Sonicate lysate at 1 Watt on ice for 5 s.
-
20.
Spin lysate at 20,000 × g for 10 min at 4°C to pellet cellular debris.
-
21.Collect supernatant in a new Eppendorf tube.
-
a.Label tubes for long-term storage.
-
a.
Pause point: Protein can be stored at −80°C.
-
22.
Conduct a protein assay to determine the protein concentration of each sample.
Note: A quick protein assay can be conducted using the Pierce™ BCA Protein assay kit (Thermo Scientific™ #23227) per manufacturer’s protocol.
-
23.
Use 20–35 μg of protein to perform Western blotting.
Expected outcomes
The expected outcome is to obtain a ≥80% knockdown efficiency with each individual siRNA, and that this knockdown efficiency is similar when both targets are knocked down. Figure 1 illustrates the decrease in PDK1 and CAMKK2 expression when the appropriate siRNAs were used. These data are quantified in Table 1.
Figure 1.
Immunoblot of PDK1, CAMKK2, and GAPDH after dual sequential knockdown of PDK1 and CAMKK2
OVCAR-3 cells were sequentially transfected with 40 μM NS or PDPK1 siRNA on Day 2 and 40 μM NS or CAMKK2 siRNA on Day 4. Obtained with permission and modified from Gocher et al.1
Table 1.
Quantification of PDK1 and CAMKK2 percent knockdown for single and combination knockdown in siRNA transfected cells
| Day | Individual siRNA |
Combination siRNA |
||
|---|---|---|---|---|
| PDK1 | CAMKK2 | PDK1 | CAMKK2 | |
| 5 | 63.6 ± 44.58 | 59.4 ± 19.5 | 56.1 ± 61.1 | 49.0 ± 54.6 |
| 7 | 83.9 ± 13.3 | 75.5 ± 17.8 | 88.5 ± 3.5 | 86.6 ± 12.3 |
| 9 | 85.1 ± 9.4 | 81.8 ± 9.0 | 93.2 ± 1.6 | 89.6 ± 5.2 |
OVCAR-3 cells were sequentially transfected with 40 μM NS or PDPK1 siRNA on Day 2 and 40 μM NS or CAMKK2 siRNA on Day 4. Values represent mean fold-change ± standard deviation relative to the NS siRNA. N=3 independent experiments. Obtained with permission from Gocher et al.1
Quantification and statistical analysis
To accurately determine if the target proteins are in a parallel or series circuitry, the knockdown efficiency of each target protein should be comparable. Herein, we achieved a 93.2 ± 1.6 percent decrease in PDK1 and an 89.6 ± 5.2 percent decrease in CAMKK2, which are comparable to one another. If the extents of knockdowns are not similar, please see the “troubleshooting” section.
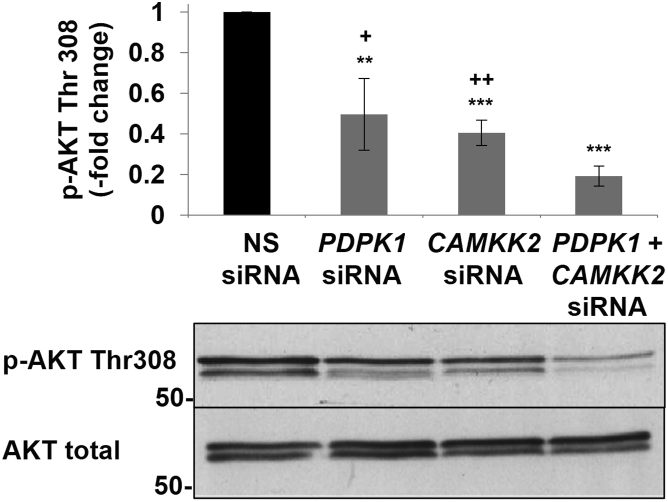
In the example shown here, Figure 2 illustrates that the double knockdown sample had a decrease in the phosphorylation of the substrate (p-AKT Thr308 was used here), that was greater than that of the single knockdowns (quantified in Table 2), and thus the circuitry is constructed in parallel. In nature, the redundancy of having multiple enzymes for a single substrate allows for activation of the substrate when one of the upstream activators is absent. In the example shown here, knockdown of PDK1 allowed for AKT to be phosphorylated at approximately 50%, since the alternative upstream activator CAMKK2 is present and can phosphorylate AKT. Similarly, knockdown of CAMKK2 allowed for AKT to be phosphorylated at approximately 50%, since the alternative upstream activator PDK1 is present and can phosphorylate AKT. When PDK1 and CAMKK2 were both knocked down, AKT phosphorylation decreased by more than half, thus producing an additive decrease in AKT activation. These data indicate that PDK1 and CAMKK2 share AKT as a substrate. As noted above under “Summary”, if the dual knockdown of the two upstream enzymes only decreased the activity of the substrate to that of the value achieved with knockdown of the penultimate kinase, the circuitry would have been concluded to be constructed in series.
