Abstract
Malaria sporozoites are transmitted from the mosquito salivary gland to host hepatocytes within minutes of an infectious bite. The circumsporozoite protein (CS), which covers the surface of Plasmodium sporozoites, functions during these minutes in the targeting of host liver cells. The protein's potentially important role in an antimalaria vaccine has spawned interest in both the host immune responses to the parasite's presence and the actual functional role of the protein in the targeting of host liver cells. Here we show that the region of CS known to elicit a cytotoxic T-lymphocyte (CTL) response to irradiated sporozoites also, somewhat ironically, mediates the receptor-ligand interaction essential to parasite invasion of the host. Hence, the structure of CS represents a balance of potentially counterdirectional forces. Polymorphism in the CTL epitope appears to be a product of this balanced state as opposed to an “arms race” as it is so often portrayed. The conceptual difference between the theories regarding the maintainance of polymorphism in CTL epitopes may have significant implication for vaccine design.
Circumsporozoite protein (CS) is an immunodominant protein present on the surface of Plasmodium sporozoites, the causative organism for malaria (39). This protein is essential to sporozoite development in the mosquito and promotes binding to liver cells (22). It has been used as a target for making antimalarial vaccines (33, 35, 37).
The main structural and antigenic properties of CS are identical in all the species of malaria sporozoites. It is made up of a secretory signal sequence at its amino terminus, a central repeat region, two conserved amino acid motifs region I and region II-plus, and an anchor sequence at its carboxyl terminus (4, 24, 34). The repeat domain is species specific and immunodominant and constitutes about one-half of the molecule (40). Region II-plus, a 18-amino-acid motif, constitutes the binding ligand of CS (4, 24, 34). The region II-plus motif is not only conserved among the CS of all malaria parasites (20), it is shared with other sporozoite surface proteins such as thrombospondin-related anonymous protein and a variety of hosts proteins such as thrombospondin or properdin (12, 17, 29). In Plasmodium falciparum it is represented by EWSPCSVTCGNGIQVRIK (4).
With regard to function of CS during invasion, it is known that basic and hydrophobic amino acids associated with region II-plus specifically interact with the negatively charged glycosaminoglycans chains of heparan sulfate proteoglycans present on the cell surface of hepatocytes (9, 25, 34). The avidity of binding relates to the degree of sulfonation of the proteoglycan and hence varies in accordance with host-related factors. Low-density lipoprotein receptor-related protein present on hepatocyte cell surface has also been shown to interact with the region II-plus of CS (32). Identification of the exact residue(s) involved in binding has yielded discrepant results (10, 28, 34). Recently, CS has also been shown to inhibit the protein synthesis in mammalian cells, but the exact mechanism is not fully understood (8, 14).
CS-specific CD8+ and CD4+ T cells are protective in murine models, and an important aim has been to identify CS T-cell epitopes recognized by malaria-exposed humans (1, 27, 31). Recently, Wang et al. demonstrated the induction of antigen-specific cytotoxic T lymphocytes (CTL) in humans by immunizing them with plasmid DNA encoding CS of P. falciparum (37). Two CTL epitopes recognized by humans have been identified in a 23-amino-acid motif (KPKDELDYANDIEKKICKMEKCS) located toward the carboxyl terminus of the protein (13, 16, 19, 30). The protein's potentially important role in eliciting host immunity sometimes overshadows interest in the functional role the protein plays in parasite development. Given the dual role of this protein in malaria infection, we investigated the structure-function relationship of a region of CS known to be involved in eliciting a protective immunological response to the sporozoite. Here we show that the region of the CS known to elicit a protective CTL response to the sporozoite is involved in the receptor-ligand interaction essential to parasite invasion of the host.
MATERIALS AND METHODS
Materials.
