Abstract
Background
To explore the feasibility of diffusion-weighted imaging (DWI) metrics to predict the histologic subtypes and genetic status of gliomas (e.g., IDH, MGMT, and TERT) noninvasively.
Methods
One hundred and eleven patients with pathologically confirmed WHO grade II-IV gliomas were recruited retrospectively. Apparent diffusion coefficient (ADC) values were measured in solid parts of gliomas on co-registered T2-weighted images and were compared with each other in terms of WHO grading and genotypes using t-tests. Receiver operating characteristic analysis was performed to assess the diagnostic performances of ADC. Subsequently, multiple linear regression was used to find independent variables, which can directly affect ADC values.
Results
The values of overall mean ADC (omADC) and normalized ADC (nADC) of high grade gliomas and IDH wildtype gliomas were lower than low grade gliomas and IDH mutated gliomas (P < 0.05). nADC values showed better diagnostic performance than omADC in identifying tumor grade (AUC: 0.787 vs. 0.750) and IDH status (AUC: 0.836 vs. 0.777). ADC values had limited abilities in distinguishing TERT status (AUC = 0.607 for nADC and 0.617 for omADC) and MGMT status (AUC = 0.651 for nADC). Only tumor grade and IDH status were tightly associated with ADC values.
Conclusion
DWI metrics can predict glioma grading and IDH mutation noninvasively, but have limited use in detecting TERT mutation and MGMT methylation.
Keywords: Diffusion-weighted imaging, Apparent diffusion coefficient, Glioma differentiation, Genetic status
Background
The 2016 World Health Organization (WHO) Classification of brain tumors integrated molecular parameters into histopathologic classification and tumor grading. Extensive analyses have been performed to study the influence of various genetic markers and glioma grading on patients’ survival and treatment with glioma. Among these genetic markers, three are noteworthy because they are common in gliomas and have great values in routine clinical treatment and prognostic prediction. The first one needs to be noted is isocitrate dehydrogenase mutation (IDH-mut), which can define glioma subtype and indicate good prognosis [1]. The second is O6-methylguanine-DNA methyltransferase promoter methylation (MGMT-m), a favorable independent prognostic biomarker, which can predict glioma patients’ response to temozolomide [1]. The third is telomerase reverse transcriptase promoter mutation (TERT-mut), which associates with a worse prognosis [2] and radiotherapy resistance [3].
Since genetic alterations and WHO grading are related to patient management and outcome, it is essential to figure out a useful method, enabling efficient and secure detection of those prognostic factors. Although the histopathologic examination is the gold standard to test genetic markers in glioma, brain surgery and autopsy are risky. Moreover, it is unable to obtain tumor samples from patients without surgical indications. Molecular detection using tumor tissue is too time-consuming to guide treatment before, during, and after operations timely.
Diffusion-weighted imaging (DWI) is a non-invasive method, which has been widely used in the diagnosis of brain tumors. The apparent diffusion coefficient (ADC) values generated from DWI can quantitatively evaluate the cellularity of tissue and movement of water molecules in vivo [4]. Conventional structural magnetic resonance imaging (MRI) features, including glioma location, glioma volume, necrosis, invasiveness, enhancement pattern, and peritumoral edema, have been used to predict the IDH, MGMT, and TERT status [5–8], however with controversies. Recently, MRI-based radiomic signatures have shown the possibility of predicting genotypes of gliomas [9, 10], while the time-consuming methodology has limited its use in routine clinical work. Advanced MRI, including arterial spin labeling imaging (ASL) [11], DWI [12–15], and dynamic susceptibility contrast perfusion imaging (DSC) [2], is used to assess MGMT and TERT status in patients with glioblastomas. However, few studies evaluate the feasibility of ADC in predicting TERT and MGMT status in WHO grade II-IV gliomas. Simultaneously, the result of several studies that correlated ADC values with WHO grading is still controversial [16–18]. Besides, how the glioma grading and genotypes impact the ADC values of gliomas remains unknown.
Therefore, this study aimed to firstly investigate the association between WHO grade and the ADC values of gliomas, secondly evaluate the predictive capability of ADC in genetic markers (e.g., IDH, MGMT, and TERT) in gliomas, thirdly confirm the parameters that affect the ADC values.
Methods
Clinical data and groupings
This retrospective study was approved by the Institutional Review Board of Peking Union Medical College Hospital. The requirement for informed consent from patients was waived. A total of 111 adult patients (mean age: 44.3 ± 12.1 years old) were enrolled in this study. They were pathologically diagnosed with primary WHO grade II-IV gliomas between August 2010 and March 2018 at Peking Union Medical College Hospital. Patients who underwent radiotherapy, chemotherapy, or invasive procedures before magnetic resonance imaging (MRI) acquisitions were excluded from this study. The details about the main clinical features, pathological diagnosis, and genetic status of the enrolled patients are listed in Table 1.
Table 1.
