Abstract
Introduction:
Apolipoprotein E (APOE) is considered the major susceptibility gene for developing Alzheimer’s disease. However, the strength of this risk factor is not well established across diverse Hispanic populations.
Methods:
We investigated the associations among APOE genotype, dementia prevalence, and memory performance (immediate and delayed recall scores) in Caribbean Hispanics (CH), African Americans (AA), Hispanic Americans (HA) and non-Hispanic White Americans (NHW). Multivariable logistic regressions and negative binomial regressions were used to examine these associations by subsample.
Results:
Our final dataset included 13,516 participants (5198 men, 8318 women) across all subsamples, with a mean age of 74.8 years. Prevalence of APOE ε4 allele was similar in CHs, HAs, and NHWs (21.8%–25.4%), but was substantially higher in AAs (33.6%; P < 0.001). APOE ε4 carriers had higher dementia prevalence across all groups.
Discussion:
APOE ε4 was similarly associated with increased relative risk of dementia and lower memory performance in all subsamples.
Keywords: admixture, Alzheimer’s disease, apolipoprotein E, Blacks, cognitive performance, dementia, Hispanics/Latinos, Non-Hispanic Whites
1 ∣. INTRODUCTION
Alzheimer’s disease (AD) is the most common cause of dementia1 and has emerged as a significant societal issue and a global priority.2 Risk of AD is influenced by both genetic and environmental factors. Genetic factors include pathogenic variants in the amyloid precursor protein(APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) genes leading to dominantly inherited AD (DIAD)3,4 and susceptibility loci in (not deterministic) genes that increase the risk of developing disease.5,6 Apolipoprotein E (APOE) is considered the major susceptibility gene for developing AD; possession of the ε4 allele leads to a significant increase in an individual’s risk of developing AD.7,8 However, disease prevalence varies according to ancestry; for example, compared to non-Hispanic Whites (NHWs) and Asian populations, the association between APOE ε4 and AD has been found in several studies to be weaker or inconsistent among African9,10 and Hispanic populations,11,12 suggesting gene by environment, and/or gene by gene interactions.13 Furthermore, within Hispanic populations, the interaction between APOE ε4 and disease has been found to vary.11,14-17
The reasons for these disparities in NHWs, African American (AAs), Hispanic Americans (HAs), and among Hispanic subgroups are still poorly understood and highly controversial. Observed diferences may be the result of gene by environment interactions,18,19 given large heterogeneity between these groups. In addition, the widely varying admixture across subgroups may also play an important role, given the unique admixtures across Amerindian, African, and European continental ancestries.20,21 Furthermore, the differential effect of APOE ε4 across populations may be confounded by unique characteristics in previous highly selective cohorts, which have often been used instead of population-based studies. Few studies of Hispanics have cross-population comparisons on the frequency and effect of APOE on dementia prevalence and cognitive performance.
To address these gaps in the literature, we used population-based data from the 10/66 Dementia Research Group (10/66) and the US Health and Retirement Study (HRS) to examine associations between APOE genotype, dementia, and memory performance in Caribbean Hispanics (CA) and US older adults by race/ethnicity (AA, HA, and NHW). We also examined the extent to which these associations may be mediated by admixture proportions on a subsample of these populations with admixture information.
2 ∣. METHODS
2.1 ∣. Data and study population
The primary analyses in this study used community-dwelling participants aged 65 years and over enrolled in the 10/66 study and in the HRS study.
2.1.1 ∣. 10/66 data
We use the baseline prevalence wave of the 10/66 study, which was a population-based sample of adults aged 65 years and over in urban areas in Cuba (Havana and Matanzas), Dominican Republic (Santo Domingo), Puerto Rico (San Juan), and Venezuela (Caracas). Participants received an assessment lasting 3 hours, including participant interview, physical examination, cognitive assessment, blood draw, and informant interview.22 Written informed consent was obtained from all participants and their study partners. Local institutional review boards, and the King’s College London Research Ethics Committee approved this project. The full protocol for the 10/66 population-based surveys is available in an open-access publication.22,23
2.1.2 ∣. HRS data
We used US data from the HRS, a biennial longitudinal panel study that has surveyed a representative sample of approximately 20,000 adults over the age of 50 in the United States since 1992.24 The HRS collects rich data on cognition, demographics, socioeconomic characteristics, and health. APOE and other genetic data were available for a subset of HRS respondents in 2008 from whom saliva was collected.
2.1.3 ∣. Study sample
We included all 10/66 respondents in the baseline surveys, conducted between 2003 and 2008, in Cuba, Dominican Republic, Puerto Rico, and Venezuela for whom APOE genotype was available (genotype data were not available to us for other 10/66 Latin American surveys). In primary analyses, we pooled respondents across the four 10/66 sites to increase statistical power. For comparability across samples, we included all HRS respondents in the 2008 wave who were aged 65 and older; had non-missing APOE genotype; and were either HA, AA, or NHW.
2.2 ∣. Measures
Participants were evaluated using the 10/66 and HRS protocols, which generate information on dementia diagnosis, physical health, demographics, non-communicable disease risk factors, disability, and socioeconomic status, among others.22 Assessments relevant to the current analyses are described in detail below.
