Abstract
Intellectual disability is a well-known hallmark of Down Syndrome (DS) that results from the triplication of the critical region of human chromosome 21 (HSA21). Major studies were conducted in recent years to gain an understanding about the contribution of individual triplicated genes to DS-related brain pathology. Global transcriptomic alterations and widespread changes in the establishment of neural lineages, as well as their differentiation and functional maturity, suggest genome-wide chromatin organization alterations in trisomy. High Mobility Group Nucleosome Binding Domain 1 (HMGN1), expressed from HSA21, is a chromatin remodeling protein that facilitates chromatin decompaction and is associated with acetylated lysine 27 on histone H3 (H3K27ac), a mark correlated with active transcription. Recent studies causatively linked overexpression of HMGN1 in trisomy and the development of DS-associated B cell acute lymphoblastic leukemia (B-ALL). HMGN1 has been shown to antagonize the activity of the Polycomb Repressive Complex 2 (PRC2) and prevent the deposition of histone H3 lysine 27 trimethylation mark (H3K27me3), which is associated with transcriptional repression and gene silencing. However, the possible ramifications of the increased levels of HMGN1 through the derepression of PRC2 target genes on brain cell pathology have not gained attention. In this review, we discuss the functional significance of HMGN1 in brain development and summarize accumulating reports about the essential role of PRC2 in the development of the neural system. Mechanistic understanding of how overexpression of HMGN1 may contribute to aberrant brain cell phenotypes in DS, such as altered proliferation of neural progenitors, abnormal cortical architecture, diminished myelination, neurodegeneration, and Alzheimer’s disease-related pathology in trisomy 21, will facilitate the development of DS therapeutic approaches targeting chromatin.
Keywords: Epigenetics, Trisomy, Neurodevelopment, Polycomb repressive complex, Chromatin remodeling, Histone modification, Neurodegeneration, Intellectual disability, Nucleosome
Introduction
Intellectual disability is perhaps the most well-known and ubiquitous feature of Down Syndrome (DS), with deficits in intellect as measured by intelligence quotient (IQ) nearly universal among individuals with the condition. The median IQ score in DS is 40, although variability exists and it can range from 10 to 70 [1]. There is often a delay in cognitive development, and individuals tend to have particular deficits in verbal working memory, executive functioning, syntactic processing, and expressive language [2–6].
Although some treatment or management options exist for other DS-associated conditions, such as congenital heart disease, addressing intellectual disability presents unique challenges since the precise mechanisms underlying the cognitive impairment have been difficult to establish. Many known neurobiological differences in individuals with DS, such as reduced brain volume [7, 8], hypocellularity [9], impairments in cellular proliferation and migration [10], delayed cortical lamination [11], and deficient dendritic arborization and synaptogenesis may contribute to this phenotype [12–14]. In addition, perturbations in glial cells in DS are manifested by precocious gliogenic shift that happens when neural progenitors in the embryonic ventricular zone start to change the fate of the cells they generate from neuronal to glial. This leads to an increased generation of astrocytes and delayed and aberrant myelination [7, 9, 12–16], as schematically shown in Fig. 1. The precise causes of these changes have remained elusive. However, spatiotemporal neurodevelopmental differences in gene expression implicate epigenetic dysregulation in DS as a possible crucial mechanism leading to global transcriptomic alterations and aberrant neural phenotypes.
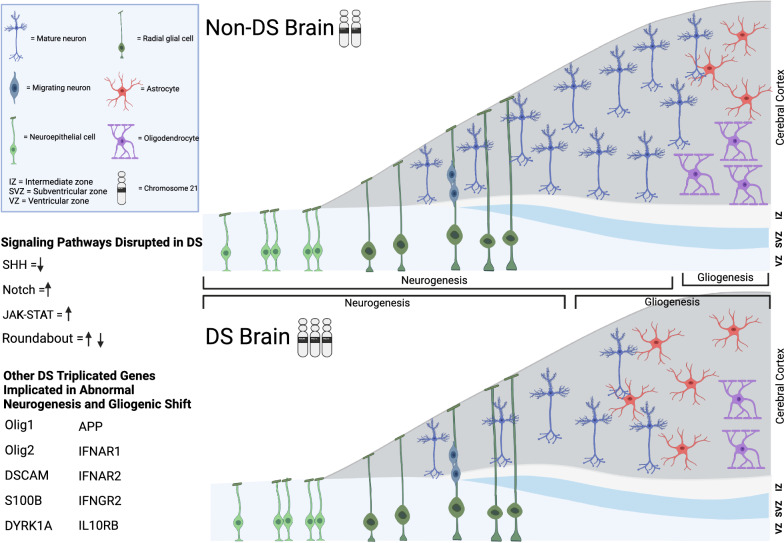
Fig. 1.
Schematic illustrating the proposed changes in neurogenesis and gliogenesis found in the DS brain. During neurodevelopment, neural progenitor cells, such as radial glial cells, generate neurons that migrate to the cortical plate and populate deep and superficial layers of the neocortex. Neurogenesis then is followed by gliogenesis, which results in the generation of glial cells, such as astrocytes and oligodendrocytes, that subsequently differentiate and mature. A premature shift from neurogenesis to gliogenesis is implicated in DS, with the potential manifestation of impaired neurogenesis, hypocellularity, an increase in the number of astrocytes, and a decrease in the number of mature oligodendrocytes. This precocious gliogenic switch can be attributed to a dosage effect of the triplicated genes (listed in the figure) resulting in a disruption of multiple signaling pathways implicated, shown in the figure. The dysregulation of these pathways can also be a result of the diminished activity of PRC2 due to increased levels of HMGN1 in DS. Created with BioRender.com
There are several genes triplicated in DS that affect the epigenetic landscape and may contribute to the described phenotypes, including High Mobility Group Nucleosome Binding Domain 1 (HMGN1), Dual specificity tyrosine-phosphorylation-regulated-kinase 1A (DYRK1A) [17–21], ETS proto-oncogene 2 (ETS2) [22–30], Bromodomain and WD repeat domain containing 1 (BRWD1) [31–33], Runt-related transcription factor 1 (RUNX1) [34–42], DNA (cytosine-5)-methyltransferase 3-like (DNMT3L) [43, 44], and SON RNA and DNA binding protein (SON) [45–50]. This review is focused on HMGN1, which is known to promote chromatin decompaction through interactions with nucleosomes [51], and its interplay with the Polycomb Repressive Complex 2 (PRC2) in DS. The disruption of the HMGN1/PRC2 balance is likely to be causatively related to a variety of DS phenotypes. We aim to highlight the consequences of HMGN1 triplication in connection to the specific transcriptomic, molecular, and cellular changes seen in DS.
Chromatin architecture and histone modifications
Before we focus our attention on HMGN1 and its functions, we will briefly review some general principles of chromatin compaction.
Genetic information is carried in an incredibly adaptable structure that can be modulated to allow for changes in gene transcription. The repeating structural unit of the human genome is the nucleosome, comprised of DNA wrapped 1.7 times around an octamer structure made of basic proteins called histones. There are two copies of each of the core histone proteins; H2A, H2B, H3, and H4, which form heterodimeric pairs [52]. There are also two linker proteins, H1 and H5, which influence chromatin structure and are known to repress transcription. For an extensive review, please refer to Talbert and Henikoff [53].
Post-translational histone modification via the addition of acetyl, methyl, phosphate, or ubiquitin groups at the N-terminal of histone tails can affect the interaction of these proteins with DNA and lead to alterations in the chromatin structure [54, 55]. This can change how DNA is read and transcribed into RNA. Therefore, histone modifications play an important role in the regulation of the transcriptome.
One important type of histone modification is the addition of an acetyl group to the lysine and arginine residues. Acetylation is present in a majority of N-terminal histone tails and primarily occurs in lysine-rich histone tails (73–80%) versus arginine-rich histone tails (36–48%) [56]. Allfrey et al. [56] demonstrated that the interaction between DNA and histones could be altered by the addition of acetyl groups and that this change in turn impacts the rate of RNA synthesis. Hebbes et al. [57] later showed the presence of acetylated core histones in transcriptionally active genes. This increase in transcriptional activity is likely due to a more open conformation of the nucleosome structure since the addition of an acetyl group to a lysine residue reduces its positive charge, and therefore decreases the strength with which the histone tail interacts with the nucleosomal DNA [58, 59]. This should allow for easier access to the DNA by transcription factors. Indeed, hyperacetylation of H4 is correlated with an elongated shape of the nucleosome core particle [60] and leads to an opening in the tetrameric particle formed by histones H3 and H4, which may allow for easier access to the DNA by transcription factors [61].
Whereas a more relaxed chromatin structure around the histone core following acetylation allows transcription factors to bind to the DNA and activate transcription, the removal of acetyl groups reestablishes the positive charge of the lysine residues and closes the nucleosomal structure, which represses transcription [57, 59, 62]. The addition of acetyl groups is carried out by enzymes known as histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs) [63, 64].
Histones can also be post-translationally modified by the addition of one, two, or three methyl groups to arginine, histidine, or lysine residues. Methylation was originally considered irreversible [65] until the discovery of the demethylases that remove methyl groups [66]. Unlike acetylation, methylation can be either transcriptionally repressive or activating, but most relevant to this review is the transcriptionally-repressive trimethylation of lysine 27 on the histone 3 tail [67, 68]. In addition to directly affecting the properties of chromatin, specific histone modifications can be recognized and bound by the “reader” proteins, which then influence chromatin organization and gene expression. This “histone code” hypothesis, which was postulated by Strahl and Allis [55], has been largely proven by numerous subsequent studies. Importantly, chromatin immunoprecipitation (ChIP) assays using antibodies recognizing specific histone modifications were combined with genomic arrays, and later with deep sequencing, which allowed a generation of genome-wide chromatin modification maps [69–73]. Thus, H3K4, H3K36, and H3K79 methylation has been correlated with transcriptionally active genes [74–78], whereas H3K9, H3K27, and H3K20 methylation is associated with transcriptional repression [67, 68, 79, 80]. Moreover, it was recognized that transcription start sites (TSS) are marked with localized enrichment in H3K4me3 [70, 76, 81, 82], and enhancer elements are marked by H3K4me1 and H3K27ac modifications [73, 83, 84].
A remarkable coincidence of H3K27 methylation domains with smaller H3K4 methylation regions was discovered in mouse embryonic stem cells (ESCs) and termed “bivalent domains” in 2006 [85]. This pattern was later found predominant in human ES cells [86]. Sequential ChIP experiments confirmed that these seemingly mutually exclusive modifications co-existed on the same histone H3 tails [85, 86]. Moreover, this bivalent feature was predominantly found on developmental genes activated by lineage-specific transcription factors that were either not expressed or expressed at low levels in ESCs. It was suggested that such genes are poised for future activation [85], and, indeed, the bivalent marks showed dynamic changes during ES cell differentiation [86]. In later sections, we will discuss the relevance of bivalent marking in neurogenesis and DS abnormalities.
To summarize, there are numerous regulators of chromatin organization, and we will focus on the high mobility group (HMG) family of proteins next.
High mobility group (HMG) proteins
High mobility group (HMG) proteins are key epigenetic regulators found in all vertebrates that exert their effect through chromatin remodeling [87, 88]. They were first extracted from calf thymus tissue and identified as a new class of non-histone proteins by Goodwin et al. [89]. Since then, they have been noted to play a role as global regulators of chromatin architecture and gene expression, mainly by reducing chromatin fiber compaction and promoting DNA accessibility for transcription and replication [51, 90–97], as well as their ability to influence modifications to histone tails [98]. There have been three families of these proteins identified and described: HMGA, HMGB, and HMGN, which are all less than 30 kDa in size and bind reversibly to DNA or nucleosomes [95, 99, 100]. HMGA proteins bind in the minor groove to AT-rich sequences [101], HMGB proteins contain 80 amino acid domains that bind with limited specificity to the minor groove of DNA [102], and HMGN proteins bind to nucleosomes, which are comprised of 146 base pairs of DNA wrapped around a central histone octamer [103]. Digestion of HMGN-associated chromatin with micrococcal nuclease results in DNA fragments that are 10 to 20 base pairs longer than core nucleosome DNA, indicating HMGN may also interact to a limited extent with linker DNA [96].
The HMGN family is made up of five different proteins: HMGN1, HMGN2, HMGN3, HMGN4, and HMGN5 [103–107]. HMGN1 and HMGN2 are expressed most ubiquitously, with HMGN3, HMGN4, and HMGN5 expression limited by tissue type or developmental stage [105, 106, 108–111]. Expression of HMGN3 is found primarily in the eye, brain, and pancreatic islet cells [105]. HMGN4 is expressed in most human tissues, but expression levels are highest in the thyroid gland, thymus, and lymph nodes [107]. This variant is also only found in primates, unlike the others which are found in all vertebrates [112]. The newest member of the family to be discovered, HMGN5, specifically interacts with the linker histone H1 and promotes its chromatin-compacting functions [113].
Similar to other HMGN proteins, HMGN1, an HSA21 triplicated gene, binds within the nucleosome core structure [114]. It has a molecular weight of approximately 10 kDa and contains three main features: a 30 amino acid nucleosome binding domain (NBD) at the N-terminus, nuclear location signals flanking the NBD, and a regulatory domain, i.e. chromatin-unfolding domain (CHUD), at the C-terminus [115]. Within the NBD, there is a sequence of amino acids that reads ‘RRSARLSA’, which confers the protein’s ability to bind to the nucleosome core particle and is a defining feature of the HMGN family [111]. In fact, HMGN proteins are the only non-histone proteins capable of binding between the DNA and the histone octamer core, basically within the nucleosome unit itself [94]. Specifically, HMGN1 binds in the major groove of the nucleosomal DNA near the nucleosome’s dyad axis [94, 103]. Furthermore, the binding of HMGN1 to nucleosomes is regulated post-translationally, occurring only in interphase and being completely abolished during mitosis when two serine residues located at position 20 and 24 in the NBD of HMGN1 are phosphorylated and negate the ability of HMGN to bind nucleosomes [116–118]. This exclusion of HMGN from the nucleosome during mitosis, when DNA is highly condensed, is in line with the proposed role of these proteins in promoting DNA accessibility for transcription and decompacting the chromatin [118].
Here we reviewed the general structure and function of different HMG family members and explained how HMGN1 binds to the nucleosome. Next, we will focus on the effect of HMGN1 on chromatin accessibility for transcription.
How can the binding of HMGN1 to nucleosomes affect transcription?
Two HMGN1 molecules bind cooperatively to the nucleosome core particle and form a homodimeric complex [119]. A majority of HMGN1 proteins are bound to nucleosomes at any given point, with a mean bind time of 4.1–24.8 s [120].
The first way HMGN1 can alter the structure of chromatin is by affecting linker histone H1’s interaction with the nucleosome since H1 promotes the compaction of chromatin [51, 103, 121]. There is evidence of overlap between the binding of HMGN1 and H1 near the nucleosome’s particle dyad axis [103]. H1 has a higher affinity for the nucleosome, and while quantities of HMGN1 are only sufficient to bind approximately 1% of the nucleosomes in a cell, the movement of HMGN proteins in the nucleus is significantly faster [122, 123]. These competitive dynamics have a direct effect on chromatin compaction, and while loss of HNGN1 enhances H1 binding to nucleosomes [124], increased levels of HMGN1 inhibit the binding of H1, decrease its residence time at nucleosomes [125], and impede its ability to effectively compact the chromatin architecture [51]. Another way HMGN1 antagonizes linker histone H1’s action is by restoring the electrostatic forces that lead to DNA repulsion and cause a more open chromatin conformation [51, 126].