Patient characteristics and genetic types of WHO II-IV gliomas
| Characteristics | LGGs | HGGs | Total | P value | |
|---|---|---|---|---|---|
| WHO II n = 36 (32.43%) |
WHO III n = 32 (28.83%) |
WHO IV n = 43 (38.74%) |
WHO II-IV n = 111 (100.00%) |
LGGs vs HGGs |
|
| Age | 42.4 | 46.5 | 57.8 | 44.3 | P < 0.0001 |
| Sex | P = 0.035 | ||||
| Male | 24 (66.67%) | 19 (59.38%) | 15 (34.88%) | 58 (52.25%) | |
| Female | 12 (33.33%) | 13 (40.62%) | 28 (65.12%) | 53 (47.75%) | |
| Genetic type | |||||
| IDH | P < 0.0001 | ||||
| Mutation | 27 (75.00%) | 15 (46.88%) | 3 (6.98%) | 45 (40.54%) | |
| Wildtype | 9 (25.00%) | 17 (53.12%) | 39 (90.70%) | 65 (58.56%) | |
| NA | 0 (0.00%) | 0 (0.00%) | 1 (2.32%) | 1 (0.90%) | |
| MGMT | P = 0.002 | ||||
| Methylated | 28 (77.78%) | 21 (65.63%) | 17 (39.53%) | 66 (59.46%) | |
| Unmethylated | 6 (16.67%) | 10 (31.25%) | 26 (60.47%) | 42 (37.84%) | |
| NA | 2 (5.55%) | 1 (3.12%) | 0 (0.00%) | 3 (2.70%) | |
| TERT | P = 0.471 | ||||
| Mutation | 17 (47.22%) | 13 (40.62%) | 25 (58.14%) | 55 (49.55%) | |
| Wildtype | 17 (47.22%) | 15 (46.88%) | 13 (30.23%) | 45 (40.54%) | |
| NA | 2 (5.56%) | 4 (12.50%) | 5 (11.63%) | 11 (9.91%) | |
Unless otherwise noted, data in the table are presented as n (%) or mean standard deviation; NA: not available
LGGs low grade gliomas; HGGs high grade gliomas
MRI data acquisition and imaging processing
MRI studies were performed preoperatively on a 3.0-T MRI scanner (Discovery MR750, GE, US). The MRI protocols included an axial T2-periodically rotated overlapping parallel lines with enhanced reconstruction (T2-PROPELLER) sequence (TR, 12507 ms; TE, 91 ms; TA, 97 s; slice thickness, 6 mm and FOV, 240 × 240 mm2) and an axial DWI sequence (TR, 3000 ms; TE, 91 ms; TA, 27 s; slice thickness, 6 mm and b value, 0 and 1000 s/mm2).
The DWI images were manually transferred to an offline workstation (Advantage Workstation, AW4.5; GE Medical Systems) supplied by the vendor. GE Functool software was further used to generate ADC maps and automatically calculated the mean ADC value for each region of interest (ROI). The solid parts of all the gliomas were confirmed by a consensus of two radiologists blinded to genetic and pathologic information. For each tumor, four ROIs were manually placed within the solid components on co-registered T2-weighted images. Necrotic, cystic, calcified, and hemorrhage areas of gliomas were avoided. Two other ROIs of each patient were selected on the contralateral normal white matter (CNWM) (Fig. 1). The area of each ROI was between 29 to 31 mm2.
Fig. 1.
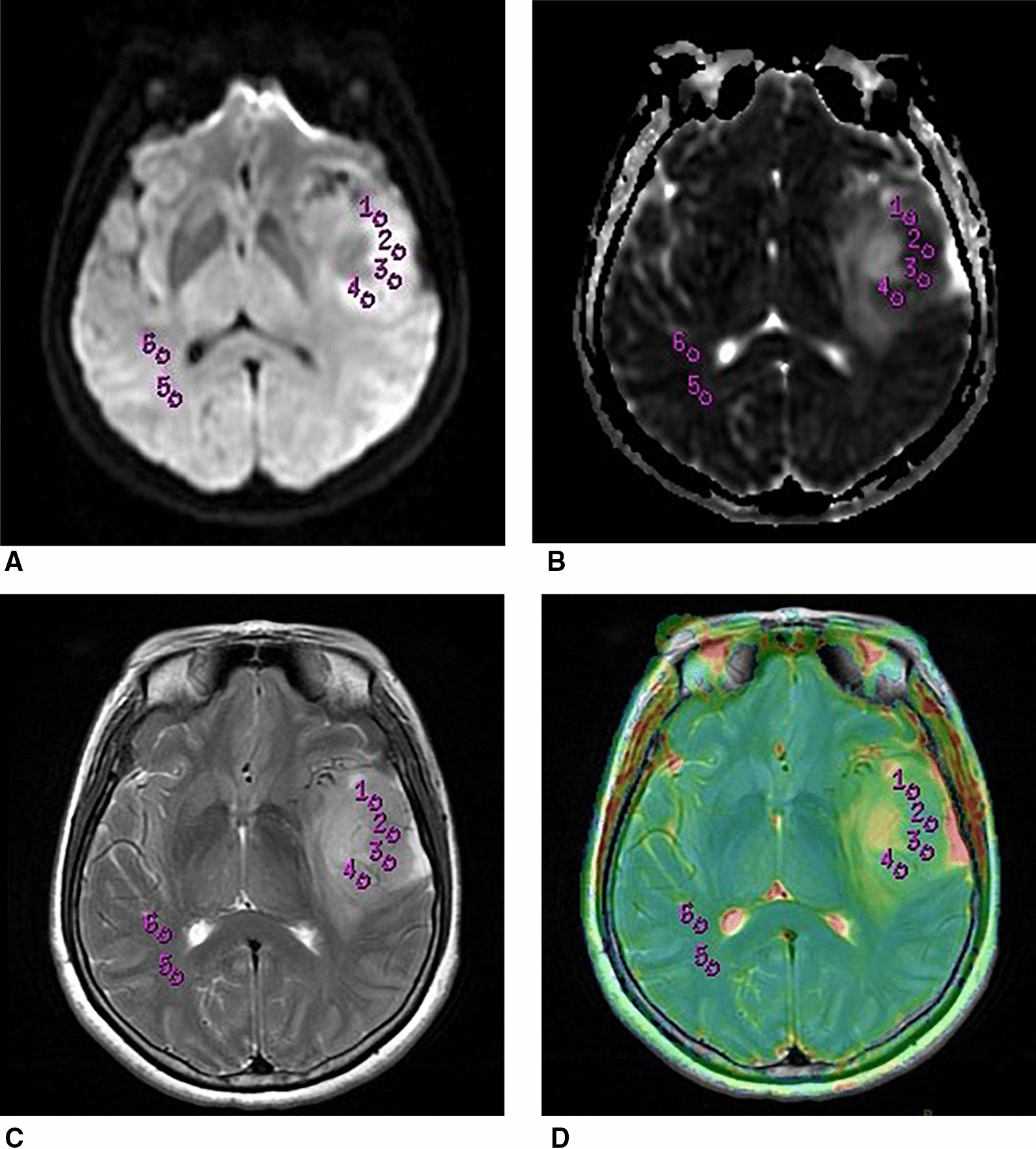
