Abstract
Mannose-binding lectin (MBL) is a collagenous serum lectin believed to be of importance in innate immunity. Genetically determined low levels of the protein are known to predispose to infections. In this study the binding of purified MBL to pathogens isolated from immunocompromised children was investigated by flow cytometry. Diverse Candida species, Aspergillus fumigatus, Staphylococcus aureus, and beta-hemolytic group A streptococci exhibited strong binding of MBL, whereas Escherichia coli, Klebsiella species, and Haemophilus influenzae type b were characterized by heterogeneous binding patterns. In contrast, beta-hemolytic group B streptococci, Streptococcus pneumoniae, and Staphylococcus epidermidis showed low levels of binding. Bound MBL was able to promote C4 deposition in a concentration-dependent manner. We conclude that MBL may be of importance in first-line immune defense against several important pathogens.
Mannose-binding lectin (MBL) is a serum protein of hepatic origin belonging to a family of Ca2+-dependent collagenous lectins, most of which are components of the innate immune system (10). MBL is able to bind through multiple sites to various carbohydrate structures (29, 37) and, on binding to its ligands, is able to activate complement in an antibody- and C1q-independent manner (12, 24, 26) using MBL-associated serine protease 1 (MASP-1) and MASP-2 (23, 34).
In humans, low levels of MBL are caused by one of three structural mutations found within exon 1 of the MBL gene (20, 22, 31). These single point mutations in codons 52, 54, and 57 result in amino acid substitutions which are believed to interfere with the stability of the protein (20, 22, 31). Individuals heterozygous for the codon 54 or 57 mutation are found at relatively high frequency within Eurasian and sub-Saharan African populations, respectively (7, 19, 20, 22, 25), and such mutations have been shown to be associated with a generalized increased risk of infections (6, 32).
MBL has the potential to express multiple biological effector functions, but a prerequisite for all such activity is primary binding to multiple arrays of sugar ligands such as those expressed on microbial surfaces. However, to date, there have been few detailed studies of the interactions between MBL and a wide range of clinically relevant microorganisms. Here, we report such a study in which, by using flow cytometry, it has been possible to detect major differences in the MBL binding capacities of different microorganisms and even differences among organisms of the same genus. In addition, we have shown that bound MBL promotes, in a concentration-dependent manner, C4 deposition on the surfaces of organisms.
MATERIALS AND METHODS
Preparation of MBL.
MBL was prepared as previously described by Kilpatrick (17). Briefly, 500 g of frozen ethanol-fractionated human plasma paste (fraction B+1, equivalent to Cohn fraction I+III; donated by C. Dash, Blood Products Laboratory, Elstree, United Kingdom) was purified by ammonium sulfate precipitation to give 42% saturation. After dialysis the solution was applied to a mannan-agarose (Sigma, Poole, United Kingdom) column (5-ml packed volume; Pharmacia Biotech, Uppsala, Sweden) and the calcium-dependent proteins were eluted with 0.01 M EDTA. The first EDTA eluate was recalcified to 0.05 M CaCl2, reapplied to the same mannan-agarose column, and eluted with 0.1 M mannose. The concentration of MBL was determined by enzyme-linked immunosorbent assay (19), and sample purity was verified by nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 3 to 10% polyacrylamide gradient gel and silver staining. Bands observed on silver staining were confirmed to be higher-order oligomers of MBL by immunoblotting and enhanced chemiluminescence detection according to the method of Lipscombe et al. (21). MBL prepared in this manner is known to be noncovalently associated with MASP (23).
Preparation of C4.
C4 was prepared from the plasma of a single donor by chromatography on Q Sepharose FF, followed by affinity chromatography using monoclonal (clone L003) anti-C4 Sepharose and anion exchange using MonoQ (Pharmacia) (2).
Organisms.