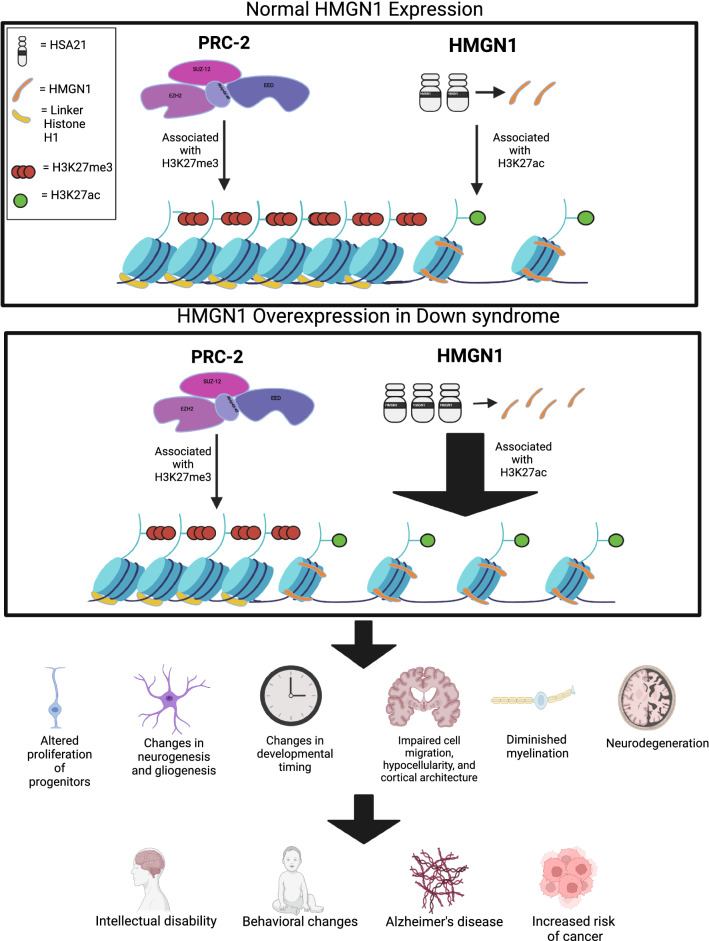
Recent studies demonstrate that the binding of HMGN1 to nucleosomes is further facilitated by the presence of an acetylation mark on H3K27 [123], which enables interactions with different transcription factors that affect gene expression. Zhang et al. [123] performed HMGN1 ChIP-seq analysis in ESCs and demonstrated that it preferentially localizes to nucleosomes containing H3K27ac marks, with no preference for other marks associated with increased transcription, such as H3K9ac and H3K4me1. Moreover, reducing acetylation levels of H3K27 with an acetyltransferase inhibitor resulted in the decreased abundance of HMGN on chromatin. Furthermore, the authors showed that the C-terminal regulatory domain of HMGN1 is not responsible for its preferential co-localization with H3K27ac and therefore cannot be responsible for H3K27ac recognition. This suggests that acetylation of H3K27 promotes nucleosome conformation that allows for easier binding of the HMGN proteins, as well as for their longer time spent bound to the nucleosome. Moreover, in vivo findings from this study using double-knockout (DKO) mice lacking both HMGN1 and HMGN2 revealed that the loss of HMGNs resulted in decreased H3K27ac marks [123]. This was accompanied by an increase in H3K27me3 marks and enhanced H1 occupancy, promoting a repressive chromatin state at the enhancer and promoter regions and interfering with the binding of transcription regulators. Furthermore, an additional study found enrichment in the occupancy of HMGN1 and HMGN2 in the areas of super-enhancers, defined as the areas containing a high density of H3K27ac and H3K4me1, in mouse ESCs, MEFs, and resting B cells [127]. In line with this, a very recent report [128] showed that HMGN1 and HMGN2 occupy compartment A (a transcriptionally active nuclear compartment) through their preferential binding to acetylated nucleosomes [128]. While HMGN1 depletion does not alter the higher-order chromatin structure, HMGNs are specifically localized to the cell-type-specific promoters and enhancers and promote transcription factor accessibility to these open chromatin areas, which form looping interactions, thus contributing to cell type-and stage-distinct gene expression [128].
In summary, HMGN1 competes with histone H1 for the same sites on the nucleosome [87, 125, 129] and preferentially associates with the H3K27ac marks of open chromatin that it helps generate. This allows the binding of transcription factors to enhancers and further leads to a more transcription-permissive conformation of chromatin. The binding of linker histone H1 to the nucleosome is itself transcriptionally repressive, but it also plays a role in the recruitment of Polycomb Repressive Complex 2 (PRC2), which imposes transcriptional silencing [124] as discussed in detail below.
The biological significance of the polycomb repressive complex 2 (PRC2)
PRC2: members, structure, and functions
Gene silencing mediated by PRC2 is believed to be based on the changes in chromatin structure, achieved through post-translational modification of histones. PRC2 is involved in transcriptional silencing through its methyltransferase (HMT) activity, catalyzing the mono-, di-, and tri-methylation of H3K27 [130, 131]. It has many roles specifically within the central nervous system, including effects on maturation [132], proliferation [133], and the identity of neural stem cells [134], migration and maturation of neurons [135], and gliogenesis [136, 137]. The proper function of PRC2 is required for the silencing of specific genes to allow the transcription of other genes that mediate alternative cellular processes.
PRC2 is comprised of three major subunits crucial for this catalytic activity: Embryonic ectoderm development (EED), Suppressor of zeste-12 (SUZ-12), either Enhancer of zeste 1 (EZH1) or EZH2, as well as the histone binding proteins RbAp46 and RbAp48 [18, 138]. EZH1 and EZH2 are proteins encoded by the mammalian homologs of the E(Z) gene originally identified in Drosophila and appear to have slightly different functions [139]. While both proteins are involved in transcriptional repression, EZH2 exerts its effect primarily as an HMT, whereas EZH1 appears to compact neighboring nucleosomal arrays [138]. The catalytic component of the PRC2 complex, EZH2, adds methyl groups to H3K27, as well as to H3K9, via its SET domain that possesses lysine-specific HMT activity [140]. Both EZH2 and EZH1 interact with the other major subunits of the PRC2 complex, SUZ-12, RbAp46/48, and EED to carry out their functions [138]. EED’s aromatic cage selectively binds to H3K27me3 marks, creating a positive feedback loop where binding to this mark propagates further binding by PRC2 and increases H3K27me3 [141, 142]. The C-terminal region of SUZ-12 is necessary for chromatin binding [143].
The setting of H3K27me3 marks by PRC2 leads to the recruitment of PRC1, an E3 ubiquitin ligase [144, 145] that is believed to silence gene transcription through the interference with RNA polymerase II activity via the mono-ubiquitination of H2AK119 [146, 147]. The crosstalk between PRC1 and PRC2 involves a positive feedback mechanism: PRC1-established H2AK119ub1 marks are recognized by PRC2, while H3K27me3 marks deposited by PRC2 recruit more PRC1 [148]. Recent studies showed that the catalytic activity of PRC1 is essential for the proper functioning of both complexes and gene silencing [149, 150].
The role of PRC2 in cell identity acquisition and establishment of neural lineages.
Beyond its global function in transcriptional repression, PRC2 plays specific roles in the process of establishing cellular identity and differentiation. Pereira et al. [134] demonstrated that PRC2 regulates a delicate balance between the self-renewal of cortical progenitor cells and neurogenesis. The researchers used mice carrying “floxed” alleles of EZH2 crossed with mice from an Emx1-Cre line, expressing Cre in the cortical pyramidal neurons [151]. Cre expression was induced starting at embryonic day 9.5 (E9.5) and resulted in the deletion of the EZH2 subunit in cortical progenitor cells. The EZH2-KO group showed increased cortical plate volume accompanied by a greater number of neurons, especially of layers V and VI, in the cortex at E14.
By the time of birth, however, EZH2-KO animals displayed a thinner cortical plate and fewer cells, as well as the compromised generation of superficial layers’ neurons (II-IV). Using Bromodeoxyuridine (BrdU) pulse labeling of the cortical progenitor cells in vivo at E13, the investigators detected a shift towards the generation of neurons, as well as basal progenitor cells (representing direct and indirect neurogenesis, respectively) at the expense of the self-renewal of cortical progenitor cells that led to premature exhaustion of their pool in EZH2-KO group, indicating that neurogenesis is temporally regulated by PRC2.
Similar dynamics were observed when another PRC2 subunit, EED, was deleted from NPCs populating the hippocampal dental gyrus (DG) and the subventricular zone (SVZ) at E13.5 [152]. This was achieved by crossing transgenic EED “floxed” mice with mice expressing Cre under the GFAP promoter. Similarly to the study performed by Pereira et al. [134], there was an initial increase in the number of postmitotic neurons in DG (at P7) followed by a drastic decrease in the numbers at P14 in EED-KO animals. This was accompanied by the reduced proliferation capacity of NPCs, suggesting once again that the ablation of the components of PRC2 leads to an impaired pattern of NPC renewal and aberrant neurogenesis.
PRC2 also regulates the acquisition and maintenance of an intact neuronal identity. Conditional deletion of the EED subunit in postmitotic dopaminergic (midbrain DA) and serotonergic (hindbrain 5HT) neurons in transgenic mice led to the diminished transcription of subtype-specific genes and derepression of non-mDA and non-5HT genes, including those coding for transcription factors involved in autoregulatory and fate-determining functions, as well as death-promoting genes [153]. These observations were similar to the finding in medium spiny neurons (MSNs) and Purkinje cells demonstrating that PRC2 activity is essential for shutting down the transcription of the death-promoting genes [154]. EED ablation also resulted in functional deficits manifested in the altered electrophysiological properties of the neurons, abnormal production of cell-specific metabolites, and pathological behavioral phenotypes in mice [153]. In agreement with this, conditional EZH2 inactivation via Cre-inducible deletion of the SET domain in glutamatergic neurons in vitro led to their altered differentiation trajectory and the switch towards a transcriptomic signature associated with GABAergic neurons [132]. The enhancement in GABAergic signature and the overproduction of interneurons following EZH2 ablation was also shown in mouse cerebellum [155], further implicating PRC2 in the establishment and maintenance of neuronal identity, as well as the balance between the different neuronal populations.
The proteins of the PRC2 complex also facilitate the appropriate migration of post-mitotic neurons in the cortex during development. The loss of EZH2 induced through intra-utero electroporation (IUE) of an EZH2-specific short hairpin RNA (shRNA) into mouse E14.5 neocortex resulted in abnormal neuronal orientation during the radial migration, with an increase in neurons in the intermediate zone (IZ) and a reduction in those that reached the upper cortical plate (CP) [135]. These findings were further attributed to the ectopically activated expression of Reelin in the migrating neurons, suggesting that silencing of Reelin in migrating post-mitotic neurons by EZH2 is essential for the proper cortical lamination [135]. Similarly, the ectopic expression of Netrin 1, due to the loss of EZH2, results in the abnormal tangential migration of precerebellar neurons in the mouse hindbrain, supporting further the essential role of PRC2-induced gene silencing in neuronal motility [156].
Deviant PRC2 expression can also affect the timing of gliogenesis, and the type of glial cell produced. The onset of gliogenesis in the developing neocortex is tightly regulated and commences at the end of neurogenesis with the production of immature astrocytes [136, 137], followed by the generation of oligodendrocyte precursor cells (OPCs) [157, 158]. Disruption in the timing of gliogenesis in PRC2 mutant mice was demonstrated by Pereira et al. [134] when they found precocious astrocyte development upon EZH2 KO, with GFAP-expressing glial cells found earlier in the cortical plate of an EZH2 KO group of mice as compared to controls. Overexpression of EZH2 in mouse embryonic neural stem cells (NSCs), on the other hand, led to a reduction in astrogenesis and an increase in oligodendrocytes [159]. The opposite was seen when the same group used shRNA to reduce EZH2 expression in mouse embryonic NSCs, with an increase in the percentage of astrocytes and a decrease in oligodendrocytes. Taken together, it is apparent that PRC2 is necessary for proper gliogenesis and balance between astrocyte and oligodendrocyte development.
An increasing body of evidence indicates that oligodendrocyte lineage development is particularly tightly regulated by PRC2, with studies highlighting the necessity of PRC2-mediated repression at different developmental stages ranging from oligodendrocyte lineage commitment to differentiation and maturation. Wang et al. [160] showed that the members of this complex are involved in the switch from the OPC to the differentiated oligodendrocyte stage. This group performed a conditional KO in transgenic mice carrying a floxed EZH2 allele using an Oligodendrocyte Transcription Factor 2 (Olig2)-Cre driver. Olig2 transcription factor governs pMN progenitor domains of the ventral spinal cord that give rise to motor neurons during neurogenesis followed by the production of oligodendrocytes and a subset of astrocytes during gliogenesis [161]. The investigators abolished EZH2 expression on day E12.5, which resulted in the absence of the H3K27me3 mark in the pMN domain at later time points [160]. No changes were observed in the OPCs produced in the absence of EZH2, indicating that this member of PRC2 is not essential for the generation of OPCs. However, a reduction in the number of myelin basic protein (MBP)-expressing oligodendrocytes by E18.5, suggests that the process of differentiation into mature OLs is delayed when EZH2 is depleted.
Similar results were obtained following a conditional KO of the EED subunit (acting in concert with both EZH1 and EZH2) in Olig2-expressing cells. Again, no change was observed in the generation of OPCs, but the number of MBP-positive oligodendrocytes was reduced, and EED-KO mice exhibited deficient myelination-associated phenotypes, including aberrant motility, seizures, and tremors, and died by P17. Noticeably, increased production of astrocytes was detected in EED KO Olig2-expressing cells, supporting the previous notion that the proteins of the PRC2 regulate the balance between the oligodendrocytes and astrocytes. The orchestrating role of the PRC2 members in the differentiation and maturation of oligodendrocyte lineage was attributed to the repressive control performed by PRC2 on the activation of Notch and Wnt signaling. This study highlights the crucial role of PRC2 in silencing the alternative pathways and allowing the expression of genes leading to functionally mature oligodendrocytes.
High levels of EZH2 are found in proliferating mouse NPCs, followed by decreased expression during the differentiation to neurons and astrocytes in vitro [159]. However, the expression of EZH2 remains high in NPCs transitioning into OPCs and immature oligodendrocytes. EZH2 overexpression in NPCs resulted in increased oligodendrocyte production, while silencing of EZH2 using shRNA led to the enhanced production of neurons and astrocytes.
Douvaras et al. [162] used human ESCs and induced pluripotent stem cell (iPSC) lines to decipher the epigenetic changes that take place during the differentiation of NSCs into OPCs and then oligodendrocytes. The authors detected the enhanced expression of EZH2 and EED mRNAs during the NSC-to-OPC transition and the pre-OPC stages suggesting that the members of PRC2 are involved in the early commitment of NPCs to the OL lineage. This contrasts with the study performed by Wang et al. [160] highlighting the requirement for the EZH2 activity at later stages of oligodendrocyte development, but still implicates PRC2 in this process. Importantly, ESCs showed consistently increasing H3K27me3 levels during the transition from the NSC stage all the way to immature oligodendrocytes, reflecting again the importance of this mark in oligodendrocyte differentiation. Interestingly, EZH1 was most highly expressed in immature oligodendrocytes, suggesting EZH1’s role in gene silencing once oligodendrocyte commitment has occurred. Taken together, this data supports the regulatory role of PRC2 in oligodendrogenesis, oligodendrocyte differentiation, and maturation.
The only study connecting PRC2-mediated control of oligodendrocytes differentiation and HMGN1, the main focus of this review, was performed by Deng et al. [124]. This work showed that the interplay between HMGN1 and PRC2 impacts Olig1 and Olig2 expression. ChIP-qPCR analysis revealed an increase in the levels of H3K27me3, as well as the EZH2 subunit of the PRC2, in the genomic locus containing Olig1 and Olig2 in HMGN−/− ESCs. Further, they showed an increase in Olig1 and Olig2 expression in vitro in ESCs that were treated with an EZH2 inhibitor, GSK126. The authors postulated that the absence of HMGN1 increased the ability of linker histone H1 to bind to the nucleosome, promoting deposition of the transcriptionally repressive mark H3K27me3 by PRC2 and reducing the expression of Olig1 and Olig2 [124]. This is an interesting finding since Olig1, Olig2, and HMGN1 are all triplicated in DS. It raises the question of how increased levels of HMGN1 further impact the levels of Olig1 and Olig2 and influence oligodendrocyte development in trisomy.
In this section, we reviewed literature reflecting the crucial role of PRC2 and its different subunits in various neurodevelopmental processes. Research to date demonstrates that PRC2 activity is essential for the maintenance of balance between the self-renewal of the cortical progenitors and their differentiation into neurons, establishment of cell identity and neuronal migration, as well as for the timely transition from neurogenesis to gliogenesis and proper differentiation and maturation of oligodendrocytes. We summarize the developmental effects of PRC2 in Table 1.
Table 1.