Images showed an example of the placement of ROIs. Axial DWI (A), ADC (B), T2WI (C), and co-registered T2WI (D) from a patient with low grade glioma. Four non-overlapping round ROIs were placed within the solid part of the glioma on co-registered T2WI (ROI 1–4), while two same-sized ROIs (ROI 5–6) were placed within the contralateral white matter to calculate the value of the mean ADC in each ROI. DWI: diffusion-weighted imaging; ADC apparent diffusion coefficient; ROI region of interest
The formula of the normalized ADC (nADC) value is listed as follows: , where mean value of the four mean ADC values within tumor and is the mean value of the two mean ADC values within CNWM.
Histopathology
IDH mutational and TERT promoter mutational analysis was performed using direct sequencing described by Horbinski et al. [19] and Chan et al. [20], respectively. MGMT-m was detected by pyrosequencing reported by Reifenberger et al.[21]. DNA extracted from formalin-fixed, paraffin-embedded tumor tissue was used to detect IDH1/2-mut, TERT-mut, and MGMT-m.
Statistics
The statistical analyses of data were performed using SPSS, version 20. The Kolmogorov–Smirnov test was used to analyze whether age and ADC data were normally distributed. Chi-square tests were performed to test distribution differences of age, sex, and genetic types between low grade gliomas (LGGs, which refer to WHO grade II gliomas) and HGGs, which refer to WHO grade III-IV gliomas. T-tests were used to compare continuous variables. Statistical significance was set at P < 0.05. Parameters with significant differences were further analyzed by receiver-operating characteristic (ROC) curve to seek the threshold nADC and omADC values to predict genetic status and assess the differentiate performances of nADC and omADC. Multiple linear regression analysis was further performed to test the association of each variable with omADC and nADC.
Results
Patient characteristics and genetic type
The detailed baseline characteristics of the 111 patients are shown in Table 1. 36 (32.43%) WHO grade II gliomas, 32 (28.83%) WHO grade III gliomas, and 43 (38.74%) WHO grade IV gliomas were enrolled in this study. IDH and TERT genotype were mutant in 45 (40.54%) and 55 (49.55%) of the 111 gliomas. Gliomas with and without MGMT promoter methylation accounted for 59.46% (66) and 37.84% (42) of all the gliomas, respectively.
Significant differences existed in age (P < 0.0001) and sex (P = 0.035) between LGGs and HGGs (Table 1). Patients in the HGGs group were significantly older than those in the LGGs group. IDH (P < 0.0001) and MGMT status (P = 0.002) were also statistically significant, with more patients in the HGGs group falling into the IDH-wildtype (IDH-wt) category and MGMT promoter unmethylation (MGMT-um) category versus the LGGs group. No significant difference in TERT status was observed.
