Abstract
The BBIBP-CorV severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) inactivated vaccine has been authorized for emergency use and widely distributed. We used single-cell transcriptome sequencing to characterize the dynamics of immune responses to the BBIBP-CorV inactivated vaccine. In addition to the expected induction of humoral immunity, we found that the inactivated vaccine induced multiple, comprehensive immune responses, including significantly increased proportions of CD16+ monocytes and activation of monocyte antigen presentation pathways; T cell activation pathway upregulation in CD8+ T cells, along with increased activation of CD4+ T cells; significant enhancement of cell-cell communications between innate and adaptive immunity; and the induction of regulatory CD4+ T cells and co-inhibitory interactions to maintain immune homeostasis after vaccination. Additionally, comparative analysis revealed higher neutralizing antibody levels, distinct expansion of naive T cells, a shared increased proportion of regulatory CD4+ T cells, and upregulated expression of functional genes in booster dose recipients with a longer interval after the second vaccination. Our research will support a comprehensive understanding of the systemic immune responses elicited by the BBIBP-CorV inactivated vaccine, which will facilitate the formulation of better vaccination strategies and the design of new vaccines.
Keywords: inactivated vaccine, COVID-19, scRNA-seq, immune response, homeostasis
Graphical abstract
Public summary
-
•
A single-cell atlas of dynamic immune responses to the SARS-CoV-2 inactivated vaccine.
-
•
The proportion of CD16+ monocytes and the antigen presentation pathway are elevated.
-
•
Both CD4+ T cells and CD8+ T cells are activated by inactivated vaccine.
-
•
Cell-cell communications between innate and adaptive immunity are enhanced.
-
•
Tregs and co-inhibitory pathways are induced to maintain immune homeostasis.
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic continues to threaten human health. The global distribution of effective vaccines has an essential role in preventing and controlling pandemics.1
The BBIBP-CorV vaccine is the first whole inactivated virus vaccine authorized by the World Health Organization.2,3 During studies of its immunogenicity and safety, the preclinical trials of BBIBP-CorV in six mammalian models and clinical trials involving healthy populations at different ages demonstrated that it induced high levels of neutralizing antibodies and efficiently protected against SARS-CoV-2 infection.4,5,6 In addition to the essential role of humoral immunity, a recent study regarding the T cell response to BBIBP-CorV vaccination suggested that cellular responses may be involved in protective immunity.7
The development of single-cell RNA sequencing (scRNA-seq) has allowed the in-depth profiling of peripheral immunity.8 Recently, several single-cell studies profiled antiviral immune responses elicited by COVID-19 vaccines, such as the expansion of plasmacytoid dendritic cells (pDCs) and the emergence of inflammatory monocytes with high interferon (IFN)-stimulated gene expression after vaccination.9,10 Studies have also revealed enhancement of the IFN-γ response in terms of cellular immunity and the activation of humoral immunity,9,11 which confirmed that the comprehensive immune response induced by a vaccine can be delineated using scRNA-seq.
In the present study, we comprehensively characterized the longitudinal changes in peripheral immune cells at single-cell resolution from vaccine recipients during a phase 1/2 trial and conducted comparative analysis of different booster vaccine strategies. Our findings indicate that the SARS-CoV-2 inactivated vaccine efficiently induced extensive immune responses in both the innate and adaptive immune systems, revealing the potential protective mechanism and providing a reference for the use of booster doses of this vaccine.
Results
Single-cell analysis of peripheral blood mononuclear cell dynamics in vaccine recipients
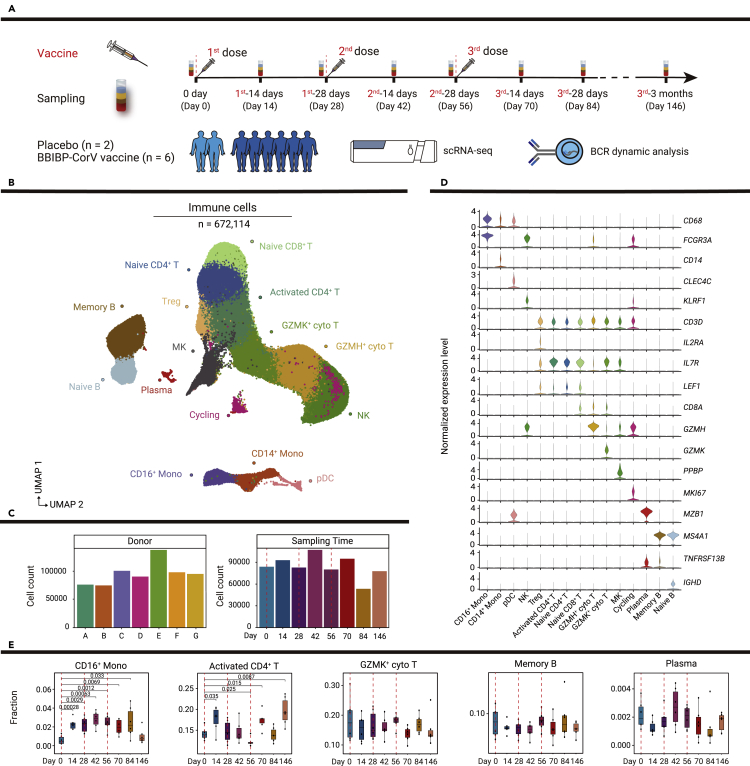
To profile dynamic immune responses to the BBIBP-CorV SARS-CoV-2 inactivated vaccine in humans, longitudinal blood samples were collected from six vaccine recipients and two placebo recipients (Table S1). All participants received three doses of vaccine (vaccine group) or placebo (placebo group). Inoculations were administered to participants on days 0, 28, and 56. Blood samples were collected from each participant on days 0, 14, 28, 42, 56, 70, 84, and 146 (Figure 1A).
Figure 1.
Study design and single-cell analysis of PBMC dynamics in vaccine and placebo recipients
(A) Flowchart of the study design. Eight participants received three doses of placebo or BBIBP-CorV vaccine, and blood samples were collected from these participants at eight time points divided before and after vaccinations. Then, scRNA-seq and B cell receptor dynamic analysis were conducted. Red dotted lines indicate inoculation days.
(B) Uniform manifold approximation and projection (UMAP) plot shows 15 cell types of 672,114 immune cells by unsupervised clustering. Each cell type is indicated by a different color.
(C) Bar plots show the number of cells in each sample and the number of cells at each time point. A, B, C, D, E, and G represent vaccine recipients, while F represents a placebo recipient.
(D) Violin plots show the normalized expression levels of canonical markers defining cell types in (B).
(E) Boxplots show the proportions of selected cells at all time points. p values < 0.05 (Student’s t test) are indicated.