Developmental effects of PRC2
| Developmental effect | Related findings | References |
|---|---|---|
| Proper timing of neurogenesis | EZH2-KO in mice leads to premature exhaustion of the pool of neural progenitor cells, decreased neuronal density at the cortical plate, and precocious astrocyte generation | Pereira et al. [134] |
| EED-KO mice experience impaired neurogenesis, growth retardation, and death. EED ablation leads to abnormal neuronal differentiation during the hippocampal dentate gyrus formation | Liu et al. [152] | |
| miR-203 is repressed by EZH2 in neural progenitor cells and negatively regulates proliferation. EZH2-KO results in the reduction of neural progenitor proliferation | Liu et al. [166] | |
| Appropriate neuronal orientation and cortical radial migration | EZH2 inhibition results in abnormal neuronal orientation reduced neuronal numbers at the cortical plate and ectopic expression of Reelin | Zhao et al. [135] |
| EZH2-mediated repression of Netrin1 is necessary for the appropriate migration of pontine neurons in the cortico-ponto-cerebellar pathway in mice. EZH2-KO results in ectopic Netrin1 induction and aberrant neuronal migration | Di Meglio et al. [156] | |
| Maintenance of neuronal identity | Deletion of EED disrupts the acquisition and maintenance of neuronal identity and functionality in differentiated dopaminergic and serotonergic neurons | Toskas et al. [153] |
| Conditional knock-out of EZH2 results in reduced proliferation of granule precursor cells, decrease in the Purkinje cell population and increase in GABAergic interneurons in the mouse cerebellum | Feng et al. [155] | |
| Conditional KO of EZH2 results in loss of H3K27me3 marks in differentiating neurons and causes changes to molecular networks that govern glutamatergic neuron differentiation, leading to a disruption in the balance of inhibitory/excitatory neurons during the development | Buontempo et al. [132] | |
| Combined EZH1 and EZH2 KO leads to a loss of H3K27me3 marks in MSNs in the striatum, down-regulation of lineage-specific and function-specific MSN genes, and upregulation of the death-promoting genes | Von Schimmelmann et al. [154] | |
| Glial cell development and fate determination | Pharmacological inhibition of EZH2 leads to increased expression of Olig1 and Olig2. Increased depositions of H3K27me3 marks are detected in the genomic loci of Olig1 and Olig2 in HMGN-KO ESCs | Deng et al. [124] |
| KO of EZH2 or EED do not affect the generation of OPC but inhibit their differentiation into mature, myelinating oligodendrocytes. PRC2 is necessary for the repression of the Notch pathway | Wang et al. [160] | |
| EZH2 regulates NSC differentiation into glial cells in mice, with high expression levels of EZH2 associated with increased oligodendrocyte production and decreased production of astrocytes while low levels of EZH2 correlate with a reduction in oligodendrocyte generation and increased numbers of astrocytes | Sher et al. [159] | |
| Increased levels of EZH2 and EED mRNAs are detected during the early stage of OPC lineage commitment and development in mouse and human ESCs and human iPSCs | Douvaras et al. [162] |
Summary of studies demonstrating the role of PRC2 in neurodevelopment
Bivalent marks and their significance in neural development
As was discussed earlier, genes activated during differentiation are marked by both “silencing”, H3K27me3, and “activating”, H3K4me3 marks in undifferentiated ESCs. Given the critical role of PRC2 in NPC differentiation, it is important to review recent insights into bivalent chromatin dynamics during this process. Importantly, bivalency is not limited to ESCs and is present in other progenitor populations defined as multipotent, such as NPCs and glial progenitors [144, 163–165]. Liu et al. [166] compared genome-wide RNA Polymerase II (Pol II), H3K4me3, and H3K27me3 landscapes between purified mouse NPCs and neurons. Remarkably, the authors found that bivalently marked genes in NPCs included those critical for cortical neuron differentiation, migration, and function. These genes acquired more prominent H3K4me3 marking in neurons, consistent with their higher expression. At the same time, genes known to express highly in OPCs, such as Olig1, Olig2, and platelet-derived growth factor receptor alpha (Pdgfr-α) were also bivalently marked in NPCs but got enriched in H3K27me3 in neurons, consistent with their repression in this lineage. The interplay between HMGN1 and PRC2 in Olig1 and Olig2 regulation shown in ESCs and discussed above, and data from the animal-extracted cells described by Liu et al. [166], strongly suggest HMGN1’s involvement in the regulation of bivalent genes during NPC differentiation in vivo.
Another important study by Yu et al. used a novel approach to define bivalent genes based on a quantitative assessment of chromatin compaction [167]. The authors purified and sequenced DNA associated with mono-nucleosomes after the treatment of chromatin with Micrococcal nuclease (MNase). They used the “time-course digestion with reduced MNase levels followed by high-throughput sequencing” (TC-rMNase-seq) approach for separately sequencing DNA fragments originating from the open chromatin regions and those coming from more compacted areas. In this manner, they defined a specific moderately compacted chromatin state, which was associated with bivalent genes present in mouse ESCs. Moreover, the authors noted several genes important for neurogenesis among those in the moderately compacted group and followed chromatin changes at these loci upon mESC differentiation into NPCs. Indeed, promoters of these genes showed higher susceptibility to MNase digest in NPCs compared to mESCs in the TC-rMNase-seq assay, consistent with their higher expression in NPCs and accumulation of higher levels of H3K4me3.
The new insight connecting moderate chromatin compaction to bivalency and its resolution during differentiation also supports the proposed role of HMGN1 in opposing PRC2 via its chromatin-opening function.
PRC2 and neurodegeneration
PRC2 deficiency has been linked to neurodegeneration in several studies [168–170]. A study performed by von Schimmelmann et al. [154], determined that PRC2 inhibition in adult neurons in vivo leads to an upregulation of death-promoting genes in MSNs in the striatum. PRC2 inactivation was achieved by the combination of a null mutation in EZH1 and the conditional ablation of the EZH2 gene in the Calcium/Calmodulin Dependent Protein Kinase II Alpha (Camk2a)-expressing neurons marking MSNs. Most of the upregulated genes detected at three and 6 months post-PRC2 ablation were identified as H3K27me3 targets, suggesting a specific regulatory role of PRC2 in adult neurons. Interestingly, the investigators pointed out redundancy in the activity of EZH1 and EZH2 in adult postmitotic MSNs, while previous studies showed that EZH1 is incompetent to substitute for the lost activity of EZH2 in dividing cells during neurodevelopment [138]. By using conditional ablation of the PRC2 members in adult neurons, the researchers deduced that PRC2 deficiency led to the downregulation of MSN-specific genes and the up-regulation of death-promoting genes previously identified as PRC2 target genes, such as Phorbol-12-Myristate-13-Acetate-Induced Protein 1 (Pmaip1), BH3 Interacting Domain Death Agonist (BID), NADPH Oxidase Activator 1 (Noxa), Cyclin-dependent kinase inhibitor 2A/B (Cdkn2a/b), and Insulin-like growth factor-binding protein 3 (Igfbp3). This was accompanied by the phenotypic signs of neurodegeneration manifested in the elevation of gamma-H2Ax, a marker of DNA damage, cytoplasmic and nuclear condensation, a decrease in the number of MSNs, and a reduction in the total brain mass.
Furthermore, altered PRC2 activity in MSNs manifested in neurodegenerative phenotypes and behavior changes, as mutant mice showed impairments in rotarod performance, difficulty in hanging on a wire top, and altered hind limb clasping followed by the termination of all voluntary eating and drinking and death at 7 months. The same approach was taken to abolish the activity of PRC2 in Purkinje cells (PC) in the cerebellum. The loss of PRC2 in PC resulted in the upregulation of death-promoting PRC2 target genes and progressive neurodegenerative phenotypes accompanied by abnormal motor behavior phenotypes [154]. Notably, the authors concluded that PRC2 silencing of the bivalent genes in particular is essential for the maintenance of cell identity in adult neurons. The upregulation of the pro-apoptotic genes was also detected in differentiated serotonergic and dopaminergic neurons upon EED deletion but did not result in the induction of cell death [153]. Altogether, these studies strongly implicate PRC2 activity in the repression of neurodegeneration-related pathways thus ensuring the survival of adult neurons [153, 168–170].
Implications of HMGN1 gene dosage effect on PRC2-mediated gene silencing in DS
The first possible link between DS and PRC2 was established by a group studying B-cell acute lymphoblastic leukemia (B-ALL) [171]. Individuals with DS are at a 20-fold risk for developing B-ALL compared to the general population [172] and 60% of B-ALL cases in DS harbor the rearrangement of cytokine receptor-like factor 2 (CRLF2) further classifying it as DS-ALL [173]. Lane et al. [171], aimed to determine what confers this increased risk using Ts1Rhr mice harboring a triplicated fragment of the mouse chromosome 16 (Mmu16) orthologous to a human chr.21q22 segment that includes HMGN1 among other 31 genes on this fragment. Ts1Rhr mice consistently developed B-ALL, and the progenitor B cells derived from the bone marrow of these mice showed an enhanced formation of colonies and increased rate of cell renewal as compared to WT, supporting a transformed phenotype of these cells.
Comparison of the transcriptomic signature of Ts1Rhr mice against human DS datasets from different cohorts done through the network enrichment analysis identified the PRC2 target genes and the H3K27me3 sites as the most highly enriched. Furthermore, differential expression of PRC2 and H3K27me3-target genes was sufficient to differentiate between DS-ALLs and other B-ALLs. Importantly, genes defined as PRC2 targets were all found to be upregulated in DS-ALL, suggesting that increased transcription and reduced silencing of PRC2 targets in DS-ALL were the root cause of these differences. Further support for this assertion was provided by mass spectrometry of H3 showing that the Ts1Rhr B-cells had a reduction in H3K27me3 peptides, and ChIP analysis revealing a global reduction in H3K27me3 marks. This data obtained from the Ts1Rhr mice provided evidence that the triplication of 31 genes orthologous to the human DS-critical region was sufficient to reduce H3K27me3 occupancy.
Remarkably, overexpression of HMGN1 alone in vivo was sufficient to reproduce many of the transcriptional and phenotypical features seen in the Ts1Rhr B cells with all 31 genes triplicated. These results support the crucial role of HMGN1 in the de-repression of the PRC2 target genes in DS. HMGN1 overexpression was further implicated in transcriptomic changes and histone modifications related to DS-ALLs in a subsequent study [174] that revealed a global increase in RNA transcripts produced per gene in pro-B cells from the Ts1Rhr cells as compared to the WT ones. Using transgenic mice overexpressing HMGN1 (HMGN1-OE) the investigators showed a genome-wide transcriptional amplification, accompanied by the global increase in H3K27ac marks in HMGN1-OE cells, similar to the Ts1Rhr cells. This supports the idea that HMGN1 is responsible for a global increase in gene expression in DS. Further gene set enrichment analysis showed that genes with enhanced expression induced by HMGN1-OE in B cell progenitors were enriched for EZH2 targets.
Remarkably, using B cell lines carrying a doxycycline-inducible HMGN1 cassette with either WT or mutated nucleosome-binding domain showed that the binding of HMGN1 to the nucleosome was essential for the enhancement of gene expression [174]. Moreover, genes showing a greater increase in expression following the induction of HMGN1-OE also displayed the enrichment in H3K27ac at the super-enhancer regulatory regions. Finally, normalizing the expression of only the HMGN1 gene in the Ts1Rhr mouse DS model rescued B cell pathological phenotypes, abolished an increase in H3K27ac marks, and mitigated mRNA expression changes, indicating that the increased dosage of HMGN1 is sufficient to induce DS-specific transcriptomic and epigenetic signatures and cellular phenotypes in B cells.
The most recent study applied HMGN1 CRISPR/Cas9 KO to the SET2 cell line edited to harbor the rearrangement of CRLF2 observed in DS-ALL patients and to the DS-ALL xenograft mouse model [175]. The authors showed that KO of HMGN1 abrogated the abnormal proliferation of the SET2 cells in vitro as well as enhanced survival of the DS-ALL xenograft model through mitigation of its disease phenotypes. Using the complementary approach of HMGN1-OE, the study concluded that HMGN1 facilitates leukemic transformation to DS-ALL due to its ability to generate a positive feedback mechanism resulting in the upregulated transcription of CRLF2 and dysregulation of the downstream signaling pathways and highlighted the crucial role of HMGN1 in DS-specific ALL.
Further support for the global genomic and epigenetic disruption in trisomy comes from a recent study conducted by Meharena et al. [176]. The authors found evidence for the disorganization in the nuclear architecture and alterations in the transcriptomic signature of human NPCs derived from DS iPSC lines compared to isogenic controls. They demonstrated an increase in H3K27ac and a decrease in H3K27me3 marks in trisomic NPCs, but not in trisomic iPSCs, which suggests developmental and stage-specific alterations of these marks in trisomy. Furthermore, transposase-accessible chromatin sequencing (ATAC-seq) showed a significant increase in differentially accessible regions caused by trisomy in NPCs compared to iPSCs (20% versus 1.6%, respectively). Consistent with this, the transcriptomic profile of trisomic NPCs showed a decrease in EZH2 and EED mRNA, as well as some histone acetyltransferases, further supporting the proposed diminished activity of PRC2 in DS, possibly due to increased dosage of HMGN1. Remarkably, transcriptional profiling of trisomic NPCs revealed senescence-associated signatures as well as upregulation of genes related to cell migration, adhesion, and inflammation. This suggests the disturbance of distinct signaling pathways due to enhanced transcription of specific genes in DS can be potentially attributed to the gene dosage effect of HMGN1. The concept that increased gene dosage of HMGN1 is necessary and sufficient for the induction of DS-specific ALL suggests the possibility that HMGN1 triplication can be detrimental in other DS-related phenotypes observed in different systems. The existing studies addressing the triplication of HMGN1 in DS are summarized in Table 2.
Table 2.
HMGN1 and DS
| DS Phenotype | Title | Related findings | References |
|---|---|---|---|
| Overexpression of HMGN1 | Chromosomal protein HMG-14 gene maps to the Down syndrome region of human chromosome 21 and is overexpressed in mouse trisomy 16 | Ts16 mouse model of DS has 1.5 times more HMGN1 protein and mRNA than WT and supports the gene dosage effect of HMGN1 triplication | Pash et al. [192] |
| Chromosomal protein HMG-14 is overexpressed in Down syndrome | Fibroblasts from DS individuals express HMGN1 at levels 1.6 higher than non-DS individuals | Pash et al. [193] | |
| Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks | HMGN1 is upregulated in the Ts1Cje mouse model of DS | Ling et al. [194] | |
| Transcriptional disruptions in Down syndrome: a case study in the Ts1Cje mouse cerebellum during post-natal development | HMGN1 was overexpressed in the cerebellum of the Ts1Cje mouse model of DS during post-natal development | Potier et al. [195] | |
| Bioinformatics analysis of biomarkers and transcriptional factor motifs in Down syndrome | Raw gene expression data from DS rat brain tissue, Ts1Cje cerebellum tissue, and adult human DS tissue analyzed using Gene Expression Omnibus and showed overexpression of HMGN1 | Kong et al. [201] | |
| Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination | Raw gene expression data analyzed in humans with DS using Gene Expression Omnibus and found HMGN1 overexpressed in the hippocampus, cerebellar cortex, and areas of the pre-frontal cortex, primary visual cortex, | Rodriguez-Ortiz et al. [203] | |
| Intellectual disability | Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium | A genome-wide association study of general cognitive function in adults found HMGN1 as the single gene-based significant association in the study | Davies et al. [288] |
| Cancer | Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation |
HMGN1 suppresses H3K27me3 and promotes B cell proliferation in B-ALL |
Lane et al. [171] |
| Trisomy of a down syndrome critical region globally amplifies transcription via HMGN1 overexpression | HMGN1 globally amplifies transcription | Mowery et al. [174] | |
| HMGN1 plays a significant role in CRLF2-driven Down Syndrome leukemia and provides a potential therapeutic target in this high-risk cohort | HMGN1 is involved in signaling pathways in CRLF2-driven DS leukemia | Page et al. [175] | |
| Behavioral changes | The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice |
HMGN1 downregulates MeCP2 and its aberrant expression produces behavioral changes in mice consistent with ASD and DS phenotypes |
Abuhatzira et al. [180] |
Summary of studies associating HMGN1 expression and changes found in DS
The gene regulatory function of HMGN1: how specific is it to neural development?
While HMGN proteins exert a global effect on chromatin compaction, an increasing body of evidence suggests that HMGN1 expression is tightly involved in developmental regulation. Since this review is targeting the potential role of HMGN1 in DS brain pathology, in this section we will be focusing on the role of HMGN1 in CNS development, which we also summarize in Table 3.
Table 3.
Role of HMGN1 in CNS Development
| Title | Major findings | References |
|---|---|---|
| Developmental role of HMGN proteins in Xenopus laevis | Altered HMGN1 levels lead to malformations in Xenopus laevis development at the post-blastula stage | Körner et al. [178] |
| High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse | HMGN1 protein is necessary for the appropriate timing of embryo development in mice; depletion leads to developmental delays | Mohamed et al. [177] |
| Binding of HMGN proteins to cell specific enhancers stabilizes cell identity | Loss of HMGN1 protein accelerates reprogramming of MEFs into iPSCs | He et al. [127] |
| High mobility group nucleosome-binding family proteins promote astrocyte differentiation of neural precursor cells | HMGN1 expression promotes astrocyte differentiation | Nagao et al. [179] |
| HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells | Loss of HMGN1 reduces the number of Nestin-positive NPCs in SVZ in mouse brain | Deng et al. [129] |
| Interplay between H1 and HMGN epigenetically regulates OLIG1 and 2 expression and oligodendrocyte differentiation |
Loss of HMGN1 reduces OLIG1 and OLIG2 expression and impairs normal oligodendrocyte differentiation Loss of HMGN1 decreases the amount of MBP and proteolipid protein (PLP) in the spinal cord of mice |
Deng et al. [124] |
Summary of studies demonstrating the importance of HMGN1 in CNS development
Early embryonic development in mice is disrupted as early as the blastocyst stage by depleted levels of HMGN1 achieved via injection of antisense oligonucleotides [177]. Studies in post-blastula X. laevis embryos exposed to microinjection of the HMGN1 recombinant protein revealed that increased HMGN1 levels resulted in profound developmental abnormalities, such as impaired blastopore closure and an inappropriate body axis establishment accompanied by aberrant head structures [178]. HMGN1 is expressed in the cells of neural lineage: it is detected as early as E12.5 and is highly expressed in the developing E18.5 mouse neocortex, in forebrain VZ and SVZ and cortical plate as well as in the postnatal NPCs and glial progenitors (P7) and some expression of HMGN1 persists in adult brains in astrocytes and to the lesser extent in postmitotic neurons [179]. Deng et al. [129] investigated the particular role of HMGN1 in neural development using ESC derived from the HMGN1-KO transgenic mouse model. In vitro differentiation of ESCs into dopaminergic neurons did not result in noticeable phenotypic alterations but was accompanied by a transcriptomic dysregulation of genes involved in organ morphogenesis, vasculature development, and tissue development. This was detected at the stages of ESCs, neural progenitor cells (NPCs), and differentiated neurons. The same study showed that in vivo HMGN1 is strongly expressed in the SVZ of two weeks old wild type mice and is co-expressed with Nestin, a marker of NPCs. HMGN1-KO mice, however, displayed a decrease in the number of Nestin-positive cells and Nestin mRNA expression in the SVZ compared to the wild type, suggesting the regulatory role of HMGN1 in the maintenance of the NPC’s pool and further neocortical development. This study also demonstrated that in ESCs and NPCs the HMGN1 protein preferentially binds to TSS of active promoters containing CpG islands and regulates the stability and positioning of nucleosomes in these regions, further supporting the role of HMGN1 in transcriptional gene regulation during development.