Correlation of the ADC values with the WHO grade
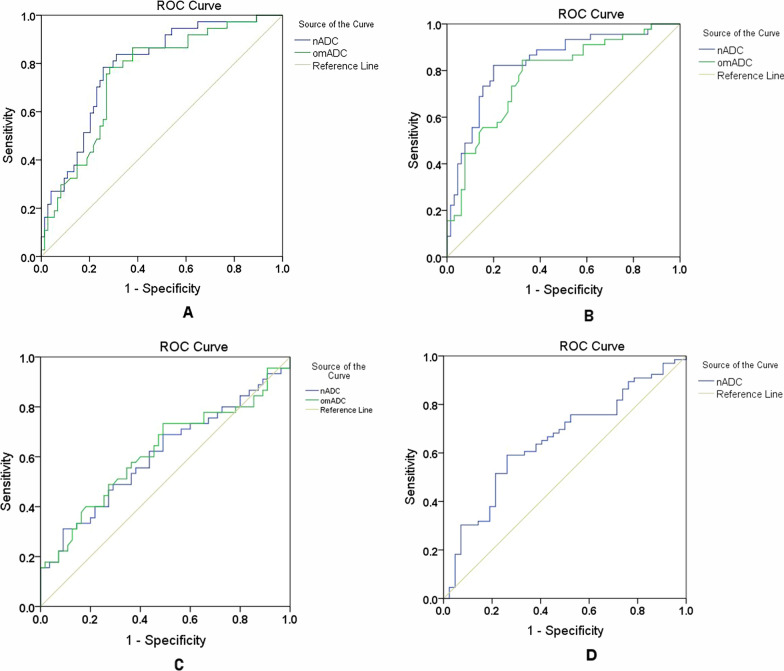
The values of nADC and omADC were significantly different between LGGs and HGGs according to WHO classification of 2007 (both P < 0.0001). ADC values in LGGs group were higher than those in HGGs group (1.83 ± 0.35 vs. 1.47 ± 0.31 for nADC, and 0.0014 ± 0.0003 vs. 0.0011 ± 0.0003 mm2/s for omADC) (Table 2). In ROC analysis, the best cutoff values for nADC and omADC to differentiate LGGs from HGGs were 1.56 (AUC: 0.787, sensitivity and specificity: 83.8% and 68.9%) and 0.0012 mm2/s (AUC: 0.750, sensitivity and specificity: 78.4% and 71.6%), respectively (Table 3 and Fig. 2A).
Table 2.
Summary of discriminant analyses
| nADC (median ± SD) | P value | omADC (median ± SD) | P value | ||
|---|---|---|---|---|---|
| Grade | Low | 1.70 ± 0.36 | < 0.0001a | 0.0014 ± 0.0003 | < 0.0001a |
| High | 1.42 ± 0.30 | 0.0011 ± 0.0003 | |||
| IDH | Mutation | 1.83 ± 0.34 | < 0.0001a | 0.0014 ± 0.0003 | < 0.0001a |
| Wildtype | 1.42 ± 0.28 | 0.0011 ± 0.0002 | |||
| MGMT | Methylation | 1.65 ± 0.37 | 0.021a | 0.0013 ± 0.0003 | 0.084 |
| Unmethylation | 1.49 ± 0.35 | 0.0012 ± 0.0003 | |||
| TERT | Mutation | 1.53 ± 0.27 | 0.046a | 0.0012 ± 0.0002 | 0.041a |
| Wildtype | 1.69 ± 0.44 | 0.0013 ± 0.0003 |
Unit of omADC: mm2/s
aSignificant at p < 0.05; this difference was significant
Table 3.
Performances of ADC in the comparison of tumor grading and genotypes
| Low grade vs High grade | IDH-mut vs IDH-wt | MGMT-m vs MGMT-um | TERT-mut vs TERT-wt | |
|---|---|---|---|---|
| nADC | ||||
| AUC | 0.787 | 0.836 | 0.651 | 0.607 |
| 95% CI | 0.701–0.874 | 0.757–0.914 | 0.546–0.757 | 0.494–0.721 |
| Cutoff value | 1.56 | 1.60 | 1.59 | 1.89 |
| Sensitivity | 83.8% | 82.2% | 59.1% | 31.0% |
| Specificity | 68.9% | 80.0% | 73.8% | 90.9% |
| omADC | ||||
| AUC | 0.750 | 0.777 | NA | 0.617 |
| 95% CI | 0.656–0.844 | 0.688–0.865 | 0.503–0.730 | |
| Cutoff value | 0.0012 | 0.0012 | 0.0012 | |
| Sensitivity | 78.4% | 88.4% | 73.3% | |
| Specificity | 71.6% | 67.7% | 50.9% | |
CI confidence interval; Sen sensitivity; Sep specificity; NA not available
Fig. 2.
Diagnostic performance of ADC values in WHO-based glioma grading and genotypes. AUC of ROC curves for discrimination between LGGs and HGGs (A), IDH-mut and IDH-wt (B), TERT-mut and TERT-wt (C), as well as between MGMT-m and MGMT-um (D), based on nADC and omADC values. ROC: receiver-operating characteristic; LGGs: low grade gliomas; HGGs: high grade gliomas
Correlation of the ADC values with genotypes
The nADC (1.42 ± 0.28) and omADC (0.0011 ± 0.0002 mm2/s) values in IDH-wt gliomas were lower than those (1.83 ± 0.34 and 0.0014 ± 0.0003 mm2/s) in IDH-mutated gliomas (both P < 0.0001) (Table 2). In ROC analysis, when the cutoffs were 1.60 and 0.0012 mm2/s, respectively, the sensitivities, specificities and AUC of nADC and omADC were 82.2% and 84.4%, 80.0% and 67.7%, and 0.836 and 0.777, respectively (Table 3 and Fig. 2B).