The organisms studied comprised isolates from the nasopharynx and blood cultures from immunocompromised children treated at Great Ormond Street Hospital for Children. The patient group included patients with oncological disease undergoing chemotherapy, children undergoing dialysis while suffering from renal failure, and patients with a clearly defined immunodeficiency. Isolates were collected and randomized by the Department of Medical Microbiology. Organisms were identified by standard laboratory techniques or classified using API20 NE and API20 Strep (BioMerieux). At least three different isolates of the same genus were collected. In addition to these clinical isolates the following organisms were also available for study: 7 defined strains (Staphylococcus aureus NCTC6571, Pseudomonas aeruginosa NCTC10662, Escherichia coli NCTC10418, Staphylococcus epidermidis ATCC 1228, Enterobacter aerogenes ATCC 13048, Streptococcus pneumoniae ATCC 6305, and Candida albicans ATCC 10231), 7 internal quality controls (S. aureus, C. albicans, Candida parapsilosis, Candida tropicalis, Candida lusitanae, and Cryptococcus neoformans [n = 2]; Department of Medical Microbiology), 10 defined S. pneumoniae vaccination strains (1, 3-6, 9V, 14, 19F, 18, and 23; kindly provided by the Public Health Laboratory Service), and 2 defined C. neoformans strains (1 encapsulated isolate, B3501, and its coisogenic acapsular mutant, B4131).
Growth and preparation of organisms.
Bacteria and fungi were cultured as described by Bridson (The Oxoid manual, 7th ed., Oxoid, Buckinghamshire, United Kingdom). Organisms were subcultured on agar once before use. Immediately before each experiment organisms were suspended in Veronal-buffered saline supplemented with 5 mM CaCl2 and 5 mM MgCl2 at 3 × 108 to 8 × 108 organisms/ml (measured as an absorbance of 1.0 at 540 nm).
Binding of MBL to organisms.
Binding of MBL to organisms was performed as previously described by Jack et al. (14). Briefly, 3 × 108 to 8 × 108 organisms/ml were incubated with 5 μg of MBL/ml for 30 min at 37°C. After the addition of 2.5 μg of fluorescein isothiocyanate (FITC)-conjugated anti-MBL (FITC-labeled clone 131-1; State Serum Institute, Copenhagen, Denmark; conjugated as described by Johnson and Holborow [15])/ml and incubation as described above, MBL binding was measured by flow cytometry performed on a FACS-Calibur at low flow rates using CellQuest software (Becton Dickinson).
The binding of MBL to the organisms was evaluated on at least three occasions and, whenever possible, compared to that for the defined strains. MBL binding to S. aureus NCTC6571 was included in each experiment as a positive control. A negative control comprising organisms processed in the same way but in the absence of MBL was included in every assay. Data were evaluated both as the percentage of positive organisms and as median fluorescence intensity (MFI). The binding of MBL to organisms was categorized arbitrarily into three levels based on MFI, namely, low (<5), moderate (5 to 20), or high (>20).
Specificity of MBL binding.
In order to evaluate whether the binding observed was mediated by C-type lectin interactions, inhibition experiments using different concentrations of monosaccharides (d-mannose and N-acetyl-d-glucosamine) as well as calcium chelating agent EDTA were performed. The monosaccharide or EDTA was added to the MBL solution 10 min prior to the addition of the MBL to six different microorganisms.
Binding of C4 to organisms.
The binding of C4 to organisms was performed as previously described by Jack et al. (14) with some modifications. Briefly, 3 × 108 to 8 × 108 organisms/ml were preincubated with MBL for 30 min at 37°C. C4 was added at a concentration of 140 μg/ml, and the organisms were incubated for a further 15 min at 37°C. A mixture of FITC-conjugated anti-MBL and biotinylated anti-C4d (Quidel) was added at a final concentration of 4 μg/ml for each, and the organisms were incubated for 20 min at 37°C. Biotin conjugation of anti-C4d was carried out using standard methodology. Streptavidin–phycoerythrin-Cy 5 (Pharmingen) at a concentration of 0.66 μg/ml was added, and the organisms were incubated as described above. MBL binding and C4 deposition were measured by two-color flow cytometry performed as described above.
RESULTS
Clinical isolates.
Over a period of 4 months 54 clinical isolates from either the nasopharynges (n = 30) or cultured blood (n = 24) of immunocompromised children were collected and investigated. These included S. aureus (n = 12), S. epidermidis (n = 6), gram-negative bacterium (n = 12), C. albicans (n = 3), Aspergillus fumigatus (n = 3), beta-hemolytic group A streptococci (n = 5), beta-hemolytic group B streptococci (n = 4), Streptococcus sanguis (n = 2), Enterococcus faecalis (n = 2), and S. pneumoniae (n = 5) isolates.
Flow cytometric analysis.
