Abstract
The mechanism of pathogenesis of Mycobacterium tuberculosis is thought to be multifactorial. Among the putative virulence factors is the antigen 85 (Ag85) complex. This family of exported fibronectin-binding proteins consists of members Ag85A, Ag85B, and Ag85C and is most prominently represented by 85A and 85B. These proteins have recently been shown to possess mycolyl transferase activity and likely play a role in cell wall synthesis. The purpose of this study was to generate strains of M. tuberculosis deficient in expression of the principal members of this complex in order to determine their role in the pathogenesis of M. tuberculosis. Constructs of fbpA and fbpB disrupted with the kanamycin resistance marker ΩKm and containing varying amounts of flanking gene and plasmid vector sequences were then introduced as linear fragments into H37Rv by electroporation. Southern blot and PCR analyses revealed disruption of the homologous gene locus in one fbpA::ΩKm transformant and one fbpB::ΩKm transformant. The fbpA::ΩKm mutant, LAa1, resulted from a double-crossover integration event, whereas the fbpB::ΩKm variant, LAb1, was the product of a single-crossover type event that resulted in insertion of both ΩKm and plasmid sequences. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis confirmed that expression of the disrupted gene was not detectable in the fbpA and fbpB mutants. Analysis of growth rates demonstrated that the fbpB mutant LAb1 grew at a rate similar to that of the wild-type parent in enriched and nutrient-poor laboratory media as well as in human (THP-1) and mouse (J774.1A) macrophage-like cell lines. The fbpA mutant LAa1 grew similarly to the parent H37Rv in enriched laboratory media but exhibited little or no growth in nutrient-poor media and macrophage-like cell lines. The targeted disruption of two genes encoding mycolyl transferase and fibronectin-binding activities in M. tuberculosis will permit the systematic determination of their roles in the physiology and pathogenesis of this organism.
It has been estimated that one-third of the world's population is infected with Mycobacterium tuberculosis, the causative agent of the disease tuberculosis (24). The incidence of tuberculosis continues to increase worldwide, particularly among groups such as the medically underserved, the immunosuppressed, and the confined (11). New concerns have been raised by the reported increase in the overall number of cases, as well as by the increase in number of drug-resistant isolates (10, 11). Investigations into potential drug targets, more-efficient vaccines, and more-effective treatment regimens are currently under way in an effort to decrease morbidity and mortality due to this disease. While numerous immunogenic antigens and putative virulence factors have been isolated, cloned, and sequenced (36), insight into the mechanisms of pathogenesis of M. tuberculosis remains elusive.
The antigen 85 (Ag85) complex is a family of fibronectin-binding proteins that are considered to be potential virulence factors. These proteins (Ag85A, Ag85B, and Ag85C, encoded by the genes fbpA, fbpB, and fbpC, respectively) garnered attention when M. tuberculosis was found to bind selectively to fibronectin and not to other purified extracellular matrix proteins tested (32). Binding of these organisms to purified fibronectin is dose dependent and can be blocked by antibodies produced either to fibronectin or to members of the Ag85 complex (31, 32). It has been clearly demonstrated that the ability to bind fibronectin and other extracellular matrix proteins enhances the virulence of pathogenic organisms, most notably staphylococci and streptococci (28). Specific binding to host extracellular matrix proteins may aid in the adherence and dissemination of organisms in tissue.
Members of the Ag85 complex are both secreted and retained in the cell wall of M. tuberculosis (1). Quantitatively, the proteins are secreted at a ratio of 3:2:1, Ag85A to Ag85B to Ag85C (15). These proteins have recently been found to possess mycolyl transferase activity (9), adding to the growing number of cell wall-synthetic enzymes identified in mycobacteria (3, 4, 6, 10). Belisle et al. (9) identified members of the Ag85 complex as enzymes responsible for the transfer of mycolic acids to α-α′-trehalose to form α-α′-trehalose monomycolate (TMM) and α-α′-trehalose dimycolate (TDM), also known as cord factor. Ag85A and Ag85C share a similar specific mycolyl transferase activity, while the specific activity of Ag85B is only about 20% of that of Ag85C. One recent study (19) has shown that a disruption mutant of a clinical isolate of M. tuberculosis deficient in Ag85C contained 40% less cell wall-bound mycolates than the parent strain. These mycolates represented not only TMM and TDM but also other molecules such as arabinogalactan, glycerol monomycolate, α-mycolates, methoxymycolates, and ketomycolates. There was no apparent change in the composition of mycolates detected, but their quantity was affected by mutation of Ag85C. It remains to be determined why mycobacteria possess three enzymes with apparently similar activities and whether the individual members of the Ag85 complex play different roles in the production of these molecules.
In this study, several constructs were utilized in an attempt to achieve homologous recombination and disruptional mutagenesis of fbpA and fbpB in the virulent M. tuberculosis strain H37Rv. PCR, Southern blot hybridization, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblot analyses confirmed the disruption and inactivation of fbpA and fbpB in individual strains of M. tuberculosis. Loss of FbpA expression was shown to inhibit the ability of H37Rv to grow in wholly synthetic media or to replicate in human or mouse macrophage-like cell lines, indicating that FbpA may play a role in the pathogenesis of M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and media.
Middlebrook media were purchased from Difco Laboratories, Detroit, Mich. The virulent laboratory strain of M. tuberculosis, H37Rv (ATCC 27294), was grown in liquid Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.25% Tween 80 (Sigma Chemical Co., St. Louis, Mo.), and albumin-dextrose complex (ADC) (consisting of 0.5% bovine serum albumin, fraction V [Sigma], 0.085% NaCl, and 0.2% glucose) or Sauton medium (25). Middlebrook 7H10 or 7H11 plates supplemented with ADC with or without 20 μg of kanamycin/ml were used for colony isolation. Cycloheximide at 50 μg/ml was added to all plates to inhibit growth of fungi during incubation. Escherichia coli DH5α1 (Stratagene, La Jolla, Calif.) was used for recombinant DNA studies and plasmid propagation. These strains were grown on solid or liquid Luria-Bertani (LB) medium (34) supplemented with 50 μg of kanamycin/ml as indicated.
Macrophage cell lines and media.
The human monocyte-like cell line THP-1 (ATCC TIB-202) and the murine monocyte-like cell line J774A1 (ATCC TIB-67) were maintained in nitrate-free RPMI 1640 medium (GIBCO BRL, Grand Island, N.Y.) supplemented with 50 mg of HEPES/liter, 200 mM glutamine, 2.2 g of sodium bicarbonate/liter, 50 mg of l-arginine/liter, 100 U of penicillin/ml, 50 μg of gentamicin/ml, and either 10% heat-inactivated human AB serum (for THP-1 cells) or 10% heat-inactivated fetal bovine serum (FBS) (for J774A1 cells). Cells for use in M. tuberculosis cocultures were expanded in media without antibiotics and harvested during log phase.
Recombinant DNA techniques.