The values of nADC and omADC were higher in TERT promoter wildtype (TERT-wt) gliomas than in TERT-mutated gliomas (P = 0.046 for nADC and 0.041 for omADC) (Table 2). However, these ADC values had limited ability in discriminating TERT status (AUC = 0.607 for nADC and 0.617 for omADC) (Table 3 and Fig. 2C).
MGMT-methylated gliomas exhibited significantly higher nADC values than MGMT-unmethylated gliomas (P = 0.021) (Table 2). However, the predictive performance of nADC was not good (AUC = 0.651, specificity = 73.8%, and sensitivity = 59.1%) (Table 3 and Fig. 2D). MGMT-m could not be detected by omADC values.
Multiple linear regression analysis of the correlation between the basic information of gliomas and ADC values
Multiple linear regression analysis, including four variables (IDH, MGMT, TERT, and WHO grade), showed that the nADC and omADC values were not statistically affected by TERT status and MGMT status. The values of nADC and omADC were significantly associated with IDH status, and nADC values were also tightly associated with the WHO grade (Table 4). However, only 60.0% of the variation in the nADC values could be explained by WHO grade and IDH status. IDH status exhibited higher nADC values of standardized coefficients than WHO grade (0.311 vs. -0.240) (Table 4), indicating that IDH status has a greater impact on nADC values than WHO grade. The analysis also revealed the trend that lower tumor grade and IDH-mutation status can increase nADC values. The students’ t-tests also demonstrated this trend.
Table 4.
Results of multiple linear regression analysis
| Variables | nADC | omADC | ||||||
|---|---|---|---|---|---|---|---|---|
| B | P value | SE | SC | B | P value | SE | SC | |
| Constant | 1.710 | < 0.0001 | 0.098 | 0.001 | < 0.0001 | < 0.0001 | ||
| Grade | − 0.204 | 0.011a | 0.079 | − 0.299 | < 0.0001 | 0.053 | < 0.0001 | − 0.213 |
| IDH-mut | 0.311 | < 0.0001a | 0.085 | 0.380 | < 0.0001 | 0.003a | < 0.0001 | 0.374 |
| MGMT-um | − 0.071 | 0.340 | 0.074 | − 0.061 | − 6.373E-005 | 0.306 | < 0.0001 | − 0.107 |
| TERT-mut | − 0.101 | 0.112 | 0.063 | − 0.167 | − 8.526E-005 | 0.109 | < 0.0001 | − 0.146 |
| Adjusted R2 | 0.600 | 0.522 | ||||||
SE standardized error, SC standardized coefficients, IDH-mut IDH mutation, MGMT-um MGMT unmethylation, TERT-mut TERT mutation
aSignificant at p < 0.05; this difference was significant
Discussion
The 2016 WHO classification of glioma emphasized the role of genetic parameters in glioma patients’ prognosis and treatment response [22]. The identification of histology and genetic status of gliomas before surgery can benefit these patients. DWI is performed as a routine preoperative method for evaluating gliomas. In this case, ADC’s discriminative abilities in histologic subtypes, IDH, MGMT, and TERT status were assessed, respectively.
In the current study, ADC values generated from DWI (b = 0 and 1000 s/mm2) decreased significantly with the WHO glioma grade, which was in accordance with previous studies [16, 23]. Cell density, mitotic activity, and vascularity play important roles in gliomas’ pathological grading [24]. For example, the increment of cell density can remarkably restrict water molecules’ movement, which can be reflected by ADC [24]. Therefore, HGGs were more prone to exhibit lower ADC values than LGGs. Louis et al. [25]discovered that HGGs also had lesser normal brain cells and more tumor cells than LGGs, which may also partly explain the lower ADC values in HGGs.
Accurate identification of IDH status is crucial because the prognosis varies greatly according to IDH status. IDH-mutated gliomas have a significantly better prognosis than IDH-wt gliomas [1]. In this study, the IDH-mut rate was 75.00% in LGGs and 24.32% in HGGs, respectively. The IDH-mut rate of HGGs was higher than the reported indices (75% for LGGs and 12% for HGGs) [26]. The ADC values for IDH-mutated gliomas were significantly higher than those for IDH-wt gliomas, which was consistent with previous research [16, 27]. This difference was more significant when high b-value (b = 3000 s/mm2) rather than standard b-value (1000 s/mm2) was used[23]. IDH may inhibit tumor growth by decreasing the level of nicotinamide adenine dinucleotide phosphate production [26] and hypoxia-inducible factor 1α [28]. This mechanism could decrease cell density and partially explain how IDH-mutated gliomas displayed higher ADC values. Besides, we found that IDH-mut had a direct and greater impact on ADC values than tumor grade, which helped to explain why IDH status could predict prognosis better than the histologic classification [29].Besides IDH, MGMT and TERT are also important genetic hallmarks in guiding clinical treatment and evaluating glioma patients’ prognosis [30, 31]. The ADC values are used as a potential marker for predicting MGMT and TERT status in glioblastomas; however, without expert consensus [2, 12–14, 32]. For WHO II-IV gliomas, we found that ADC values had less accuracy and reliability in discriminating MGMT and TERT status, which limited the use of DWI metrics in predicting these two genotypes. Multiple linear regression analysis also revealed that MGMT and TERT status were not independent parameters for ADC values. We hypothesized that coexisting factors or interactions between variables might induce the increment of ADC in TERT-wt and MGMT-m gliomas. For example, in this study, HGGs were more likely to have MGMT-um and IDH-wt than LGGs (P = 0.002 and P < 0.0001, respectively), and consequently, the ADC values in MGMT-unmethylated gliomas might be affected by the tumor grading and concurrent IDH-wt. In accordance with our results, no significant relationship between ADC values in glioblastomas and TERT [2, 15] and MGMT status [32] was reported. However, several studies [12–14] showed that ADC values were significantly higher in glioblastomas with MGMT-m than with MGMT-um. These conflicting results may be partly due to the difference in ROI selection and subject recruitment [14]. Unlike previous studies [2, 12–15] that only included glioblastomas, this study recruited patients with WHO II-IV gliomas. Besides, we placed ROIs on the solid part of the tumor, which is different from the previous study where ROIs were placed on the contrast-enhanced part of the tumor [25]. The predictive value of ADC values still needs to be verified by further large-scale comparative studies.