A more recent study [127] demonstrated that HMGNs stabilize the epigenetic landscape in specific cell types and allow for the proper establishment and maintenance of cell identity through their clustering at the areas of super-enhancers. The researchers utilized MEFs, ESCs, and neurons derived from HMGN1 and HMGN2 DKO mice and observed enhanced efficiency of pluripotency reprogramming during the process of transformation of MEF cells into iPSCs using Yamanaka factors. They also found that further differentiation of ESCs into neurons was accelerated in the absence of both HMGN1 and HMGN2. This suggests that their loss results in reduced accessibility of enhancer regions to the transcription factors maintaining cell type specificity, while their presence stabilizes cell identity. HMGN1 not only regulates the differentiation of NPCs into neurons but is also involved in the switch from neurogenesis to gliogenesis and the generation of astrocytes and oligodendrocytes. HMGN1 overexpression in mouse forebrain NPCs in vitro resulted in enhanced production of astrocytes and decreased production of neurons, but no change in the generation of oligodendrocytes [179]. Conversely, the knock-down of HMGN1 expression resulted in a decrease in the production of astrocytes and an increase in the fraction of neurons. Similar results were observed in vivo when HMGN1 was overexpressed through IUE into the fetal mouse neocortical NPCs. This led to the greater numbers of astrocytes accompanied by a decreased generation of superficial layer neurons at P7 suggesting that HMGN1 regulates the generation of astrocytes in vitro and at perinatal stages in vivo. Remarkably, the knockdown of the HMGN1 expression postnatally (at P0), decreased the fraction of cortical astrocytes at P7 and led to the over-production of the immature neurons. Together, these findings indicate that HMGN1 plays a role in astrocyte differentiation prenatally as well as postnatally in vivo.
HMGN1 has also been implicated in oligodendrocyte development. Deng et al. [124] differentiated ESCs derived from HMGN1 and HMGN2 DKO mice into embryoid bodies (EBs) and towards the oligodendrocyte lineage and observed a decrease in the expression of Olig1 and Olig2 transcription factors throughout the process. A decrease in Olig2-positive cells was also shown in vivo in the spinal cord of the DKO mice and was accompanied by the decreased expression of oligodendrocyte lineage markers across the differentiation stages. Specifically, PDGFR-α was downregulated in the OPCs, and 2ʹ,3ʹ-cyclic-nucleotide 3ʹ-phosphodiesterase (CNPase), proteolipid protein (PLP), and MBP were downregulated in the more mature oligodendrocytes. These results implicate the HMGN proteins in the generation, differentiation, and maturation of oligodendrocytes.
Taken together, the existing studies strongly support the regulatory role of HMGN1 in the CNS starting from the early embryonic development and continuing through the later stages of CNS development and cell maturation. HMGN1 has been shown to stabilize the cell identity of the neural lineages and to play an important role in the maintenance of the pool of NPCs and controlling their differentiation into neurons, astrocytes, and oligodendrocytes.
HMGN1 and changes in behavior
HMGN1 dysregulation may lead to various developmental abnormalities beyond DS through its global epigenetic regulatory function and subsequent downstream effects on the expression of other genes. Abuhatzira et al. [180] showed for the first time that altered HMGN1 levels in mice lead to behavioral changes and autism-related features. This group also showed that HMGN1 controls the expression of methyl CpG-binding protein 2 (MeCP2), dysregulation of which has been linked to the behavioral patterns associated with autism spectrum disorder (ASD) [181]. Abnormal MeCP2 expression is correlated with cognitive disabilities [181], and transgenic mice expressing 50% reduced levels of MeCP2 demonstrated significant impairments in learning and memory tasks [182].
MeCP2 is thought to participate in transcriptional repression due to its preferential association with methylated DNA [183] as well as with histones carrying the H3K27me3 mark via its methylation binding domain (MBD) [184–187]. Similar to the action of the linker histone H1, MeCP2 can induce compaction of the chromatin fiber and promote transcriptional repression [184]. Reducing H3K27me3 with GSK23, an EZH2-specific inhibitor, results in a decrease in MeCP2 levels on chromatin, as determined by ChIP-seq analysis, indicating that the recruitment of MeCP2 to genomic loci is dependent on H3K27me3 [186]. This is further supported by the co-localization of MeCP2 and H3K27me3 at TSS [183, 186] resulting in a cooperative regulation of transcriptional repression.
Abuhatzira et al. [180] showed that HMGN1 binds to the promoter and the first exon of MeCP2 in the human and mouse brain cells and regulates its expression. In line with this, in DS human brains, the 50% increase in transcript levels of HMGN1 is inversely correlated with MeCP2 transcript levels, which are decreased by 30%. This group also showed that overexpression of HMGN1 in vivo or in vitro using MEFs from HMGN1-KO mice transfected with an HMGN1-OE vector resulted in a decrease in mRNA and protein levels of MeCP2, while KO of HMGN1 led to a significant increase in the mRNA and protein levels of MeCP2 [180]. The most profound findings of this work led to the discovery that changes in the level of HMGN1 can lead to autism-related behaviors. Through the battery of behavioral tests to evaluate social memory and preference, the study showed that both groups with either upregulation or downregulation of HMGN1 demonstrated a lack of preference in a novel social interaction and novel object recognition, behaviors linked to autistic-like phenotypes [188] and DS-associated cognitive deficits [189]. Notably, this work did not show that the behavioral changes induced by misexpression of HMGN1 were mediated by direct changes to MeCP2 but instead recognized the global effect of HMGN1 expression on chromatin accessibility. It is possible, however, that reduced MeCP2 expression due to the high levels of HMGN1 in trisomy can potentially promote further decompaction of the chromatin.
To the best of our knowledge, the study described above is the only investigation directly showing that altered levels of HMGN1 are causatively linked to the behavioral features associated with the autistic and DS-related phenotypes.
HMGN1 expression in mouse and human DS brain tissue
DS is caused by the presence of all or part of an extra copy of chromosome 21 [190], and the region of q22.1 to q22.3 is considered to be critical for DS [188, 191]. The HMGN1 gene was mapped to a 21q22.3 region of human chromosome 21 by Pash et al. [192, 193]. The gene dosage effect of HMGN1 was observed in several mouse models of DS. The Ts16 mouse model (harboring the triplication of Mmu16) shows a 1.6- to 3.3-fold increase in the HMGN1 mRNA in the brain and whole trunk of Ts16 fetuses, respectively [192]. The HMGN1 mRNA was found to be increased in the cortex, hippocampus, and cerebellum of the Ts1Cje mice [194, 195], representing another mouse model of DS that contains a partial triplication of Mmu16 [196], and in the embryonic brain tissue of Ts65Dn [197], the most widely used mouse model of DS, containing an extra-small chromosome resulting from a fusion of the region of Mmu16 orthologous to Hsa21 with the centromeric region of Mmu17 as well as the additional extra segment of non-DS-related genes [198]. Increased hippocampal HMGN1 mRNA expression in adult mice has also been confirmed in the Dp1Tyb mouse model that contains an additional copy of 63% of mouse genes orthologous to the HSA21 genes [199]. The HMGN1 mRNA levels are also increased in the forebrain tissue at postnatal day 1 in a novel non-mosaic DS model, TcMAC21, which incorporates 93% of HSA21q protein coding genes as a segregated chromosome [200]. The correlation between the gene dosage imbalance and the HMGN1 expression upregulation across different DS mouse models is of extreme importance since a large part of our understanding of DS pathology and the underlying molecular mechanisms are based on the high utility of mouse models of trisomy 21.
Importantly, the bioinformatic analysis of the data obtained through Gene Expression Omnibus (GEO) concluded that HMGN1 is at least 1.5-fold upregulated in the human trisomic tissue in an orthologous comparison to the mouse trisomic tissue [201]. Human cultured DS fibroblasts also exhibit a 1.6-fold increase in HMGN1 mRNA expression, and increased protein levels of HMGN1 are detected in human embryonic DS brains compared to control samples [192, 193]. In addition, HMGN1 mRNA expression is upregulated in the cerebellum and dorsal prefrontal cortex across multiple developmental periods in DS human postmortem brain tissue compared to controls [202].
Rodriguez-Ortiz et al. [203] conducted a study to determine the distribution of HMGN1 expression in the brain of individuals with DS. The authors accessed raw gene expression data collected from individuals with and without DS from GEO, focusing on a microarray experiment that compared gene expression in multiple brain areas. They found that HMGN1 is overexpressed in DS brains compared to control in the hippocampus, cerebellar cortex, and primary visual cortex, as well as areas of the prefrontal cortex (PFC), including the dorso- and ventrolateral, and orbital PFC. This study was one of the first to look at the distribution of HMGN1 expression in the DS brain and demonstrate differences in specific areas.
In summary, the studies mentioned above describe a strong concordance between the increased mRNA and protein levels of HMGN1 and trisomy. This was measured across different mouse models as well as human tissue and cellular systems (Table 4).
Table 4.
Levels of HMGN1 expression in different mouse models and cell lines
| Mouse model | Triplicated regions/genes | Expression of HMGN1 | References |
|---|---|---|---|
| Ts16 | Harbors triplication of Mmu16 | 1.6- fold increase in HMGN1 mRNA in the brain and a 3.3-fold increase in the whole trunk of fetuses | Pash et al. [192] |
| Ts65Dn | Contains a fusion of the region of Mmu16 orthologous to Hsa21 and the centromeric region of Mmu17, containing a segment of non-DS-related genes | Increase in HMGN1 mRNA expression found in E15.5 embryonic forebrain tissue | Guedj et al. [197] |
| Ts1Cje | Harbors partial triplication of Mmu16 (Sod1-Zbtb21 region) | Analysis of raw gene expression data from the Gene Expression Omnibus for Ts1Cje and euploid cerebellar tissue revealed upregulation of HMGN1 mRNA | Kong et al. [201] |
| HMGN1 was identified as one of 18 differentially expressed genes in the cerebellum of Ts1Cje mice versus disomic mice | Ling et al. [194] | ||
| HMGN1 mRNA increase in the cerebellum at P0, P15, and P30 | Potier et al. [195] | ||
| Dp1Tyb | Contains an additional copy of 63% of Mmu16 genes, orthologous to HSA21 genes | Increased HMGN1 mRNA expression in the hippocampus of adult mice | Lana-Elola et al. [199] |
| TcMAC21 | Incorporates 93% of HSA21 protein coding genes as a separate chromosome | Increase in HMGN1 mRNA in forebrain tissue at P1 | Kazuki et al. [200] |
| Gene expression omnibus of the national center for biotechnology information data from human DS brain tissue | Triplication of HSA21 | HMGN1 mRNA overexpressed in the hippocampus, cerebellar cortex, primary visual cortex, and pre-frontal cortex | Rodriguez-Ortiz et al. [203] |
| HMGN1 mRNA upregulated 1.5 times in human DS tissue | Kong et al. [201] | ||
| Human DS fibroblasts | Triplication of HSA21 | 1.6 fold increase in HMGN1 mRNA and 1.8 fold increase in HMGN1 protein levels in human DS fibroblasts | Pash et al. [193] |
| Post-mortem DS tissue | Triplication of HSA21 | 1.6-fold increase in HMGN1 mRNA expression in the cerebellum and dorsal prefrontal cortex across multiple developmental periods | Olmos-Serrano et al. [202] |
Summary of the relevant studies discussed in this review
DS phenotypes and possible correlation with gene dosage effect of HMGN1
Neurodevelopmental phenotypes and HMGN1
Although altered HMGN1 expression has been linked to neurological disorders [180], it is not clear whether the neurological phenotypes seen in DS individuals are caused by elevated HMGN1 levels. Though a direct connection remains elusive, there is a strong basis to suggest that the pathological cellular phenotypes in DS can be associated with enhanced HMGN1 expression in DS brain cells.
Individuals with DS have an 18% reduction in brain volume as compared to those without DS, and this decrease can be detected as early as the second trimester [8, 204, 205]. This is recapitulated in the iPSC-based three-dimensional models (organoids and spheroids), which show a decrease in the size of DS-derived cortical organoids [206] and in our own work utilizing cortical spheroids [207] as well as in the Ts65Dn mouse model [208]. It is noteworthy that in the mouse and human cerebellum, there is also a decrease in the number of granule and Purkinje cells [10, 200, 209, 210]. For the updated and comprehensive review describing DS-related developmental brain changes across different mouse models and cellular systems, please refer to Klein and Haydar [211].
The diminished brain volume in DS is thought to be related to hypocellularity, as reductions in neuronal numbers have been found in multiple areas of the brain, including the neocortex, hippocampus, dentate gyrus, inferior temporal gyrus, fusiform gyrus and cerebellum [9, 210, 212–214]. The hypocellularity of the DS brain is often attributed to the decrease in proliferating NPCs observed as early as 17–21 gestational weeks in human brain tissue [9, 10, 15, 215, 216] and is replicated in NPCs generated from DS-derived iPSCs [206, 217]. Deranged neurogenesis is considered to be a significant factor in the reduced number of neurons in DS brains, as it has been observed both during human fetal development in DS and DS animal models [9, 218], accompanied by delayed cortical lamination and aberrant axonal and dendritic arborization [16], as well as abnormal synaptogenesis [12–14, 16].
Many of these abnormal cellular phenotypes have been linked to other HSA21 genes, such as DYRK1A [219], amyloid precursor protein (APP) [220–224], DS cell adhesion molecule (DSCAM), S100 Calcium Binding Protein B (S100B) [225–227], OLIG2 [228–230], and more. However, it is possible that similar neurodevelopmental and neurodegenerative phenotypes result from the triplication of different genes on HSA21. For example, the enhanced production of APP and the products of its cleavage were shown to induce transcriptomic changes through deficient retrograde trafficking of neurotrophins (mediated through activation of Rab proteins) [220, 221], and to attenuate signaling mediated via Sonic hedgehog (SHH) [222–224]. The triplication of APP and DYRK1A in DS was also shown to interrupt Notch-mediated signaling [219, 222, 231]. The dysregulation of these signaling pathways has been implicated in abnormalities in neurogenesis [232], neuronal migration [233], and cortical lamination [234]. Importantly, many of the genes driving these pathways are targeted by the PRC2 family members. For example, the delta-like canonical Notch ligand 3 (DLL3) and basic helix-loop-helix (bHLH) HES genes that encode the Notch-HES pathway components are the targets for SUZ-12 binding [68, 80] and are affected by EZH2 knockdown [235], and some of the RAB genes have binding sites for the PRC2 proteins [68, 235, 236].
Similarly, HSA21 genes DSCAM and DYRK1A have been implicated in the dysregulated neuronal migration and cortical lamination in DS [206, 219]. Deficient neuronal motility and the dysregulation of the “roundabout” signaling pathway have been widely observed in DS [176, 207, 237]. Importantly, genes, such as ROBO2, ROBO4, and SLIT2, which mediate neuronal migration and axon guidance, appear to be targeted by EZH2 and SUZ-12 [68, 235, 236]. This suggests that triplicated HMGN1 can lead to developmental phenotypes similar to those induced by other triplicated genes, such as but not limited to APP, DSCAM, and DYRK1A. While direct evidence for the causal link between the triplication of HMGN1 and the DS-associated cellular processes has not yet been found, the tight regulation of these processes by the members of PRC2 and the global effect of HMGN1 on PRC2 target genes’ transcription and H3K27me3 levels could be the potential missing link.