2.2.1 ∣. 10/66 dementia status
Dementia was diagnosed using the cross-culturally validated 10/66 dementia diagnosis algorithm, for which strong concurrent and predictive validity has been demonstrated.25,26 Dementia diagnosis was established following: (1) a structured clinical interview; (2) a cognitive test battery including (a) the Community Screening Instrument for Dementia (CSI-D),27 (b) a verbal fluency task, and (c) the modified Consortium to Establish a Registry for Alzheimer’s Disease 10 word list learning task with delayed recall;28 and (3) an informant interview (CSI-D)27 for evidence of cognitive and functional decline. This dementia measure has been previously used29 for cross-national comparisons to the Langa-Weir dementia measure in the US HRS.
2.2.2 ∣. HRS dementia status
We assigned dementia status among HRS respondents using the validated Langa-Weir method.30 The Langa-Weir method relies on an individual’s Modified Telephone Interview for Cognitive Status (TICS-M) score, which ranges from 0 to 27 and is based on the following components: an immediate and delayed 10-noun free recall test, a serial 7 subtraction test, and a backward count from 20 test. A respondent was classified as having dementia if their TICS-M score was below 7. The method uses an analogous algorithm that relies on proxy responses for respondents who could not answer the survey for themselves.31 We used the Langa-Weir method, as alternative algorithms32,33 explicitly incorporated education or race/ethnicity in calculating dementia probability, whereas we aimed to make comparisons in dementia and its determinants across race/ethnicity subgroups.
2.2.3 ∣. Memory performance
Memory performance is among the cognitive measures best harmonized across the 10/66 and HRS assessments. To complement the analysis of dementia status, we study the APOE relationship with memory performance. Specifically, both the 10/66 and HRS include a 10 word list learning task with delayed recall.28 We used the scores from the 10 word list learning immediate, and from the delayed recall, as the two dependent variables to study memory performance.
2.2.4 ∣. APOE genotype
Our key independent variable of interest was APOE genotype. In the primary analysis, we used an indicator for whether the respondent carried any APOE ε4 allele, as well as an indicator for the carriage of APOE ε2. In secondary analysis, we examined the full APOE genotype including six indicators for all combinations of the APOE alleles (ε2, ε3, and ε4).
2.2.5 ∣. Covariates
Age was ascertained using documented age, or an event calendar as provided by participant or informant report. Educational attainment was measured differently between 10/66 and the HRS, due in part to contextual differences across regions, following the approach of prior research.29 For 10/66 respondents, we categorized educational attainment as (1) not completing primary school, (2) completed primary school, or (3) secondary school or above. For HRS respondents, we categorized educational attainment as (1) no high school degree, (2) high school degree or equivalent, or (3) some college or above. Ethnicity in the HRS was ascertained using self-report by participant or informant as follows: NHW, AA, and HA. Self-reported ethnicity is not available in 10/66 data; for ease of description we refer to all as “Caribbean Hispanic” (CH).
Biochemical analysis
DNA was extracted, quantitated, and archived for Cuba at the National Centre for Medical Genetics in Havana; for Venezuela at the immunogenetics section of the Instituto Venezolano de Investigaciones Cientificas; and for Dominican Republic and Puerto Rico at KBioscience, UK. In Cuba and Venezuela, APOE ε4 genotype was determined using Hhal digestion of amplified products. For the Dominican Republic and Puerto Rico we applied a method based on the observation that APOE ε2, ε3, and ε4 alleles differ in amino acid sequence at C112R, single nucleotide polymorphism (SNP) rs429358 and R158C, SNP rs7412. Admixture data were available only in a Caribbean subsample (Cuba and Dominican Republic). Genetic admixture was determine by using 60 SNPs, which has been shown to be sufficient to estimate three-way individual admixture proportions with a standard error less than 0.1.14,34,35 Genotyping was performed by KBiosciences (http://kbioscience.co.uk), using the KAS-Par chemistry allele specific polymerase chain reaction SNP genotyping system (http://www.kbioscience.co.uk/genotyping/genotyping_chemistry.htm). The ADMIXMAP program36,37 (http://homepages.ed.ac.uk/pmckeigu/admixmap/) generated posterior means of individual admixture from ancestry informative marker data.
In the HRS, saliva samples were collected from HRS participants beginning in 2006. HRS respondents were genotyped using the Illumina HumanOnmi2.5 array and the Illumina HumanExome-12v1 array.
2.3 ∣. Statistical analysis
Descriptive statistics were calculated for all variables. For continuous variables, these included means and standard deviations. For categorical variables, these included counts and proportions in each category. First, we described the APOE genotype distribution and allele frequency by sample and race/ethnicity. We then examined associations between carriage of any APOE ε4 allele versus none, and outcome variables (dementia prevalence and immediate and delayed recall scores) with crude and adjusted prevalence ratios (PR). Associations were estimated separately by race/ethnicity (HA, AA, and NHW) among the HRS sample and pooled across the 10/66 sites (CH). All adjusted analyses controlled for age, sex, and level of education.