Advances in radiomics [9, 10] and MRI techniques, including ASL [11, 16, 33, 34], DSC [2, 33, 35], and diffusion tensor imaging [36, 37], have been used in evaluating glioma grade or genotypes. Several studies [14, 16, 34] have shown that, compared with perfusion parameters, ADC values have a better predictive effect on tumor grade and genotypes. In this study, only ADC values were assessed because DWI is a commonly used sequence and can be performed in all hospitals. Besides, the postprocessing method of DWI is simple and time-saving.
This study’s strength was that it evaluated the discriminative ability of ADC values in WHO glioma grade and various genetic status in the same study. Therefore, an overall assessment of the predictive power of DWI metrics was available. Accessing various genetic features in one study also helped us identify the valuable genotypes which directly affected ADC values. Since higher ADC values were associated with a more favorable prognosis [12, 38], it was crucial to find out meaningful genotypes that were tightly associated with patients’ outcomes.
Besides the intrinsic limitations of retrospective researches, the other four limitations of this study should be noted. Firstly, biopsy samples used in this study were not acquired by ADC-guided biopsy. Because the ROI-based method cannot assess the direct correlation between histopathology and ADC values, some bias can be produced, especially in more heterogeneous gliomas like HGGs. Secondly, the ROIs did not include peri-tumor areas that may also be infiltrated by glioma cells and contain information reflecting tumor genotypes. Thirdly, the sample size was small. Thus, a larger cohort of patients is needed to verify our conclusions. Fourthly, the genotypes evaluated in this study were limited.
Conclusion
DWI metrics, including nADC and omADC from the solid part of the glioma, have a potential ability to predict tumor grade and IDH-mut, but have limited use in the prediction of TERT-mut and MGMT-m.
Acknowledgements
The authors thank American Journal Experts for providing language help.
Abbreviations
- DWI
Diffusion-weighted imaging
- IDH-mut
Isocitrate dehydrogenase mutation
- IDH-wt
Isocitrate dehydrogenase wildtype
- MGMT-m
O6-methylguanine-DNA methyltransferase promoter methylation
- MGMT-um
O6-methylguanine-DNA methyltransferase promoter unmethylation
- TERT-mut
Telomerase reverse transcriptase promoter mutation
- TERT-wt
Telomerase reverse transcriptase promoter wildtype
- ADC
Apparent diffusion coefficient
- omADC
Overall mean apparent diffusion coefficient
- nADC
Normalized apparent diffusion coefficient
- WHO
World Health Organization
- T2-PROPELLER
T2-periodically rotated overlapping parallel lines with enhanced reconstruction
- ROI
Region of interest
- ROC
Receiver-operating characteristic
- CNWM
Contralateral normal white matter
- HGGs
High grade gliomas
- LGGs
Low grade gliomas
Author contributions
I confirm that all authors have made substantial contributions to all of the following: (1) the conception and design of the study (LSR, ZYW, YH, MWB, and FF), or acquisition of data (KZR, JCD, and WY), or analysis and interpretation of data (LSR, ZYW, and ZDC), (2) drafting the article (LSR), (3) final approval of the version to be submitted (LSR, ZYW, KZR, JCD, WY, ZDC, YH, MWB, and FF). All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This retrospective study was approved by the Institutional Review Board of Peking Union Medical College Hospital. The requirement for informed consent from patients was waived.
Consent for publication
Not applicable.
Competing interests
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yiwei Zhang and Ziren Kong are co-first authors.
Contributor Information
Sirui Liu, Email: lsr0303@126.com.
Yiwei Zhang, Email: zhangyiweilucy@126.com.
Ziren Kong, Email: kongziren@pumc.edu.cn.
Chendan Jiang, Email: jiangchendan@gmail.com.
Yu Wang, Email: ywang@pumch.cn.
Dachun Zhao, Email: dachunzhao@126.com.
Hui You, Email: you_hui@hotmail.com.
Wenbin Ma, Email: mawb2001@hotmail.com.