To isolate chromosomal DNA, M. tuberculosis cells were grown to confluence on Löwenstein-Jensen slants at 37°C under 9.5% CO2. Cells were scraped from the surface of the slant and resuspended in 500 μl of TE buffer (10 mM Tris–1 mM EDTA [pH 8]) in a 1.5-ml microcentrifuge tube. Two milligrams of achromopeptidase (Sigma) per milliliter was added, and the samples were incubated for 1 h at 37°C. At that time, 120 μg of proteinase K/ml and 1.5% SDS were added, and samples were incubated for 10 min at 65°C. N-cetyl-N,N,N-trimethyl ammonium bromide (1.3%, vol/vol) was added, and the samples were incubated for another 10 min at 65°C. These samples were then extracted twice with phenol-chloroform (1:1) and once with chloroform-isoamyl alcohol (24:1) and were ethanol precipitated. DNA was pelleted and resuspended in an appropriate volume of TE buffer. Molecular cloning and restriction endonuclease digestion were performed by standard techniques (34). Cloning vectors used were pBluescript KS(+) (Stratagene) and pNEB193 (New England Biolabs, Beverly, Mass.). Restriction endonucleases and other enzymes (New England Biolabs; Promega, Madison, Wis.) were used according to the manufacturers' instructions. The ΩKm cassette in plasmid pHP45ΩKm (14) was graciously provided by the laboratory of Malcolm Winkler, Department of Microbiology and Molecular Genetics, University of Texas—Houston Medical School.
Generation of transforming plasmids.
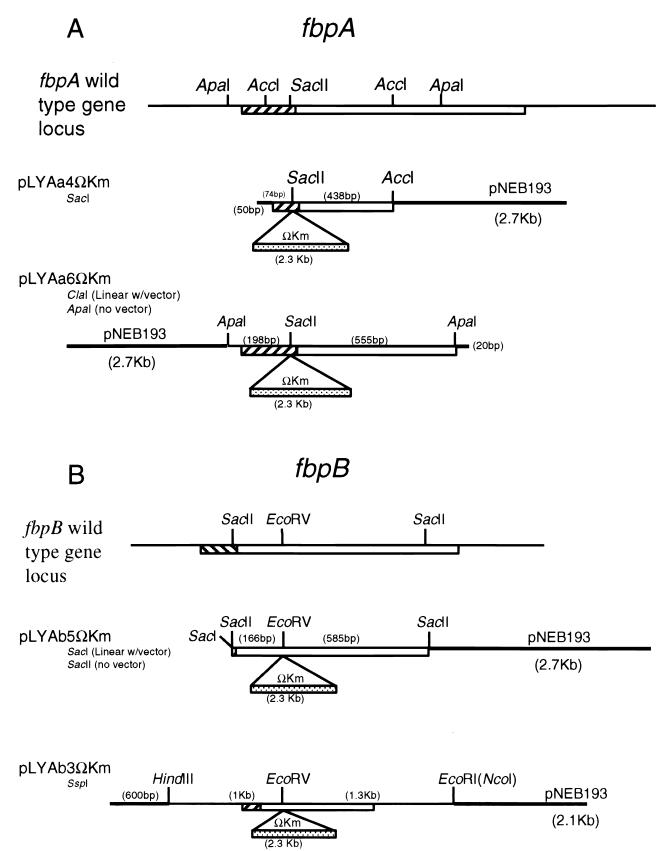
A 9.4-kb BamHI/ClaI fragment containing the H37Rv fbpA gene was cloned into pBluescript KS by standard methods (34) and identified by Southern blot hybridization using a PCR-generated fragment of fbpA as a probe. An internal 750-bp ApaI fragment was subcloned into pBluescript KS(+), excised by using the vector KpnI and EcoRI sites, and then ligated into pNEB193 linearized with the same two enzymes. The resultant plasmid, pLYAa6, was linearized at the unique SacII site within fbpA (see Fig. 1) and treated with the Klenow fragment of DNA polymerase I to provide blunt ends. A blunt-ended ΩKm cassette was prepared by liberating the ΩKm fragment from pHP45ΩKm with EcoRI and treating the resultant fragment with the Klenow fragment. The blunt-ended pLYAa6 and ΩKm DNA fragments were ligated to generate plasmid pLYAa6ΩKm, containing an fbpA::ΩKm gene disruption (see Fig. 1). An internal 500-bp AccI fragment was subcloned directly into the AccI site of pNEB193 and likewise interrupted with the ΩKm cassette at the SacII site to generate the transforming plasmid pLYAa4ΩKm (see Fig. 1).
FIG. 1.
Plasmids generated for transformation of M. tuberculosis H37Rv. Open boxes, fbpA and fbpB open reading frames; hatched boxes, signal sequences; heavy lines, vector sequences. Numbers in parentheses are distances between restriction sites. (A) fbpA::ΩKm constructs. A 500-bp AccI fragment and a 750-bp ApaI fragment were cloned from fbpA into pNEB193, a ColE1 plasmid, and mutated by the addition of an ΩKm cassette at the SacII site. Plasmid pLYAa4ΩKm was digested with SacI and introduced into H37Rv as a linear fragment, while pLYAa6ΩKm was electroporated into H37Rv associated with (by digestion with ClaI) and liberated from (by digestion with ApaI) the vector pNEB193. (B) fbpB::ΩKm constructs. A 750-bp ApaI fragment and a 2.3-kb HindIII/NcoI fragment were cloned from fbpB into pNEB193 and mutated by the addition of an ΩKm cassette at the EcoRV site. Plasmid pLYAb5ΩKm was introduced into H37Rv associated with (by SacI digestion) and liberated from (by SacII digestion) the vector pNEB193.
For fbpB::ΩKm gene disruptions, a 6.6-kb EcoRI/HindIII fragment containing the fbpB gene of H37Rv was cloned into the EcoRI and HindIII sites of pBluescript KS. An internal 750-bp SacII fragment was subcloned into pBluescript KS and then into pNEB193 by using the vector SacI and XbaI sites to generate pLYAb5. This plasmid was linearized with EcoRV and ligated with a blunt-ended ΩKm cassette, prepared as described above, to generate plasmid pLYAb5ΩKm (Fig. 1). A 2.3-kb HindIII/NcoI fragment containing fbpB was subcloned into pNEB193 to generate pLYAb3. This plasmid was mutated by addition of a blunt-ended ΩKm cassette at the unique EcoRV site within fbpB to generate pLYAb3ΩKm (Fig. 1).
To prepare the DNA for transformation of M. tuberculosis, plasmids pLYAa4ΩKm and pLYAb5ΩKm were linearized with SacI and pLYAa6ΩKm was linearized with ClaI (Fig. 1). In addition, pLYAa6ΩKm and pLYAb5ΩKm were treated with ApaI and SacII, respectively, to yield the mutated genes without vector sequences.
The shuttle plasmid pLYAspk was generated by cloning the 3-kb KpnI/EcoRV origin of replication fragment of the Mycobacterium fortuitum plasmid pAL5000 from the recombinant plasmid pYUB18 (20) into pBluescript KS and cloning the ΩKm marker into the unique BamHI site. This plasmid is able to replicate in both E. coli and M. tuberculosis and to confer kanamycin resistance.
Electroporation of M. tuberculosis H37Rv.