We performed scRNA-seq of 56 peripheral blood mononuclear cell (PBMC) samples from the participants; we also performed bulk RNA-seq of 64 PBMC samples. After quality filtering, we profiled 672,114 single cells from six vaccine recipients and one placebo recipient (Figures 1B and S1A). The number of cells in each sample ranged from 74,759 to 137,446; the number of cells at each time point ranged from 53,343 to 107,013 (Figure 1C). Unsupervised clustering was performed and 15 cell populations were identified on the basis of canonical markers (Figures 1B and 1D). Then, we examined the single-cell dynamics of PBMCs in these vaccine recipients at eight time points before and after the three vaccinations. The proportions of CD16+ monocytes, activated CD4+ T cells, and regulatory CD4+ T cells dramatically increased after vaccination, whereas the proportions of other cell types did not significantly change (Figures 1E, S1B, and S1C). These findings indicated that scRNA-seq could robustly measure dynamic peripheral immune responses induced by the inactivated vaccine.
Vaccine-mediated stimulation of CD16+ monocyte-dominated innate immune responses
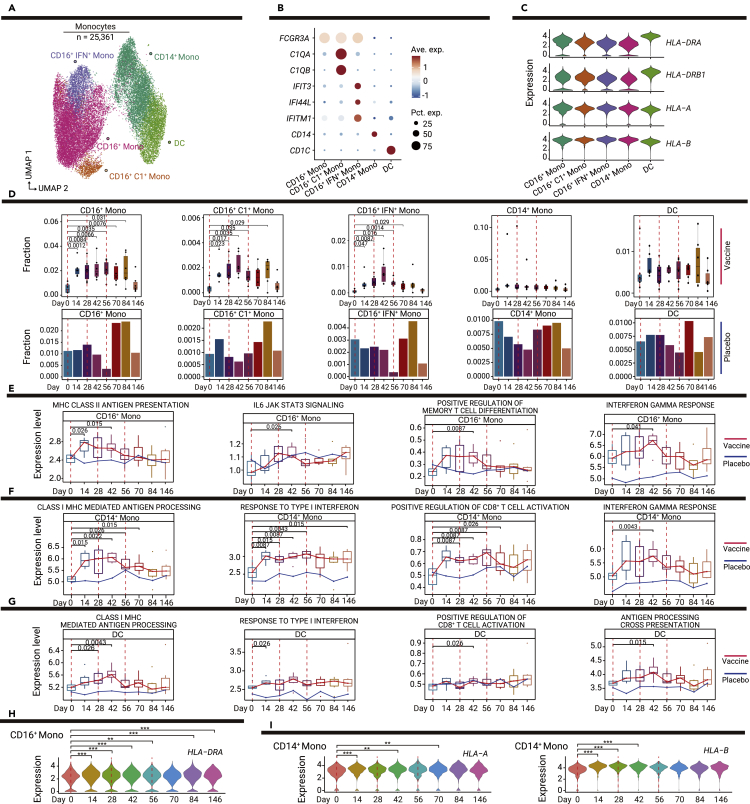
Vaccine-mediated induction of antigen-specific B and T cell responses requires activation of the innate immune system, particularly with respect to antigen-presenting cells (APCs).12 We subclustered monocytes (n = 25,361) to analyze the dynamic response of APCs to vaccination (Figure 2A). Re-clustering revealed five clusters: non-classical CD16+ monocytes (FCGR3A+; CD16+ mono), CD16+ complement-associated monocytes (FCGR3A+C1QA+C1QB+; CD16+C1+ mono), CD16+IFN+ monocytes (FCGR3A+IFIT3+; CD16+IFN+ mono), CD14+ monocytes (CD14+; CD14+ mono), and dendritic cells (CD1C+; DCs) (Figure 2B). Cells in these clusters expressed high levels of MHC-II (HLA-DRA, HLA-DRB1) and MHC-I (HLA-A, HLA-B) genes, indicating their antigen presentation capabilities (Figure 2C).
Figure 2.
Dynamic changes in innate immune responses among myeloid cell subsets
(A) UMAP plot shows the results of sub-clustering involving 25,361 monocytes.
(B) Dot plot shows the expression levels of selected genes in the five subsets of myeloid cells. Dot color indicates mean expression level and dot size indicates the percentage of the cell expressing the gene.
(C) Violin plot shows the expression levels of MHC-II and MHC-I genes in myeloid cell subsets.
(D) Boxplots show the proportions of myeloid cell subsets in vaccine recipients at all time points; bar plots show the proportions of myeloid cell subsets in one placebo recipient at all time points. p values < 0.05 (Student’s t test) are indicated.
(E–G) Changes in functional pathways over time in monocyte subsets, the red line indicates the median expression level of vaccine recipients, the blue line indicates the expression level of the placebo recipient, including CD16+ monocytes (E), CD4+ monocytes (F), and DCs (G). p values < 0.05 (Wilcoxon rank-sum test) are indicated.
(H) Violin plot shows the dynamic changes in MHC-II expression levels over time in CD16+ monocytes. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
(I) Violin plots show dynamic changes in MHC-II and MHC-I expression levels over time in CD14+ monocytes. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
We first examined compositional changes in response to the vaccine. The proportions of three CD16+ monocyte states (CD16+, CD16+C1+, and CD16+IFN+) dramatically increased after the first vaccination (Figure 2D). By contrast, we did not observe significant increases in CD14+ monocytes or DCs. CD16+IFN+ monocytes mainly expressed genes related to the IFN response; cells in these clusters may be directly related to the IFN response (Figure S2A). The increased proportion of CD16+IFN+ monocytes indicated that vaccination had caused the activation of IFN-mediated antiviral immunity.
We also used pathway analyses to investigate changes in the functional status of each subpopulation of myeloid cells after vaccination. The expression levels of genes in the MHC-II antigen presentation pathway in CD16+ monocytes were significantly upregulated after the first vaccination (Figure 2E). Analysis of differentially expressed genes at dynamic time points (Table S2) also showed that the expression of the MHC-II molecule HLA-DRA began to significantly increase in CD16+ monocytes at 14 days after injection of the vaccine (Figure 2H). The expression levels of genes in the MHC-I-mediated antigen processing and presentation pathways were significantly upregulated in CD14+ monocytes and DCs (Figures 2F and 2G); the expression levels of MHC-I molecules (HLA-A and HLA-B) in CD14+ monocytes were enhanced after the first vaccination (Figure 2I). The activities of several other immune response-related pathways and genes were also significantly enhanced by vaccination (Figures 2E–2G, S2B–S2F, S3A, and S3B). The pathway activity of positive regulation of memory T cell differentiation by CD16+ monocytes (Figure 2E) and CD8+ T cell activation by CD14+ monocytes and DCs (Figures 2F and 2G) were upregulated after vaccination, indicating that the abilities of APCs to stimulate T cell activation and differentiation were enhanced by the vaccine.