Figure 2.
CAMKK2 and PDK1 regulate AKT phosphorylation in a parallel fashion
OVCAR-3 cells were transfected with NS, PDPK1, and CAMKK2 siRNAs singly or in combination, and phosphorylation of AKT at Thr308 was quantified. Values represent mean fold-change ± standard deviation relative to the NS siRNA. ∗∗, p < 0.01; ∗∗∗, p < 0.001 relative to NS siRNA. +, p < 0.05; ++, p < 0.01 for individual siRNAs relative to combined siRNAs. N=3 independent experiments. Obtained with permission and modified from Gocher et al.1
Table 2.
Quantification of p-AKT Thr308 for single and combination knockdown in siRNA transfected cells
| siRNA(s) | % Decrease in p-AKT Thr308 |
|---|---|
| PDPK1 | 50.3 ± 17.6 |
| CAMKK2 | 59.4 ± 6.2 |
| PDPK1 + CAMKK2 | 80.7 ± 4.9 |
Values represent mean fold-change ± standard deviation relative to the NS siRNA. N=3 independent experiments. Obtained with permission and modified from Gocher et al.1
Limitations
This dual sequential knockdown approach has only been validated in three cells lines (OVCAR-3, SK-OV-3 and Caov-3 cells) with one combination of two targeting siRNAs (CAMKK2 and PDPK1). Although we achieved a robust knockdown of CAMKK2 or PDPK1 when singly transfecting HeLa, A431, A549, N87 and LNCaP cell, we have not tested this dual sequential knockdown approach on additional cell lines nor additional gene targets.8
A potential limitation of this procedure is if knockdown of the target gene promotes cell death or cell cycle arrest. This can make it difficult to carry out the 5-day dual transfection procedure. Higher seeding number may be needed in this case.
It can be very challenging if there is not an assay available to quantitatively measure the change in activity of the substrate and/or if the substrate is not expressed abundantly or is difficult to detect, a series versus parallel circuitry of the network is unable to be determined.
Knockdowns using siRNA are not 100% complete, thus resulting in low levels of the targeted gene to still be detected. Knockdowns using siRNA are not long-lived, therefore repeated transfections of cell lines in independent experiments must be conducted to validate the results.
Troubleshooting
Problem 1
Knockdown efficiency is low.
Potential solution
-
•
Preliminary experiments should be used to establish the optimal concentrations of the siRNAs used for transfection.
-
•
Preliminary experiments should be used to determine the times required for maximal deletion of each protein and the time window that includes maximal knockdown efficacy of both siRNAs.
-
•
Some cells such as leukemic cells are difficult to transfect and thus yield a low knockdown efficiency with lipid-based transfection approaches. An alternative approach using electroporation should be used for cells that are difficult-to-transfect, and are advised to refer to Beyer et al.9
Problem 2
Knockdown efficiency is adequate but short lived.
Potential solution
-
•
Short hairpin RNAs (shRNA) can be used instead of siRNAs since shRNAs typically have longer durations of action than siRNAs. However, lentiviral transductions with shRNAs require cells to undergo a selection process with antibiotics for long-term cultures. Additionally, sometimes methylation-induced silencing occurs within regions of the lentivirus (promoters) that can result in antibiotic resistance, yet the gene of interest is not knocked down.