We conducted several secondary analyses. First, to assess the representativeness of the subset of HRS respondents with genetic data, we compared the sociodemographic and cognitive variables between subsamples of HRS respondents with and without genetic data. We also examined associations between specific APOE genotypes and dementia prevalence by sample. We further focused on a subsample with ancestry data in both 10/66 and HRS to examine whether admixture mediates the associations among APOE genotype, dementia, and cognitive performance; this also allows us to test the extent to which the APOE relationships with our outcome variables would be different when controlling for ancestry. Only a subset of respondents in the Cuba and Dominican Republic sample had complete ancestry data, whereas all our HRS sample had data on the first 10 principal components. We first reported the frequency of APOE genotype by categories of African and Native American ancestry proportions (> 60%, 30%–60%, < 30%) in the combined Cuba and Dominican Republic subsample. We then examined whether ancestry mediates the effect of APOE ε4 on cognitive performance or dementia in a series of multivariable regression analyses analogous to our main regressions; these regressions controlled for African and Native American ancestry for Dominican Republic and Cuba subsamples, and indicators for the 10 principal components by race/ethnicity for the HRS sample.
All analyses were performed using Stata 17 MP (College Station, TX). All P-values were from two-sided tests and results were deemed statistically significant at P < 0.05. The study protocol was reviewed and approved by the institutional review boards of University of California, Berkeley and Weill Cornell Medical College.
3 ∣. RESULTS
3.1 ∣. Sample characteristics
Our final dataset included 13,516 participants (5198 men, 8318 women), with a mean age of 74.8 years. Summary statistics including sample size, sex, age, education, cognitive performance, and distribution of the APOE alleles by ethnicity are shown in Table 1. Mean age 74.8 (±7.1) was similar across all cohorts. Levels of education were higher in NHWs, with 45.6% completing college or above, compared to 29.0% in AA, 16.6% in HAs, and 36.7% in CHs who completed secondary schooling and above. In the HRS sample the prevalence of dementia was 3.1% in NHWs, 10.6% in HAs, and 12.5% in AAs. The low dementia prevalence particularly in the HRS NHW group could be partially due to selection into genetic data collection. We showed that HRS respondents by ethnicity with and without genetic information were broadly similar in demographic characteristics, except that those with genetic information were slightly younger and better educated than their counterparts (Table S1 in supporting information). However, Table S1 also shows that HRS participants with dementia were substantially less likely to have genetic information in the data; nevertheless, these data have been widely used for genetic analyses; thus, we proceed while noting this caveat. In the 10/66 sample, prevalence of dementia was 9.5% (6.9% in Venezuela, 10.6% in Cuba, 11.7% in Puerto Rico, and 11.8% in Dominican Republic). The cognitive healthy cohort included 7182 in the US-HRS (AA = 795, NHW = 5874, HA = 513) and 5383 in the Caribbean cohort. Summary statistics of the Caribbean cohort by country/island are shown in Table S2 in supporting information.
TABLE 1.
Summary statistics of study sample
| United States | ||||
|---|---|---|---|---|
| Caribbean Hispanics N = 5947 |
Hispanic Americans N = 576 |
African Americans N = 917 |
Non-Hispanic White Americans N = 6070 |
|
| Demographics | ||||
| Female n (%) | 3959 (66.6) | 341 (55.6) | 575 (62.4) | 3440 (56.8) |
| Age mean (SD) | 74.8 (7.1) | 73.0 (6.7) | 73.8 (6.9) | 74.9 (7.4) |
| Education | ||||
| Less than high school (US)/less than primary school (Carib) n(%) | 1935 (32.5) | 386 (67.6) | 417 (45.0) | 1210 (19.2) |
| High-school graduate (US)/completing primary school (Carib) n (%) | 1830 (30.8) | 104 (15.8) | 244 (26.0) | 2159 (35.3) |
| Some college and above (US)/Secondary schooling and above (Carib) n (%) | 2182 (36.7) | 86 (16.6) | 256 (29.0) | 2701 (45.6) |
| APOE genotype | ||||
| Number of APOE ε4 alleles n (%) | ||||
| 0 n (%) | 4651 (78.2) | 445 (77.1) | 605 (66.4) | 4525 (74.6) |
| 1 n (%) | 1192 (20.0) | 126 (22.4) | 280 (30.7) | 1447 (23.9) |
| 2 n (%) | 104 (1.7) | 5 (0.6) | 32 (2.9) | 98 (1.5) |
| Any ε4 allele | 1296 (21.8) | 131 (22.9) | 312 (33.6) | 1545 (25.4) |
| Cognitive status | ||||
| Dementia n (%) | 564 (9.5) | 63 (10.6) | 122 (12.5) | 196 (3.1) |
| Immediate recall, mean (SD) | 4.8 (1.6) | 4.6 (1.7) | 4.6 (1.7) | 5.3 (1.6) |
| Delayed recall, mean (SD) | 4.5 (2.2) | 3.6 (2.0) | 3.2 (2.0) | 4.2 (2.0) |
Summary statistics for the Caribbean Hispanics sample are unweighted.
Total number of observations and number of observations in each cell of categories for US subsamples are unweighted. Percentages for categorical variables and means and standard deviations for continuous variables for US subsamples are weighted using person-level analytic weights available in the Health and Retirement Study. All P-values for differences across groups were significant at 0.1%.
Abbreviations: APOE, apolipoprotein E; SD, standard deviation.
3.2 ∣. APOE genotype and allele distributions by ethnic group
APOE ε4 allele frequencies were similar in NHWs, HAs, and CHs, but ε4 frequency was substantially higher in AAs (P < 0.001; Table 1). Further stratification among the Caribbean sample show frequencies of APOE ε4 by country (Table S2). In Cuba, 413 (16.5%) had one or more ε4 alleles, in Dominican Republic 336 (32.2%), in Venezuela 200 (21.3%), in Puerto Rico 347 (23.8%). Genetic ancestry was available in Dominican Republic and Cuba. Mean African admixture was higher in Dominican Republic relative to Cuba (52% vs. 18%, P < 0.001). Pooling Dominican Republic and Cuba we found that mean African admixture increased from 50% among those with no ε4 alleles to 55% with one and 56% for those with two ε4 alleles (P = 0.016).