M. tuberculosis H37Rv was prepared for electroporation as previously described (20). Briefly, cells were grown in Middlebrook 7H9 medium–ADC–Tween 80 with gentle shaking to an optical density at 600 nm of 0.6 to 1.0, washed three times in 1/50 volume of cold 10% glycerol, resuspended in 10% glycerol at a concentration of ∼1011 cells/ml, and stored at −70°C until needed. The linearized plasmids (2 to 4 μg of DNA) diagrammed in Fig. 1 were electroporated at 0°C into 1010 electrocompetent H37Rv cells by using an Electroporator 2510 (Eppendorf North America, Madison, Wis.) at a setting of 1,250 V; under these conditions, the pulse time was 4 to 5 ms. One milliliter of 7H9–ADC broth without antibiotics was added immediately, and the bacteria were incubated at 37°C for 2.5 h with agitation. Transformants were then plated on 7H10–ADC–kanamycin plates and incubated at 37°C for 3 weeks under 9.3% CO2. Individual kanamycin-resistant colonies were subcultured onto fresh 7H10–ADC–kanamycin plates and grown an additional 2 to 3 weeks prior to further evaluation.
Hybridization analysis.
Fluorescein conjugation of the DNA fragments used as probes and chemiluminescent detection of hybridization were carried out according to the GeneImages procedure (Amersham Life Science Inc., Arlington Heights, Ill.). For initial screening of transformants, chromosomal DNA liberated from boiled M. tuberculosis colonies was immobilized onto a Hybond N+ nylon membrane (Amersham Life Science Inc.) by using a slot blot apparatus. Immobilized DNA was hybridized with a fluorescein-labeled 700-bp fragment from the ΩKm cassette obtained by digestion with PvuII, agarose gel separation, and purification using the PCR Cleanup kit (Promega). In addition, intact chromosomal DNA was digested with the enzymes indicated in Fig. 3, electrophoresed in 0.7% agarose, and transferred to a nylon membrane by using the alkaline transfer procedure (34). The membrane was then hybridized with a fluorescein-labeled probe (either the internal 750-bp ApaI fragment from fbpA or the internal 750-bp SacII fragment from fbpB).
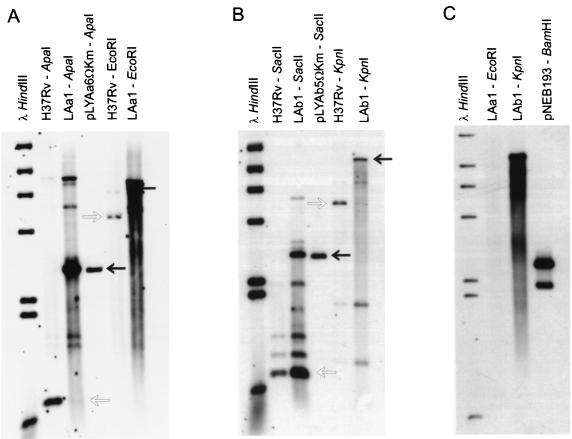
FIG. 3.
Southern blot analysis of H37Rv and fbpA::ΩKm and fbpB::ΩKm transformants. In each panel, open arrows indicate the locations of hybridizing bands corresponding to wild-type H37Rv sequences, whereas solid arrows indicate bands containing the ΩKm disruption. (A) Chromosomal digest of H37Rv and the fbpA::ΩKm transformant LAa1, which has a disruption of the 5′ region of fbpA. The probe is the 750-bp ApaI fragment of fbpA. (B) Chromosomal digest of H37Rv and the fbpB::ΩKm transformant LAb1 with a 5′-region disruption of fbpB by PCR analysis. The probe is the 750-bp SacII fragment of fbpB (Fig. 1). The transformant possesses both the wild-type SacII fragment and the fbpB::ΩKm fragment. (C) Chromosomal digest of fbpA::ΩKm and fbpB::ΩKm transformants hybridized with the vector pNEB193. LAa1 lacks the vector, while LAb1 retains the vector. Additional bands observed in some lanes are due to cross-hybridization of the fbpA or fbpB probe with other fbp genes.
PCR.
Primers used in this study are listed in Table 1. PCR was performed to screen for disruptions in fbpA (see Fig. 2) and fbpB (data not shown). Two primers were unique to the upstream regions of fbpA (5′A2) and fbpB (5′B2), and one primer was common to both fbpA and fbpB (3′AB). The primers 5′ΩKm and 3′ΩKm were used to screen for the presence of the ΩKm cassette. Cells (104 to 106 per reaction) from transformants were boiled in TE buffer for 10 min to liberate chromosomal DNA, which was used as a template. PCR conditions were 10 mM Tris (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 200 μM deoxynucleoside triphosphates, 0.5 μM each primer, and 2 U of Thermalase polymerase (Amresco, Solon, Ohio) per 100 μl of reaction mixture. Denaturation, annealing, and extension temperatures (and times) were as follows: 1 cycle of 96°C for 2 min; 5 cycles of 94°C for 40 s, 56°C for 40 s, and 72°C for 1.5 min; 30 cycles of 94°C for 40 s, 68°C for 40 s, and 72°C for 1.5 min; and a final extension of 72°C for 10 min.
TABLE 1.
PCR primers used for analysis of fbpA and fbpB mutants
| Primer | Sequence |
|---|---|
| 5′A2 | 5′-TTGATGCGGGTGGACGCC-3′ |
| 3′Aexp | 5′-GATCAAGCTTGTTCGGAGCTAGGCGCCC-3′ |
| 5′B2 | 5′-GCGGCTGTAGTCCTTCC-3′ |
| 3′Bexp | 5′-GATCAAGCTTTCAGCCGGCGCCTAACG-3′ |
| 3′AB | 5′-CGGCAGCTCGCTGGTCAGGA-3′ |
| 5′ΩKm | 5′-TCTCACCTTGCTCCTGCC-3′ |
| 3′ΩKm | 5′-GGTCCGCCACACCCAGCC-3′ |
| 5′ΩKm2 | 5′-GAAGTAATCGCAACATCCGC-3′ |
| 3′ΩKm2 | 5′-CAAGGGCTCCAAGGATCGGG-3′ |
| pNEB1 | 5′-GGTGTTGGCGGGTGTCGGGG-3′ |
FIG. 2.
PCR analysis of M. tuberculosis H37Rv transformed with constructs pLYAa4ΩKm treated with SacI (a4-SacI), pLYAa6ΩKm treated with ClaI (a6-ClaI), and pLYAa6ΩKm treated with ApaI (a6-ApaI). (A) Initial screening strategy. The locations of the primers used are indicated in the diagram. DNA from pLYAa4ΩKm- and pLYAa6ΩKm-transformed Kmr colonies was used as a template in a PCR using three primers: one unique to the upstream region of fbpA (5′A2), one unique to the upstream region of fbpB (5′B2), and one common to both fbpA and fbpB (3′AB). The PCR conditions used allowed complete amplification of ∼1 kb of DNA template (the product for fbpA is 600 bp and that for fbpB is 300 bp), but not of larger products with ΩKm inserts. Several transformants had a 300-bp PCR product representing fbpB but no 600-bp product representing intact fbpA. (B) PCR verification and characterization of fbpA::ΩKm mutants. The locations of the primers used are shown in the diagram. A single pLYAa6ΩKm transformant (starred; lane a6-ClaI) lacked a wild-type 5′ fbpA (5′A2→3′AB) (left panel), while its 3′ region was intact (middle panels). This clone was also found to possess the ΩKm cassette used for selection (right panel). The fbpB::ΩKm transformants were screened in an identical fashion, and a single fbpB::ΩKm transformant with a single-crossover insertion was identified (data not shown).