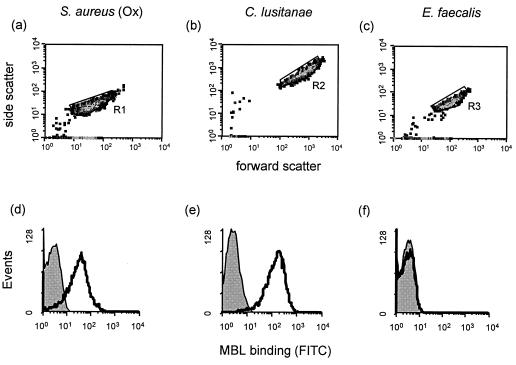
Distinct populations of all of the above organisms could be visualized on the basis of size (forward scatter) and granularity (side scatter) by flow cytometry. Representative population density plots for three of these organisms (S. aureus NCTC6571, C. lusitanae, and E. faecalis) are shown in Fig. 1a to c. The levels of binding of MBL to each of these three organisms also differed markedly and are shown in Fig. 1d to f.
FIG. 1.
Representative population density plots of three organisms (S. aureus NCTC6571, C. lusitanae, and E. faecalis) detected by a combination of forward scatter and side scatter and gated as regions R1, R2, and R3, respectively (a to c). The corresponding flow cytometric profiles of MBL binding to these organisms using the protein at a final concentration of 5 μg/ml are shown (d to f). The control profiles (shaded) show organisms incubated with FITC-conjugated anti-MBL alone. Experimental samples (open profiles) were obtained after incubation of organisms with MBL and FITC-conjugated anti-MBL.
The use of MBL at a final concentration of 5 μg/ml resulted in strong binding of the lectin to S. aureus NCTC6571 and also to C. lusitanae. In contrast, a low level of binding to E. faecalis was observed.
Binding of MBL to microorganisms.
The commonest isolate studied was S. aureus. Eight isolates were collected from the nasopharynx, and four isolates were from blood culture. All isolates of S. aureus (n = 12), including three methicillin-resistant isolates, were found to bind MBL in the moderate and high categories. Similar binding patterns were also seen with C. albicans (n = 3) and A. fumigatus (n = 3). In addition, all beta-hemolytic group A streptococci (n = 5), three isolates collected from the nasopharynx and two isolates from blood culture, showed a distinct pattern of MBL deposition, with 60 to 80% of the organisms binding the protein. In contrast, all beta-hemolytic group B streptococci (n = 4), all S. pneumoniae isolates (n = 5), and two isolates each of E. faecalis and S. sanguis showed low binding. The same results were obtained when selected S. pneumoniae isolates were grown in log phase. Similarly S. epidermidis (n = 6) generally demonstrated low binding activity except for one isolate, which showed moderate binding. Most gram-negative isolates (n = 12) bound little or no MBL, apart from one isolate each of E. coli, Klebsiella aerogenes, and Haemophilus influenzae type b showing high or moderate MBL binding. The E. coli and K. aerogenes strains were isolated from blood cultures obtained from the same patient, who was subsequently shown to be heterozygous for the codon 54 mutation of the MBL gene.
In many analyses it was noted that a minority of organisms (up to 20%) bound MBL whereas the majority showed no such binding. In Fig. 2a a typical example of such a pattern is shown. Further analysis of the binding population (Fig. 2c) failed to demonstrate any distinctive features (e.g., in size and granularity) compared to the population as a whole, shown in Fig. 2b, indicating that the binding was not due to clumping or major disruption of the organisms. However, the intensity of the binding was always low, so, compared to background staining (in the absence of MBL) little difference was observed.
FIG. 2.
Representative cytometric profile of MBL binding to K. aerogenes (a). A typical tail is seen in the experimental sample, shown as an open profile, but not in the control profile (shaded). The forward scatter-versus-side scatter density plot of the entire population (b) is shown. The size and granularity characteristics of the subpopulation (c) did not differ markedly from those shown in panel b.
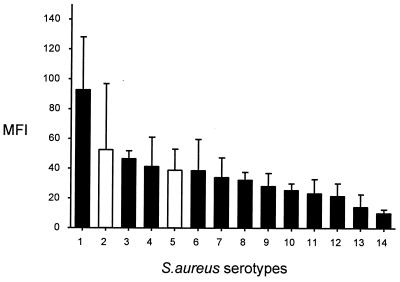
When MBL binding to organisms was expressed as the MFI rather than as the percentage of positive organisms, differences not only between different genera but also within one species were observed. This is illustrated in Fig. 3, where S. aureus isolates have been ranked in order of binding.