Vector pET11a and Escherichia coli strain BL21(λDE3) were obtained from Novagen (Madison, Wis.). All restriction and modifying enzymes were either from Life Technologies or Boehringer Mannheim. RPMI 1640, fetal bovine serum, trypsin, and l-glutamine were obtained from Life Technologies (Gaithersburg, Md.). Hepatoma cell line HepG2 was obtained from The American Type Culture Collection (Manassas, Va.). Paraformaldehyde was obtained from Electron Microscopy Sciences (Washington, Pa.). Anti-mouse antibody-alkaline phosphatase conjugate was obtained from Pierce Chemical Co. (Rockford, Ill.). A heparin Sepharose column was obtained from Amersham-Pharmacia Biotech (Pricataway, N.J.). Monoclonal antibody 2A10 directed against the (NANP)n repeat domain was kindly provided by Robert Wirtz (Centers for Disease Control, Atlanta, Ga.). Plasmid pCS27IVC was kindly provided by Photini Sinnis (New York University Medical Center, New York).
Peptide synthesis.
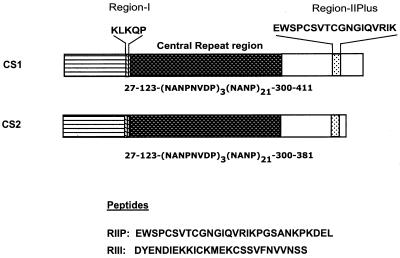
Peptides were synthesized by using solid-phase Fmoc chemistry on chlorotrityl resins. They were purified by reversed-phase chromatography in a high-performance liquid chromatography system. Purity of the peptides was verified by mass spectrometry. Peptides RIIP and RIII were composed of 29 and 26 amino acids, respectively (Fig. 1).
FIG. 1.
Schematic representation of pCS1 and pCS2. pCS1 encodes the full-length protein, while pCS2 represents a truncated version of CS where the DNA encoding amino acids 382 to 411 has been deleted.
Construction of pCS1 and pCS2.
Plasmid pCS27IVC (36) was used as template to PCR amplify the DNA encoding CS. pCS27IVC contains the DNA sequence encoding amino acid sequence 27–123[NANPNVDP]3[NANP]21300–411 of CS fused to a six-amino-acid histidine tag at the carboxyl terminus. It represents the complete P. falciparum CS sequence from the T4 isolate except that the hydrophobic clusters of amino- and carboxyl-terminal amino acids 1 to 26 and 412 to 424 have been deleted. Primers were designed to introduce NdeI and BamHI restriction sites at the 5′ and 3′ ends of the amplified fragments, respectively. A BamHI site present near the 5′ end of the CS DNA was also changed without changing the encoding amino acid. The amplified fragment was cloned in E. coli expression vector pET11a that was digested with the same enzymes. This gave rise to plasmid pCS1 (27–123[NANPNVDP]3[NANP]21300–411). Plasmid pCS2 (27–123[NANPNVDP]3[NANP]21300–381) is essentially the same as pCS1 except that it lacks DNA encoding the last 30 amino acids from the carboxyl terminus.
Expression and localization of recombinant proteins.
Bacterial strain BL21(λDE3), containing T7 RNA polymerase gene under control of the lac promoter was used for the expression of recombinant proteins. Cells were transformed with the desired construct and grown in Super Broth (pH 7.2) containing 100 μg of ampicillin per ml at 37°C with shaking. At an optical density at 600 nm of 1.0, the cells were induced with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and after 4 h they were harvested by centrifugation at 4,000 × g for 20 min. To localize the protein, the cell pellet was gently resuspended in 20% sucrose in 50 mM Tris (pH 7.4) and incubated on ice for 10 min. Suspended cells were spun at 8,000 × g for 20 min at 4°C, and the pellet was saved and resuspended in chilled distilled water. After incubation on ice for 10 min, the cell suspension was centrifuged at 15,000 × g for 10 min at 4°C. The supernatant, obtained after centrifugation, contained the periplasmic fraction, while the pellet represented the spheroplast.