Sequence analysis.
The PCR primer pairs 5′A2 and 3′ΩKm2 and 5′ΩKm2 and 3′Aexp (see Fig. 2) were used to amplify the 5′ and 3′ regions, respectively, of the fbpA gene locus in LAa1. Primers 3′ΩKm2 and 5′ΩKm2 are specific for sequences within the ΩKm cassette. PCR primers 5′B2 and 3′ΩKm2 were used to amplify the 5′ region of the fbpB locus of LAb1. Southern blot analysis revealed that the transforming vector pNEB193 had integrated into the fbpB locus. Primer pNEB1, which is specific for pNEB193, was used with 3′Bexp to amplify the 3′ region of LAb1. PCR products were purified, desalted, and used directly as templates for sequencing. DNA sequencing was performed by using an ABI 377 automatic DNA sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.) at the DNA Core Laboratory, Department of Microbiology and Molecular Genetics, University of Texas–Houston Medical School. Sequences were analyzed with the GAP and BESTFIT programs (Genetics Computer Group, Madison, Wis.).
SDS-PAGE and Western blot analysis.
The parent strain, H37Rv, and the fbpA::ΩKm and fbpB::ΩKm mutants were grown in stationary cultures (without agitation) in Middlebrook 7H9 broth without additional supplements, except 20 μg of kanamycin/ml for selection of the two mutant strains. Five milliliters of 11-day cultures were filtered twice through a 22-μm-pore-size filter. Two milliliters of each filtrate was concentrated to ∼150 μl by using a Centricon-10 membrane apparatus (Amicon, Beverly, Mass.). The retentates were resuspended in an equal volume of solubilization buffer (2% SDS, 5% 2-mercaptoethanol, 10% glycerol). Proteins were electrophoresed in 8-to-20% polyacrylamide gradient gels and stained with Coomassie blue or transferred to polyvinyl difluoride (PVDF) membranes as previously described (27). Western blot analysis was carried out by incubating PVDF membranes with a 1:100 dilution of hybridoma culture supernatant containing the monoclonal antibody HYT27 (1), kindly provided by T. M. Shinnick, Centers for Disease Control and Prevention, Atlanta, Ga. This antibody has been shown to react with FbpA, FbpB, and FbpC. Second antibody incubation and detection were carried out according to the GeneImages procedure (Amersham).
In vitro growth studies.
The parent strain, H37Rv, and the fbpA and fbpB mutants were washed with saline and added at 104 CFU/ml to the enriched Middlebrook 7H9–ADC and synthetic Sauton media and incubated with gentle mixing at 37°C for 14 days. On days 3, 5, 7, 10, and 14, aliquots were removed from each broth culture and plated in 10-fold dilutions on 7H10–ADC plates with or without kanamycin for colony counts.
Infection of macrophages.
Macrophage infections were carried out as previously described (21, 22). Briefly, THP-1 cells were washed three times in the RPMI 1640 assay medium described above but with 2% AB serum, 1 μg of tetrahydrobiopterin/ml, and no antibiotics. Suspensions of M. tuberculosis H37Rv or the fbpA or fbpB mutant were dispersed by gentle sonication and mixed with 108 THP-1 cells at a concentration of 1010 CFU in 5 ml of assay medium. Phagocytosis was allowed to occur for 4 h with gentle mixing at 37°C. Cells were then washed by low-speed centrifugation and resuspension in assay medium six times, diluted to 106 cells/ml, and plated at 1 ml per well in 24-well culture plates. For infection of J774A1 cell cultures, adherent monolayers were established in 24-well plates by using RPMI 1640 assay medium with 10% FBS. Monolayers were infected at a CFU/macrophage ratio of 1:1 for 4 h with gentle mixing at 37°C. The monolayers were then washed extensively with warm assay medium. Twenty-four-well plates with infected THP-1 and J774A1 cells were incubated at 37°C under 5% CO2, and 1 ml of fresh assay medium was added to each well on day 3. Aliquots of THP-1 cells and supernatants were aspirated from wells, pelleted, and lysed with 0.05% SDS in saline, while J774A1 cells were lysed in situ on days 0 (baseline CFU), 3, 5, and 7 (three replicates per time point). SDS lysates were neutralized by the addition of sterile 15% bovine serum albumin in saline, and lysates were diluted in sterile saline. Serial 10-fold dilutions of the diluents were plated out on 7H11 agar for CFU counts. Controls for extracellular growth of M. tuberculosis were obtained by incubating 104 CFU of M. tuberculosis H37Rv/ml in 1 ml of assay medium containing a sonicated lysate of 106 THP-1 macrophages which had been passed through a 0.22-μm-pore-size filter to remove cell debris. Heat-inactivated human AB serum does not support M. tuberculosis growth, nor does M. tuberculosis grow in RPMI 1640 assay medium containing macrophage lysate and 5% heat-inactivated AB serum (data not shown).
Electron microscopic examination of M. tuberculosis-infected THP-1 cells.
On day 7 post-macrophage infection, aliquots of THP-1 cells infected with H37Rv or LAa1 were aspirated from wells, pelleted, and washed with phosphate-buffered saline three times. The cells were then prepared for electron microscopy as previously described (16).
RESULTS
Transformation of M. tuberculosis.
H37Rv was transformed by electroporation with 2 to 4 μg of several linearized constructs containing the fbpA and fbpB genes interrupted by the ΩKm cassette (Fig. 1). The transforming fragments contained between 74 and 585 bp of the M. tuberculosis sequences on either side of the ΩKm insert; in some cases the vector sequences were retained to protect the construct from potential exonuclease activity. These constructs lacked an origin of replication active in M. tuberculosis; therefore, kanamycin resistance could be conferred only if part or all of the transforming fragment was integrated into the M. tuberculosis chromosome. Plasmid pLYAspk, which contains the ΩKm cassette and is capable of replication in both M. tuberculosis and E. coli, was used as a positive control for transformation. Following electroporation, the transformation mixtures (originally containing ∼1010 M. tuberculosis cells) were plated on 7H10–ADC–kanamycin plates to select for kanamycin-resistant (Kmr) colonies. To assess the rate of Kmr due to spontaneous mutation versus integration or recombination events, transformants were subjected to slot blot analysis using a 700-bp fragment of ΩKm as the probe.