FIG. 3.
MBL binding to S. aureus isolates ranked in order of binding. Results are expressed as median fluorescence, with each bar representing the mean of three to six experiments; error bars, standard errors of the means. Solid bars, clinical isolates; open bars, defined strains.
Serum MBL levels are determined by both structural gene mutations and promoter region polymorphisms, which differ in frequency in the general population. Because of this heterogeneity, it was considered important to determine the concentration dependence of MBL binding, and Fig. 4 shows the results of an experiment in which S. aureus NCTC6571 was incubated with the protein.
FIG. 4.
MBL binding to S. aureus NCTC6571 at different concentrations. Binding of MBL is expressed as median fluorescence. Each point represents the mean of three experiments ± the standard error of the mean. Both structural gene mutations and promoter polymorphisms determine the serum MBL level and may influence the degree of MBL binding to the organisms in vivo. The approximate MBL concentration ranges of individuals with and without mutations are indicated by arrows.
In British Caucasians lacking MBL mutations, the protein is present at a median concentration of 1.63 μg/ml. Strong binding to S. aureus NCTC6571 was observed at a concentration close to this median. Minimal or poor binding was seen at MBL concentrations typically found in individuals heterozygous for the codon 54 mutation (0.358 μg/ml) and the codon 52 mutation (0.6 μg/ml). The median fluorescence of staining increased with increasing MBL concentrations and did not reach a plateau.
In order to study the influence of bacterial concentration on MBL binding, suspensions of S. aureus NCTC6571 were prepared over a range of concentrations from 108/ml to 7.25 × 105/ml. No differences in MBL binding were observed (in three separate experiments performed on three independent occasions; data not shown).
Specificity of MBL binding.
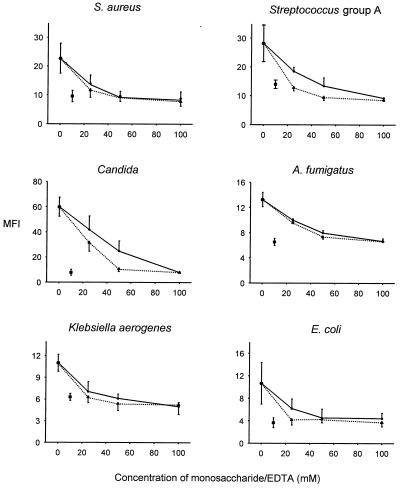
MBL binding to six representative organisms was markedly reduced in the presence of 10 mM EDTA, demonstrating the divalent-cation-dependent nature of the observed interactions (Fig. 5). In addition mannose and N-acetylglucosamine inhibited binding of MBL in a dose-dependent manner.
FIG. 5.
Inhibition of MBL binding to six representative microorganisms using two monosaccharides (solid line, d-mannose; dotted line, N-acetylglucosamine) at different concentrations and 10 mM EDTA (■). The monosaccharide or EDTA was added to the MBL solution 10 min prior to the addition of the MBL to the organisms. Each point represents the mean ± standard error of the mean of the MFI for three independent experiments.
Overall summary of observed MBL binding patterns.
In total 80 microorganisms were studied (clinical isolates and defined strains). The results, expressed as the ratio of median fluorescence using 5 μg of MBL/ml relative to the appropriate control, are illustrated in Fig. 6. Nearly half of the organisms bound MBL. However, three patterns of binding were observed. All isolates from S. aureus, C. albicans, A. fumigatus, and beta-hemolytic group A streptococci showed MBL binding. For several organisms a single isolate was observed to exhibit binding (E. coli, K. aerogenes, H. influenzae type b, S. epidermidis, and nonencapsulated C. neoformans). For six of the organisms (beta-hemolytic group B streptococci, S. pneumoniae, S. sanguis, E. faecalis, non-type b H. influenzae, and P. aeruginosa) there was no evidence of binding with any of the isolates tested.
FIG. 6.
MBL binding to 80 microorganisms. Data are expressed as the ratios of median fluorescence of the experimental samples (incubated with MBL and FITC-conjugated anti-MBL) to the median fluorescence of the control samples (incubated with FITC-conjugated anti-MBL alone). Each circle represents the mean of three to six experiments. The solid circles represent clinical isolates, whereas the open circles show either defined National Collection of Type Cultures strains or internal quality controls. The dashed line represents a ratio of one. β-haem. Strep. A indicates beta-hemolytic group A streptococcus.