Purification of recombinant proteins.
Periplasm, containing the recombinant protein in water, was added to 25 mM Tris (pH 7.4) and loaded onto a heparin Sepharose affinity column equilibrated in the same buffer. The protein was sequentially eluted with a step gradient with 0.125, 0.25, and 0.5 M NaCl in 25 mM Tris (pH 7.4) on a fast protein liquid chromatography system. CS-containing fractions were pooled, concentrated, and purified to homogeneity by gel filtration chromatography. The protein runs as a single band on polyacrylamide electrophoresis. The amino terminus of the protein has been sequenced and yields the single, expected sequence with little background, thus indicating that it is essentially a pure product.
Cell binding assay.
The hepatoma cell line HepG2, maintained in RPMI 1640 containing 2 mM glutamine and 10% heat-inactivated fetal bovine serum, was used, and the assay was performed as described earlier (3). Briefly, cells were seeded at a density of 100,000 cells per well in a 96-well plate and allowed to adhere for at least 24 h. For the assay, cells were fixed with 4% paraformaldehyde, followed by blocking with Tris-buffered saline containing 2.5% bovine serum albumin. This was followed by the addition of different concentrations of the proteins and incubation at 37°C for 1 h. Unbound protein was removed by washing, and the cells were incubated with anti-CS monoclonal antibody (1 μg/ml) for 45 min. Unbound monoclonal antibody was removed by washing, followed by the addition of and further incubation with alkaline phosphatase-coupled anti-mouse antibody for 30 min. Unbound conjugate was removed, and 0.1 ml of 1 mM 4-methylumbelliferyl was added as substrate. After 15 min the fluorescence was measured in a fluorometer with excitation at 350 nm and emission at 460 nm. For competition analysis, cells after blocking were incubated with different concentrations of the peptides for 2 h before the addition of recombinant protein. Each experiment was repeated four separate times, and the binding profile, as measured by fluorescence, remained the same. We saw less than 10% variation in any quantitative measure.
RESULTS AND DISCUSSION
We have studied CS protein with or without the CTL epitope (CS1 or CS2, respectively) in an attempt to correlate the absence of this region with a measurable biological affect (Fig. 1). For the sake of comparison with work from other laboratories, the CS was produced from the plasmid construct used by others who study structure-function relationships of the protein (3, 9, 32, 34, 38). In all comparative studies, therefore, a signal sequence and a small anchor is missing from the protein (23). Proteins were isolated and purified according to the binding affinity for the receptor. For this purpose, both proteins were separated by using a heparin Sepharose column and later purified to homogeneity by gel filtration chromatography.
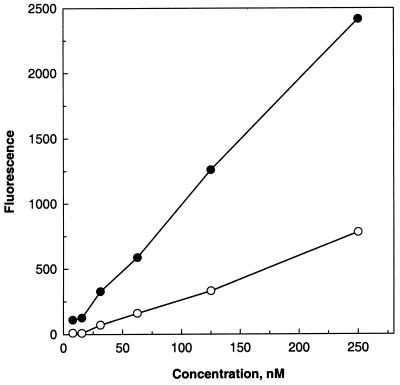
The CS binds to hepatocytes through a conserved 18-amino-acid binding motif known as region II-plus. To check if the deletion of the CTL epitopes makes a difference in the binding efficiency of the protein, its activity was tested on HepG2 cells. Even when the region II-plus (binding domain) was totally intact, deletion of the last 30 amino acids encompassing the CTL epitopes resulted in a significant loss (70%) of binding activity of CS2 (Fig. 2). This study demonstrates that the region containing CTL epitopes plays a significant role in the hepatocyte invasion and that an intact carboxyl terminus is essential for the binding activity of CS.
FIG. 2.
Binding activity of CS1 and CS2 on HepG2 cells. Cells were incubated with different concentrations of CS1 (●) and CS2 (○) for 1 h, followed by successive addition of anti-CS monoclonal antibody and anti-mouse antibody-alkaline phosphatase conjugate. Fluorescence was measured as described in the text.