Feng Feng, Email: cjr.fengfeng@vip.163.com.
References
- 1.Cagney DN, Sul J, Huang RY, Ligon KL, Wen PY, Alexander BM. The FDA NIH biomarkers, endpoints, and other tools (BEST) resource in neuro-oncology. Neuro Oncol. 2018;20(9):1162–1172. doi: 10.1093/neuonc/nox242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanidze J, Lum M, Pisapia D, Magge R, Ramakrishna R, Kovanlikaya I, Fine HA, Chiang GC. MRI features associated with TERT promoter mutation status in glioblastoma. J Neuroimaging . 2019;29(3):357–363. doi: 10.1111/jon.12596. [DOI] [PubMed] [Google Scholar]
- 3.Gao K, Li G, Qu Y, Wang M, Cui B, Ji M, Shi B, Hou P. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7(8):8712–8725. doi: 10.18632/oncotarget.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chawla S, Krejza J, Vossough A, Zhang Y, Kapoor GS, Wang S, O'Rourke DM, Melhem ER, Poptani H. Differentiation between oligodendroglioma genotypes using dynamic susceptibility contrast perfusion-weighted imaging and proton MR spectroscopy. AJNR Am J Neuroradiol. 2013;34(8):1542–1549. doi: 10.3174/ajnr.A3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellingson BM, Cloughesy TF, Pope WB, Zaw TM, Phillips H, Lalezari S, Nghiemphu PL, Ibrahim H, Naeini KM, Harris RJ, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage. 2012;59(2):908–916. doi: 10.1016/j.neuroimage.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 6.Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed. 2017;140:249–257. doi: 10.1016/j.cmpb.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Shu C, Wang Q, Yan X, Wang J. The TERT promoter mutation status and MGMT promoter methylation status, combined with dichotomized MRI-derived and clinical features, predict adult primary glioblastoma survival. Cancer Med. 2018;7(8):3704–3712. doi: 10.1002/cam4.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel SH, Poisson LM, Brat DJ, Zhou Y, Cooper L, Snuderl M, Thomas C, Franceschi AM, Griffith B, Flanders AE, et al. T2-FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res. 2017;23(20):6078–6085. doi: 10.1158/1078-0432.CCR-17-0560. [DOI] [PubMed] [Google Scholar]
- 9.Jiang C, Kong Z, Zhang Y, Liu S, Liu Z, Chen W, Liu P, Liu D, Wang Y, Lyu Y, et al. Conventional magnetic resonance imaging-based radiomic signature predicts telomerase reverse transcriptase promoter mutation status in grade II and III gliomas. Neuroradiology. 2020;62(7):803–813. doi: 10.1007/s00234-020-02392-1. [DOI] [PubMed] [Google Scholar]
- 10.Jiang C, Kong Z, Liu S, Feng S, Zhang Y, Zhu R, Chen W, Wang Y, Lyu Y, You H, et al. Fusion Radiomics Features from Conventional MRI Predict MGMT Promoter Methylation Status in Lower Grade Gliomas. Eur J Radiol. 2019;121:108714. doi: 10.1016/j.ejrad.2019.108714. [DOI] [PubMed] [Google Scholar]
- 11.Yoo RE, Yun TJ, Hwang I, Hong EK, Kang KM, Choi SH, Park CK, Won JK, Kim JH, Sohn CH. Arterial spin labeling perfusion-weighted imaging aids in prediction of molecular biomarkers and survival in glioblastomas. Eur Radiol. 2020;30(2):1202–1211. doi: 10.1007/s00330-019-06379-2. [DOI] [PubMed] [Google Scholar]
- 12.Romano A, Calabria LF, Tavanti F, Minniti G, Rossi-Espagnet MC, Coppola V, Pugliese S, Guida D, Francione G, Colonnese C, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol. 2013;23(2):513–520. doi: 10.1007/s00330-012-2601-4. [DOI] [PubMed] [Google Scholar]
- 13.Kanazawa T, Minami Y, Jinzaki M, Toda M, Yoshida K, Sasaki H. Predictive markers for MGMT promoter methylation in glioblastomas. Neurosurg Rev. 2019 doi: 10.1007/s10143-018-01061-5. [DOI] [PubMed] [Google Scholar]
- 14.Han Y, Yan LF, Wang XB, Sun YZ, Zhang X, Liu ZC, Nan HY, Hu YC, Yang Y, Zhang J, et al. Structural and advanced imaging in predicting MGMT promoter methylation of primary glioblastoma: a region of interest based analysis. BMC Cancer. 2018;18(1):215. doi: 10.1186/s12885-018-4114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita K, Hatae R, Hiwatashi A, Togao O, Kikuchi K, Momosaka D, Yamashita Y, Kuga D, Hata N, Yoshimoto K, et al. Predicting TERT promoter mutation using MR images in patients with wild-type IDH1 glioblastoma. Diagn Interv Imaging. 2019;100(7–8):411–419. doi: 10.1016/j.diii.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Cheng G, Kang X, Xi Y, Zhu Y, Wang K, Sun C, Ye J, Li P, Yin H. Noninvasively evaluating the grading and IDH1 mutation status of diffuse gliomas by three-dimensional pseudo-continuous arterial spin labeling and diffusion-weighted imaging. Neuroradiology. 2018;60(7):693–702. doi: 10.1007/s00234-018-2021-5. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y, Choi SH, Kim YJ, Kim KG, Sohn CH, Kim JH, Yun TJ, Chang KH. Gliomas: histogram analysis of apparent diffusion coefficient maps with standard- or high-b-value diffusion-weighted MR imaging–correlation with tumor grade. Radiology. 2011;261(3):882–890. doi: 10.1148/radiol.11110686. [DOI] [PubMed] [Google Scholar]