Vaccine-mediated induction of activated and regulatory CD4+ T cells and activation of CD8+ cytotoxic T cells
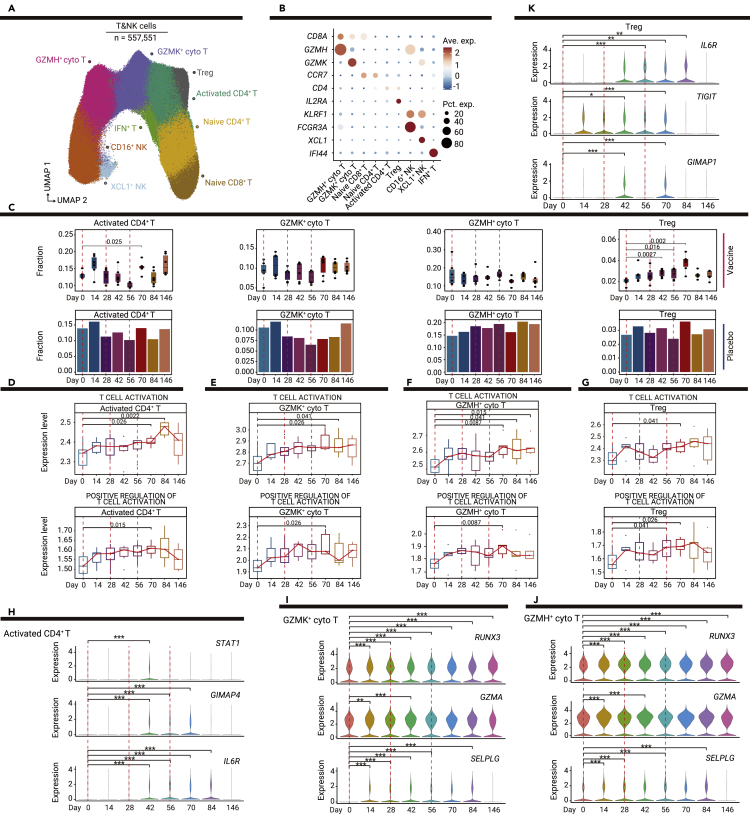
T cell-mediated cellular immunity is critical for protection against SARS-CoV-2 infection.13 Therefore, to investigate the T cell and natural killer (NK) cell responses elicited by the vaccine, we re-clustered T cells and NK cells with more granularity (Figure 3A). Seven clusters of T cells and two clusters of NK cells were identified: CD8+GZMH+ cytotoxic T cells (CD8A+GZMH+; GZMH+ cyto T), activated CD4+ T cells (CD4+; activated CD4+ T), CD8+ GZMK+ cytotoxic T cells (CD8A+GZMK+; GZMK+ cyto T), regulatory CD4+ T cells (CD4+IL2RA+; Treg), naive CD4+ T cells (CD4+CCR7+; naive CD4+ T), naive CD8+ T cells (CD8A+CCR7+; naive CD8+ T), IFN+ T cells (IFI44+; IFN+ T), CD16+ NK cells (FCGR3A+; CD16+ NK), and XCL1+ NK cells (XCL1+; XCL1+ NK) (Figure 3B).
Figure 3.
Vaccine-mediated induction of T cell responses in T and NK cell subsets
(A) UMAP plot shows the results of sub-clustering involving 557,551 T and NK cells.
(B) Dot plot shows the expression levels of selected genes in the nine subsets of T and NK cells. Dot color indicates mean expression level and dot size indicates the proportion of cells expressing the gene.
(C) Boxplots show the proportions of T and NK cell subsets in vaccine recipients at all time points; bar plots show the proportions of T and NK cell subsets in one placebo recipient at all time points. p values < 0.05 (Student’s t test) are indicated.
(D–G) Changes in T cell activation and positive regulation of T cell activation pathways over time in T cell subsets; the red line indicates the median expression level of vaccine recipients, including activated CD4+ T cells (D), GZMK+ cytotoxic T cells (E), GZMH+ cytotoxic T cells (F), and Treg cells (G). p values < 0.05 (Wilcoxon rank-sum test) are indicated.
(H–K) Violin plots show dynamic changes in the expression levels of selected genes over time in T cell subsets, including activated CD4+ T cells (H), GZMK+ cytotoxic T cells (I), GZMH+ cytotoxic T cells (J), and Treg cells (K). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
The proportions of naive T cells, cytotoxic T cells and NK cells did not significantly change after vaccination (Figures 3C and S4). However, vaccine-mediated induction of activated and regulatory CD4+ T cells was observed. The proportion of activated CD4+ T cells and T cell activation pathway activity significantly increased 14 days after the third vaccination, indicating that the third vaccination could induce the proliferation of activated CD4+ T cells (Figures 3C, 3D, and S5A).
Regulatory CD4+ T cell expansion was detected 14 days after the second vaccination, and the third vaccination further increased the proportion of regulatory CD4+ T cells (Figure 3C). Activation of regulatory CD4+ T cells was also observed after vaccination (Figures 3G and S5D). These findings suggested that the SARS-CoV-2 inactivated vaccine could elicit robust production of regulatory CD4+ T cells. Regulatory CD4+ T cells have a critical role in the maintenance of immune homeostasis14; regulatory CD4+ T cells induced by the vaccine may contribute to the suppression of excessive immune responses and the prevention of allergic responses. The expression of IL6R was significantly induced after the second vaccination in both activated and regulatory CD4+ T cells (Figures 3H and 3K; Table S3); thus IL6-IL6R signaling may play a critical role in regulating the balance between effector and regulatory CD4+ T cells in response to the SARS-CoV-2 inactivated vaccine.
Although the proportion of CD8+ cytotoxic T cells did not significantly change (Figure 3C), the expression levels of genes in T cell activation pathways significantly increased in these CD8+ T cells after the third vaccination (Figures 3E, 3F, S5B, and S5C). In cytotoxic T cells (GZMK+ cyto T, GZMH+ cyto T) and CD16+ NK cells, the expression level of RUNX3, a transcription factor that regulates the cytolytic activity of T cells,15 was significantly enhanced by the first vaccination (Figures 3I, 3J, and S6D). The expression levels of cytotoxicity-related genes (GZMA, PRF1, GZMK, and NKG7) were also significantly upregulated after the first vaccination, and their expression was enhanced by the second or third vaccinations (Figures 3I, 3J, and S6B–S6D). These results indicate that CD8+ T cell responses are elicited by the SARS-CoV-2 inactivated vaccine.