Problem 3
The sequential knockdown approach may promote adaptive signaling rewiring after knocking down the first targeted gene, and prior to knocking down the second targeted gene. However, these knockdown studies are short-term and transient, therefore rewiring is likely to be a rare event.
Potential solution
-
•
Time course studies should we conducted in single transfected cells to observe the impact that the knockdown of first targeted gene has on the signaling cascade over-time. This will allow for the observation of a potential adaptive signaling rewiring event that may occur prior to knocking down the second targeted gene.
Problem 4
Knockdown of the two upstream kinases resulted in an additive decrease in activity of the substrate however, some basal activity of the substrate remained.
Potential solution
-
•
There is incomplete knockdown of the two presumed upstream kinases. Evaluate how much residual protein of each upstream kinase is expressed after single and dual knockdown. Refer to “problem 1”.
-
•
There are more than two upstream kinases that regulate the activity of the substrate.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the technical and lead contact, Angela Gocher-Demske, agocher@pitt.edu.
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by the National Institutes of Health (F32 CA247004 to A.M.G.D. and T32 CA082083 to A.M.G.D.) and the Department of Defense (Ovarian Cancer Research Program Grant OC150368 to A.M.E.).
Author contributions
Conceptualization, A.M.D.G., A.M.E.; investigation and testing, A.M.G.D.; writing original draft, A.M.G.D.; review and editing, A.M.G.D., S.D., A.M.E.; funding acquisition, A.M.E.
Declaration of interests
The authors declare no competing interests.
Data and code availability
The published article includes all datasets generated and analyzed in this study. https://doi.org/10.1074/jbc.M117.778464.
References
- 1.Gocher A.M., Azabdaftari G., Euscher L.M., Dai S., Karacosta L.G., Franke T.F., Edelman A.M. Akt activation by Ca(2+)/calmodulin-dependent protein kinase kinase 2 (CaMKK2) in ovarian cancer cells. J. Biol. Chem. 2017;292:14188–14204. doi: 10.1074/jbc.M117.778464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang M., Huang Y. siRNA modification and delivery for drug development. Trends Mol. Med. 2022;28:892–893. doi: 10.1016/j.molmed.2022.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Ong J.Y. Synonymous Mutation Generator: a web tool for designing RNAi-resistant sequences. bioRxiv. 2021 doi: 10.1101/2021.01.02.425100. Preprint at. [DOI] [Google Scholar]
- 4.Holmes K., Lantz L.M., Fowlkes B.J., Schmid I., Giorgi J.V. Preparation of cells and reagents for flow cytometry. Curr. Protoc. Immunol. 2001;Chapter 5 doi: 10.1002/0471142735.im0503s44. Unit 5.3. [DOI] [PubMed] [Google Scholar]
- 5.Bookout A.L., Cummins C.L., Mangelsdorf D.J., Pesola J.M., Kramer M.F. High-throughput real-time quantitative reverse transcription PCR. Curr. Protoc. Mol. Biol. 2006;Chapter 15 doi: 10.1002/0471142727.mb1508s73. Unit 15.18. [DOI] [PubMed] [Google Scholar]
- 6.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dar S., Chhina J., Mert I., Chitale D., Buekers T., Kaur H., Giri S., Munkarah A., Rattan R. Bioenergetic adaptations in chemoresistant ovarian cancer cells. Sci. Rep. 2017;7:8760. doi: 10.1038/s41598-017-09206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai S., Venturini E., Yadav S., Lin X., Clapp D., Steckiewicz M., Gocher-Demske A.M., Hardie D.G., Edelman A.M. Calcium/calmodulin-dependent protein kinase kinase 2 mediates pleiotropic effects of epidermal growth factor in cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2022;1869:119252. doi: 10.1016/j.bbamcr.2022.119252. [DOI] [PubMed] [Google Scholar]
- 9.Beyer M., Krämer O.H. RNA interference protocol to silence oncogenic drivers in leukemia cell lines. STAR Protoc. 2022;3:101512. doi: 10.1016/j.xpro.2022.101512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets generated and analyzed in this study. https://doi.org/10.1074/jbc.M117.778464.