- 18.Latysheva A, Emblem KE, Brandal P, Vik-Mo EO, Pahnke J, Roysland K, Hald JK, Server A. Dynamic susceptibility contrast and diffusion MR imaging identify oligodendroglioma as defined by the 2016 WHO classification for brain tumors: histogram analysis approach. Neuroradiology. 2019;61(5):545–555. doi: 10.1007/s00234-019-02173-5. [DOI] [PubMed] [Google Scholar]
- 19.Horbinski C, Kofler J, Kelly LM, Murdoch GH, Nikiforova MN. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68(12):1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 20.Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, Shi Z, Chan DT, Poon WS, Zhou L, et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Modern Pathol. 2015;28(2):177–186. doi: 10.1038/modpathol.2014.94. [DOI] [PubMed] [Google Scholar]
- 21.Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, Westphal M, Wick W, Pietsch T, Loeffler M, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131(6):1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 22.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 23.Han H, Han C, Wu X, Zhong S, Zhuang X, Tan G, Wu H. Preoperative grading of supratentorial nonenhancing gliomas by high b-value diffusion-weighted 3 T magnetic resonance imaging. J Neurooncol. 2017;133(1):147–154. doi: 10.1007/s11060-017-2423-y. [DOI] [PubMed] [Google Scholar]
- 24.Togao O, Hiwatashi A, Yamashita K, Kikuchi K, Mizoguchi M, Yoshimoto K, Suzuki SO, Iwaki T, Obara M, Van Cauteren M, et al. Differentiation of high-grade and low-grade diffuse gliomas by intravoxel incoherent motion MR imaging. Neuro Oncol. 2016;18(1):132–141. doi: 10.1093/neuonc/nov147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol. 2011;12(1):83–91. doi: 10.1016/S1470-2045(10)70053-X. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Choi SH, Ryoo I, Yoon TJ, Kim TM, Lee SH, Park CK, Kim JH, Sohn CH, Park SH, et al. Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neurooncol. 2015;121(1):141–150. doi: 10.1007/s11060-014-1614-z. [DOI] [PubMed] [Google Scholar]
- 28.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler-Wirth H, Kaulich K, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129(5):679–693. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 30.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M, Shimizu S, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang P, Cai J, Yan W, Zhang W, Wang Y, Chen B, Li G, Li S, Wu C, Yao K, et al. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neuro Oncol. 2016;18(8):1099–1108. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi YS, Ahn SS, Kim DW, Chang JH, Kang SG, Kim EH, Kim SH, Rim TH, Lee SK. Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology. 2016;281(1):175–184. doi: 10.1148/radiol.2016151913. [DOI] [PubMed] [Google Scholar]
- 33.Ma H, Wang Z, Xu K, Shao Z, Yang C, Xu P, Liu X, Hu C, Lu X, Rong Y. Three-dimensional arterial spin labeling imaging and dynamic susceptibility contrast perfusion-weighted imaging value in diagnosing glioma grade prior to surgery. Exp Ther Med. 2017;13(6):2691–2698. doi: 10.3892/etm.2017.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leu K, Ott GA, Lai A, Nghiemphu PL, Pope WB, Yong WH, Liau LM, Cloughesy TF, Ellingson BM. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II-III diffuse gliomas. J Neurooncol. 2017;134(1):177–188. doi: 10.1007/s11060-017-2506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita N, Wang S, Chawla S, Poptani H, Melhem ER. Dynamic susceptibility contrast perfusion weighted imaging in grading of nonenhancing astrocytomas. J Magn Reson Imaging JMRI. 2010;32(4):803–808. doi: 10.1002/jmri.22324. [DOI] [PubMed] [Google Scholar]
- 36.Tan Y, Zhang H, Wang X, Qin J, Wang L, Yang G, Yan H. Comparing the value of DKI and DTI in detecting isocitrate dehydrogenase genotype of astrocytomas. Clin Radiol. 2019;74(4):314–320. doi: 10.1016/j.crad.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Aliotta E, Nourzadeh H, Batchala PP, Schiff D, Lopes MB, Druzgal JT, Mukherjee S, Patel SH. Molecular subtype classification in lower-grade glioma with accelerated DTI. AJNR Am J Neuroradiol. 2019;40(9):1458–1463. doi: 10.3174/ajnr.A6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Y, Ma L, Chen X, Zhang Z, Jiang H, Lin S. Lower apparent diffusion coefficients indicate distinct prognosis in low-grade and high-grade glioma. J Neurooncol. 2014;119(2):377–385. doi: 10.1007/s11060-014-1490-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.