The data related to the underproduction or overproduction of the inhibitory neurons in DS is controversial, however, the reports related to this disbalance further expose the disturbances in the acquisition of neuronal identity. Several studies have found a decrease in excitatory synapses [238] and an increase in cortical and hippocampal inhibitory synapses in mouse models of DS [239, 240]. Ts65Dn mice have decreased excitatory neuronal density in the neocortex noticeable by P8, and an increase in parvalbumin-(Pvalb) and somatostatin-(SST) positive inhibitory neurons at the same time frame in both the neocortex and the CA1 region of the hippocampus [228]. These disruptions were attributed to the increased dosage effect of Olig2 in trisomy [228–230], as normalizing Olig2 expression to euploid levels was enough to rescue the phenotype and reduce the number of Pvalb+ and SST+ interneurons generated in the mouse medial ganglionic eminence (MGE). Similar findings were described in DS iPSC-derived organoids, showing a significant overabundance of the GABAergic neurons and dysregulation of genes related to the establishment of interneuron fate and migration [230]. Other studies, however, found fewer cortical interneurons in DS fetal tissue and adult DS cortex [241]. The underproduction of GABAergic neurons and their abnormal migration was also demonstrated in an in vitro model derived from DS iPSCs and attributed to the increased levels of p21-activated kinase 1 (PAK1) [242], a binding target for SUZ12 [68]. Moreover, the commitment and lineage development of inhibitory neurons are PRC2-dependent [132, 243] and while OLIG2 is triplicated in DS, it is also known to be highly regulated by HMGN1 [121], suggesting that despite the above discrepancy between the studies, there is a consistency in reported deficits in the interneuron motility and the disbalance between the excitatory and inhibitory neurons that can be potentially caused by the gene dosage of HMGN1 and subsequent dysregulation of PRC2 targets.
Collectively, neuronal perturbations observed in DS are manifested in hypocellularity, delayed cortical lamination, diminished axonal and dendritic arborization, deficient neurogenesis and synaptogenesis, and disbalance between the inhibitory and excitatory neurons. Extensive research linked these alterations to the gene dosage effect of different triplicated genes or their combinations, as well as to the global genomic and epigenetic dysregulation in DS, as shown in Fig. 1. In this section, we provided evidence that many of the genes coding for the signaling molecules involved in intracellular and intercellular communication, such as SHH, Notch-HES, and “roundabout”, are targets for the transcriptional repression by PRC2 and therefore can be dysregulated due to antagonizing the action of HMGN1 on PRC2 target genes. We also pointed out an additional level of complexity arising from the fact that increased levels of HMGN1 can further potentiate the expression of other HSA21 genes, such as OLIG1, OLIG2, and APP, amplifying the deranged neuronal biology in trisomy.
Neuronal cell death in DS and a possible link to PRC2 dysregulation
An enhanced rate of apoptosis has also been found in certain regions of the DS brain, including the hippocampus [9], cerebral cortex [14, 244], cerebellum [244], as well as in the DS iPSC-derived NPCs [217], and cortical spheroids [207]. Increased neuronal cell death was detected in DS fetal brain [244] and the brains of middle-aged DS individuals [245]. However, the extent to which cell death contributes to the overall reduction in the number of neurons remains contested, and evidence related to the role of apoptosis and neurodegeneration in DS is somewhat controversial [10, 218, 227]. The recently demonstrated inhibition of neurodegenerative genes by PRC2 and antagonism between PRC2 and HMGN1 can potentially contribute to the upregulation of the pro-apoptotic genes in DS and enhanced neurodegeneration. The pro-apoptotic gene upregulation in DS has not been previously attributed to the triplication of HMGN1.
Glial cells in DS and the possible link to the increased gene dosage of HMGN1
In addition to the abnormal proliferation of NPCs, aberrant neurogenesis, and increased cell death, the reduction in neuronal numbers detected in DS can also be causatively linked to an aberrant shift from neurogenesis to gliogenesis [9, 227, 246]. Significantly more astrocytes were detected in human DS frontal lobe brain tissue compared to samples from age-matched controls across different developmental time points [247]. Similar findings have been noted in the hippocampus and frontal cortex of individuals with DS [226]. These findings were attributed previously to the gene dosage effect of the triplicated genes, such as DYRK1A acting through the enhanced STAT activity [248], APP leading to decreased SHH signaling [249], and more.
Since increased levels of HMGN1 can intensify the generation of astrocytes at the late embryonic neocortex at the expense of neurogenesis [179], one can speculate that the gene dosage effect of HMGN1 in trisomy can be another contributing factor to the gliogenic shift in DS. This is further supported by the regulatory role of the PRC2-deposited H3K27me3 chromatin mark in defining cell identity during the shift from neurogenesis to gliogenesis [134, 159]. Furthermore, enhanced astrocyte production and diminished generation of oligodendrocytes in DS reflect the aberrant transition from astrogenesis to oligodendrogenesis, as has been described previously [222] (Fig. 1).
Indeed, impaired development of oligodendrocytes and aberrant patterns of myelination in DS are well-documented [250]. Children with DS exhibit delayed myelination in the frontotemporal lobes from 2 months to 6 years old, developmentally lagging age-matched controls by almost 12 months. They also display specific delays in myelination in the hippocampus [250, 251]. Overall, people with DS have a decreased density of myelinated fibers and lower levels of myelin basic protein (MBP) and myelin-associated glycoprotein (MAG) [202, 250]. Due to the necessity of myelination for the appropriate and efficient signal propagation, these findings may be significant for explaining the intellectual deficits seen in DS.
Relevant to our studies, PRC2 deficiency is associated with impaired progression from OPCs to myelinating OLs [160]. Even though the dysregulation of the SHH pathway and enhanced expression of the PTCH1 receptor in DS [209, 223, 252, 253] has been attributed, at least partially, to the triplication of APP [252], both SHH and PTCH1 are targets for transcriptional repression by PRC2 members [68, 236], and dysregulation of the SHH pathway can be caused by increased transcription of key pathway genes due to the increased dosage of HMGN1 and alleviation of PRC2 repression. Similarly to neuronal dysregulation, the perturbation in glial cells and diminished maturation of oligodendrocytes can be potentially linked to a deficient silencing of the genes necessary for proper lineage development and functional maturity.
Neuroinflammation in DS and HMGN1 gene dosage effect
The role of neuroinflammation in DS has been widely acknowledged. Previous studies demonstrated increased activation of microglia, astrogliosis, dysregulation of inflammation-related genes, as well as the enhanced formation of complement complexes detected in DS brain tissue [189, 254–257]. The frontal cortex of young DS individuals displays an enhanced neuroinflammatory signature of microglia represented by a high ratio of the soma size to the length of the processes and the appearance of the pathological rod-like microglial phenotypes accompanied by enhanced production of pro-inflammatory cytokines [255]. Remarkably, these inflammation-related changes precede the fully blown AD pathology in DS persons [255].
In addition to the increased generation of astrocytes at the expanse of neurons (aka gliogenic shift, as we mentioned earlier), there is an abnormal astrocyte biology. Impaired Ca2+ homeostasis and signaling [258], and mitochondrial dysfunction, including altered mitochondrial membrane potential, have been demonstrated in DS-derived astrocytes [259, 260]. In fact, mitochondrial networks are more fragmented and are associated with increased production of reactive oxygen species (ROS) in trisomic astrocytes leading to enhanced oxidative stress and neuronal injury in DS [260]. In addition to the gene dosage effect of APP, DYRK1A, and S100B, implicated previously in enhanced gliosis in DS, other HSA21 genes can play a role in neuroinflammation associated with trisomy. Among these genes are ubiquitin-specific protease 25 (USP25), which regulates microglial activation, T-cell lymphoma invasion and metastasis 1 (TIAM1), and Superoxide dismutase 1 (SOD1), which both participate in a response to the ROS, as well as the interferon family members: interferon α, β, and ω receptor 1 (IFNAR1), IFNAR2, interferon γ receptor 2 (IFNGR2), and interleukin-10 receptor subunit beta (IL10RB) [261, 262]. Altogether, an increase in the products of these genes can contribute to the neuroinflammatory signature observed in DS. For instance, overexpression of APP and S100B results in an enhanced expression of interleukin 1 (IL-1) and induction of astrocytic and microglial activation [257, 261, 263].
Notably, recent studies showed that aberrant PRC2 activity related to the HMGN1 gene dosage effect could further exacerbate the inflammatory profile associated with trisomy. It was shown that IFN signaling is a target of EZH2 suppression in cancer: EZH2 represses IFNγ target genes in prostate [264], colorectal [265], and melanoma [266] cancer cells, while pharmacological inhibition of EZH2 in melanoma results in the upregulated profile of IFNγ and IFN-α downstream targets [266]. In addition, previous studies have linked HMGN1 to a variety of inflammation-related processes [267–269] that have been harnessed for anti-cancer therapeutic approaches. These studies showed that HMGN1 activates and promotes the migration of dendritic cells, participates in an antigen-specific immune response, positively regulates CD8+ T cell activation, and acts as an alarmin [269–271]. On the other hand, the ability of HMGN1 to potentiate the immune response can further contribute to the neuroinflammatory phenotypes seen in DS that can further aggravate the development of AD pathology.
Alzheimer’s disease (AD)-related pathology in DS (DS-AD) and HMGN1
Premature development of AD pathology is common in DS, with most individuals displaying AD-related pathology, such as amyloid-β (Aβ) plaques, accumulation of hyper-phosphorylated tau (p-tau), markers of oxidative stress, neuroinflammation, and neurodegeneration by age 40 [257, 261, 263, 272–274]. The density of plaques and tangles in DS-AD is higher compared to that seen in non-DS AD, but the arrangement is similar [275]. There is variability in the age of onset and the level of dementia, despite the near guarantee of someone with DS developing AD during their lifetime [276].
The data related to post-translational histone modifications in AD show patterns of altered histone acetylation, methylation, and phosphorylation [277, 278]. The available information supporting the role of HMGN1 in DS-AD through the decrease in H3K27me3 and increase in H3K27ac is somewhat controversial since some studies of human postmortem AD brain tissue show different H3K27 modification and based on our knowledge, no genome-wide studies focused on H3K27 marks in DS-AD human brains were performed. However, the existing data obtained from human post-mortem AD brain tissue highlights epigenetic dysregulation in AD pathology and implicates changes in both acetylation and trimethylation histone marks in the disease. Decreased levels of histone H3 (K18/K23) acetylation have been found in the temporal lobe of post-mortem human AD tissue [279]. A recent study, looking specifically at the genome-wide distribution of H3 marks in the entorhinal cortex of post-mortem human tissue from severe cases of AD identified diminished levels of H3K4me3 and enhanced levels of H3K27me3 in association with the disease that was more prominent in males [280]. A genome-wide acetylome study in the human entorhinal cortex of AD patients uncovered a redistributed pattern of H3K27 acetylation and identified 4,162 differentially modified peaks (both hypo- and hyper-acetylated) as compared to matched controls [281]. The investigators found alterations in the H3K27ac patterns near the following genes involved in the pathological generation of Aβ: APP, microtubule-associated protein tau (MAPT), presenilin-1 (PSEN1), and PSEN2 [282–285]. Gene ontology analysis revealed that hyperacetylated genes were enriched in categories associated with Aβ metabolic processes and tau interactome, while hypoacetylated genes were enriched in categories associated with neurotransmitter function, neuronal transmission, and synapses.
Additionally, increased levels of acetylated histone H3 and H4 were found in the pyramidal neurons in the inferior temporal gyrus of post-mortem AD tissue and were significantly correlated with both tau and β-amyloid load [286]. A recent multiome study further confirmed the previously shown reconfiguration of acetylation patterns in the lateral temporal lobe from AD patients and revealed an increase in H3K27ac and H3K9ac marks [287]. Remarkably, the gain of these acetylation marks was linked to disease-specific AD pathways, and increasing H3K27ac and H3K9ac levels in Drosophila, performed by the same group, exacerbated Aβ42-induced neurodegeneration.
Altogether, these studies show the functional relevance between the posttranslational modification of histones and the classical features of AD pathology in mouse and human brains. The changes in the epigenetic landscape induced by HMGN1 in DS may contribute to the development of AD pathology. Notably, a meta-analysis of cognition-related genome-wide association studies yielded HMGN1 as only one significantly associated gene [288]. Both AD and DS pathology have been connected to changes at H3K27 acetylation/trimethylation marks but future studies are needed to provide a causative link between the specific changes to the epigenetic environment induced by overexpression of HMGN1 in DS and the development of AD pathology.
Conclusion
In this review, we discussed a possible mechanistic connection between the HMGN1 triplication in DS and downstream events leading the neurodevelopmental and neurodegenerative pathology in trisomy. We specifically hypothesized that increased HMGN1 dosage promotes global chromatin accessibility for transcription, thus leading to the derepression of PRC2 target genes, causatively linking the triplication of HMGN1 in trisomy to several DS-related cellular phenotypes. We present a summary of this hypothesis in Fig. 2.
Fig. 2.
Schematic illustrating the effects of increased dosage of HMGN1 in DS through the antagonizing action on PRC2. The epigenetic changes induced by increased levels of PRC2 lead to diverse transcriptomic and phenotypic changes that can potentially lead to the pathological features observed in DS. Created with BioRender.com
Abbreviations
- DS
Down syndrome
- IQ
Intelligence quotient
- B-ALL
B-cell acute lymphoblastic leukemia
- HMGN1
High mobility group binding domain 1
- DYRK1A
Dual specificity tyrosine-phosphorylation-regulated-kinase 1A
- ETS2
ETS proto-oncogene 2
- BRWD1
Bromodomain and WD repeat domain containing 1
- RUNX1
Runt-related transcription factor 1
- DNMT3L
DNA (cytosine-5)-metyltransferase 3-like
- SON
SON RNA and DNA binding protein
- PRC2
Polycomb repressive complex 2
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- ChIP
Chromatin immunoprecipitation
- TSS
Transcription start site
- HMG
High mobility group
- CHUD
Chromatin unfolding domain
- MEF
Mouse embryonic fibroblast
- ESC
Embryonic stem cell
- NBD
Nuclear-binding domain
- DKO
Double-knock-out
- HMT
Histone methyltransferase
- EED
Embryonic ectoderm development
- EZH2
Enhancer of zeste
- SUZ-12
Suppressor of zeste
- BrdU
Bromodeoxyuridine
- DG
Dentate gyrus
- SVZ
Subventricular zone
- DA
Dopamine
- 5HT
Serotonin
- MSN
Medium spiny neuron
- IUE
Intra-utero electroporation
- shRNA
Short-hairpin RNA
- CP
Cortical plate
- OPC
Oligodendrocyte precursor cell
- NSC
Neural stem cell
- Olig2
Oligodendrocyte transcription factor 2
- MBP
Myelin basic protein
- iPSC
Induced pluripotent stem cell
- PolII
RNA polymerase II
- Pdgfr-α
Platelet-derived growth factor receptor alpha
- MNase
Micrococcal nuclease
- TC-rMNase-seq
Time-course digestion with reduced MNase levels followed by high-throughput sequencing
- Camk2a
Calcium/calmodulin dependent protein kinase II alpha
- Pmaip1
Phorbol-12-myristate-13-acetate-induced protein 1
- BID
BH3 interacting domain death agonist
- Noxa
NADPH oxidase activator 1
- Cdkn2a/b
Cyclin-dependent kinase inhibitor 2A/B
- Igfbp3
Insulin-like growth factor-binding protein 3
- CRLF2
Cytokine receptor-like factor 2
- Mmu16
Mouse chromosome 16
- ATAC-seq
Transposase-accessible chromatin sequencing
- EB
Embryoid body
- CNPase
2ʹ,3ʹ-Cyclic-nucleotide 3ʹ-phosphodiesterase
- PLP
Proteolipid protein
- MeCP2
Methyl CpG-binding protein 2
- GEO
Gene expression omnibus
- PFC
Prefrontal cortex
- APP
Amyloid precursor protein
- DSCAM
DS cell adhesin molecule
- SHH
Sonic hedgehog
- bHLH
Basic helix-loop-helix
- Pvalb
Paralbumin
- Sst
Somatostatin
- MGE
Medial ganglionic eminence
- PAK1
P21 activated kinase 1
- MAPT
Microtubule-associated protein tau
- PSEN1
Presenilin-1
- JAK-STAT
Janus tyrosine kinase-signal transducer and activator of transcription
Author contributions
EZ and AG conceived the idea. SJF, AG, and EZ performed the screen of the literature and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the National Institutes of Health, NINDS, 1R21NS125469-01A1, Award RNS125469A by NICHD to E.Z.
Availability of data and materials
Not applicable.
Declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alla Grishok and Ella Zeldich have contributed equally to this work and share the last authorship
References
- 1.Gueant JL. Homocysteine and related genetic polymorphisms in Down’s syndrome IQ. J Neurol Neurosurg Psychiatry. 2005;76(5):706–709. doi: 10.1136/jnnp.2004.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baburamani AA, Patkee PA, Arichi T, Rutherford MA. New approaches to studying early brain development in Down syndrome. Dev Med Child Neurol. 2019;61(8):867–879. doi: 10.1111/dmcn.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanfranchi S, Jerman O, Dal Pont E, Alberti A, Vianello R. Executive function in adolescents with Down syndrome: executive function in Down syndrome. J Intellect Disabil Res. 2010;54(4):308–319. doi: 10.1111/j.1365-2788.2010.01262.x. [DOI] [PubMed] [Google Scholar]
- 4.Vicari S, Bellucci S, Carlesimo GA. Visual and spatial long-term memory: differential pattern of impairments in Williams and Down syndromes. Dev Med Child Neurol. 2005;47(5):305–311. doi: 10.1017/S0012162205000599. [DOI] [PubMed] [Google Scholar]
- 5.Startin CM, Hamburg S, Hithersay R, Davies A, Rodger E, Aggarwal N, et al. The LonDownS adult cognitive assessment to study cognitive abilities and decline in Down syndrome. Wellcome Open Res. 2016;15(1):11. doi: 10.12688/wellcomeopenres.9961.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lott IT, Dierssen M. Cognitive deficits and associated neurological complications in individuals with Down’s syndrome. Lancet Neurol. 2010;9(6):623–633. doi: 10.1016/S1474-4422(10)70112-5. [DOI] [PubMed] [Google Scholar]
- 7.Pinter JD, Eliez S, Schmitt JE, Capone GT, Reiss AL. Neuroanatomy of Down’s syndrome: a high-resolution MRI study. AJP. 2001;158(10):1659–1665. doi: 10.1176/appi.ajp.158.10.1659. [DOI] [PubMed] [Google Scholar]
- 8.Pinter JD, Brown WE, Eliez S, Schmitt JE, Capone GT, Reiss AL. Amygdala and hippocampal volumes in children with Down syndrome: a high-resolution MRI study. Neurology. 2001;56(7):972–974. doi: 10.1212/WNL.56.7.972. [DOI] [PubMed] [Google Scholar]
- 9.Guidi S, Bonasoni P, Ceccarelli C, Santini D, Gualtieri F, Ciani E, et al. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome: hippocampal hypocellularity in down fetuses. Brain Pathol. 2007;18(2):180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidi S, Ciani E, Bonasoni P, Santini D, Bartesaghi R. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with Down syndrome: down syndrome fetal cerebellum. Brain Pathol. 2011;21(4):361–373. doi: 10.1111/j.1750-3639.2010.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Utagawa EC, Moreno DG, Schafernak KT, Arva NC, Malek-Ahmadi MH, Mufson EJ, et al. Neurogenesis and neuronal differentiation in the postnatal frontal cortex in Down syndrome. Acta Neuropathol Commun. 2022;10(1):86. doi: 10.1186/s40478-022-01385-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petit TL, LeBoutillier JC, Alfano DP, Becker LE. Synaptic development in the human fetus: a morphometric analysis of normal and Down’s syndrome neocortex. Exp Neurol. 1984;83(1):13–23. doi: 10.1016/0014-4886(84)90041-4. [DOI] [PubMed] [Google Scholar]
- 13.Becker LE, Armstrong DL, Chan F. Dendritic atrophy in children with Down’s syndrome. Ann Neurol. 1986;20(4):520–526. doi: 10.1002/ana.410200413. [DOI] [PubMed] [Google Scholar]
- 14.Takashima S, Ieshima A, Nakamura H, Becker LE. Dendrites, dementia and the Down syndrome. Brain Dev. 1989;11(2):131–133. doi: 10.1016/S0387-7604(89)80082-8. [DOI] [PubMed] [Google Scholar]
- 15.Contestabile A, Fila T, Ceccarelli C, Bonasoni P, Bonapace L, Santini D, et al. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus. 2007;17(8):665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- 16.Golden JA, Hyman BT. Development of the superior temporal neocortex is anomalous in trisomy 21. J Neuropathol Exp Neurol. 1994;53(5):513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Becker W, Joost HG. Structural and Functional characteristics of dyrk, a novel subfamily of protein kinases with dual specificity. Prog Nucleic Acid Res Mol Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- 18.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang EJ, Ahn YS, Chung KC. Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J Biol Chem. 2001;276(43):39819–39824. doi: 10.1074/jbc.M104091200. [DOI] [PubMed] [Google Scholar]
- 20.Lepagnol-Bestel AM, Zvara A, Maussion G, Quignon F, Ngimbous B, Ramoz N, et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum Mol Genet. 2009;18(8):1405–1414. doi: 10.1093/hmg/ddp047. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Xu C, Fu Y, Lei PJ, Yao Y, Yang W, et al. DYRK1A interacts with histone acetyl transferase p300 and CBP and localizes to enhancers. Nucleic Acids Res. 2018;46(21):11202–11213. doi: 10.1093/nar/gky754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, et al. p300/cAMP-responsive element-binding protein interactions with Ets-1 and Ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem. 1999;274(24):17342–17352. doi: 10.1074/jbc.274.24.17342. [DOI] [PubMed] [Google Scholar]
- 23.Sun HJ, Xu X, Wang XL, Wei L, Li F, Lu J, et al. Transcription factors Ets2 and Sp1 Act synergistically with histone acetyltransferase p300 in activating human interleukin-12 p40 promoter. ABBS. 2006;38(3):194–200. doi: 10.1111/j.1745-7270.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- 24.Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/S0065-230X(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 25.Tymms MJ, Kola I. Regulation of gene expression by transcription factors Ets-1 and Ets-2. Mol Reprod Dev. 1994;39(2):208–214. doi: 10.1002/mrd.1080390214. [DOI] [PubMed] [Google Scholar]
- 26.Seth A, Watson DK, Blair DG, Papas TS. c-ets-2 protooncogene has mitogenic and oncogenic activity. Proc Natl Acad Sci USA. 1989;86(20):7833–7837. doi: 10.1073/pnas.86.20.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sevilla L, Aperlo C, Dulic V, Chambard JC, Boutonnet C, Pasquier O, et al. The Ets2 transcription factor inhibits apoptosis induced by colony-stimulating factor 1 deprivation of macrophages through a Bcl-xL-dependent mechanism. Mol Cell Biol. 1999;19(4):2624–2634. doi: 10.1128/MCB.19.4.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson DK, Robinson L, Hodge DR, Kola I, Papas TS, Seth A. FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene. 1997;14(2):213–221. doi: 10.1038/sj.onc.1200839. [DOI] [PubMed] [Google Scholar]
- 29.Wolvetang EJ. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12(3):247–255. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
- 30.Heru Sumarsono S, Wilson TJ, Tymms MJ, Venter DJ, Corrick CM, Kola R, et al. Down’s syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379(6565):534–537. doi: 10.1038/379534a0. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Rambaldi I, Daniels E, Featherstone M. Expression of theWdr9 gene and protein products during mouse development. Dev Dyn. 2003;227(4):608–614. doi: 10.1002/dvdy.10344. [DOI] [PubMed] [Google Scholar]
- 32.Mandal M, Hamel KM, Maienschein-Cline M, Tanaka A, Teng G, Tuteja JH, et al. Histone reader BRWD1 targets and restricts recombination to the Igk locus. Nat Immunol. 2015;16(10):1094–1103. doi: 10.1038/ni.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal M, Maienschein-Cline M, Maffucci P, Veselits M, Kennedy DE, McLean KC, et al. BRWD1 orchestrates epigenetic landscape of late B lymphopoiesis. Nat Commun. 2018;9(1):3888. doi: 10.1038/s41467-018-06165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, et al. The runt domain identifies a new family of heterometric transcriptional regulators. Trends Genet. 1993;9(10):338–341. doi: 10.1016/0168-9525(93)90026-E. [DOI] [PubMed] [Google Scholar]
- 35.Kitabayashi I. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17(11):2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakford PC, James SR, Qadi A, West AC, Ray SN, Bert AG, et al. Transcriptional and epigenetic regulation of the GM-CSF promoter by RUNX1. Leuk Res. 2010;34(9):1203–1213. doi: 10.1016/j.leukres.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 37.Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, Xie J, Lutterbach B, Durst-Goodwin KL, et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25(42):5777–5786. doi: 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- 38.Bowers SR, Calero-Nieto FJ, Valeaux S, Fernandez-Fuentes N, Cockerill PN. Runx1 binds as a dimeric complex to overlapping Runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids Res. 2010;38(18):6124–6134. doi: 10.1093/nar/gkq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103(6):2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 40.Lacaud G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100(2):458–466. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- 41.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MFTR. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22(2):158–168. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- 42.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117(8):2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 43.Chédin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99(26):16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laufer BI, Gomez JA, Jianu JM, LaSalle JM. Stable DNMT3L overexpression in SH-SY5Y neurons recreates a facet of the genome-wide Down syndrome DNA methylation signature. Epigenetics Chromatin. 2021;14(1):13. doi: 10.1186/s13072-021-00387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berdichevskiĭ FB, Chumakov IM, Kiselev LL. Decoding of the primary structure of the son3 region in human genome: identification of a new protein with unusual structure and homology with DNA-binding proteins. Mol Biol. 1988;22(3):794–801. [PubMed] [Google Scholar]
- 46.Mattioni T, Hume CR, Konigorski S, Hayes P, Osterweil Z, Lee JS. A cDNA clone for a novel nuclear protein with DNA binding activity. Chromosoma. 1992;101(10):618–624. doi: 10.1007/BF00360539. [DOI] [PubMed] [Google Scholar]
- 47.Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, et al. SON Controls Cell-Cycle Progression by Coordinated Regulation of RNA Splicing. Mol Cell. 2011;42(2):185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA. 2008;105(44):17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa T, Tanji E, Kuboki Y, Hatori T, Yamamoto M, Shimizu K, et al. Targeting of MAPK-associated molecules identifies SON as a prime target to attenuate the proliferation and tumorigenicity of pancreatic cancer cells. Mol Cancer. 2012;11(1):88. doi: 10.1186/1476-4598-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huen MSY, Sy SMH, Leung KM, Ching YP, Tipoe GL, Man C, et al. SON is a spliceosome-associated factor required for mitotic progression. Cell Cycle. 2010;9(13):2679–2685. doi: 10.4161/cc.9.13.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17(10):5843–5855. doi: 10.1128/MCB.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997. 389(6648):251–60. [DOI] [PubMed]
- 53.Talbert PB, Henikoff S. Histone variants at a glance. J Cell Sci. 2021;134(6):jcs244749. doi: 10.1242/jcs.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lennartsson A, Ekwall K. Histone modification patterns and epigenetic codes. Biochim Biophys Acta. 2009;1790(9):863–868. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 56.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesi. Proc Natl Acad Sci USA. 1964;51(5):786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bode J, Gomez-Lira MM, Schroter H. Nucleosomal particles open as the histone core becomes hyperacetylated. Eur J Biochem. 1983;130(3):437–445. doi: 10.1111/j.1432-1033.1983.tb07170.x. [DOI] [PubMed] [Google Scholar]
- 59.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. doi: 10.1016/0092-8674(93)90051-Q. [DOI] [PubMed] [Google Scholar]
- 60.Oliva R, Bazett-Jones DP, Locklear L, Dixon GH. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucl Acids Res. 1990;18(9):2739–2747. doi: 10.1093/nar/18.9.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morales V, Richard-Foy H. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol Cell Biol. 2000;20(19):7230–7237. doi: 10.1128/MCB.20.19.7230-7237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392(6678):831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 63.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384(6610):641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 64.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–959. doi: 10.1016/S0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 65.Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148(2):558–567. doi: 10.1016/0003-9861(72)90174-9. [DOI] [PubMed] [Google Scholar]
- 66.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 68.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robyr D, Grunstein M. Genomewide histone acetylation microarrays. Methods. 2003;31(1):83–89. doi: 10.1016/S1046-2023(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 70.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103(43):15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kondo Y, Shen L, Yan PS, Huang THM, Issa JPJ. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci USA. 2004;101(19):7398–7403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huebert DJ, Kamal M, O’Donovan A, Bernstein BE. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods. 2006;40(4):365–369. doi: 10.1016/j.ymeth.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 73.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 75.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280(18):17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 76.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 77.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 78.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24(12):5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Sung-wook J, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16(7):890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang G, Lin JCY, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3–K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101(19):7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436(7052):876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 84.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 86.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 87.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goodwin GH, Shooter KV, Johns EW. Interaction of a non-histone chromatin protein (high-mobility group protein 2) with DNA. Eur J Biochem. 1975;54(2):427–433. doi: 10.1111/j.1432-1033.1975.tb04153.x. [DOI] [PubMed] [Google Scholar]
- 89.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38(1):14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 90.Crippa MP, Trieschmann L, Alfonso PJ, Wolffe AP, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993;12(10):3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding HF, Rimsky S, Batson SC, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265(5173):796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 92.Paranjape SM, Krumm A, Kadonaga JT. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9(16):1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 93.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14(7):1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trieschmann L, Postnikov YV, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995;15(12):6663–6669. doi: 10.1128/MCB.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bustin M, Trieschmann L, Postnikov YV. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin Cell Biol. 1995;6(4):247–255. doi: 10.1006/scel.1995.0033. [DOI] [PubMed] [Google Scholar]
- 96.Tremethick DJ, Hyman L. High mobility group proteins 14 and 17 can prevent the close packing of nucleosomes by increasing the strength of protein contacts in the linker DNA. J Biol Chem. 1996;271(20):12009–12016. doi: 10.1074/jbc.271.20.12009. [DOI] [PubMed] [Google Scholar]
- 97.Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17(23):6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, et al. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15(4):573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 99.Levinger L, Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci USA. 1982;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004;255(3):332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- 101.Garabedian A, Fouque KJD, Chapagain PP, Leng F, Fernandez-Lima F. AT-hook peptides bind the major and minor groove of AT-rich DNA duplexes. Nucleic Acids Res. 2022;50(5):2431–2439. doi: 10.1093/nar/gkac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dintilhac A, Bernués J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J Biol Chem. 2002;277(9):7021–7028. doi: 10.1074/jbc.M108417200. [DOI] [PubMed] [Google Scholar]
- 103.Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236(1):189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 104.Shirakawa H, Landsman D, Postnikov YV, Bustin M. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem. 2000;275(9):6368–6374. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- 105.West KL, Ito Y, Birger Y, Postnikov Y, Shirakawa H, Bustin M. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J Biol Chem. 2001;276(28):25959–25969. doi: 10.1074/jbc.M101692200. [DOI] [PubMed] [Google Scholar]
- 106.Lehtonen S, Lehtonen E. HMG-17 is an early marker of inductive interactions in the developing mouse kidney. Differentiation. 2001;67(4–5):154–163. doi: 10.1046/j.1432-0436.2001.670407.x. [DOI] [PubMed] [Google Scholar]
- 107.Birger Y, Ito Y, West KL, Landsman D, Bustin M. HMGN4, a newly discovered nucleosome-binding protein encoded by an intronless gene. DNA Cell Biol. 2001;20(5):257–264. doi: 10.1089/104454901750232454. [DOI] [PubMed] [Google Scholar]
- 108.Furusawa T, Lim JH, Catez F, Birger Y, Mackem S, Bustin M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol Cell Biol. 2006;26(2):592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ito Y, Bustin M. Immunohistochemical localization of the nucleosome-binding protein HMGN3 in mouse brain. J Histochem Cytochem. 2002;50(9):1273–1275. doi: 10.1177/002215540205000914. [DOI] [PubMed] [Google Scholar]