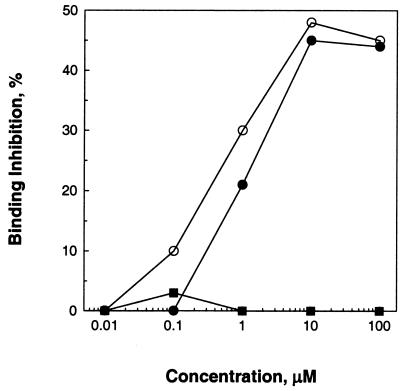
Both CS1 and CS2 utilize region II-plus as the primary binding ligand, since a peptide encoding region II-plus competes with the binding of both CS1 and CS2 in a dose-dependent manner yielding similar profiles (Fig. 3). In contrast, peptide RIII, representing the deleted region, failed to compete with CS1 for binding to the cells (Fig. 3). This indicates that the deleted domain does not bind directly with the protein receptor but acts to modulate the interaction of region II-plus with the receptor. It also suggests that the residues recognized as a CTL epitope also provide a proper structural conformation to the protein for its interaction with hepatocyte receptors. Given that the region plays dual roles, the results of molecular change must be considered to potentially alter at least two potentially opposing forces.
FIG. 3.
Inhibition of binding activity. Cells were preincubated with different concentrations of peptide RII (circles) or RIII (square), followed by the addition of CS1 (○, ■) or CS2 (●) and incubation for 1 h. The percent inhibition is decrease in fluorescence compared to the control where no peptide was added.
How and why are substantial amounts of amino acid polymorphism maintained in a pathogen's dominant surface protein and why are these exclusively concentrated in its immunodominant T-cell epitopes? De La Cruz et al. (5, 6) were the first to study polymorphism in CS genes, and they concluded that (i) there is a correlation between the small number of T epitopes in the protein and the regions of polymorphism and (ii) there is a biased ratio of nonsynonymous to synonymous mutations resulting in change at the protein level. These authors proceeded to speculate that the ratio of nonsynonymous to synonymous mutation was an indicator (not proof) of positive selection for variation at the site and that variation may be driven by immune selection. For better or worse, considerable work and thought since that time have focused on evaluating those ideas. To review the arguments here, both pro and con, would serve no purpose, and thus we only acknowledge those who questioned the interpretation (2), those that developed experimental approaches that led to alternative interpretations (3, 34), and those that used statistical approaches and a growing database of CS sequences to assess the confidence levels of putative indicators (15).
The relationship between host and parasite is not an “arms race” or even a Faustian chess game, depending upon individual move and countermove, as it is portrayed by the popular press and in many journal articles. At least in the case of CS, the host and parasite have come to a balanced state(s). For example, the most effective way for sporozoites to evade a directed immune response to CS would be to rid the protein of sequence motifs that lead to an antiparasite response. The CTL epitope has not been eliminated and therefore appears to be essential to parasite survival. Hence, one already suspects what we have shown here: the immune system recognizes a sequence motif that is essential to the function of the parasite. This balance would suggest limitations on the number of acceptable forms of the epitope that are biologically successful. This hypothesis is supported by the observed homoplasy in the CTL epitopes (21), which often indicates a high rate of mutation in the region but a low number of possible character states. A vaccine including only dominant polymorphic forms would therefore focus the immune response on character states that best balance the necessity to invade liver cells and to evade the immune system. Since these features are tied, surviving minor variants genotypes would represent outliers with either reduced potential for transmission or an increased facility to elicit an immune response. Reduction in parasite numbers would disproportionately affect the fittest combinations.