Binding of C4.
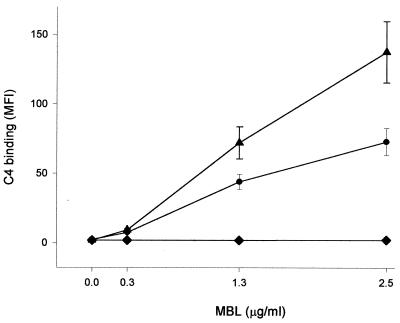
In order to investigate the ability of MBL and MASP bound to different organisms to activate the complement system, purified C4 was added to suspensions of three selected organisms. MBL at a range of concentrations (0 to 2.5 μg/ml) was preincubated with these organisms before exposure to C4 (140 μg/ml). As shown in Fig. 7, MBL concentration-dependent C4 binding was observed with S. aureus NCTC6571 and one strain of Klebsiella whereas no C4 was bound by another strain of Klebsiella, which had previously been shown not to bind MBL.
FIG. 7.
C4 deposition on three representative organisms previously incubated with different concentrations of MBL. S. aureus NCTC6571 (●) and one strain of Klebsiella (▴) were selected for their known ability to bind MBL, whereas another strain of Klebsiella (⧫) did not bind to the protein. Each point represents the mean ± the standard error of the mean of the MFI for three independent experiments.
DISCUSSION
The acquisition of detailed knowledge of the fine structure of MBL has been paralleled by a series of publications reporting significant binding of this collectin to particular microorganisms. Such reports have presented evidence of MBL binding to individual pathogens, including human immunodeficiency virus type 1 and influenza A virus (4, 8, 9), and to the yeasts C. albicans and C. neoformans (28, 33). There have also been studies of MBL binding to various bacteria such as Salmonella enterica serovar Montevideo (18), E. coli (16), Mycobacterium avium (27), and Neisseria meningitidis (14). To the best of our knowledge only one other study has attempted a more comprehensive investigation of binding. This was the report by van Emmerik et al. (36), which described studies using radiolabeled MBL and which focused on organisms known to cause meningeal disease (e.g., Listeria monocytogenes, streptococci, H. influenzae type b, and Neisseria species).
In the present study we have used a flow cytometric methodology developed and refined in a previous study (14) to investigate a wide range of clinically relevant microorganisms. In addition to being a nonisotopic procedure, this has the advantage that single organisms can be visualized and specificity controls can be readily incorporated.
We were particularly concerned that misleading results could arise if surface lectins expressed by the bacteria were able to interact with sugar groups on the MBL molecule. We have attempted to control for this by performing calcium chelation and dose-dependent inhibition experiments using N-acetylglucosamine and mannose with each organism exhibiting strong binding of MBL. The concordance observed suggests that the binding was indeed between the C-type lectin domain of MBL and sugar groups expressed on the surfaces of organisms.
While the binding of MBL to simple sugars is relatively well understood (3, 13, 29, 37, 38), the binding to complex microbial structures and surfaces has not been extensively studied. Although the variation in expression of mannose structures has been shown to alter MBL binding (18, 27), it is likely that in many instances pathogens also utilize other structures to evade MBL binding. The expression of capsular polysaccharide has been shown to decrease the binding of MBL to N. meningitidis serogroup B and H. influenzae type b (36) and to C. neoformans (28). The structure and composition of bacterial endotoxins have also been shown to have a major influence on MBL binding (11, 14). These structures appear to be able to mask ligands to which MBL could bind (e.g., mannose and N-acetylglucosamine) or may alter the sugar conformation or density to prevent MBL binding.
For certain microorganisms every isolate studied bound MBL unequivocally, although there was considerable variation in the amount deposited on the microbial surface as determined by measuring the MFI of binding. Examples of such organisms included S. aureus, C. albicans, A. fumigatus, and beta-hemolytic group A streptococcus. However, one of the most striking findings in this study was the identification of heterogeneity in MBL binding for certain organisms such as E. coli, Klebsiella species, and H. influenzae type b. This emphasizes the need to study several isolates of a particular organism before concluding that MBL either binds or does not bind.