The results obtained are summarized in Table 2. The transformation efficiency, calculated by transformation with the autologously replicating plasmid, pLYAspk, was ∼105 transformants/μg of DNA. In this experiment, 57 to 389 Kmr colonies were obtained in the transformations utilizing fbpA::ΩKm and fbpB::ΩKm constructs. However, 17 Kmr colonies were identified in the negative control without transforming DNA, indicating the occurrence of spontaneous mutations leading to Kmr. Indeed, only a small proportion of the Kmr colonies representing organisms transformed with fbpA::ΩKm or fbpB::ΩKm constructs contained ΩKm, as determined by slot blot analysis (Table 2). Two constructs, pLYAa6ΩKm and pLYAb5ΩKm, integrated into the chromosome in 3 of 10 (30%) and 9 of 26 (34.6%) of the Kmr colonies screened (Table 2). Recombinants containing the ΩKm cassettes were not detected in the limited number of clones examined from the other transformation groups.
TABLE 2.
Results obtained from electroporation of M. tuberculosis H37Rv with different fbpA::Km and fbpB::Km constructs
| Transforming DNA (restriction enzyme used for linearization) | Gene construct (± vector sequences) | No. of Kmr colonies | No. of colonies with ΩKm/total no. screened | Disruption mutants identified |
|---|---|---|---|---|
| No DNA | 17 | Not done | ||
| pLYAa4ΩKm (SacI) | fbpA::ΩKm (+ vector) | 68 | 0/10 | |
| pLYAa6ΩKm (ClaI) | fbpA::ΩKm (+ vector) | 53 | 3/10 | 1 |
| pLYAa6ΩKm (ApaI) | fbpA::ΩKm (− vector) | 254 | 0/14 | |
| pLYAb5ΩKm (SacI) | fbpB::ΩKm (+ vector) | 57 | 9/26 | 1 |
| pLYAb5ΩKm (SacII) | fbpB::ΩKm (− vector) | 389 | 0/30 | |
| pLYAa4ΩKm (SspI) | fbpB::ΩKm (+ vector) | 366 | 0/28 | |
| pLYAspk | (Control plasmid) | 9.09 × 104 | 1/1 |
PCR screening.
To identify H37Rv clones with the desired fbpA::ΩKm and fbpB::ΩKm gene disruptions, a PCR strategy was devised to amplify the region where the targeted disruption was to occur. Each PCR mixture contained a primer unique to the upstream region of fbpA (5′A2), a primer unique to the upstream region of fbpB (5′B2), and a primer that was common to both (3′AB) (Fig. 2A). Under the PCR amplification conditions used, an integrative or recombinative event at one or the other gene locus would result in a corresponding loss of product. The amplification of both genes thus provided an internal PCR control.
Ninety-six Kmr colonies each from the fbpA::ΩKm and fbpB::ΩKm transformations were screened in this manner. The results of several of the fbpA::ΩKm transformants are shown in Fig. 2A. Most of the amplification reactions resulted in an intact fbpA product (600 bp) and an intact fbpB product (300 bp). Transformants that contained the fbpB PCR product but lacked the fbpA PCR product were chosen for further analysis. Additional primers specific for ΩKm and different regions of fbpA (Fig. 2B) were used to further characterize the mutants by PCR. One fbpA::ΩKm transformant was found to have a disruption in the 5′ region of the gene, while the 3′ region was intact. By using primers to ΩKm sequences, this transformant was found to contain the ΩKm cassette (Fig. 2B, rightmost panel). The ΩKm cassette was used as a positive control in this case. Four fbpB::ΩKm transformants were analyzed in a similar manner, and one was found by PCR to contain a 5′ disruption of fbpB, an intact 3′ region, and ΩKm sequences (data not shown). Screening was discontinued when single fbpA::ΩKm and fbpB::ΩKm mutants were obtained. These two disruption mutants were analyzed further.
Southern blot analysis of fbpA::ΩKm and fbpB::ΩKm mutants.
Digestion of wild-type H37Rv chromosomal DNA with the restriction endonuclease ApaI generates a 750-bp fbpA gene fragment (Fig. 1), while digestion with EcoRI generates a 5-kb fragment with the 1,059-bp fbpA gene located near the center (12). When chromosomal DNA from the fbpA::ΩKm mutant was digested with ApaI and probed with a PCR-generated fbpA fragment, it was found to lack the 750-bp native gene fragment. Instead, it now contained a 3-kb fragment identical in size to the transforming DNA fragment (Fig. 3A). Digestion of DNA from this transformant with EcoRI revealed that the locus had undergone an increase in size roughly equivalent to the size of the ΩKm cassette (∼2.3 kb). In addition, EcoRI-digested DNA from the transformant was probed with the labeled transforming vector, pNEB193, and vector sequences were found to be absent (Fig. 3C). These data are consistent with the occurrence of a double-crossover recombination event at the fbpA locus.
The fbpB gene locus in the fbpB::ΩKm mutant was evaluated in a similar manner. Digestion of H37Rv chromosomal DNA with the restriction endonuclease SacII generates an fbpB gene fragment of approximately 750 bp, and digestion with KpnI generates a ∼7-kb fragment containing fbpB. Analysis of the fbpB::ΩKm mutant by digestion with SacII and KpnI and Southern blot hybridization with the labeled 750-bp SacII fbpB gene fragment revealed the presence of the native gene fragment in addition to the transforming fragment (Fig. 3B). This result is consistent with an integrative event in which both fbpB::ΩKm and vector sequences were inserted at the native fbpB site. This interpretation was corroborated by the fact that the 7-kb KpnI fragment increased to ∼12 kb in the fbpB::ΩKm mutant. Thus, the pLYAb5ΩKm fragment was inserted at the native gene locus through a single-crossover event near the 5′ end, as supported by the presence of vector sequences at this site (Fig. 3C). The fbpA::ΩKm and fbpB::ΩKm mutants were designated LAa1 and LAb1, respectively.
Sequence analysis.
Sequence analysis was performed on LAa1 and LAb1 to characterize the recombination events that had occurred. Analysis of the 5′ and 3′ regions of the fbpA gene sequences flanking the ΩKm cassette in LAa1 revealed that the sequences were identical to those of the wild-type gene in these regions (data not shown). These results were consistent with a double-crossover, homologous-recombination event within the fbpA gene (Fig. 4A). Sequence analysis of the 5′ region of LAb1 also revealed sequence identity to wild-type fbpB in this region. The sequence downstream of the ΩKm cassette was identical to the end of the construct pLYAb5ΩKm (including the segment of pNEB193), indicating that this region was integrated in its entirety into the fbpB::ΩKm gene locus. Surprisingly, the 3′ end of the inserted sequence terminated at the SacI site used for linearization of the transforming fragment (Fig. 4C); therefore, no degradation of the end of the insert occurred before integration. Following the insert sequence, the wild-type fbpB gene resumed at bp 107 of the open reading frame, marking the exact point of insertion. A model of the integrative event occurring in LAb1 is shown in Fig. 4B.
FIG. 4.
Results of the analysis of the M. tuberculosis H37Rv transformants LAa1 and LAb1B. (A) Model of the fbpA mutant, LAa1; (B) model of the fbpB mutant, LAb1; (C) sequences from the 3′ “joint” region of the fbpB::ΩKm mutant, LAb1. Comparisons to the transforming vector sequence (pNEB193) and the wild-type fbpB sequence are shown. Only one difference of 1 bp (corresponding to bp 108 in the fbpB coding region) was observed in the joint region.