The overall expression levels of genes in type I IFN pathways and the expression levels of specific genes in those pathways (ISG20, XAF1, and IRF3) were significantly increased in activated CD4+ T cells and CD8+ cytotoxic T cells after vaccination, indicating that vaccination induced a broad IFN response in T cells (Figures S6A–S6C and S7). Furthermore, the expression level of SELPLG, an immune checkpoint regulator that promotes T cell exhaustion,16 was significantly enhanced in CD8+ cytotoxic T cells (Figures 3I and 3J); this finding suggested that negative regulators of T cell responses may be induced by the vaccine to prevent the excessive activation of effector T cells.
Taken together, these results indicate that the SARS-CoV-2 inactivated vaccine induced increased production of activated and regulatory CD4+ T cells, along with increased activation of CD8+ cytotoxic T cells; these changes may contribute to vaccine-induced T cell memory.
Dynamic changes in B cell functional status after vaccination
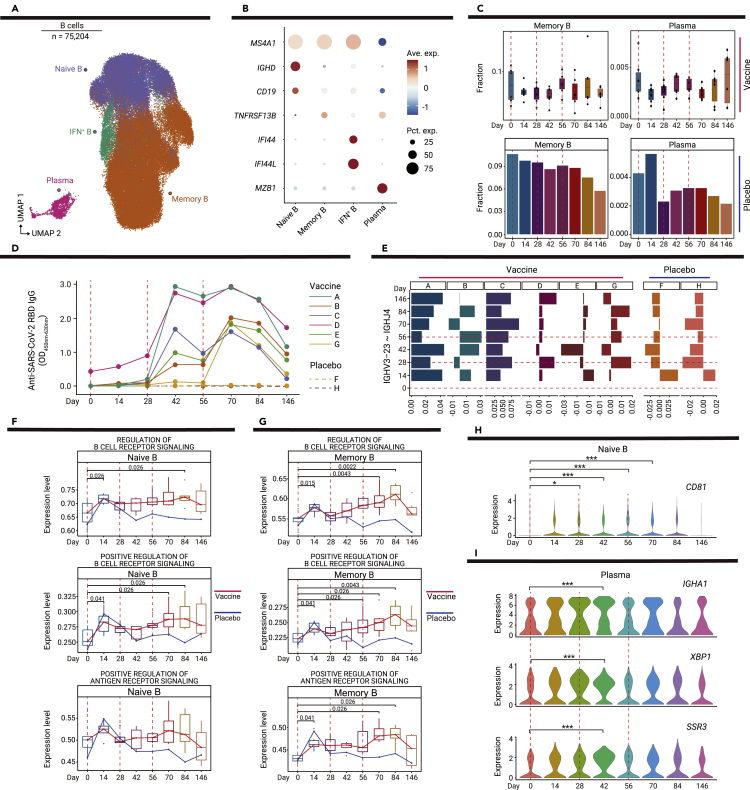
Long-term humoral immunity mediated by memory B cells and plasma cells is critical for protection against SARS-CoV-2.17 Through sub-clustering, we identified four B cell subsets: naive B cells (MS4A1+IGHD+CD19+; naive B), memory B cells (MS4A1+IGHD−; memory B), IFN+ B cells (MS4A1+IFI44+IFI44L+; IFN+ B), and plasma cells (MZB1+; plasma) (Figures 4A and 4B). The proportions of B cell subsets did not significantly change before and after vaccination (Figures 4C and S8A).
Figure 4.
Dynamic changes in functional status among B cell subsets
(A) UMAP plot shows the results of sub-clustering involving 75,204 B cells.
(B) Dot plot shows the expression level of selected genes in four subsets of B cells. Dot color indicates mean expression level and dot size indicates the proportion of cells expressing the gene.
(C) Boxplots show the proportions of memory B and plasma cells in six vaccine recipients at all time points; bar plots show the proportions of memory B and plasma cells in one placebo recipient at all time points. p values < 0.05 (Student’s t test) are indicated.
(D) Line chart shows the dynamic changes in anti-receptor-binding domain IgG levels in six vaccine recipients and two placebo recipients, as determined by enzyme-linked immunosorbent assays.
(E) Bar plot shows the proportion of IGHV3-23-IGHJ4 pairs in six vaccine recipients and two placebo recipients. The proportion of IGHV3-23-IGHJ4 pairs in each time point is minus the proportion on day 0.
(F and G) Changes in pathways regulating B cell receptor signaling over time in memory B cells (F) and plasma cells (G); the red line indicates the median expression level of vaccine recipients, the blue line indicates the expression level of the placebo recipient. p values < 0.05 (Wilcoxon rank-sum test) are indicated.
(H) Violin plot shows the expression level CD81 at dynamic time points in naive B cells. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
(I) Violin plots show dynamic changes in the expression levels of IGHA1, XBP1, and SSR3 over time in plasma cells. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
We measured the anti-receptor-binding domain IgG levels of all recipients on the basis of the longitudinally collected plasma samples by enzyme-linked immunosorbent assays (Figure 4D). Although the IgG antibody level exhibited minor elevation after the first vaccination, the level significantly increased in most vaccine recipients after the second vaccination. Furthermore, the third vaccination maintained the IgG antibody levels; thus, a high level of antibody remained detectable until 146 days after the initial vaccination. However, the plasma IgG levels in two placebo recipients remained undetectable (Figure 4D).
On the basis of the results of bulk RNA sequencing, we analyzed changes in B cell receptors before and after vaccination. Analysis of VDJ gene usage showed dynamic changes in the IGHV-IGHJ pairing after vaccination. In previous studies, IGHV3-23-IGHJ4 was identified as the most frequent pairing among COVID-19 patients with early recovery.18 Interestingly, IGHV3-23-IGHJ4 was detected as one of the main clone types in all recipients (Figure S9). In the vaccine recipients, an elevated IGHV3-23-IGHJ4 proportion was constantly observed in recipients A and C, while decreased proportions were observed at some time points in other recipients (Figures 4E and S9), reflecting individual differences in vaccine responses.
Pathway analyses were conducted to analyze vaccine-driven changes in B cell functional status. After vaccination, a series of pathways that regulated B cell receptor signaling and type I IFN regulatory pathways were significantly upregulated in naive B cells and memory B cells (Figures 4F, 4G, and S8B–S8D). These results indicated that vaccination had a significant impact on B cell functional status (i.e., the pathways responsible for the regulation of B cell receptor signaling were activated), which could contribute to the generation of B cell memory responses.