3.3 ∣. APOE genotype, dementia prevalence, and cognitive performance
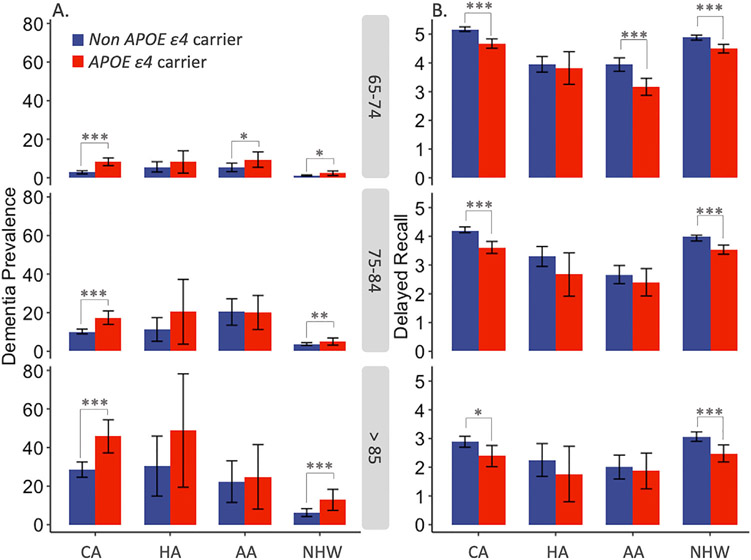
Dementia prevalence by APOE genotypes is shown in Table 2. Further stratification among the Caribbean sample is shown in Table S3 in supporting information. Across all ethnic groups, APOE ε4 carriers had higher dementia prevalence. Figure 1 shows dementia prevalence and 10-word list delayed recall scores separately for those with and without APOE ε4 for each of our sample (CH sample and the US-HRS sample by ethnicity), stratified by age group (65–74, 75–84, 85 and above). Differences in dementia prevalence by carriage of APOE ε4 were most pronounced among the oldest age group (85 and above); they visually appeared smaller in magnitude among AAs compared to other groups, though the wide confidence intervals in this group make it impossible to reject magnitudes similar to those of other race/ethnic groups. Those with APOE ε4 generally had lower 10-word list delayed recall scores than those without APOE ε4 across ethnic groups and age categories.
TABLE 2.
Dementia prevalence by number of APOE ε4 alleles and race/ethnicity
| Number of APOE alleles | Dementia prevalence n/N (%) | |||
|---|---|---|---|---|
| Caribbean Hispanicsa |
Hispanic Americansb |
Blacks/African Americansb |
Non-Hispanic White Americansb |
|
| 0 | 372/4651 (8.0) | 44/445 (8.9) | 73/605 (11.7) | 123/4525 (2.6) |
| 1 | 171/1192 (14.3) | N/Ac | 45/280 (14.6) | 65/1447 (4.1) |
| 2 | 21/104 (20.2) | N/Ac | 4/32 (10.8) | 8/98 (9.4) |
| 1 or 2 | 192/1296 (14.8) | 19/131 (16.1) | 49/312 (14.2) | 73/1545 (4.4) |
Dementia prevalence and number of observations with dementia in each cell for Caribbean Hispanics subsamples are unweighted.
Number of observations with dementia is in each cell for US subsamples are unweighted. Dementia prevalence for US subsamples are weighted using person-level analyticweights available in the Health and Retirement Study.
Numbers censored due to small cell size.
Abbreviation: APOE, apolipoprotein E.
FIGURE 1.
Dementia prevalence and 10-word delayed recall scores by ethnic group and age category. Notes: Bars in Panel A show dementia prevalence with 95% confidence intervals. Panel B shows mean number of 10-word delayed recall with 95% confidence intervals. CA, Caribbean Hispanic; HA, Hispanic Americans; AA, African American;NHW, Non-Hispanic White. Red color indicates APOE ε4 carriers (4/ε4, 3/ε4, ε2/ε4), Blue color indicates non APOE ε4 carriers (ε2/ε2, ε2/ε3, ε3/ε3) *P < 0.05, **P < 0.01, ***P < 0.001
Table 3 shows the adjusted associations between the APOE alleles and cognitive outcomes across all groups. Carriage of one or more APOE ε4 alleles was independently associated with increased prevalence of dementia, with prevalence ratios significant for CHs (1.92, 95% confidence interval [CI] 1.62, 2.22) and NHWs (1.83, 95% CI 1.26, 2.40). They were positive though non-significant for HAs (1.64, 95% 0.81, 2.47) and AAs (1.26, 95% CI 0.81, 1.71), which could be due to smaller sample sizes. Carriage of APOE ε4 was consistently associated with incident rate ratios (IRRs) less than one for both immediate and delayed recall scores, though they were non-significant for HAs for both outcomes. There was no evidence of a protective effect of ε2/ε2 or ε2/ε3 genotypes across all dependent variables and ethnic groups, although the confidence intervals were wide enough to prevent rejecting reasonably side protective effects. Overall, results across groups were similar with overlapping 95% CIs for all estimates. Analogous regressions examining the associations between the full APOE genotype and dementia prevalence show similar results (Table S4 in supporting information).