SDS-PAGE and Western blot analysis.
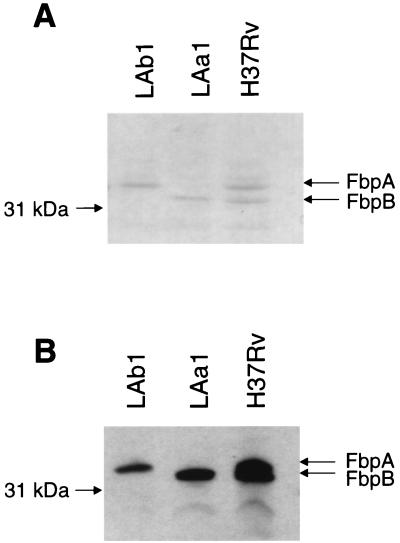
To verify that the chromosomal mutations in LAa1 and LAb1 disrupted synthesis of FbpA and FbpB, respectively, concentrated supernatant proteins and whole-cell lysates from wild-type H37Rv and the two mutants were analyzed by SDS-PAGE. Results for supernatant and lysate proteins were the same. As demonstrated in Fig. 5A, the parent, H37Rv, had two prominent protein bands at 32 kDa (representing FbpA) and 31 kDa (representing FbpB). The FbpA band was absent in LAa1, while the other visible protein bands were all intact. Likewise, the protein band representing FbpB was absent in LAb1. Western blot analysis with monoclonal antibody HYT27 confirmed these results (Fig. 5B). HYT27 is reactive with all members of the Ag85 complex and bound specifically to the FbpA and FbpB bands in H37Rv, the FbpB band only in LAa1, and the FbpA band in LAb1. The quantity of FbpC was apparently too low in these preparations to be detected by these techniques.
FIG. 5.
SDS-PAGE and Western blot analysis of supernatant proteins from H37Rv, LAa1, and LAb1. (A) Coomassie blue-stained SDS-PAGE gel of concentrated supernatant proteins from H37Rv, LAa1, and LAb1 revealed that LAa1 lacks the FbpA protein band and LAb1 lacks the FbpB protein band. Both protein bands were present in the parent, H37Rv. (B) Western blot analysis of supernatant proteins detected with monoclonal antibody HYT27.
In vitro growth studies.
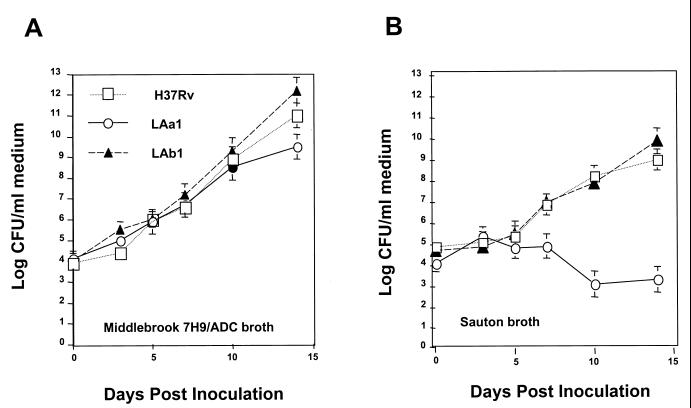
Since the Ag85 complex has been identified as having mycolyl transferase activity, the fbpA and fbpB mutants were anticipated to display differences during in vitro culture. In order to test this hypothesis, the parent and mutant strains were cultured in enriched Middlebrook 7H9–ADC broth or Sauton broth, a commonly used synthetic medium. While all three strains grew in the Middlebrook 7H9–ADC broth at very similar rates, the fbpA mutant, LAa1, was unable to grow in the synthetic Sauton medium (Fig. 6).
FIG. 6.
Effect of medium composition on the growth of fbpA- and fbpB-deficient M. tuberculosis mutants. Saline-washed M. tuberculosis H37Rv or an fbpA or fbpB mutant was inoculated into enriched liquid Middlebrook 7H9 medium or wholly synthetic Sauton medium. All strains grew similarly in the enriched Middlebrook 7H9–ADC medium. The fbpA mutant, LAa1, was unable to grow in the Sauton medium, which lacks the ADC supplement.
Macrophage infection studies.
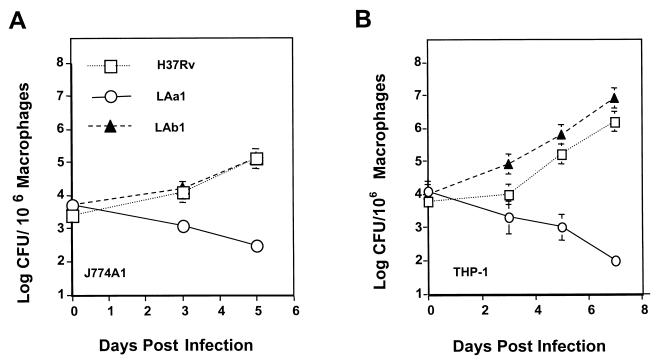
The macrophage cell line culture method can be used to measure differences in capacity for intracellular growth among strains of mycobacteria (21, 22). Human THP-1 macrophage cultures were infected with H37Rv and with the fbpA and fbpB mutants LAa1 and LAb1 to determine intracellular growth. In this system, the parent strain, H37Rv, increased in number from 104 to ≥106 mycobacteria per culture over a 7-day incubation period (Fig. 7). These results are similar to those obtained with M. tuberculosis Erdman in the same system (22). The fbpB mutant, LAb1, exhibited a similar growth curve. However, the fbpA mutant, LAa1, actually decreased in number under the same incubation conditions, indicating an inability to survive and replicate in macrophages (Fig. 7B). Monolayers of the J774A1 murine macrophage cell line also supported the growth of the parent strain, H37Rv, and the fbpB mutant, whereas the fbpA mutant did not replicate (Fig. 7A). Examination by electron microscopy (Fig. 8A) revealed that infection of THP-1 cells by H37Rv resulted in high numbers of intact, intracellular mycobacteria. In contrast, LAa1 infection resulted in fewer mycobacteria that were rarely intact and exhibited varying degrees of degradation. LAa1-infected cells also had many extensions indicative of membrane ruffling and cell activation (Fig. 8B), whereas H37Rv-infected cells appeared relatively quiescent.
FIG. 7.
Growth of H37Rv and the fbpA and fbpB mutants in monocyte-like human THP-1 and murine J774A1 cell cultures. J774A1 (A) or THP-1 (B) cells were infected with the parental H37Rv strain, the fbpA mutant LAa1, or the fbpB mutant LAb1 for 4 h and plated at 106 cells/culture, as described in Materials and Methods. On days 0, 3, 5, and 7, the cell cultures were harvested, lysed, and plated for mycobacterial CFU counts on 7H11 agar with appropriate antibiotics. Mean CFU ± standard deviations for representative experiments are shown.
FIG. 8.