- 110.Lucey MM, Wang Y, Bustin M, Duncan MK. Differential expression of the HMGN family of chromatin proteins during ocular development. Gene Expr Patterns. 2008;8(6):433–437. doi: 10.1016/j.gep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28(9):2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.González-Romero R, Eirín-López JM, Ausió J. Evolution of high mobility group nucleosome-binding proteins and its implications for vertebrate chromatin specialization. Mol Biol Evol. 2015;32(1):121–131. doi: 10.1093/molbev/msu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell. 2009;35(5):642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mardian JKW, Paton AE, Bunick GJ, Olins DE. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- 115.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci USA. 1998;95(10):5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell. 2008;19(5):1816–1824. doi: 10.1091/mbc.e07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, et al. Mitotic phosphorylation prevents the binding of hmgn proteins to chromatin. Mol Cell Biol. 2001;21(15):5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prymakowska-Bosak M, Hock R, Catez F, Lim JH, Birger Y, Shirakawa H, et al. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol Cell Biol. 2002;22(19):6809–6819. doi: 10.1128/MCB.22.19.6809-6819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Postnikov YV, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252(4):423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 120.Phair RD, Scaffidi P, Elbi C, Vecerová J, Dey A, Ozato K, et al. Global Nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24(14):6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Postnikov YV, Shick VV, Belyavsky AV, Khrapko KR, Brodolin KL, Nikolskaya TA, et al. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucl Acids Res. 1991;19(4):717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19(8):5237–5246. doi: 10.1128/MCB.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang S, Postnikov Y, Lobanov A, Furusawa T, Deng T, Bustin M. H3K27ac nucleosomes facilitate HMGN localization at regulatory sites to modulate chromatin binding of transcription factors. Commun Biol. 2022;5(1):159. doi: 10.1038/s42003-022-03099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deng T, Postnikov Y, Zhang S, Garrett L, Becker L, Rácz I, et al. Interplay between H1 and HMGN epigenetically regulates OLIG1 and 2 expression and oligodendrocyte differentiation. Nucleic Acids Res. 2017;45(6):3031–3045. doi: 10.1093/nar/gkw1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3(8):760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990;211(4):883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 127.He B, Deng T, Zhu I, Furusawa T, Zhang S, Tang W, et al. Binding of HMGN proteins to cell specific enhancers stabilizes cell identity. Nat Commun. 2018;9(1):5240. doi: 10.1038/s41467-018-07687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He B, Zhu I, Postnikov Y, Furusawa T, Jenkins L, Nanduri R, et al. Multiple epigenetic factors co-localize with HMGN proteins in A-compartment chromatin. Epigenetics Chromatin. 2022;15(1):23. doi: 10.1186/s13072-022-00457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, et al. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol Cell Biol. 2013;33(16):3377–3389. doi: 10.1128/MCB.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Højfeldt JW, Laugesen A, Willumsen BM, Damhofer H, Hedehus L, Tvardovskiy A, et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol. 2018;25(3):225–232. doi: 10.1038/s41594-018-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15(10):942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 132.Buontempo S, Laise P, Hughes JM, Trattaro S, Das V, Rencurel C, et al. EZH2-mediated H3K27me3 targets transcriptional circuits of neuronal differentiation. Front Neurosci. 2022;12(16):814144. doi: 10.3389/fnins.2022.814144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu PP, Tang GB, Xu YJ, Zeng YQ, Zhang SF, Du HZ, et al. MiR-203 Interplays with polycomb repressive complexes to regulate the proliferation of neural stem/progenitor cells. Stem Cell Reports. 2017;9(1):190–202. doi: 10.1016/j.stemcr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107(36):15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhao L, Li J, Ma Y, Wang J, Pan W, Gao K, et al. Ezh2 is involved in radial neuronal migration through regulating Reelin expression in cerebral cortex. Sci Rep. 2015;5(1):15484. doi: 10.1038/srep15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 137.Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, et al. Timing of CNS cell generation. Neuron. 2000;28(1):69–80. doi: 10.1016/S0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 138.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16(11):3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci USA. 2010;107(45):19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 Is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 144.Corley M, Kroll KL. The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 2015;359(1):65–85. doi: 10.1007/s00441-014-2011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9(12):1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 147.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA Polymerase II transcriptional elongation. Mol Cell. 2008;29(1):69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Healy E, Mucha M, Glancy E, Fitzpatrick DJ, Conway E, Neikes HK, et al. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol Cell. 2019;76(3):437–4526. doi: 10.1016/j.molcel.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 149.Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. PRC1 catalytic activity is central to polycomb system function. Mol Cell. 2020;77(4):857–874.e9. doi: 10.1016/j.molcel.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sugishita H, Kondo T, Ito S, Nakayama M, Yakushiji-Kaminatsui N, Kawakami E, et al. Variant PCGF1-PRC1 links PRC2 recruitment with differentiation-associated transcriptional inactivation at target genes. Nat Commun. 2021;12(1):5341. doi: 10.1038/s41467-021-24894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chan CH, Godinho LN, Thomaidou D, Tan SS, Gulisano M, Parnavelas JG. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cereb Cortex. 2001;11(12):1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- 152.Liu PP, Xu YJ, Dai SK, Du HZ, Wang YY, Li XG, et al. Polycomb protein EED regulates neuronal differentiation through targeting SOX11 in hippocampal dentate gyrus. Stem Cell Reports. 2019;13(1):115–131. doi: 10.1016/j.stemcr.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Toskas K, Yaghmaeian-Salmani B, Skiteva O, Paslawski W, Gillberg L, Skara V, et al. PRC2-mediated repression is essential to maintain identity and function of differentiated dopaminergic and serotonergic neurons. Sci Adv. 2022;8(34):eabo1543. doi: 10.1126/sciadv.abo1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.vonSchimmelmann M, Feinberg PA, Sullivan JM, Ku SM, Badimon A, Duff MK, et al. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat Neurosci. 2016;19(10):1321–1330. doi: 10.1038/nn.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Feng X, Juan AH, Wang HA, Ko KD, Zare H, Sartorelli V. Polycomb Ezh2 controls the fate of GABAergic neurons in the embryonic cerebellum. Development. 2016;143(11):1971–1980. doi: 10.1242/dev.132902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.DiMeglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, et al. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339(6116):204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jakovcevski I, Filipovic R, Mo Z, Rakic S, Zecevic N. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front Neuroanat. 2009;3:5. doi: 10.3389/neuro.05.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89(1):37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sher F, Rößler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 160.Wang W, Cho H, Kim D, Park Y, Moon JH, Lim SJ, et al. PRC2 acts as a critical timer that drives oligodendrocyte fate over astrocyte identity by repressing the notch pathway. Cell Rep. 2020;32(11):108147. doi: 10.1016/j.celrep.2020.108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Masahira N, Takebayashi H, Ono K, Watanabe K, Ding L, Furusho M, et al. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev Biol. 2006;293(2):358–369. doi: 10.1016/j.ydbio.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 162.Douvaras P, Rusielewicz T, Kim K, Haines J, Casaccia P, Fossati V. Epigenetic modulation of human induced pluripotent stem cell differentiation to oligodendrocytes. IJMS. 2016;17(4):614. doi: 10.3390/ijms17040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4(1):80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 166.Liu J, Wu X, Zhang H, Pfeifer GP, Lu Q. Dynamics of RNA polymerase II pausing and bivalent histone H3 methylation during neuronal differentiation in brain development. Cell Rep. 2017;20(6):1307–1318. doi: 10.1016/j.celrep.2017.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu J, Xiong C, Zhuo B, Wen Z, Shen J, Liu C, et al. Analysis of local chromatin states reveals gene transcription potential during mouse neural progenitor cell differentiation. Cell Rep. 2020;32(4):107953. doi: 10.1016/j.celrep.2020.107953. [DOI] [PubMed] [Google Scholar]
- 168.Li J, Hart RP, Mallimo EM, Swerdel MR, Kusnecov AW, Herrup K. EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat Neurosci. 2013;16(12):1745–1753. doi: 10.1038/nn.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Seong IS, Woda JM, Song JJ, Lloret A, Abeyrathne PD, Woo CJ, et al. Huntingtin facilitates polycomb repressive complex 2. Hum Mol Genet. 2010;19(4):573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Södersten E, Feyder M, Lerdrup M, Gomes AL, Kryh H, Spigolon G, et al. Dopamine signaling leads to loss of polycomb repression and aberrant gene activation in experimental parkinsonism. PLoS Genet. 2014;10(9):e1004574. doi: 10.1371/journal.pgen.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lane AA, Chapuy B, Lin CY, Tivey T, Li H, Townsend EC, et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet. 2014;46(6):618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rabin KR, Whitlock JA. Malignancy in children with trisomy 21. Oncologist. 2009;14(2):164–173. doi: 10.1634/theoncologist.2008-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mullighan CG, Collins-Underwood JR, Phillips LAA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor-and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41(11):1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mowery CT, Reyes JM, Cabal-Hierro L, Higby KJ, Karlin KL, Wang JH, et al. Trisomy of a Down syndrome critical region globally amplifies transcription via HMGN1 overexpression. Cell Rep. 2018;25(7):1898–1911.e5. doi: 10.1016/j.celrep.2018.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Page EC, Heatley SL, Eadie LN, McClure BJ, de Bock CE, Omari S, et al. HMGN1 plays a significant role in CRLF2 driven Down syndrome leukemia and provides a potential therapeutic target in this high-risk cohort. Oncogene. 2022;41(6):797–808. doi: 10.1038/s41388-021-02126-4. [DOI] [PubMed] [Google Scholar]
- 176.Meharena HS, Marco A, Dileep V, Lockshin ER, Akatsu GY, Mullahoo J, et al. Down-syndrome-induced senescence disrupts the nuclear architecture of neural progenitors. Cell Stem Cell. 2022;29(1):116–130.e7. doi: 10.1016/j.stem.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mohamed OA, Bustin M, Clarke HJ. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev Biol. 2001;229(1):237–249. doi: 10.1006/dbio.2000.9942. [DOI] [PubMed] [Google Scholar]
- 178.Körner U, Bustin M, Scheer U, Hock R. Developmental role of HMGN proteins in Xenopus laevis. Mech Dev. 2003;120(10):1177–1192. doi: 10.1016/j.mod.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 179.Nagao M, Lanjakornsiripan D, Itoh Y, Kishi Y, Ogata T, Gotoh Y. High mobility group nucleosome-binding family proteins promote astrocyte differentiation of neural precursor cells. Stem Cells. 2014;32(11):2983–2997. doi: 10.1002/stem.1787. [DOI] [PubMed] [Google Scholar]
- 180.Abuhatzira L, Shamir A, Schones DE, Schäffer AA, Bustin M. The chromatin-binding protein HMGN1 regulates the expression of Methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice. J Biol Chem. 2011;286(49):42051–42062. doi: 10.1074/jbc.M111.300541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28(3):205–211. doi: 10.1016/S0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 182.Samaco RC, Fryer JD, Ren J, Fyffe S, Chao HT, Sun Y, et al. A partial loss of function allele of Methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum Mol Genet. 2008;17(12):1718–1727. doi: 10.1093/hmg/ddn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–481. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 184.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. J Biol Chem. 2003;278(34):32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 185.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010;30(19):4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee W, Kim J, Yun JM, Ohn T, Gong Q. MeCP2 regulates gene expression through recognition of H3K27me3. Nat Commun. 2020;11(1):3140. doi: 10.1038/s41467-020-16907-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lorincz MC, Schübeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 Recruitment and localized histone H3 deacetylation. Mol Cell Biol. 2001;21(23):7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Korbel JO, Tirosh-Wagner T, Urban AE, Chen XN, Kasowski M, Dai L, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci USA. 2009;106(29):12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pinto B, Morelli G, Rastogi M, Savardi A, Fumagalli A, Petretto A, et al. Rescuing over-activated microglia restores cognitive performance in juvenile animals of the Dp(16) mouse model of down syndrome. Neuron. 2020;108(5):887–904.e12. doi: 10.1016/j.neuron.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gardiner K, Herault Y, Lott IT, Antonarakis SE, Reeves RH, Dierssen M. Down syndrome: from understanding the neurobiology to therapy. J Neurosci. 2010;30(45):14943–14945. doi: 10.1523/JNEUROSCI.3728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Theodoropoulos DS, Cowan JM, Elias ER, Cole C. Physical findings in 21q22 deletion suggest critical region for 21q- phenotype in q22. Am J Med Genet. 1995;59(2):161–163. doi: 10.1002/ajmg.1320590209. [DOI] [PubMed] [Google Scholar]
- 192.Pash J, Popescu N, Matocha M, Rapoport S, Bustin M. Chromosomal protein HMG-14 gene maps to the Down syndrome region of human chromosome 21 and is overexpressed in mouse trisomy 16. Proc Natl Acad Sci USA. 1990;87(10):3836–3840. doi: 10.1073/pnas.87.10.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pash J, Smithgall T, Bustin M. Chromosomal protein HMG-14 is overexpressed in Down syndrome. Exp Cell Res. 1991;193(1):232–235. doi: 10.1016/0014-4827(91)90562-9. [DOI] [PubMed] [Google Scholar]
- 194.Ling KH, Hewitt CA, Tan KL, Cheah PS, Vidyadaran S, Lai MI, et al. Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks. BMC Genomics. 2014;15(1):624. doi: 10.1186/1471-2164-15-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Potier MC, Rivals I, Mercier G, Ettwiller L, Moldrich RX, Laffaire J, et al. Transcriptional disruptions in Down syndrome: a case study in the Ts1Cje mouse cerebellum during post-natal development. J Neurochem. 2006;97:104–109. doi: 10.1111/j.1471-4159.2005.03624.x. [DOI] [PubMed] [Google Scholar]
- 196.Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA. 1998;95(11):6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guedj F, Pennings JL, Massingham LJ, Wick HC, Siegel AE, Tantravahi U, et al. An integrated human/murine transcriptome and pathway approach to identify prenatal treatments for Down syndrome. Sci Rep. 2016;2(6):32353. doi: 10.1038/srep32353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11(2):177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 199.Lana-Elola E, Cater H, Watson-Scales S, Greenaway S, Müller-Winkler J, Gibbins D, et al. Comprehensive phenotypic analysis of the Dp1Tyb mouse strain reveals a broad range of Down syndrome-related phenotypes. Dis Model Mech. 2021;14(10):dmm049157. doi: 10.1242/dmm.049157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kazuki Y, Gao FJ, Li Y, Moyer AJ, Devenney B, Hiramatsu K, et al. A non-mosaic transchromosomic mouse model of down syndrome carrying the long arm of human chromosome 21. Elife. 2020;29(9):e56223. doi: 10.7554/eLife.56223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kong XD, Liu N, Xu XJ. Bioinformatics analysis of biomarkers and transcriptional factor motifs in Down syndrome. Braz J Med Biol Res. 2014;47(10):834–841. doi: 10.1590/1414-431X20143792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, et al. Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron. 2016;89(6):1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rodríguez-Ortiz A, Montoya-Villegas JC, García-Vallejo F, Mina-Paz Y. Spatial and temporal expression of high-mobility-group nucleosome-binding (HMGN) genes in brain areas associated with cognition in individuals with Down syndrome. Genes. 2021;12(12):2000. doi: 10.3390/genes12122000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Patkee PA, Baburamani AA, Kyriakopoulou V, Davidson A, Avini E, Dimitrova R, et al. Early alterations in cortical and cerebellar regional brain growth in Down Syndrome: an in vivo fetal and neonatal MRI assessment. NeuroImage Clin. 2020;25:102139. doi: 10.1016/j.nicl.2019.102139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schmidt-Sidor B, Wisniewski KE, Shepard TH, Sersen EA. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin Neuropathol. 1990;9(4):181–190. [PubMed] [Google Scholar]
- 206.Tang XY, Xu L, Wang J, Hong Y, Wang Y, Zhu Q, et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J Clin Invest. 2021;131(12):135763. doi: 10.1172/JCI135763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li Z, Klein JA, Rampam S, Kurzion R, Patel Y, Haydar TF, et al. Asynchronous excitatory neuron development in an isogenic cortical spheroid model of Down syndrome. Front Neurosci. 2022;16:932384. doi: 10.3389/fnins.2022.932384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chakrabarti L, Galdzicki Z, Haydar TF. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27(43):11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Roper RJ, Baxter LL, Saran NG, Klinedinst DK, Beachy PA, Reeves RH. Defective cerebellar response to mitogenic Hedgehog signaling in Down’s syndrome mice. Proc Natl Acad Sci USA. 2006;103(5):1452–1456. doi: 10.1073/pnas.0510750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rotmensch S, Goldstein I, Liberati M, Shalev J, Ben-Rafael Z, Copel JA. Fetal transcerebellar diameter in Down syndrome. Obstet Gynecol. 1997;89(4):534–537. doi: 10.1016/S0029-7844(97)00076-8. [DOI] [PubMed] [Google Scholar]