TABLE 3.
Associations between APOE allele and cognitive function
| Caribbean Hispanicsc N = 5947 |
Hispanic Americansd N = 576 |
Blacks/African Americansd N = 917 |
Non-Hispanic White Americansd N = 6070 |
|
|---|---|---|---|---|
| Panel A: Dementia status a | ||||
| Any ε4 | 1.92 [1.62–2.22] | 1.64 [0.81–2.47] | 1.26 [0.81–1.71] | 1.83 [1.26–2.40] |
| Any ε2 | 0.87 [0.65–1.09] | 0.82 [0.07–1.56] | 1.26 [0.78–1.74] | 1.01 [0.60–1.42] |
| Panel B: Immediate recall b | ||||
| Any ε4 | 0.94 [0.92–0.96] | 0.95 [0.88–1.03] | 0.96 [0.92–1.01] | 0.95 [0.93–0.96] |
| Any ε2 | 1.00 [0.98–1.03] | 1.04 [0.94–1.16] | 0.98 [0.93–1.03] | 1.00 [0.98–1.02] |
| Panel C: Delayed recall b | ||||
| Any ε4 | 0.90 [0.87–0.93] | 0.91 [0.81–1.03] | 0.84 [0.76–0.93] | 0.91 [0.88–0.93] |
| Any ε2 | 1.00 [0.96–1.04] | 1.10 [0.96–1.25] | 0.98 [0.88–1.09] | 1.01 [0.98–1.05] |
Coefficients reported in Panel A are prevalence ratios (PRs) estimated from logistic regressions with dementia status as the dependent variable. PRs are interpreted as the ratios of estimated dementia prevalence between those with a particular APOE allele (e.g., ε4) to those without the allele in each sample. Ninety-five percent confidence intervals of the PRs are in brackets. All regressions adjusted for age, age squared, sex, and education.
Coefficients reported in Panels B and C are incidence rate ratios (IRRs) estimated from Poisson regressions with number of correct words recalled in 10-word immediate recall (Panel B) and 10-word delayed recall (Panel C) as the dependent variable. IRRs are interpreted as the ratios of expected number of words recalled between those with a particular APOE allele to those without the allele in each sample. Ninety-five percent confidence intervals of the IRRs are in brackets. All regressions adjusted for age, age squared, sex, and education.
Country indicators were additionally adjusted for the Caribbean Hispanics sample.
Prevalence ratios, incidence rate ratios, and their confidence intervals for US samples are from weighted regressions using person-level analytic weights. Number of observations for each sample is unweighted.
Abbreviation: APOE, apolipoprotein E.
3.4 ∣. Admixture, APOE genotype, and dementia prevalence.
Genetic ancestry was available in Cuban (n = 585) and Dominican Republic (n = 1055) subsamples. We examined the effect of admixture on dementia prevalence and cognitive performance and whether admixture mediates the effect of APOE ε4 on dementia prevalence and cognitive performance. African admixture was not associated with increased risk for dementia or overall cognitive performance in the Caribbean subsample. Associations between APOE ε4 carriage and cognitive outcomes were very similar before and after controlling for admixture variables across all subsamples (Table S6 in supporting information).
4 ∣. DISCUSSION
In the present study we contribute to a growing body of research examining the extent to which associations among APOE genotype, dementia prevalence, and cognitive performance may be heterogeneous across different ethno-racial groups and/or contexts. In these population-based samples of CHs, HAs, AAs, and NHWs, we found that carriage of one or more APOE ε4 alleles was associated with prevalent dementia and lower memory performance, and furthermore that these associations were of similar magnitudes across racial and ethnic groups. This is despite varying admixtures and life course environments. In addition, we found no evidence that admixture mediates the associations between APOE genotype and cognitive outcomes across racial and ethnic groups.
The association between APOE genotype and risk of AD is well established in White populations; however, a number of studies have suggested a weaker or inconsistent association between ε4 and AD in AAs.38-40 Similarly, previous studies investigating the association of APOE ε4 and AD in HAs have found inconsistent results. Due to the high heterogeneity in Hispanic populations (e.g., admixture, diet, environment, vascular risk factors), it is possible that genetic susceptibility related to the APOE ε4 allele is inconsistently expressed, varying according to population origin. Nevertheless, in our study we found no significant differences in the effect of APOE ε4 across ethnic cohorts.
Furthermore, we note that previous studies have shown that associations between ε4 and AD have been substantially larger when estimated in clinic-based samples versus community-based populations (odds ratio [OR] = 8.6 vs. 2.8, respectively).41 Our associations are smaller still. Among factors that may explain this is our use of a relative elderly cohort. Prior evidence suggests that the association between ε4 and dementia is stronger in younger age groups;42,43 for example, a similar study in the Framingham cohort suggests a low AD positive predictive value of ε4 in older population-based cohorts.44 Furthermore, older population cohorts are more subject to mortality bias, particularly for ε4 homozygotes, due to cardiovascular disease, in which APOE ε4 is known to play a role.45,46
In relation to the associations between APOE ε4 and memory performance, previous studies have found that ε4 is associated with lower cognitive performance and faster decline on episodic memory.47 Our results provide further confirmation of this association, with notably similar magnitudes across diverse US and CH samples.