Poor survival and growth of the fbpA disruption mutant LAa1 in THP-1 cells, as revealed by electron microscopy. THP-1 cells were inoculated with either the wild-type progenitor, H37Rv (A), or LAa1 (B) and were processed for electron microscopy 7 days postinfection. The H37Rv-infected cells contained many intact, electron-dense mycobacteria (solid arrows), whereas LAa1-infected cells contained few intact mycobacteria and many vacuoles containing apparent mycobacterial cell debris (open arrows).
DISCUSSION
While homologous recombination with gene replacement has been demonstrated readily in fast-growing, nonpathogenic mycobacteria, such as Mycobacterium smegmatis (17), generation of defined mutations in the genomes of slow-growing mycobacteria initially proved difficult. Attempts to achieve homologous recombination in M. tuberculosis and Mycobacterium bovis BCG revealed a high rate of illegitimate recombination, with random insertion of linear DNA fragments into the chromosome (23, 35). Despite this finding, intraplasmic (8, 26) and interplasmic (8) recombination experiments have demonstrated that homologous recombination does occur in slow-growing mycobacteria, although at a significantly lower rate than illegitimate recombination. Aldovini et al. (2) obtained recombination without gene replacement at the chromosomal uraA site of M. bovis BCG, producing transformants that had undergone single-crossover homologous recombination at only one end of the transforming fragment. These results indicated that disruption could be achieved by using fragments internal to the coding region of a gene. Other studies involving the use of linear DNA fragments of single-gene length (4, 33), linear DNA fragments of 40 to 50 kb (5), transposon delivery systems (7, 29, 30), and SacB counterselection plasmids (3, 30) have proven successful with increasing frequency. Despite the increased proficiency at mutagenesis of the M. tuberculosis chromosome, the list of identified potential virulence factors remains brief.
In this study, several constructs were utilized in our attempts to disrupt the fbpA and fbpB chromosomal loci in M. tuberculosis (Fig. 1). The size of the M. tuberculosis DNA insert ranged from 0.5 to 2.3 kb, and the amount of M. tuberculosis DNA flanking the ΩKm cassette ranged from 74 bp to 1.3 kb. Linear constructs with and without flanking vector sequences were used. Each of the six constructs tested was modeled after prior attempts to disrupt genes in M. tuberculosis (2, 23, 26). We reasoned that using several constructs would increase the likelihood of gene disruption.
Insertional mutants were obtained for both fbpA and fbpB. Of the six constructs utilized for transformation in this study, only two yielded H37Rv clones containing ΩKm cassettes (Table 2). Both of these (pLYAa6ΩKm, treated with ClaI, and pLYAb5ΩKm, treated with SacI) had relatively small M. tuberculosis DNA inserts (∼750 bp) and contained flanking vector DNA. Although the number of clones examined was too small for statistical analysis, we can conclude that insertional mutagenesis of M. tuberculosis genes can be achieved with small inserts and may be aided by the presence of flanking vector sequences. Our efforts were focused on the single fbpA::ΩKm and fbpB::ΩKm mutants described here, but it is likely that additional site-specific mutations could be identified with additional screening.
In the case of fbpA, mutagenesis occurred by double-crossover homologous recombination, resulting in an insertion of ΩKm into the chromosomal fbpA site. This result was confirmed by PCR, Southern blot, and sequence analyses. PCR results using primers within fbpA and ΩKm and primers outside the region used for transformation were consistent with a double-crossover event without addition of extraneous DNA. By Southern blot analysis, there is an increase in size at the native fbpA gene locus equal to the size of the ΩKm cassette. Sequence data confirmed addition of the ΩKm cassette at the SacII site of fbpA without addition or deletion of base pairs. With regard to fbpB, Southern blot analysis revealed that the entire transforming fragment, including the vector pNEB193, had integrated into the chromosome. Sequencing of this region demonstrated the occurrence of homologous recombination at the 5′ end of the gene with a double-stranded break and insertion of the nonhomologous vector sequence adjacent to the fbpB chromosomal sequences at the 3′ end.
Results of previous attempts to mutate individual genes in slow-growing mycobacteria using chromosomal fragments mutated with a kanamycin resistance marker have suggested a predominance of illegitimate over legitimate recombination. This was based on the presence of a high number of antibiotic-resistant colonies in the absence of the desired gene disruption. Our studies suggest that there is a high rate of spontaneous kanamycin resistance in these organisms and that a large number of the Kmr colonies reported previously were due to spontaneous mutation rather than illegitimate recombination. Of the 118 Kmr colonies that we screened (Table 2), 106 (90%) were spontaneous kanamycin mutants, in that slot blot analysis revealed the absence of the kanamycin resistance marker. Of the remaining 12 transformants that were found to have integrated the ΩKm cassette into their chromosomes, only 2 (17%) had undergone integration at the homologous gene loci. Previous studies have involved screening of transformants for the desired mutation by looking for a phenotypic change (i.e., auxotrophy or urease activity) and have not addressed the rate of spontaneous mutation. We have found that the rate of spontaneous mutation is somehow increased by the electroporation of DNA but not by the act of electroporation itself (unpublished data). As an example, the rate of spontaneous mutation was lower under control conditions where no DNA was used during electroporation (Table 2).
Members of the Ag85 complex have been found to possess mycolyl transferase activity. Since disruption of members of the complex could potentially affect cell wall synthesis, we examined the abilities of the two mutants, LAa1 and LAb1, to grow in routine laboratory media, one ADC-containing medium and one minimal medium composed of basic salts with no supplements. There was no significant difference in the growth rates of the mutant strains compared to that of the parent strain in media containing ADC (Fig. 6A). In contrast, the fbpA mutant, LAa1, exhibited little growth in a minimal medium that lacked ADC (Fig. 6B). Albumin-containing enrichments are added to the growth media of mycobacteria primarily to bind toxic lipid byproducts produced during routine growth in the presence of lipids such as Tween 80. The albumin-oleate complex also acts as an additional nutrient source for the organisms. The lack of growth of the fbpA mutant in minimal medium may indicate an increased dependence on lipids or other compounds associated with albumin in the enriched medium. In macrophage-like cell line infection models with both human and mouse cell lines, disruption of Ag85B again had no obvious effect on growth compared to that of the parent strain while the Ag85A mutant, LAa1, was severely hampered in its growth (Fig. 7). The number of CFU of LAa1 actually decreased during macrophage cell line infection, and killing of the mycobacteria by macrophages was verified by electron microscopy (Fig. 8A). Poor survival of LAa1 in macrophages may reflect an alteration in phagosome processing such that the mycobacteria are exposed to lysosomal contents or phagosome acidification (13). Alternatively, the increased dependence on nutritional compounds observed in broth cultures could result in decreased survival and growth in the intracellular compartment. These possibilities will be addressed in subsequent studies.
It has been postulated that the members of the Ag85 complex, though closely related, are not coordinately regulated (15). In keeping with this hypothesis, we found no evidence in our mutants that expression of the other genes of the complex increased or decreased quantitatively in response to the loss of one of the members (Fig. 5). The same study also demonstrated that the genes are transcribed as monocistronic messages, decreasing the likelihood that the effects seen in this study are due to a polar effect on genes downstream of the fbpA gene locus.