Another hypothesis regarding the maintenance of CS polymorphism proposes that variation itself is selected and maintained because of the advantage given coinfection by antagonistic pairs (26). It also proposes limitations on the extent of T-cell epitope variation and hence is also supported by the observation of homoplasy in the CS gene (21). Selective advantage, in this case, relates to the concordance of pairs by way of mutual interference with human T-cell responses. It implies that “inclusion of all allelic peptide variants in such vaccines may not offer a solution to the well-recognized problem of antigenic polymorphism in malaria: inclusion of a variant epitope might even lead to antagonism of naturally acquired immunity of other variants, resulting in increased susceptibility” (26). This is an appealing hypothesis and has some experimental support. It does, however, depend upon the assumption that polymorphism is maintained solely as a response to the immune system and ignores the biological role of the immune target (7, 18). Further, the implications for vaccine design are directly opposite those indicated by the above results. Attributing this type of polymorphism solely to immune selective pressure is, at best, an incomplete explanation.
ACKNOWLEDGMENTS
We thank Photini Sinnis, New York University Medical Center, New York, for providing plasmid pCS27IVC. We also thank Robert Wirtz, Center for Disease Control, Atlanta, Ga., for providing anti-CS monoclonal antibody.
REFERENCES
- 1.Aggarwal A, Kumar S, Jaffe R, Hone D, Gross M, Sadoff J. Oral Salmonella: malaria circumsporozoite recombinants induce specific CD8+ cytotoxic T cells. J Exp Med. 1990;172:1083–1090. doi: 10.1084/jem.172.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnot D E. Possible mechanisms for the maintenance of polymorphisms in Plasmodium populations. Acta Leiden. 1991;60:29–35. [PubMed] [Google Scholar]
- 3.Cerami C, Frevert U, Sinnis P, Takacs B, Clavijo P, Santos M J, Nussenzweig V. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell. 1992;70:1021–1033. doi: 10.1016/0092-8674(92)90251-7. [DOI] [PubMed] [Google Scholar]
- 4.Dame J B, Williams J L, McCutchan T F, Weber J L, Wirtz R A, Hockmeyer W T, Maloy W L, Haynes J D, Schneider I, Roberts D, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 5.De La Cruz V F, Maloy W L, Miller L H, Good M F, McCutchan T F. The immunologic significance of variation within malaria circumsporozoite protein sequences. J Immunol. 1989;142:3568–3575. [PubMed] [Google Scholar]
- 6.De La Cruz V F, Maloy W L, Miller L H, Lal A A, Good M F, McCutchan T F. Lack of cross-reactivity between variant T cell determinants from malaria circumsporozoite protein. J Immunol. 1988;141:2456–2460. [PubMed] [Google Scholar]
- 7.Doolan D L, Saul A J, Good M F. Geographically restricted heterogeneity of the Plasmodium falciparum circumsporozoite protein: relevance for vaccine development. Infect Immun. 1992;60:675–682. doi: 10.1128/iai.60.2.675-682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frevert U, Galinski M R, Hugel F U, Allon N, Schreier H, Smulevitch S, Shakibaei M, Clavijo P. Malaria circumsporozoite protein inhibits protein synthesis in mammalian cells. EMBO J. 1998;17:3816–3826. doi: 10.1093/emboj/17.14.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frevert U, Sinnis P, Cerami C, Shreffler W, Takacs B, Nussenzweig V. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J Exp Med. 1993;177:1287–1298. doi: 10.1084/jem.177.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantt S M, Clavijo P, Bai X, Esko J D, Sinnis P. Cell adhesion to a motif shared by the malaria circumsporozoite protein and thrombospondin is mediated by its glycosaminoglycan-binding region and not by CSVTCG. J Biol Chem. 1997;272:19205–19213. doi: 10.1074/jbc.272.31.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert S C, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood B M, Whittle H C, Hill A V. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 1998;279:1173–1177. doi: 10.1126/science.279.5354.1173. [DOI] [PubMed] [Google Scholar]