It was noteworthy that in many of the analyses reported here a significant proportion of organisms (up to 20%) were observed to bind MBL (visualized typically as a tail in the fluorescence histogram) while the majority of organisms showed no such binding. Initially it was considered that this might be a technical artifact. However, the binding population failed to demonstrate any gross distinctive features (size and granularity) compared to the population as a whole. This suggests that the binding was not due to clumping or major disruption of the organisms. An alternative explanation is that it represents organisms expressing sugar arrays differing from those of the nonbinding majority. Such differences could reflect experimental damage or simply natural variation.
At the time of primary contact with any microorganism the role of the innate immune system, including MBL, is paramount. For this reason individuals who are homozygous for the structural gene mutations are believed to be at increased risk of infection, and several studies support this (5, 6, 32). However, heterozygous individuals also manifest significant reductions in protein level (typically 0.3 to 0.6 μg/ml) (20), and MBL binding to S. aureus at these concentrations was shown to be markedly impaired in this study.
We were also able to demonstrate a close relationship between C4 deposition and initial MBL concentration for both S. aureus NCTC6571 and one strain of Klebsiella. At an MBL concentration of 0.3 μg/ml there was minimal C4 deposition, but at 1.3 μg of MBL/ml significant levels of C4 could be detected. It is reasonable to extrapolate from these findings and conclude that C3b deposition would parallel the C4 results. The latter may therefore be regarded as a surrogate for MBL-mediated opsonophagocytosis.
It was of interest that two of the clinical isolates of organisms showing heterogeneity in MBL binding (K. aerogenes and E. coli) were from the same patient and that that particular individual was heterozygous for the codon 54 mutation of the MBL gene. There is, at present, no clear evidence about the minimal levels of MBL which might be protective against particular pathogens. The levels of MBL required may be dependent upon the nature of the infectious agent and/or variations in host defense mechanisms. Arriving at such estimates will pose formidable difficulties until we have more data on the relative importance of the various clearance mechanisms involved. In some cases simple steric hindrance by MBL may effectively inhibit the spread of organisms by blocking access to appropriate receptors. In other cases direct opsonic adherence or, more likely, complement activation and the deposition of C3b opsonins will be the critical routes of elimination. Recent evidence has also indicated that MBL can modulate the host inflammatory response to infections (1, 30). All of these aspects should be the target of further research and will help to identify the clinical situations in which MBL replacement or adjunctive therapy (35) might play a positive role.
ACKNOWLEDGMENTS
O.N. was supported by the Dr. Mildred Scheel Stiftung für Krebsforschung, Germany. D.L.J. is supported by the Wellcome Trust, United Kingdom.
We are grateful for the help and assistance of members of the Microbiology Department, Great Ormond Street Hospital For Children. We also thank D. Goldblatt for providing selected isolates of S. pneumoniae and G. Bancroft for providing the acapsular mutant of C. neoformans.
REFERENCES
- 1.Chaka W, Verheul A F M, Vaishnav V V, Cherniak R, Scharringa J, Verhof J, Snippe H, Hoepelman A I M. TNF-alpha in human peripheral blood mononuclear cells by the mannoprotein of Cryptococcus neoformans involves human mannose binding protein. J Immunol. 1997;159:2979–2985. [PubMed] [Google Scholar]
- 2.Dodds A W. Small scale preparation of complement components C3 and C4. Methods Enzymol. 1993;223:46–61. doi: 10.1016/0076-6879(93)23037-n. [DOI] [PubMed] [Google Scholar]
- 3.Drickamer K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature. 1992;360:183–186. doi: 10.1038/360183a0. [DOI] [PubMed] [Google Scholar]