Regarding genetic admixture and APOE ε4 associations with dementia, our study is not consistent with prior research showing that they may vary according to admixture.48,49 However, we cannot reject that the similar associations found in our different samples are due to offsetting factors. Further research is needed to understand this relationship in diverse populations, including the role of education, access to health care, socioeconomic status, and other environmental factors that may make ancestry factors difficult to interpret. In particular, education level and quality of education may explain differences in dementia prevalence across populations.29,50,51
The present study is not without limitations, and several aspects of our research need to be kept in mind when interpreting these results. First, the classification procedure for assigning dementia status and cognitive assessment differed in the HRS and 10/66, which could influence the comparability of our risk estimations between dementia and APOE status. Previous studies have shown that cross-cohort administration, scoring, and procedural differences are frequent and may impact data interpretation.52 Importantly, our dementia outcome did not involve neuroimaging, disease-specific biomarker measures, or trained dementia specialist assessment, therefore etiological diagnosis (e.g., AD dementia, vascular dementia, Lewy body dementia) was not available. The use of dementia as a broad category may reduce our power to detect specific interactions between APOE genotype and dementia etiology. Second, ethnic minority samples included in the HRS were relatively small; thus, these results must be interpreted cautiously. Third, the subset of HRS respondents for whom genetic data were available may produce different associations than had genetic data been available in the full HRS sample, though we attempted to account for this by controlling for sex, age, and education in all adjusted analyses. Fourth, genetic ancestry was only available for a random subsample of the 10/66; thus, analyses controlling for ancestry were only possible in this subsample of the CHs. Future studies will be needed to better understand the role of APOE and admixture in affecting cognitive performance and the development of dementia. Latino populations, a uniquely admixed group, may provide the ideal environment to estimate such interactions.
Despite these limitations, the present study includes a large population-based sample with a high response rate, which alleviates concerns of possible confounders related to highly selective cohorts derived from memory-based clinics and minimizes selection bias. In addition, only a limited number of APOE ε4 and dementia research studies have included Hispanic populations, which are typically included as monolithic without taking into consideration the diversity of the Hispanic community. Our study included a diverse ethnic representation including CHs, AAs, NHWs, and Hispanics living in the United States.
Future studies in larger and more diverse data sets are needed to develop a better understanding of the role of APOE in the development of dementia in Latinos, and evaluate the complex relationships among ancestry, APOE, dementia, and environmental risk factors.
Supplementary Material
RESEARCH IN CONTEXT.
1. Systematic Review:
A literature review was completed using traditional sources and databases. Several studies suggest that in populations of West African ancestry, and Hispanics, the association of apolipoprotein E (APOE) ε4 genotype and Alzheimer’s disease (AD) is weaker. However, these studies have been rare in Latin American population-based samples.
2. Interpretation:
We found similar associations between APOE ε4 genotype, dementia prevalence, and memory performance across diverse populations of Caribbean Hispanics and US ethnic groups. These associations remained similar after we controlled for admixture proportions.
3. Future Directions:
Future studies are needed to better understand the interaction between admixture and APOE genotype in modifying risk for dementia and AD, especially in highly admixed populations. Latin American populations, where many populations have three-way admixture (European, African, and Native American) might be ideal to further explore such interactions.
ACKNOWLEDGMENTS
We acknowledge the altruism of the participants and their families, and the 10/66 research group for their contributions to this study. We thank Shao-Pang Wang for assistance in data analysis. The work was supported by a grant from the National Institute on Aging of the National Institutes of Health under Award Number R01AG064778. Jorge J. Llibre-Guerra is supported by grants from the Alzheimer’s Association under award number AARFD-21-851415 and SG-20-690363. Jing Li is supported by NIH/NIA grant K01AG066946. The 10/66 Dementia Research Group’s research has been funded by the Welcome Trust Health Consequences of Population Change Program (GR066133–Prevalence phase in Cuba and Brazil; GR080002–Incidence phase in Peru, Mexico, Argentina, Cuba, Dominican Republic, Venezuela, and China). The content is solely the responsibility of the authors and does not represent the official views of the NIA or Wellcome Trust.