- 12.Goundis D, Reid K B. Properdin, the terminal complement components, thrombospondin and the circumsporozoite protein of malaria parasites contain similar sequence motifs. Nature. 1988;335:82–85. doi: 10.1038/335082a0. [DOI] [PubMed] [Google Scholar]
- 13.Hill A V, Elvin J, Willis A C, Aidoo M, Allsopp C E, Gotch F M, Gao X M, Takiguchi M, Greenwood B M, Townsend A R, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. 1992;360:434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- 14.Hugel F U, Pradel G, Frevert U. Release of malaria circumsporozoite protein into the host cell cytoplasm and interaction with ribosomes. Mol Biochem Parasitol. 1996;81:151–170. doi: 10.1016/0166-6851(96)02701-6. [DOI] [PubMed] [Google Scholar]
- 15.Hughes A L. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics. 1991;127:345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Miller L H, Quakyi I A, Keister D B, Houghten R A, Maloy W L, Moss B, Berzofsky J A, Good M F. Cytotoxic T cells specific for the circumsporozoite protein of Plasmodium falciparum. Nature. 1988;334:258–260. doi: 10.1038/334258a0. [DOI] [PubMed] [Google Scholar]
- 17.Lawler J, Hynes R O. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J Cell Biol. 1986;103:1635–1648. doi: 10.1083/jcb.103.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockyer M J, Marsh K, Newbold C I. Wild isolates of Plasmodium falciparum show extensive polymorphism in T cell epitopes of the circumsporozoite protein. Mol Biochem Parasitol. 1989;37:275–280. doi: 10.1016/0166-6851(89)90159-x. [DOI] [PubMed] [Google Scholar]
- 19.Malik A, Egan J E, Houghten R A, Sadoff J C, Hoffman S L. Human cytotoxic T lymphocytes against the Plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci USA. 1991;88:3300–3304. doi: 10.1073/pnas.88.8.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCutchan T F, Kissinger J C, Touray M G, Rogers M J, Li J, Sullivan M, Braga E M, Krettli A U, Miller L H. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci USA. 1996;93:11889–11894. doi: 10.1073/pnas.93.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCutchan T F, Lal A A, do Rosario V, Waters A P. Two types of sequence polymorphism in the circumsporozoite gene of Plasmodium falciparum. Mol Biochem Parasitol. 1992;50:37–45. doi: 10.1016/0166-6851(92)90242-c. [DOI] [PubMed] [Google Scholar]
- 22.Menard R, Sultan A A, Cortes C, Altszuler R, van Dijk M R, Janse C J, Waters A P, Nussenzweig R S, Nussenzweig V. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. 1997;385:336–340. doi: 10.1038/385336a0. [DOI] [PubMed] [Google Scholar]
- 23.Nussenzweig V, Nussenzweig R S. Circumsporozoite proteins of malaria parasites. Cell. 1985;42:401–403. doi: 10.1016/0092-8674(85)90093-5. [DOI] [PubMed] [Google Scholar]
- 24.Ozaki L S, Svec P, Nussenzweig R S, Nussenzweig V, Godson G N. Structure of the plasmodium knowlesi gene coding for the circumsporozoite protein. Cell. 1983;34:815–822. doi: 10.1016/0092-8674(83)90538-x. [DOI] [PubMed] [Google Scholar]
- 25.Pancake S J, Holt G D, Mellouk S, Hoffman S L. Malaria sporozoites and circumsporozoite proteins bind specifically to sulfated glycoconjugates. J Cell Biol. 1992;117:1351–1357. doi: 10.1083/jcb.117.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plebanski M, Lee E A, Hannan C M, Flanagan K L, Gilbert S C, Gravenor M B, Hill A V. Altered peptide ligands narrow the repertoire of cellular immune responses by interfering with T-cell priming. Nat Med. 1999;5:565–571. doi: 10.1038/8444. [DOI] [PubMed] [Google Scholar]