Differentially expressed gene analysis (Table S4) revealed that the expression level of CD81, which is reportedly important for B cell activation and antibody production,19 was significantly enhanced in naive B cells after vaccination (Figure 4H). Moreover, the expression levels of antibody-encoding genes (IGHA1 and IGHA2), a transcriptional factor required for plasma cell differentiation (XBP1), and antibody secretion-related genes (SSR1, SSR2, SSR3, SSR4, SEC11C, SEC61A1, and SEC61G) were significantly upregulated; these findings were consistent with the heightened antibody response after the second vaccination (Figures 4I and S8E). The significant upregulation of TLR10, TFEB, and IGHM was observed in memory B cells (Figure S8F).
In summary, inoculation using the SARS-CoV-2 inactivated vaccine elicited activation of the B cell receptor pathway in B cells, secretion of SARS-CoV-2-specific antibodies, and generation of long-term B cell immune memory.
Dynamic interactions between innate and adaptive immunity after vaccination
Crosstalk between innate and adaptive immunity is critical for generating vaccine-induced immunological memory and protective immune responses to pathogens.12 To assess changes in the interactions between innate and adaptive immunity after vaccination, we used the CellPhoneDB to identify potential ligand-receptor pairs among all cell types (Figure S10).20
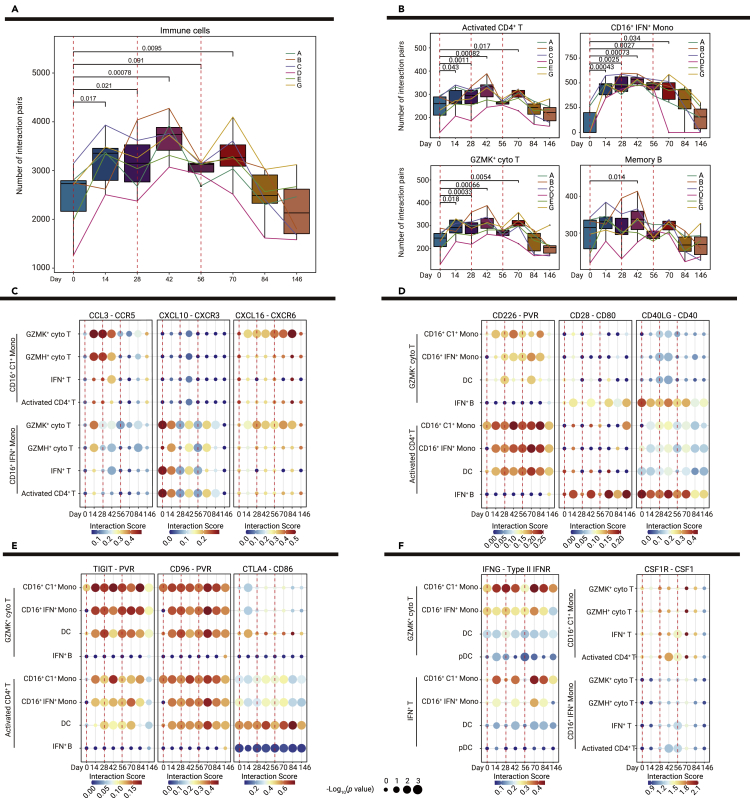
We observed a significantly increased number of ligand-receptor pairs among total immune cells after the first vaccination and further enhancement by the second vaccination in five out of six donors (Figure 5A), the slight reduction followed by the delayed elevation in the sixth donor may reflect individual differences in vaccine-induced immune responses. As we further calculated the number of ligand-receptor pairs in immune cell subsets, similar enhancements in activated CD4+ T, GZMK+ cyto T, and CD16+IFN+ monocytes were observed, though the increased tendency was not significant in memory B cells until 14 days after the second inoculation (Figure 5B). Accordingly, the overall rise in the number of interaction pairs indicated that the inactivated vaccine induced extensive cell-cell communication among immune cells (Figures 5A and 5B; Table S5).
Figure 5.
Dynamic changes in interactions between innate and adaptive immunity
(A) Boxplots show the numbers of interaction pairs among overall immune cells over time in six vaccine recipients; sample points from the same recipient are connected via lines in the same color. p values < 0.05 (Student' s t test) are indicated.
(B) Bar and line plots show the numbers of interaction pairs in selected cell subsets over time in six vaccine recipients; sample points from the same recipient are connected via lines in the same color. p values < 0.05 (Student' s t test) are indicated.
(C–F) Dot plots show the potential ligand-receptor pairs among innate immune cell subsets and adaptive immune cell subsets over time, including chemokine-receptor pairs indicative of interactions between monocyte and T cell subsets (C), co-stimulatory ligand-receptor pairs indicative of interactions between T cell and APC subsets (D), co-inhibitory ligand-receptor pairs indicative of interactions between T cell and APCs subsets (E), and IFNG-type II IFNR and CSF1-CSR1R pairs (F). Dot colors show the interaction scores calculated using the CellPhoneDB, while dot sizes indicate p values.
We observed that cellular interactions between innate and adaptive immunity were significantly enhanced in response to vaccination through ligand-receptor pairs of chemokines, cytokines, and co-stimulatory and co-inhibitory pathways. The proportions of CD16+C1+ monocytes and CD16+IFN+ monocytes dramatically increased after vaccination (Figure 2D); their interactions with several subsets of T cells (GZMK+ cyto T, GZMH+ cyto T, IFN+ T, and activated CD4+ T) were also significantly enhanced through chemokine–receptor pairing (CCL3-CCR5, CXCL10-CXCR3, and CXCL16-CXCR6) (Figure 5C). These results suggested that chemokine production in innate immune cells was stimulated by vaccination; the increased levels of chemokines may promote the recruitment and activation of T cells.
Co-stimulatory pathways are essential for T cell activation by APCs.21 Multiple co-stimulatory ligand-receptor interactions (CD226-PVR, CD28-CD80, CD40LG-CD40, and ICOS-ICOSLG) between T cells (GZMK+ cyto T and activated CD4+ T) and APCs (CD16+C1+ monocytes, CD16+IFN+ monocytes, DCs, and IFN+ B cells) were significantly enhanced after vaccination, indicating that sustained co-stimulatory pathway activity had been induced by vaccination (Figures 5D and S11). The balance of co-stimulatory and co-inhibitory pathways is critical for regulating T cell responses.21 Notably, several ligand-receptor interactions of co-inhibitory pathways (TIGIT-PVR, CD96-PVR, CTLA4-CD86, and CTLA4-CD80) between T cells and APCs were increased after vaccination (Figures 5E and S11), which implies a negative control mechanism to prevent vaccine-mediated induction of excessive T cell responses.