- 4.Ezekowitz R A B, Kuhlman M, Groopman J E, Byrn R A. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J Exp Med. 1989;169:185–196. doi: 10.1084/jem.169.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garred P, Madsen H O, Balsev U, Hofmann B, Pedersen C, Gerstoft J, Svejgaard A. Susceptibility to HIV infection and progression of AIDS in relation to variant alleles of mannose-binding lectin. Lancet. 1997;349:236–240. doi: 10.1016/S0140-6736(96)08440-1. [DOI] [PubMed] [Google Scholar]
- 6.Garred P, Madsen H O, Hofmann B, Svejgaard A. Increased frequency of homozygosity of abnormal mannan-binding protein alleles in patients with suspected immunodeficiency. Lancet. 1995;346:941–943. doi: 10.1016/s0140-6736(95)91559-1. [DOI] [PubMed] [Google Scholar]
- 7.Garred P, Madsen H O, Kurtzhals J A, Lamm L U, Thiel S, Hey A S, Svejgaard A. Diallelic polymorphism may explain variations of the blood concentration of mannan-binding protein in Eskimos, but not in black Africans. Eur J Immunogenet. 1992;19:403–412. doi: 10.1111/j.1744-313x.1992.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 8.Hartshorn K L, Sastry K, White M R, Anders E M, Super M, Ezekowitz R A, Tauber A I. Human mannose-binding protein functions as an opsonin for influenza A viruses. J Clin Investig. 1993;91:1414–1420. doi: 10.1172/JCI116345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haurum J S, Thiel S, Jones I M, Fischer P B, Laursen S B, Jensenius J C. Complement activation upon binding of mannan-binding protein to HIV envelope glycoproteins. AIDS. 1993;7:1307–1313. doi: 10.1097/00002030-199310000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Holmskov U, Malhotra R, Sim R B, Jensenius J C. Collectins: collagenous C-type lectins of the innate immune defense system. Immunol Today. 1994;15:67–74. doi: 10.1016/0167-5699(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 11.Ihara I, Harada Y, Ihara S, Kawakami M. A new complement-dependent bactericidal factor found in nonimmune mouse sera: specific binding to polysaccharide of Ra chemotype Salmonella. J Immunol. 1982;128:1256–1260. [PubMed] [Google Scholar]
- 12.Ikeda K, Sannoh T, Kawasaki N, Kawasaki T, Yamashina I. Serum lectin with known structure activates complement through the classical pathway. J Biol Chem. 1987;262:7451–7454. [PubMed] [Google Scholar]
- 13.Iobst S T, Drickamer K. Binding of sugar ligands to Ca2+-dependent animal lectins. II. Generation of high-affinity galactose binding by site-directed mutagenesis. J Biol Chem. 1994;269:15512–15519. [PubMed] [Google Scholar]
- 14.Jack D L, Dodds A W, Anwar N, Ison C A, Law A, Frosch M, Turner M, Klein N J. Activation of complement by mannose-binding lectin on isogenic mutants of Neisseria meningitidis serogroup B. J Immunol. 1998;160:1346–1353. [PubMed] [Google Scholar]
- 15.Johnson G D, Holborow E J. Immunofluorescence. In: Weir D M, editor. Handbook of experimental immunology. 2nd ed. Oxford, England: Blackwell Scientific Publications Ltd.; 1973. pp. 18.1–18.20. [Google Scholar]
- 16.Kawasaki N, Kawasaki T, Yamashina I. A serum lectin (mannan-binding protein) has complement-dependent bactericidal activity. J Biochem. 1989;106:483–489. doi: 10.1093/oxfordjournals.jbchem.a122878. [DOI] [PubMed] [Google Scholar]
- 17.Kilpatrick D C. Isolation of human mannan binding lectin, serum amyloid P component and related factors from Cohn fraction III. Transfus Med. 1997;7:289–294. doi: 10.1046/j.1365-3148.1997.d01-40.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuhlman M, Joiner K, Ezekowitz R A. The human mannose-binding protein functions as an opsonin. J Exp Med. 1989;169:1733–1745. doi: 10.1084/jem.169.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipscombe R J, Beatty D W, Ganczakowski M, Goddard E A, Jenkins T, Lau Y-L, Spurdle A B, Sumiya M, Summerfield J A, Turner M W. Mutations in the human mannan binding lectin gene: frequencies in several population groups. Eur J Hum Genet. 1996;4:13–19. doi: 10.1159/000472164. [DOI] [PubMed] [Google Scholar]
- 20.Lipscombe R J, Sumiya M, Hill A V, Lau Y L, Levinsky R J, Summerfield J A, Turner M W. High frequencies in African and non-African populations of independent mutations in the mannose binding protein gene. Hum Mol Genet. 1992;1:709–715. doi: 10.1093/hmg/1.9.709. . (Erratum, 2:342, 1993.) [DOI] [PubMed] [Google Scholar]