- 27.Renia L, Grillot D, Marussig M, Corradin G, Miltgen F, Lambert P H, Mazier D, Del Giudice G. Effector functions of circumsporozoite peptide-primed CD4+ T cell clones against Plasmodium yoelii liver stages. J Immunol. 1993;150:1471–1478. [PubMed] [Google Scholar]
- 28.Rich K A, George F W T, Law J L, Martin W J. Cell-adhesive motif in region II of malarial circumsporozoite protein. Science. 1990;249:1574–1577. doi: 10.1126/science.2120774. [DOI] [PubMed] [Google Scholar]
- 29.Robson K J, Hall J R, Jennings M W, Harris T J, Marsh K, Newbold C I, Tate V E, Weatherall D J. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature. 1988;335:79–82. doi: 10.1038/335079a0. [DOI] [PubMed] [Google Scholar]
- 30.Romero P, Maryanski J L, Corradin G, Nussenzweig R S, Nussenzweig V, Zavala F. Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature. 1989;341:323–326. doi: 10.1038/341323a0. [DOI] [PubMed] [Google Scholar]
- 31.Sadoff J C, Ballou W R, Baron L S, Majarian W R, Brey R N, Hockmeyer W T, Young J F, Cryz S J, Ou J, Lowell G H, et al. Oral Salmonella typhimurium vaccine expressing circumsporozoite protein protects against malaria. Science. 1988;240:336–338. doi: 10.1126/science.3281260. [DOI] [PubMed] [Google Scholar]
- 32.Shakibaei M, Frevert U. Dual interaction of the malaria circumsporozoite protein with the low density lipoprotein receptor-related protein (LRP) and heparan sulfate proteoglycans. J Exp Med. 1996;184:1699–1711. doi: 10.1084/jem.184.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y P, Hasnain S E, Sacci J B, Holloway B P, Fujioka H, Kumar N, Wohlhueter R, Hoffman S L, Collins W E, Lal A A. Immunogenicity and in vitro protective efficacy of a recombinant multistage Plasmodium falciparum candidate vaccine. Proc Natl Acad Sci USA. 1999;96:1615–1620. doi: 10.1073/pnas.96.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinnis P, Clavijo P, Fenyo D, Chait B T, Cerami C, Nussenzweig V. Structural and functional properties of region II-plus of the malaria circumsporozoite protein. J Exp Med. 1994;180:297–306. doi: 10.1084/jem.180.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoute J A, Slaoui M, Heppner D G, Momin P, Kester K E, Desmons P, Wellde B T, Garcon N, Krzych U, Marchand M. A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. N Engl J Med. 1997;336:86–91. doi: 10.1056/NEJM199701093360202. [DOI] [PubMed] [Google Scholar]
- 36.Takacs B J, Girard M F. Preparation of clinical grade proteins produced by recombinant DNA technologies. J Immunol Methods. 1991;143:231–240. doi: 10.1016/0022-1759(91)90048-k. [DOI] [PubMed] [Google Scholar]
- 37.Wang R, Doolan D L, Le T P, Hedstrom R C, Coonan K M, Charoenvit Y, Jones T R, Hobart P, Margalith M, Ng J, Weiss W R, Sedegah M, de Taisne C, Norman J A, Hoffman S L. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998;282:476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 38.Ying P, Shakibaei M, Patankar M S, Clavijo P, Beavis R C, Clark G F, Frevert U. The malaria circumsporozoite protein: interaction of the conserved regions I and II-plus with heparin-like oligosaccharides in heparan sulfate. Exp Parasitol. 1997;85:168–182. doi: 10.1006/expr.1996.4134. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida N, Nussenzweig R S, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980;207:71–73. doi: 10.1126/science.6985745. [DOI] [PubMed] [Google Scholar]
- 40.Zavala F, Cochrane A H, Nardin E H, Nussenzweig R S, Nussenzweig V. Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J Exp Med. 1983;157:1947–1957. doi: 10.1084/jem.157.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]