Although no significant changes were observed in the proportions of IFN+ T cells, we found that interactions between IFN+ T cells and APCs (CD16+C1+ monocytes, CD16+IFN+ monocytes, and DCs) were significantly enhanced after each vaccination via IFNG-type II IFNR (Figure 5F). Furthermore, the CSF1-CSR1R interaction score was enhanced by the second vaccination among IFN+ T cells, activated CD4+ T cells, CD16+C1+ monocytes, and CD16+IFN+ monocytes (Figure 5F). These results suggest that IFN+ T cells and activated CD4+ T cells may provide cytokine feedback signals to modulate innate immune cells in response to vaccination.
Taken together, our cell-cell communication analysis indicated that vaccination elicited extensive interactions between innate and adaptive immunity; these interactions may be critical for establishing long-term immunological memory and immune homeostasis in response to the vaccine.
Validation of the enhancement of vaccine-elicited immune responses by in vitro experiments
To confirm the findings from our scRNA-seq data, we performed in vitro experiments on longitudinal plasma from validation cohort 1, which enrolled six vaccine recipients who had received injections at the same time points as the previous discovery cohort (Table S1). A similar dynamic tendency of IgG antibody levels with the previous discovery cohort was observed in validation cohort 1 (Figures 4D and S12A). We further examined antibody function by a pseudotyped virus neutralization assay, and the neutralizing antibody titers were not significantly increased until the second and third doses (Figure S12B). The highest titer was at 14 days after the third vaccination (Figure S12B), which suggested the limited protection of the first vaccination and the necessity of prime-boost vaccine strategies. We then tested whether vaccine-induced antibodies were influenced by sex or age in these two cohorts (n = 12), but no significant differences were observed at any time points (Figures S12C, S12D, S13A, and S13B).
To verify the functional pathways engaged in the vaccine-induced immune response, we determined the plasma levels of cytokines and chemokines among the 12 volunteers before vaccination (0 days) and every 14 days after the vaccine injection (14, 42, and 70 days) (Figures S12E–S12G and S13E). First, we discovered a significant elevation in the concentration of IL-2Rα 14 days after the first inoculation (Figure S12E), which may correlate with the high levels of Treg cells induced by vaccines.22 The concentrations of MIP-1α (CCL3), M-CSF (CSF1), and IP-10 (CXCL10) were also significantly increased at 14 days, which was in line with enhancement of the CCL3-CCR5, CSF1R-CSF1, and CXCL10-CXCR3 pairs in the cell-cell communication results (Figure S12F). There were no significant variations in these concentrations between different sexes or ages (Figures S12D, S13C and S13D). In addition, the plasma level of IL-1β has been reported to be higher in patients with COVID-19 infection,23 and MCP-1, which is a vital cytokine for the activation and transition of APCs, increased after mRNA vaccine inoculation in mice,24 and both of these factors were significantly increased after BBIBP-CorV inactivated vaccine inoculation (Figure S12G). TNF-β was also elevated after vaccination (Figure S12G). All of these results confirmed the vaccine-induced enhancement of the immune response.
Comparison of the immune responses led by different booster dosing strategies
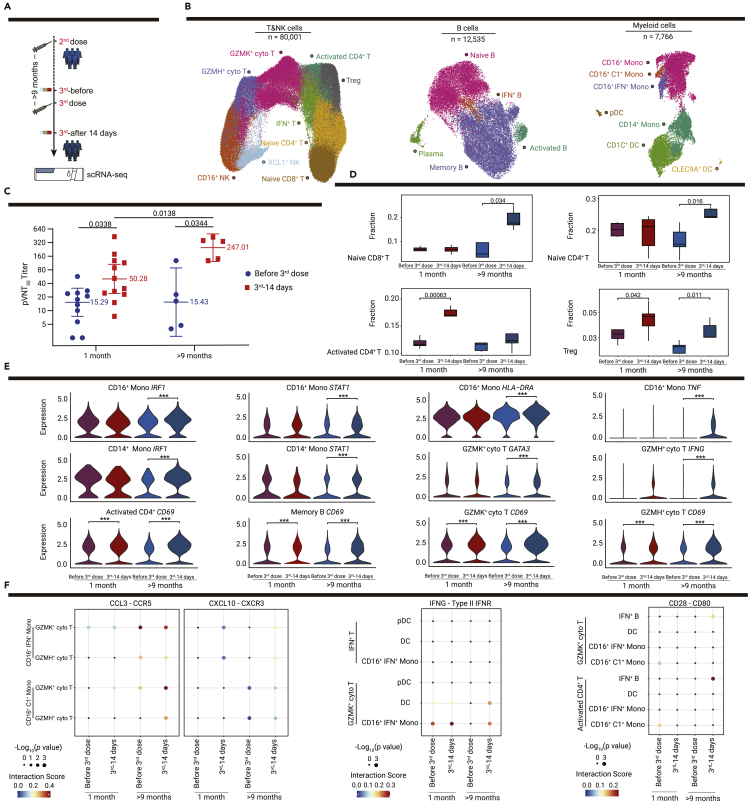
To further understand the immune responses to different vaccination strategies for the third dose, we recruited validation cohort 2 enrolling six volunteers who received the third dose more than 9 months after the second vaccination (Figure 6A; Table S1). Analyses by scRNA-seq of PBMCs and a pseudotyped virus neutralization assay of plasma were also conducted in five individuals from validation cohort 2 (Figures 6B and 6C). We integrated the scRNA-seq data of the validation cohort 2 with the discovery cohort on days 56 and 70, who received a booster dose 1 month after the second dose, and then re-clustered the T and NK cells, and B cells and myeloid cells, and further annotated their subsets (Figure 6B). Although a shared significant increase in neutralizing antibody titers 14 days after the booster dose was observed in the previous two cohorts (n = 12, vaccine recipients in the discovery cohort and validation cohort 1, who received the third dose 1 month after the second injection) and validation cohort 2 (Figure 6C), a substantially higher titer in validation cohort 2 was noted (Figure 6C), which was consistent with the results of recent booster dose studies.25,26
Figure 6.
Changes in the immune response after the third dose vaccination using different booster dosing strategies
(A) Flowchart of the sampling design for validation cohort 2 (n = 6, who received the third dose more than 9 months after the second injection).
(B) UMAP plots show immune cells of the discovery cohort (n = 6) and validation cohort 2 (n = 5) before and after the third dose vaccination, including subsets of T and NK cells, B cells and myeloid cells by unsupervised clustering. Each cell type is indicated by a different color.
(C) Comparison of neutralizing antibody titers before and after the third dose vaccination with 1 month or more than 9 month intervals between the second and third vaccinations, blue dots show the neutralizing antibody titers of vaccine recipients before the third dose vaccination, and red squares show the neutralizing antibody titers 14 days after the third injections. Median titers of groups at each time point are indicated. p values < 0.05 (paired t test and Student’s t test) are indicated.