- 211.Klein JA, Haydar TF. Neurodevelopment in Down syndrome: concordance in humans and models. Front Cell Neurosci. 2022;15(16):941855. doi: 10.3389/fncel.2022.941855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Guidi S, Giacomini A, Stagni F, Emili M, Uguagliati B, Bonasoni MP, et al. Abnormal development of the inferior temporal region in fetuses with Down syndrome. Brain Pathol. 2018;28(6):986–998. doi: 10.1111/bpa.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Larsen KB, Laursen H, Græm N, Samuelsen GB, Bogdanovic N, Pakkenberg B. Reduced cell number in the neocortical part of the human fetal brain in Down syndrome. Ann Anat-Anatomischer Anzeiger. 2008;190(5):421–427. doi: 10.1016/j.aanat.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 214.Sylvester PE. The hippocampus in Down’s syndrome. J Ment Defic Res. 1983;27(Pt 3):227–236. doi: 10.1111/j.1365-2788.1983.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 215.Chan WY, Lorke DE, Tiu SC, Yew DT. Proliferation and apoptosis in the developing human neocortex. Anat Rec. 2002;267(4):261–276. doi: 10.1002/ar.10100. [DOI] [PubMed] [Google Scholar]
- 216.Stagni F, Giacomini A, Emili M, Uguagliati B, Bonasoni MP, Bartesaghi R, et al. Neuroanatomical alterations in higher-order thalamic nuclei of fetuses with Down syndrome. Clin Neurol Neurosurg. 2020;194:105870. doi: 10.1016/j.clineuro.2020.105870. [DOI] [PubMed] [Google Scholar]
- 217.Hibaoui Y, Grad I, Letourneau A, Sailani MR, Dahoun S, Santoni FA, et al. Modelling and rescuing neurodevelopmental defect of D own syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol Med. 2014;6(2):259–277. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Contestabile A, Greco B, Ghezzi D, Tucci V, Benfenati F, Gasparini L. Lithium rescues synaptic plasticity and memory in Down syndrome mice. J Clin Invest. 2013;123(1):348–361. doi: 10.1172/JCI64650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fernandez-Martinez J, Vela EM, Tora-Ponsioen M, Ocaña OH, Nieto MA, Galceran J. Attenuation of Notch signalling by the Down-syndrome-associated kinase DYRK1A. J Cell Sci. 2009;122(Pt 10):1574–1583. doi: 10.1242/jcs.044354. [DOI] [PubMed] [Google Scholar]
- 220.Chen X, Salehi A, Pearn ML, Overk C, Nguyen PD, Kleschevnikov AM, et al. Targeting increased levels of APP in Down syndrome: Posiphen-mediated reductions in APP and its products reverse endosomal phenotypes in the Ts65Dn mouse model. Alzheimer’s Dement. 2021;17(2):271–292. doi: 10.1002/alz.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chen XQ, Sawa M, Mobley WC. Dysregulation of neurotrophin signaling in the pathogenesis of Alzheimer disease and of Alzheimer disease in Down syndrome. Free Radical Biol Med. 2018;114:52–61. doi: 10.1016/j.freeradbiomed.2017.10.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Reiche L, Küry P, Göttle P. Aberrant oligodendrogenesis in Down syndrome: shift in gliogenesis? Cells. 2019;8(12):1591. doi: 10.3390/cells8121591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Klein JA, Li Z, Rampam S, Cardini J, Ayoub A, Shaw P, et al. Sonic hedgehog pathway modulation normalizes expression of Olig2 in rostrally patterned NPCs with trisomy 21. Front Cell Neurosci. 2022;4(15):794675. doi: 10.3389/fncel.2021.794675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zakaria M, Ferent J, Hristovska I, Laouarem Y, Zahaf A, Kassoussi A, et al. The Shh receptor Boc is important for myelin formation and repair. Development. 2019;146(9):dev172502. doi: 10.1242/dev.172502. [DOI] [PubMed] [Google Scholar]
- 225.Becker LE, Mito T, Takashima S, Onodera K, Friend WC. Association of phenotypic abnormalities of Down syndrome with an imbalance of genes on chromosome 21. APMIS Suppl. 1993;40:57–70. [PubMed] [Google Scholar]
- 226.Mito T, Becker LE. Developmental changes of S-100 protein and glial fibrillary acidic protein in the brain in Down syndrome. Exp Neurol. 1993;120(2):170–176. doi: 10.1006/exnr.1993.1052. [DOI] [PubMed] [Google Scholar]
- 227.Lu J, Esposito G, Scuderi C, Steardo L, Delli-Bovi LC, Hecht JL, et al. S100B and APP promote a gliocentric shift and impaired neurogenesis in Down syndrome neural progenitors. PLoS ONE. 2011;6(7):e22126. doi: 10.1371/journal.pone.0022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chakrabarti L, Best TK, Cramer NP, Carney RSE, Isaac JTR, Galdzicki Z, et al. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13(8):927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jakovcevski I. Olig Transcription factors are expressed in oligodendrocyte and neuronal cells in human fetal CNS. J Neurosci. 2005;25(44):10064–10073. doi: 10.1523/JNEUROSCI.2324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xu R, Brawner AT, Li S, Liu JJ, Kim H, Xue H, et al. OLIG2 drives abnormal neurodevelopmental phenotypes in human iPSC-based organoid and chimeric mouse models of down syndrome. Cell Stem Cell. 2019;24(6):908–926.e8. doi: 10.1016/j.stem.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fischer DF. Activation of the Notch pathway in Down syndrome: cross-talk of Notch and APP. FASEB J. 2005;19(11):1451–1458. doi: 10.1096/fj.04-3395.com. [DOI] [PubMed] [Google Scholar]
- 232.Jang J, Byun SH, Han D, Lee J, Kim J, Lee N, et al. Notch intracellular domain deficiency in nuclear localization activity retains the ability to enhance neural stem cell character and block neurogenesis in mammalian brain development. Stem Cells Dev. 2014;23(23):2841–2850. doi: 10.1089/scd.2014.0031. [DOI] [PubMed] [Google Scholar]
- 233.Zhou ZW, Kirtay M, Schneble N, Yakoub G, Ding M, Rüdiger T, et al. NBS1 interacts with Notch signaling in neuronal homeostasis. Nucleic Acids Res. 2020;48(19):10924–10939. doi: 10.1093/nar/gkaa716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Edri R, Yaffe Y, Ziller MJ, Mutukula N, Volkman R, David E, et al. Analysing human neural stem cell ontogeny by consecutive isolation of Notch active neural progenitors. Nat Commun. 2015;6(1):6500. doi: 10.1038/ncomms7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nuytten M, Beke L, Van Eynde A, Ceulemans H, Beullens M, Van Hummelen P, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27(10):1449–1460. doi: 10.1038/sj.onc.1210774. [DOI] [PubMed] [Google Scholar]
- 236.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Palmer CR, Liu CS, Romanow WJ, Lee MH, Chun J. Altered cell and RNA isoform diversity in aging Down syndrome brains. Proc Natl Acad Sci USA. 2021;118(47):e2114326118. doi: 10.1073/pnas.2114326118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kurt MA, Davies DC, Kidd M, Dierssen M, Flórez J. Synaptic deficit in the temporal cortex of partial trisomy 16 (Ts65Dn) mice. Brain Res. 2000;858(1):191–197. doi: 10.1016/S0006-8993(00)01984-3. [DOI] [PubMed] [Google Scholar]
- 239.Belichenko PV, Kleschevnikov AM, Masliah E, Wu C, Takimoto-Kimura R, Salehi A, et al. Excitatory-inhibitory relationship in the fascia dentata in the Ts65Dn mouse model of down syndrome. J Comp Neurol. 2009;512(4):453–466. doi: 10.1002/cne.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pérez-Cremades D, Hernández S, Blasco-Ibáñez JM, Crespo C, Nacher J, Varea E. Alteration of inhibitory circuits in the somatosensory cortex of Ts65Dn mice, a model for Down’s syndrome. J Neural Transm. 2010;117(4):445–455. doi: 10.1007/s00702-010-0376-9. [DOI] [PubMed] [Google Scholar]
- 241.Giffin-Rao Y, Sheng J, Strand B, Xu K, Huang L, Medo M, et al. Altered patterning of trisomy 21 interneuron progenitors. Stem Cell Reports. 2022;17(6):1366–1379. doi: 10.1016/j.stemcr.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Huo HQ, Qu ZY, Yuan F, Ma L, Yao L, Xu M, et al. Modeling Down syndrome with patient iPSCs reveals cellular and migration deficits of GABAergic neurons. Stem Cell Reports. 2018;10(4):1251–1266. doi: 10.1016/j.stemcr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mona B, Uruena A, Kollipara RK, Ma Z, Borromeo MD, Chang JC, et al. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife. 2017;6:e25787. doi: 10.7554/eLife.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Seidl R, Bidmon B, Bajo M, Yoo PC, Cairns N, LaCasse EC, et al. Evidence for apoptosis in the fetal Down syndrome brain. J Child Neurol. 2001;16(6):438–442. doi: 10.1177/088307380101600610. [DOI] [PubMed] [Google Scholar]
- 245.Mann DM, Yates PO, Marcyniuk B, Ravindra CR. Loss of neurones from cortical and subcortical areas in Down’s syndrome patients at middle age. Quantitative comparisons with younger Down’s patients and patients with Alzheimer’s disease. J Neurol Sci. 1987;80(1):79–89. doi: 10.1016/0022-510X(87)90223-1. [DOI] [PubMed] [Google Scholar]
- 246.Contestabile A, Fila T, Cappellini A, Bartesaghi R, Ciani E. Widespread impairment of cell proliferation in the neonate Ts65Dn mouse, a model for Down syndrome. Cell Prolif. 2009;42(2):171–181. doi: 10.1111/j.1365-2184.2009.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zdaniuk G, Wierzba-Bobrowicz T, Szpak GM, Stępień T. Astroglia disturbances during development of the central nervous system in fetuses with Down’s syndrome. Folia Neuropathol. 2011;49(2):109–114. [PubMed] [Google Scholar]
- 248.Kurabayashi N, Nguyen MD, Sanada K. DYRK1A overexpression enhances STAT activity and astrogliogenesis in a Down syndrome mouse model. EMBO Rep. 2015;16(11):1548–1562. doi: 10.15252/embr.201540374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Vorobyeva AG, Saunders AJ. Amyloid-β interrupts canonical Sonic hedgehog signaling by distorting primary cilia structure. Cilia. 2018;7:5. doi: 10.1186/s13630-018-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ábrahám H, Vincze A, Veszprémi B, Kravják A, Gömöri É, Kovács GG, et al. Impaired myelination of the human hippocampal formation in Down syndrome. Int j dev neurosci. 2012;30(2):147–158. doi: 10.1016/j.ijdevneu.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 251.Wisniewski KE, Schmidt-Sidor B. Postnatal delay of myelin formation in brains from Down syndrome infants and children. Clin Neuropathol. 1989;8(2):55–62. [PubMed] [Google Scholar]
- 252.Trazzi S, Mitrugno VM, Valli E, Fuchs C, Rizzi S, Guidi S, et al. APP-dependent up-regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum Mol Genet. 2011;20(8):1560–1573. doi: 10.1093/hmg/ddr033. [DOI] [PubMed] [Google Scholar]
- 253.Currier DG, Polk RC, Reeves RH. A Sonic hedgehog (Shh) response deficit in trisomic cells may be a common denominator for multiple features of Down syndrome. Prog Brain Res. 2012;197:223–236. doi: 10.1016/B978-0-444-54299-1.00011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Head E, Azizeh BY, Lott IT, Tenner AJ, Cotman CW, Cribbs DH. Complement association with neurons and beta-amyloid deposition in the brains of aged individuals with Down Syndrome. Neurobiol Dis. 2001;8(2):252–265. doi: 10.1006/nbdi.2000.0380. [DOI] [PubMed] [Google Scholar]
- 255.Flores-Aguilar L, Iulita MF, Kovecses O, Torres MD, Levi SM, Zhang Y, et al. Evolution of neuroinflammation across the lifespan of individuals with Down syndrome. Brain. 2020;143(12):3653–3671. doi: 10.1093/brain/awaa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wilcock DM, Hurban J, Helman AM, Sudduth TL, McCarty KL, Beckett TL, et al. Down syndrome individuals with Alzheimer’s disease have a distinct neuroinflammatory phenotype compared to sporadic Alzheimer’s disease. Neurobiol Aging. 2015;36(9):2468–2474. doi: 10.1016/j.neurobiolaging.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, et al. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci USA. 1989;86(19):7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bambrick LL, Yarowsky PJ, Krueger BK. Altered astrocyte calcium homeostasis and proliferation in theTs65Dn mouse, a model of Down syndrome. J Neurosci Res. 2003;73(1):89–94. doi: 10.1002/jnr.10630. [DOI] [PubMed] [Google Scholar]
- 259.Shukkur EA, Shimohata A, Akagi T, Yu W, Yamaguchi M, Murayama M, et al. Mitochondrial dysfunction and tau hyperphosphorylation in Ts1Cje, a mouse model for Down syndrome. Hum Mol Genet. 2006;15(18):2752–2762. doi: 10.1093/hmg/ddl211. [DOI] [PubMed] [Google Scholar]
- 260.Helguera P, Seiglie J, Rodriguez J, Hanna M, Helguera G, Busciglio J. Adaptive downregulation of mitochondrial function in Down syndrome. Cell Metab. 2013;17(1):132–140. doi: 10.1016/j.cmet.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wilcock DM, Griffin WST. Down’s syndrome, neuroinflammation, and Alzheimer neuropathogenesis. J Neuroinflammation. 2013;16(10):84. doi: 10.1186/1742-2094-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kong XF, Worley L, Rinchai D, Bondet V, Jithesh PV, Goulet M, et al. Three copies of four interferon receptor genes underlie a mild type I interferonopathy in Down syndrome. J Clin Immunol. 2020;40(6):807–819. doi: 10.1007/s10875-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Royston MC, McKenzie JE, Gentleman SM, Sheng JG, Mann DM, Griffin WS, et al. Overexpression of s100beta in Down’s syndrome: correlation with patient age and with beta-amyloid deposition. Neuropathol Appl Neurobiol. 1999;25(5):387–393. doi: 10.1046/j.1365-2990.1999.00196.x. [DOI] [PubMed] [Google Scholar]
- 264.Wee ZN, Li Z, Lee PL, Lee ST, Lim YP, Yu Q. EZH2-mediated inactivation of IFN-γ-JAK-STAT1 signaling is an effective therapeutic target in MYC-driven prostate cancer. Cell Rep. 2014;8(1):204–216. doi: 10.1016/j.celrep.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 265.Du W, Frankel TL, Green M, Zou W. IFNγ signaling integrity in colorectal cancer immunity and immunotherapy. Cell Mol Immunol. 2022;19(1):23–32. doi: 10.1038/s41423-021-00735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Tiffen J, Gallagher SJ, Filipp F, Gunatilake D, Emran AA, Cullinane C, et al. EZH2 cooperates with DNA methylation to downregulate key tumor suppressors and IFN gene signatures in melanoma. J Invest Dermatol. 2020;140(12):2442–2454.e5. doi: 10.1016/j.jid.2020.02.042. [DOI] [PubMed] [Google Scholar]
- 267.Yang D, Postnikov YV, Li Y, Tewary P, de la Rosa G, Wei F, et al. High-mobility group nucleosome-binding protein 1 acts as an alarmin and is critical for lipopolysaccharide-induced immune responses. J Exp Med. 2012;209(1):157–171. doi: 10.1084/jem.20101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wei F, Yang D, Tewary P, Li Y, Li S, Chen X, et al. The Alarmin HMGN1 contributes to antitumor immunity and is a potent immunoadjuvant. Cancer Res. 2014;74(21):5989–5998. doi: 10.1158/0008-5472.CAN-13-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yang D, Bustin M, Oppenheim JJ. Harnessing the alarmin HMGN1 for anticancer therapy. Immunotherapy. 2015;7(11):1129–1131. doi: 10.2217/imt.15.76. [DOI] [PubMed] [Google Scholar]
- 270.Chen CY, Ueha S, Ishiwata Y, Yokochi S, Yang D, Oppenheim JJ, et al. Combined treatment with HMGN1 and anti-CD4 depleting antibody reverses T cell exhaustion and exerts robust anti-tumor effects in mice. J Immunother Cancer. 2019;7(1):21. doi: 10.1186/s40425-019-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Alam MM, Yang D, Trivett A, Meyer TJ, Oppenheim JJ. HMGN1 and R848 synergistically activate dendritic cells using multiple signaling pathways. Front Immunol. 2018;9:2982. doi: 10.3389/fimmu.2018.02982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mann DMA, Esiri MM. The pattern of acquisition of plaques and tangles in the brains of patients under 50 years of age with Down’s syndrome. J Neurol Sci. 1989;89(2–3):169–179. doi: 10.1016/0022-510X(89)90019-1. [DOI] [PubMed] [Google Scholar]
- 273.Schupf N, Kapell D, Nightingale B, Rodriguez A, Tycko B, Mayeux R. Earlier onset of Alzheimer’s disease in men with Down syndrome. Neurology. 1998;50(4):991–995. doi: 10.1212/WNL.50.4.991. [DOI] [PubMed] [Google Scholar]
- 274.Perluigi M, Butterfield DA. Oxidative stress and Down syndrome: a route toward Alzheimer-like dementia. Curr Gerontol Geriatr Res. 2012;2012:724904. doi: 10.1155/2012/724904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hof PR. Age-related distribution of neuropathologic changes in the cerebral cortex of patients with Down’s syndrome: quantitative regional analysis and comparison With Alzheimer’s disease. Arch Neurol. 1995;52(4):379. doi: 10.1001/archneur.1995.00540280065020. [DOI] [PubMed] [Google Scholar]
- 276.Lai F, Kammann E, Rebeck GW, Anderson A, Chen Y, Nixon RA. APOE genotype and gender effects on Alzheimer disease in 100 adults with Down syndrome. Neurology. 1999;53(2):331–331. doi: 10.1212/WNL.53.2.331. [DOI] [PubMed] [Google Scholar]
- 277.Lithner CU, Lacor PN, Zhao WQ, Mustafiz T, Klein WL, Sweatt JD, et al. Disruption of neocortical histone H3 homeostasis by soluble Aβ: implications for Alzheimer’s disease. Neurobiol Aging. 2013;34(9):2081–2090. doi: 10.1016/j.neurobiolaging.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 278.Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Transl Psychiatry. 2012;2(7):e132–e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhang K, Schrag M, Crofton A, Trivedi R, Vinters H, Kirsch W. Targeted proteomics for quantification of histone acetylation in Alzheimer’s disease. Proteomics. 2012;12(8):1261–1268. doi: 10.1002/pmic.201200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Persico G, Casciaro F, Amatori S, Rusin M, Cantatore F, Perna A, et al. Histone H3 lysine 4 and 27 trimethylation landscape of human Alzheimer’s disease. Cells. 2022;11(4):734. doi: 10.3390/cells11040734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Marzi SJ, Leung SK, Ribarska T, Hannon E, Smith AR, Pishva E, et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat Neurosci. 2018;21(11):1618–1627. doi: 10.1038/s41593-018-0253-7. [DOI] [PubMed] [Google Scholar]
- 282.Cruchaga C, Chakraverty S, Mayo K, Vallania FLM, Mitra RD, Faber K, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s Disease families. PLoS ONE. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391(6665):387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 284.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 285.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, et al. Secreted amyloid β–protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2(8):864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 286.Narayan PJ, Lill C, Faull R, Curtis MA, Dragunow M. Increased acetyl and total histone levels in post-mortem Alzheimer’s disease brain. Neurobiol Dis. 2015;74:281–294. doi: 10.1016/j.nbd.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 287.Nativio R, Lan Y, Donahue G, Sidoli S, Berson A, Srinivasan AR, et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat Genet. 2020;52(10):1024–1035. doi: 10.1038/s41588-020-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53 949) Mol Psychiatry. 2015;20(2):183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.