ROLE OF THE FUNDER/SPONSOR
The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
CONFLICTS OF INTEREST
The authors report no disclosures relevant to this manuscript.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Brayne C, Miller B, Robinson L, et al. Dementia and aging populations—A global priority for contextualized research and health policy. PLOS Med. 2017;14:e1002275. 10.1371/journal.pmed.1002275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prince M, Comas-Herrera A, Knapp M, Guerchet M, Karagiannidou M, World Alzheimer Report 2016 improving healthcare for people living with dementia. Coverage, Quality and costs now and in the future 2016:1–140. [Google Scholar]
- 3.Bateman RJ, Xiong C, Benzinger TLS, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llibre-Guerra JJ, Li Y, Allegri RF, et al. Dominantly inherited Alzheimer’s disease in Latin America: genetic heterogeneity and clinical phenotypes. Alzheimer’s Dement 2020:alz.12227. 10.1002/alz.12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med. 2016;18:421–430. 10.1038/gim.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrer LA. Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease. Jama. 1997;278:1349. 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- 8.Emrani S, Arain HA, DeMarshall C, Nuriel T. APOE4 is associated with cognitive and pathological heterogeneity in patients with Alzheimer’s disease: a systematic review. Alzheimers Res Ther. 2020;12:141. 10.1186/s13195-020-00712-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrie HC, Murrell J, Baiyewu O, et al. APOE ε4 and the risk for Alzheimer’s disease and cognitive decline in African Americans and Yoruba. Int Psychogeriatrics. 2014;26:977–985. 10.1017/S1041610214000167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gureje O, Ogunniyi A, Baiyewu O, et al. APOE epsilon4 is not associated with Alzheimer’s disease in elderly Nigerians. Ann Neurol. 2006;59:182–185. 10.1002/ana.20694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llibre-Guerra JJ, Li Y, Allen IE, et al. Race, genetic admixture, and cognitive performance in the cuban population. Journals Gerontol Ser A. 2021. 10.1093/gerona/glab063 [DOI] [PubMed] [Google Scholar]
- 12.Granot-hershkovitz E, Daviglus M, Tarraf W, et al. APOE alleles’ association with cognitive function differs across Hispanic /Latino groups and genetic ancestry in the study of Latinos-investigation of neurocognitive aging. HCHS/SOL. 2020:1–9. 10.1002/alz.12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrie HC, Murrell J, Gao S, Unverzagt FW, Ogunniyi A, Hall KS. International studies in dementia with particular emphasis on populations of African origin. Alzheimer Dis Assoc Disord. 2011;20:S42–6. 10.1097/00002093-200607001-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teruel BM, Rodríguez JJL, McKeigue P, et al. Interactions between genetic admixture, ethnic identity, APOE genotype and dementia prevalence in an admixed Cuban sample; a cross-sectional population survey and nested case-control study. BMC Med Genet. 2011;12:43. 10.1186/1471-2350-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajabli F, Feliciano BE, Celis K, et al. Ancestral origin of ApoE ε4 Alzheimer’s disease risk in Puerto Rican and African American populations. PLoS Genet. 2018;14:e1007791. 10.1371/journal.pgen.1007791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Bryant SE, Johnson L, Balldin V, et al. Characterization of Mexican Americans with Mild Cognitive Impairment and Alzheimer’s Disease. J Alzheimer’s. Dis 2012;33:373–379. 10.3233/JAD-2012-121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haan MN, Mungas DM, Gonzalez HM, Ortiz TA, Acharya A, Jagust WJ. Prevalence of dementia in older Latinos: the influence of type 2 diabetes mellitus, stroke and genetic factors. J Am Geriatr Soc. 2003;51:169–177. 10.1046/j.1532-5415.2003.51054.x [DOI] [PubMed] [Google Scholar]
- 18.Chin AL, Negash S, Hamilton R. Diversity and disparity in dementia: the impact of ethnoracial differences in Alzheimer disease. Alzheimer Dis Assoc Disord. 2011;25:187–195. 10.1097/WAD.0b013e318211c6c9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González HM, Tarraf W, Jian X, et al. Apolipoprotein E genotypes among diverse middle-aged and older Latinos: study of Latinos-Investigation of Neurocognitive Aging results (HCHS/SOL). Sci Reports. 2018;8:1–6. 10.1038/s41598-018-35573-3.812018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homburger JR, Moreno-Estrada A, Gignoux CR, et al. Genomic Insights into the Ancestry and Demographic History of South America. PLoS Genet. 2015;11:e1005602. 10.1371/journal.pgen.1005602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Estrada A, Gravel S, Zakharia F, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. 10.1371/journal.pgen.1003925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prince M, Ferri CP, Acosta D, et al. The protocols for the 10/66 dementia research group population-based research programme. BMC Public Health. 2007;7:165. 10.1186/1471-2458-7-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prina AM, Acosta D, Acostas I, et al. Cohort Profile: the 10/66 study. Int J Epidemiol. 2016;46:dyw056. 10.1093/ije/dyw056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnega A, Faul JD, Ofstedal MB, Langa KM, Phillips JWR, Weir DR. Cohort Profile: the Health and Retirement Study (HRS). Int J Epidemiol. 2014;43:576–585. 10.1093/IJE/DYU067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prince M, Acosta D, Chiu H, Scazufca M, Varghese M. Dementia diagnosis in developing countries: a cross-cultural validation study. Lancet. 2003;03:12772–12779. 10.1016/S0140-6736 [DOI] [PubMed] [Google Scholar]
- 26.Prince MJ, De Rodriguez JL, Noriega L, et al. The 10/66 Dementia Research Group’s fully operationalised DSM-IV dementia computerized diagnostic algorithm, compared with the 10/66 dementia algorithm and a clinician diagnosis: a population validation study. BMC Public Health. 2008. 10.1186/1471-2458-8-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall KS, Hendrie HC, Rodgers DD, et al. The development of a dementia screening interview in two distinct languages. Int J Methods Psychiatry. 1993. [Google Scholar]