(D) Boxplots show the proportions of T cell subsets for the discovery cohort and validation cohort 2 before and after the third dose vaccination. p values < 0.05 are indicated; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Student’s t test).
(E) Violin plot shows the expression levels of selected genes in selected cell types before and after the third dose vaccination in the discovery cohort and validation cohort 2. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Wilcoxon rank-sum test).
(F) Dot plots show the potential ligand-receptor pairs among APCs and T cell subsets before and after the third dose vaccination in the discovery cohort and validation cohort 2. Dot colors show the interaction scores calculated using the CellPhoneDB, while dot sizes indicate the p values.
Then, we compared the differences in immune subsets elicited by the third dose between the discovery cohort and validation cohort 2. Notably, the significantly elevated proportions of naive CD4+ T cells and naive CD8+ T cells identified in validation cohort 2 with a more than 9 month interval after the second dose, were not observed in the previous discovery cohort with a 1-month interval (Figure 6D). This suggested that immunization after a longer interval may induce expansion of naive T cells in the circulation or greater release from the thymus into the circulation. The shared expansion of Treg cells after the third vaccination was identified in the two cohorts, which revealed that the regulation of immune homeostasis mediated by Treg cells was also induced by the booster dose (Figure 6D). The upregulation of IFN pathway genes IRF1 and STAT1 in CD16+ mono and CD14+ mono cells, as well as the antigen-presenting-related gene HLA-DRA and TNF in CD16+ mono cells was more pronounced in validation cohort 2 than in the discovery cohort, which indicated the more active state of APCs following the third immunization with a longer interval (Figure 6E). The significantly increased expression of the GATA3 gene, an important transcription factor involved in the maintenance of cytotoxic T cells and the production of IFN-γ,27 was observed in the GZMK+ cyto T cells of validation cohort 2 (Figure 6E), which showed the strengthened function of cytotoxic T cells in recipients with a 9-month interval, together with the upregulated expression of IFNG in GZMH+ cyto T cells. In addition, elevated expression levels of activation marker CD69 were observed in memory B cells, activated CD4+ T cells, GZMK+ cyto T cells, and GZMH+ cyto T cells in the two cohorts after the booster dose, which highlighted the activation of adaptive immune responses induced by vaccines (Figure 6E). Crosstalk between some subsets of innate immune cells and adaptive immune cells were also intensified in validation cohort 2. The interactions between CD16+ C1+ monocytes and two T cell subsets (GZMK+ cyto T and GZMH+ cyto T) through CCL3-CCR5 were enhanced after the booster dose vaccination with a 9-month interval, while enhancement of the interactions between CD16+ IFN+ monocytes and these two T cell subsets through CXCL10-CXCR3 were observed in the two cohorts (Figure 6F). Interestingly, the expression of IFNG-type II IFNR between IFN+ T cells and GZMK+ cyto T with APCs (CD16+C1+ monocytes and DCs) was depleted 9 months after the second vaccination, but the booster dose drastically induced its expression again in validation cohort 2 (Figure 6F). The co-stimulatory pairing of CD28−CD80 between IFN+ B cells with activated CD4+ T cells and GZMK+ cyto T cells were also enhanced in validation cohort 2 (Figure 6F). Collectively, we evaluated and compared the diverse immune responses led by different booster dosing strategies, and revealed that a higher level of neutralizing antibodies, distinct expansion of naive T cells, and shared elevation of the Treg proportion, together with significant upregulation of functional genes and cellular interactions were evident in individuals with a longer interval between the second and third vaccinations.
Discussion
In recent studies of the COVID-19 vaccine, vaccine-induced immune responses have been widely reported, such as the enhancement of innate immunity, generation of CD4+ and CD8+ memory T cell subsets, as well as the development of antigen-specific memory B cells.10,28,29 In our study, the specific features of the immune dynamics elicited by BBIBP-CorV were investigated and are detailed below.
The BBIBP-CorV SARS-CoV-2 inactivated vaccine induced a series of immune regulatory mechanisms that suppressed excessive responses by immune cells, which could aid the maintenance of immune homeostasis after vaccination. First, the proportion of CD4+ regulatory T cells was significantly increased. Second, the expression of the immune checkpoint regulator SELPLG was upregulated in CD8+ cytotoxic T cells. Finally, co-inhibitory interactions between T cells and APCs were significantly enhanced. These results indicate that inactivated vaccine activates the immune system, while providing a negative feedback regulation mechanism to ensure that immune homeostasis is maintained and excessive immunity is prevented during the establishment of immune protection. Thus, methods to enhance the protective effects of vaccines while maintaining immune homeostasis may be essential for the future development of vaccines with greater efficacy and fewer side effects.
Regarding concerns about the influence of booster doses on the immune response, a comparative analysis of individuals administered different booster dosing strategies was conducted. In the interest of brevity, our study found that longer intervals between the second and third inoculation could trigger higher levels of neutralizing antibodies, which was consistent with a recent report on inactivated vaccine CoronaVac.25 We further discovered the distinct expansion of naive T cells and the shared elevation of the Treg proportion in individuals with a longer interval between the second and third vaccinations. The stronger active signatures of functional genes and cellular interaction pairs were also observed. Therefore, a suitable interval prior to the third dose must be established to improve the protective effects of the vaccine.
In summary, our study revealed that innate and adaptive immune cells were activated after the administration of the SARS-CoV-2 inactivated vaccine; our findings also demonstrated extensive crosstalk between innate immune cells and adaptive immune cells after vaccination. Furthermore, our results showed a potential role for immune regulatory mechanisms in the maintenance of immune homeostasis after vaccination. Our findings support a comprehensive understanding of the immune responses elicited by the SARS-CoV-2 inactivated vaccine; they also provide insights that will be useful in the optimization of vaccination strategies.
Data availability
Raw scRNA-seq data were deposited in the CNGB Nucleotide Sequence Archive (CNSA) (Database: CNP0002118) of the China National GeneBank DataBase (CNGBdb). Other relevant data are available from the corresponding author upon reasonable request.