Much speculation has been made about the roles of the individual members of the Ag85 complex. Although they all have mycolyl transferase activity, there is clear evidence that the efficiency at which this reaction is carried out differs among the three proteins (9). Studies have shown that naked DNA vectors containing the Ag85A gene injected into mice afforded a promising degree of protection as a vaccine against M. tuberculosis infection (18), which would seem to indicate a role for the Ag85 complex in the immunology of this process. Mutation of individual members of this complex will aid in determining the individual roles of these proteins in the pathogenesis and immunology of mycobacterial infection as well as in the synthesis of the mycobacterial cell wall.
ACKNOWLEDGMENTS
We thank T. M. Shinnick, Centers for Disease Control and Prevention, for providing monoclonal antibodies, M. E. Winkler, Department of Microbiology and Molecular Genetics, University of Texas—Houston Medical School, for supplying plasmid pHP45ΩKm, and W. R. Jacobs, Department of Microbiology and Immunology, Howard Hughes Medical Institute, Albert Einstein College of Medicine, Bronx, N.Y., for providing plasmid pYUB18. We also thank G. M. Weinstock, H. B. Kaplan, E. M. Walker, and S. Mueller for invaluable advice and discussions, Betty Boulet and Emem Akpaffiong for technical assistance, and Patricia Navarro for electron microscopy.
REFERENCES
- 1.Abou-Zeid C, Ratliff T L, Wiker H G, Harboe M, Bennedsen J, Rook G A. Characterization of fibronectin-binding antigens released by Mycobacterium tuberculosis and Mycobacterium bovis BCG. Infect Immun. 1988;56:3046–3051. doi: 10.1128/iai.56.12.3046-3051.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Husson R N, Young R A. The uraA locus and homologous recombination in Mycobacterium bovis BCG. J Bacteriol. 1993;175:7282–7289. doi: 10.1128/jb.175.22.7282-7289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad A K, Sirakova T D, Fernandes N D, Kolattukudy P E. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 4.Azad A K, Sirakova T D, Rogers L M, Kolattukudy P E. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc Natl Acad Sci USA. 1996;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 7.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam R A, Bloom B R, Hatfull G F, Jacobs W R., Jr Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baulard A, Kremer L, Locht C. Efficient homologous recombination in fast-growing and slow-growing mycobacteria. J Bacteriol. 1996;178:3091–3098. doi: 10.1128/jb.178.11.3091-3098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard J S. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem. 1996;65:215–239. doi: 10.1146/annurev.bi.65.070196.001243. [DOI] [PubMed] [Google Scholar]
- 11.Bloom B R, Murray C J L. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 12.Borremans M, de Wit L, Volckaert G, Ooms J, de Bruyn J, Huygen K, van Vooren J P, Stelandre M, Verhofstadt R, Content J. Cloning, sequence determination, and expression of a 32-kilodalton-protein gene of Mycobacterium tuberculosis. Infect Immun. 1989;57:3123–3130. doi: 10.1128/iai.57.10.3123-3130.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens D L. Characterization of the Mycobacterium tuberculosis phagosome. Trends Microbiol. 1996;4:113–118. doi: 10.1016/0966-842X(96)81528-9. [DOI] [PubMed] [Google Scholar]
- 14.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 15.Harth G, Lee B, Wang J, Clemens D L, Horwitz M A. Novel insights into the genetics, biochemistry, and immunocytochemistry of the 30-kilodalton major extracellular protein of Mycobacterium tuberculosis. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayat M. Principles and techniques of electron microscopy: biological applications. Vol. 1. 1986. Litton Educational Publishing, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 17.Husson R N, James B E, Young R A. Gene replacement and expression of foreign DNA in mycobacteria. J Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huygen K, Content J, Denis O, Montgomery D L, Yawman A M, Deck R R, DeWitt C M, Orme I M, Baldwin S, Drowart D S C A, Lozes E, Vandenbussche P, Van Vooren J P, Liu M A, Ulmer J B. Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat Med. 1996;2:893–898. doi: 10.1038/nm0896-893. [DOI] [PubMed] [Google Scholar]
- 19.Jackson M, Raynaud C, Laneelle M A, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffe M. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999;31:1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 21.Jagannath, C., J. A. Actor, and R. L. Hunter. 1998. Induction of nitric oxide in human monocytes and monocyte cell lines by Mycobacterium tuberculosis. Nitric oxide 2:174–186. [DOI] [PubMed]
- 22.Jagannath C, Allaudeen H S, Hunter R L. Activities of poloxamer CRL8131 against Mycobacterium tuberculosis in vitro and in vivo. Antimicrob Agents Chemother. 1995;39:1349–1354. doi: 10.1128/aac.39.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalpana G V, Bloom B R, Jacobs W R., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci USA. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kochi A. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle. 1991;72:1–12. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 25.Kubica G, Wayne L, editors. The Mycobacteria: a sourcebook. Vol. 15. New York, N.Y: Marcel Dekker, Inc.; 1984. [Google Scholar]
- 26.Norman E, Dellagostin O A, McFadden J, Dale J W. Gene replacement by homologous recombination in Mycobacterium bovis BCG. Mol Microbiol. 1995;16:755–760. doi: 10.1111/j.1365-2958.1995.tb02436.x. [DOI] [PubMed] [Google Scholar]
- 27.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 29.Pavelka M S, Jr, Jacobs W R., Jr Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guérin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J Bacteriol. 1999;181:4780–4789. doi: 10.1128/jb.181.16.4780-4789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratliff T L, McCarthy R, Telle W B, Brown E J. Purification of a mycobacterial adhesin for fibronectin. Infect Immun. 1993;61:1889–1894. doi: 10.1128/iai.61.5.1889-1894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratliff T L, McGarr J A, Abou-Zeid C, Rook G A, Stanford J L, Aslanzadeh J, Brown E J. Attachment of mycobacteria to fibronectin-coated surfaces. J Gen Microbiol. 1988;134:1307–1313. doi: 10.1099/00221287-134-5-1307. [DOI] [PubMed] [Google Scholar]
- 33.Reyrat J M, Berthet F X, Gicquel B. The urease locus of Mycobacterium tuberculosis and its utilization for the demonstration of allelic exchange in Mycobacterium bovis bacillus Calmette-Guerin. Proc Natl Acad Sci USA. 1995;92:8768–8772. doi: 10.1073/pnas.92.19.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Wilson T, Wards B J, White S J, Skou B, de Lisle G W, Collins D M. Production of avirulent Mycobacterium bovis strains by illegitimate recombination with deoxyribonucleic acid fragments containing an interrupted ahpC gene. Tuber Lung Dis. 1997;78:229–235. doi: 10.1016/s0962-8479(97)90003-4. [DOI] [PubMed] [Google Scholar]
- 36.Young D B, Kaufmann S H E, Hermans P W M, Thole J E R. Mycobacterial protein antigens: a compilation. Mol Microbiol. 1992;6:133–145. doi: 10.1111/j.1365-2958.1992.tb01994.x. [DOI] [PubMed] [Google Scholar]