- 28.Sosa AL, Albanese E, Prince M, et al. Population normative data for the 10/66 Dementia Research Group cognitive test battery from Latin America, India and China: a cross-sectional survey. BMC Neurol. 2009;9:48. 10.1186/1471-2377-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Llibre-Guerra JJ, Harrati A, et al. Associations between education and dementia in the Caribbean and the United States: an international comparison. Alzheimer’s Dement Transl Res Clin Interv. 2021;7. 10.1002/TRC2.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crimmins EM, Kim JK, Langa KM, Weir DR. Assessment of Cognition using surveys and neuropsychological assessment: the health and retirement study and the aging, demographics, and memory study. J Gerontol Ser B. 2011;66B:i162–71. 10.1093/GERONB/GBR048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.AF J A short form of the informant questionnaire on cognitive decline in the elderly (IQCODE): development and cross-validation. Psychol Med. 1994;24:145–153. 10.1017/S003329170002691X [DOI] [PubMed] [Google Scholar]
- 32.Gianattasio KZ, Wu Q, Glymour MM, Power MC. Comparison of methods for algorithmic classification of dementia status in the health and Retirement Study. Epidemiology. 2019;30:291–302. 10.1097/EDE.0000000000000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gianattasio KZ, Ciarleglio A, Power MC. Development of algorithmic dementia ascertainment for racial/ethnic disparities research in the US Health and Retirement Study. Epidemiology. 2020;31:126–133. 10.1097/EDE.0000000000001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith MW, Patterson N, Lautenberger JA, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004. 10.1086/420856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith MW, Lautenberger JA, Shin HD, et al. Markers for mapping by admixture linkage disequilibrium in African American and hispanic populations. Am J Hum Genet. 2001. 10.1086/323922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004. 10.1086/420855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shriner D. Overview of admixture mapping n.d. 10.1002/0471142905.hg0123s76 [DOI] [PubMed] [Google Scholar]
- 38.Maestre G, Ottman R, Stern Y, et al. Apolipoprotein E and Alzheimer’s disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37:254–259. 10.1002/ana.410370217 [DOI] [PubMed] [Google Scholar]
- 39.Tang MX, Maestre G, Tsai WY, et al. Effect of age, ethnicity, and head injury on the Association between APOE Genotypes and Alzheimer’s Disease. Ann N Y Acad Sci. 1996;802:6–15. 10.1111/J.1749-6632.1996.TB32593.X [DOI] [PubMed] [Google Scholar]
- 40.Sahota A, Yang M, Gao S, et al. Apolipoprotein E—associated risk for Alzheimer’s disease in the African-American population is genotype dependent. Ann Neurol. 1997;42:659–661. 10.1002/ANA.410420418 [DOI] [PubMed] [Google Scholar]
- 41.Gianattasio KZ, Bennett EE, Wei J, et al. Generalizability of findings from a clinical sample to a community-based sample: a comparison of ADNI and ARIC. Alzheimer’s Dement. 2021;17:1265–1276. 10.1002/ALZ.12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slooter AJC, Cruts M, Kalmijn S, et al. Risk estimates of dementia by apolipoprotein e genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–968. 10.1001/ARCHNEUR.55.7.964 [DOI] [PubMed] [Google Scholar]
- 43.van der Lee SJ, Wolters FJ, Ikram MK, et al. The effect of APOE and other common genetic variants on the onset of Alzheimer’s disease and dementia: a community-based cohort study. Lancet Neurol. 2018;17:434–444. 10.1016/S1474-4422(18)30053-X [DOI] [PubMed] [Google Scholar]
- 44.Myers RH, Schaefer EJ, Wilson PWF, et al. Apolipoprotein E epsilon4 association with dementia in a population-based study: the Framingham study. Neurology. 1996;46:673–677. 10.1212/WNL.46.3.673 [DOI] [PubMed] [Google Scholar]
- 45.Heffernan AL, Chidgey C, Peng P, Masters CL, Roberts BR. The Neurobiology and Age-Related Prevalence of the ε4 Allele of Apolipoprotein E in Alzheimer’s Disease Cohorts. J Mol Neurosci. 2016;60:316–324. 10.1007/S12031-016-0804-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolters FJ, Yang Q, Biggs ML, et al. The impact of APOE genotype on survival: results of 38537 participants from six population-based cohorts (E2-CHARGE). PLoS One. 2019;14:e0219668. 10.1371/JOURNAL.PONE.0219668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes LL, Arvanitakis Z, Yu L, Kelly J, De Jager PL, Bennett DA. Apolipoprotein E and change in episodic memory in blacks and whites. Neuroepidemiology. 2013;40:211–219. 10.1159/000342778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blue EE, Horimoto ARVR, Mukherjee S, Wijsman EM, Thornton TA. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement. 2019;15:1524–1532. 10.1016/j.jalz.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohman TJ, Cooke-Bailey JN, Reitz C, et al. Global and local ancestry in African-Americans: implications for Alzheimer’s disease risk. Alzheimers Dement. 2016;12:233–243. 10.1016/j.jalz.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross AL, Mungas DM, Crane PK, et al. Effects of education and race on cognitive decline: an integrative study of generalizability versus study-specific results. Psychol Aging. 2015;30:863–880. 10.1037/pag0000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manly JJ, Schupf N, Tang M-X, Stern Y. Cognitive decline and literacy among ethnically diverse elders. J Geriatr Psychiatry Neurol. 2005;18:213–217. 10.1177/0891988705281868 [DOI] [PubMed] [Google Scholar]
- 52.Briceño EM, Gross AL, Giordani BJ, et al. Pre-Statistical considerations for harmonization of cognitive instruments: harmonization of ARIC, CARDIA, CHS, FHS, MESA, and NOMAS. J Alzheimer’s Dis. 2021;83:1803–1813. 10.3233/JAD-210459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.