Acknowledgments
This work was supported by the National Key Research and Development Program of China, China (2021YFC2301400); the Project of International Cooperation and Exchanges National Natural Science Foundation of China, China (NSFC, China) (82161148008); the Shenzhen Key Laboratory of Single-Cell Omics (ZDSYS20190902093613831); the Special Major Application Research Project for COVID-19 Prevention and Control in Universities, Department of Education of Guangdong, Provincial Program of Innovation and Strengthening School, Guangdong, China (2020KZDZX1093); and the Special Project for COVID-19 Prevention and Treatment of Shantou Science and Technology Bureau, Guangdong, China (2020-1-61). W.J.L. was supported by the Excellent Young Scientist Program of the NSFC, China (81822040). X.X. was supported by the Guangdong Provincial Key Laboratory of Genome Read and Write (2017B030301011). We thank Ryan Chastain-Gross, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Author contributions
W.J.L., X.Y., Chuanyu Liu, X.J., G.F.G., G.W., and Longqi Liu designed and supervised the study. W.J.L., X.Y., Chuanyu Liu, X.J., G.F.G., G.W., and Yuntao Zhang conceived the project. Yingze Zhao, Yunkai Yang, Yanxia Wang, Z.X., B.Y., Y.G., W.L., Lei Li, J.T., J.G., H.W., P.L., Hao Liang and W.W. collected clinical samples and analyzed the clinical and treatment data. Yingze Zhao, Yaling Huang, Zhifei Wang, X.L., W.Z., S.W., Chang Liu, X.W., X.S., Zifei Wang, Yang Wang, Ya Liu, M.Y., Yue Yuan, Ying Liu, Yunting Huang, and Haorong Lu performed the experiments. J.Y., Yingze Zhao, Z.Z., F.H., Yuhui Zheng, X.W., P.M., Z.H., Y.S., W.M., and Chuanyu Liu analyzed the data. Yong Hou and X.X. contributed to fruitful discussions and key ideas. J.Y., Z.Z., Yingze Zhao, and W.J.L. wrote the manuscript. J.Y., Yingze Zhao, Chuanyu Liu, and W.J.L. participated in the manuscript editing and discussion.
Declaration of interests
The authors declare no competing interests.
Published Online: December 05, 2022
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2022.100359.
Contributor Information
Yuntao Zhang, Email: zhangyuntao@sinopharm.com.
Guizhen Wu, Email: wugz@chinacdc.cn.
George F. Gao, Email: gaof@im.ac.cn.
Xin Jin, Email: jinxin@genomics.cn.
Chuanyu Liu, Email: liuchuanyu@genomics.cn.
Xiaoming Yang, Email: yangxiaoming@sinopharm.com.
William J. Liu, Email: liujun@ivdc.chinacdc.cn.
Lead contact website
Supplemental information
References
- 1.Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadarangani M., Marchant A., Kollmann T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021;21:475–484. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotez P.J., Bottazzi M.E. Whole inactivated virus and protein-based COVID-19 vaccines. Annu. Rev. Med. 2022;73:55–64. doi: 10.1146/annurev-med-042420-113212. [DOI] [PubMed] [Google Scholar]
- 4.Wang H., Zhang Y., Huang B., et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia S., Zhang Y., Wang Y., et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia S., Zhang Y., Wang Y., et al. Safety and immunogenicity of an inactivated COVID-19 vaccine, BBIBP-CorV, in people younger than 18 years: a randomised, double-blind, controlled, phase 1/2 trial. Lancet Infect. Dis. 2022;22:196–208. doi: 10.1016/S1473-3099(21)00462-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Y., Li Y., Yang R., Tan W. SARS-CoV-2-specific T cell immunity to structural proteins in inactivated COVID-19 vaccine recipients. Cell. Mol. Immunol. 2021;18:2040–2041. doi: 10.1038/s41423-021-00730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noé A., Cargill T.N., Nielsen C.M., et al. The application of single-cell RNA sequencing in vaccinology. J. Immunol. Res. 2020;2020:8624963. doi: 10.1155/2020/8624963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Q., Wu S., Xiao C., et al. Integrated single-cell analysis revealed immune dynamics during Ad5-nCoV immunization. Cell Discov. 2021;7:64. doi: 10.1038/s41421-021-00300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arunachalam P.S., Scott M.K.D., Hagan T., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Wang J., Xu J., et al. Comprehensive investigations revealed consistent pathophysiological alterations after vaccination with COVID-19 vaccines. Cell Discov. 2021;7:99. doi: 10.1038/s41421-021-00329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulendran B., Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Jarjour N.N., Masopust D., Jameson S.C. T cell memory: understanding COVID-19. Immunity. 2021;54:14–18. doi: 10.1016/j.immuni.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Diao H., Getzler A.J., et al. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity. 2018;48:659–674.e6. doi: 10.1016/j.immuni.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tinoco R., Carrette F., Barraza M.L., et al. PSGL-1 is an immune checkpoint regulator that promotes T cell exhaustion. Immunity. 2016;44:1470. doi: 10.1016/j.immuni.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Quast I., Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54:205–210. doi: 10.1016/j.immuni.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen W., Su W., Tang H., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maecker H.T., Levy S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 1997;185:1505–1510. doi: 10.1084/jem.185.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen L., Flies D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindqvist C.A., Christiansson L.H., Simonsson B., et al. T regulatory cells control T-cell proliferation partly by the release of soluble CD25 in patients with B-cell malignancies. Immunology. 2010;131:371–376. doi: 10.1111/j.1365-2567.2010.03308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McElvaney O.J., McEvoy N.L., McElvaney O.F., et al. Characterization of the inflammatory response to severe COVID-19 illness. Am. J. Respir. Crit. Care Med. 2020;202:812–821. doi: 10.1164/rccm.202005-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebre M.S., Rauch S., Roth N., et al. mRNA vaccines induce rapid antibody responses in mice. NPJ Vaccines. 2022;7:88. doi: 10.1038/s41541-022-00511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng G., Wu Q., Pan H., et al. Immunogenicity and safety of a third dose of CoronaVac, and immune persistence of a two-dose schedule, in healthy adults: interim results from two single-centre, double-blind, randomised, placebo-controlled phase 2 clinical trials. Lancet Infect. Dis. 2022;22:483–495. doi: 10.1016/S1473-3099(21)00681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X., Zhang R., Qiao S., et al. Omicron SARS-CoV-2 neutralization from inactivated and ZF2001 vaccines. N. Engl. J. Med. 2022;387:277–280. doi: 10.1056/NEJMc2206900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tindemans I., Serafini N., Di Santo J.P., Hendriks R.W. GATA-3 function in innate and adaptive immunity. Immunity. 2014;41:191–206. doi: 10.1016/j.immuni.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z., Mateus J., Coelho C.H., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Zeng Q., Deng C., et al. Robust induction of B cell and T cell responses by a third dose of inactivated SARS-CoV-2 vaccine. Cell Discov. 2022;8:10. doi: 10.1038/s41421-022-00373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw scRNA-seq data were deposited in the CNGB Nucleotide Sequence Archive (CNSA) (Database: CNP0002118) of the China National GeneBank DataBase (CNGBdb). Other relevant data are available from the corresponding author upon reasonable request.