- 21.Lipscombe R J, Sumiya M, Summerfield J A, Turner M W. Distinct physicochemical characteristics of human mannose binding protein expressed by individuals of differing genotype. Immunology. 1995;85:660–667. [PMC free article] [PubMed] [Google Scholar]
- 22.Madsen H O, Garred P, Kurtzhals J A, Lamm L U, Ryder L P, Thiel S, Svejgaard A. A new frequent allele is the missing link in the structural polymorphism of the human mannan-binding protein. Immunogenetics. 1994;40:37–44. doi: 10.1007/BF00163962. [DOI] [PubMed] [Google Scholar]
- 23.Matsushita M, Fujita T. Activation of the classical complement pathway by mannose-binding protein in association with a novel C1s-like serine protease. J Exp Med. 1992;176:1497–1502. doi: 10.1084/jem.176.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita M, Takahashi A, Hatsuse H, Kawakami M, Fujita T. Human mannose-binding protein is identical to a component of Ra-reactive factor. Biochem Biophys Res Commun. 1992;183:645–651. doi: 10.1016/0006-291x(92)90531-o. [DOI] [PubMed] [Google Scholar]
- 25.Mead R, Jack D, Pembrey M, Tyfield L, Turner M the ALSPAC Study Team. Mannose-binding lectin alleles in a prospectively recruited UK population. Lancet. 1997;349:1669–1670. doi: 10.1016/s0140-6736(05)62635-9. [DOI] [PubMed] [Google Scholar]
- 26.Ohta M, Okada M, Yamashina I, Kawasaki T. The mechanism of carbohydrate-mediated complement activation by the serum mannan-binding protein. J Biol Chem. 1990;265:1980–1984. [PubMed] [Google Scholar]
- 27.Polotsky V Y, Belisle J T, Mikusova K, Ezekowitz R A B, Joiner K A. Interaction of human mannose-binding protein with Mycobacterium avium. J Infect Dis. 1997;175:1159–1168. doi: 10.1086/520354. [DOI] [PubMed] [Google Scholar]
- 28.Schelenz S, Malhotra R, Sim R B, Holmskov U, Bancroft G J. Binding of host collectins to the pathogenic yeast Cryptococcus neoformans: human surfactant protein D acts as an agglutinin for acapsular yeast cells. Infect Immun. 1995;63:3360–3366. doi: 10.1128/iai.63.9.3360-3366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheriff S, Chang C Y, Ezekowitz R A B. Human mannose-binding protein carbohydrate recognition domain trimerizes through a triple α-helical coiled-coil. Nat Struct Biol. 1994;1:789–794. doi: 10.1038/nsb1194-789. [DOI] [PubMed] [Google Scholar]
- 30.Soell M, Lett E, Holveck F, Scholler M, Wachsmann D, Klein J P. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-alpha release. J Immunol. 1995;154:851–860. [PubMed] [Google Scholar]
- 31.Sumiya M, Super M, Tabona P, Levinsky R J, Arai T, Turner M W, Summerfield J A. Molecular basis of opsonic defect in immunodeficient children. Lancet. 1991;337:1569–1570. doi: 10.1016/0140-6736(91)93263-9. [DOI] [PubMed] [Google Scholar]
- 32.Summerfield J A, Sumiya M, Levin M, Turner M W. Association of mutations in mannose binding protein gene with childhood infections in consecutive hospital series. Br Med J. 1997;314:1229–1232. doi: 10.1136/bmj.314.7089.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabona P, Mellor A, Summerfield J A. Mannose-binding protein is involved in first-line host-defense—evidence from transgenic mice. Immunology. 1995;85:153–159. [PMC free article] [PubMed] [Google Scholar]
- 34.Thiel S, Vorup-Jensen T, Stover C M, Schwaeble W, Laursen S B, Poulsen K, Willis A C, Eggleton P, Hansen S, Holmskov U, Reid K B M, Jensenius J. A second serine protease associated with mannan-binding lectin that activates complement. Nature. 1997;386:506–510. doi: 10.1038/386506a0. [DOI] [PubMed] [Google Scholar]
- 35.Valdimarsson H, Stefansson M, Vikingsdottir T, Arason G J, Koch C, Thiel S, Jensenius J. Reconstitution of opsonizing activity by infusion of mannan-binding lectin (MBL) to MBL-deficient humans. Scand J Immunol. 1998;48:116–123. doi: 10.1046/j.1365-3083.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- 36.van Emmerik L C, Kuijper E J, Fijen C A, Dankert J, Thiel S. Binding of mannan-binding protein to various bacterial pathogens of meningitis. Clin Exp Immunol. 1994;97:411–416. doi: 10.1111/j.1365-2249.1994.tb06103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weis W I, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 38.Weis W I, Drickamer K, Hendrickson W A. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]