Abstract
Background
TP53 is the most frequently mutated gene in the human cancer, and the awareness of its mutational status is useful in the diagnosis and treatment of cancer patients. In the present study, we investigated the association between TP53 gene mutations and p53 immunohistochemical staining (IHC) patterns and non-genetic effect of MDM2 as a negative regulator of p53.
Methods
A total of 135 solid cancer cases with next generation sequencing data were subjected to p53 IHC and classified as overexpression, null type or usual pattern.
Results
TP53 mutation was observed in 104 out of 135 cases (77.0%). When the TP53 mutations were annotated into DISRUPTED (truncations, frameshifts, splice site mutations, and deep deletions) and IF-DBD (in-frame mutations in the DNA binding domain), the null type p53 IHC pattern was associated with DISRUPTED mutations (sensitivity 86.2%, specificity 97.2%) while the overexpression pattern was associated with IF-DBD mutations (sensitivity 100%, specificity 81.7%). The specificity of p53 IHC usual pattern predicting wild type TP53 was also as high as 100%. Regardless of MDM2 amplification, p53 IHC pattern showed a perfect association with TP53 mutation pattern.
Conclusions
p53 IHC pattern (overexpression, null type, usual) reasonably predicted TP53 mutational status (DISRUPTED, IF-DBD), and MDM2 amplification status did not have any impact on the p53 IHC pattern.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13000-022-01273-w.
Keywords: TP53 gene, Immunohistochemistry, Mutation, p53 protein, Association
Background
TP53, functioning as a cellular stress sensor for physiologic or oncogenic activation, is the most frequently mutated gene in human cancer [1–4]. When there is oncogenic activation, DNA replication stress disrupts physiologic function of p53, resulting in cell death evasion and thus, TP53 mutation is selected during tumor progression [1, 5–7]. Because the p53 is so essential for tumorigenesis and tumor progression, there have been several attempts to elicit tumor cell death by reactivation of mutated TP53 [8–11]. For instance, a conceptual basis for alleviating the effect of inactivating TP53 mutation through artificial second-site mutation has been proposed [12, 13]. Furthermore, pathogenic TP53 mutations are known to be associated with poor survival or chemo-resistance in various tumor types [12, 14–21]. Therefore, it is very important to check the TP53 mutational status to predict the patient’s prognosis and treatment responsiveness.
Sanger sequencing is known as a traditional gold standard method to confirm the mutational status of TP53, but it’s labor intensive and time consuming especially because the TP53 gene is quite big. Because of these practical problems, it’s difficult to use Sanger sequencing in actual practice, and instead, immunohistochemical staining (IHC) is the most widely used way to infer TP53 mutational status. Several attempts have been made to infer TP53 mutation type through p53 IHC pattern, but most previous studies were limited to one specific organ, and the methods of interpreting the p53 IHC results varied, such as pattern-based or positive cell proportions [22–30].
To better understand the association between TP53 gene mutations and p53 IHC patterns, we used a set of tumor samples having TP53 gene mutation data across entire TP53 exons obtained by a clinical grade next-generation sequencing (NGS) test. Through annotation of TP53 mutations according to the predicted effects on p53 protein function and classification of p53 IHC patterns according to the presence of overexpression or complete loss of expression, we investigated the association between the TP53 mutation status and p53 IHC pattern. When the p53 IHC pattern and expected TP53 mutation status were discordant, we analyzed the reasons for the discrepancies. Secondly, we investigated p53 IHC pattern in MDM2-amplified cases to determine how p53 IHC appears through the non-genetic effect of MDM2, a negative regulator of p53.
Methods
Case selection
After the approval (protocol number, 2018–0976) from the institutional review board, we retrospectively collected 135 adult solid cancer samples that have clinical next-generation sequencing (NGS) data from the records of the Department of Pathology at the Asan Medical Center, University of Ulsan College of Medicine. Among them, 26 cases were obtained after neoadjuvant treatment and 12 cases showed unequivocal MDM2 amplification.
Mutational analysis
The clinical NGS cancer panel test was performed as described previously [31]. Briefly, the targeted NGS used the MiSeq platform (Illumina, San Diego, CA) and the gene panel, OncoPanel AMC version 3, was designed in house through SureDesign (Agilent Technologies, Santa Clara, CA) using GRCh37 reference version. This 1.2 Mbp-sized panel included 33,524 probes targeting a total of 382 genes, including entire exons of 199 genes, 184 hot spots, and partial introns for eight genes often rearranged in cancer. Of course, the entire TP53 and MDM2 exonic regions were included.
We classified the TP53 mutations according to the type of nucleotide changes: single nucleotide variation (SNV), in-frame insertion or deletion (In-frame indel), frameshift insertion of deletion (frameshift indel), premature stop codon (truncation), splice site mutation (splice), and copy-number (CN) loss. We classified the TP53 mutations into two major categories, in-frame alterations across the DNA binding domain (IF-DBD) and alterations involving significant disruption of protein coding sequences (DISRUPTED). The IF-DBD mutations included SNVs and In-frame indel across DNA binding domain. The DISRUPTED mutations included frameshift indel, truncation, splice, and CN loss. The MDM2 amplifications were detected through our bioinformatics pipeline using CNVkit v0.9.6 [32]. Copy numbers of tumors were called against a panel of normal. Copy number segments were inferred using circular binary segmentation(CBS) method. Absolute copy number of given segments was estimated using ‘call’ module of CNVkit and pathologist’s tumor cellularity. We used pathologist’s tumor purity estimates to infer copy-number more accurately. This involved solving an equation: (measured copy number) = (actual copy number) * (tumor purity) + 2 * (1-tumor purity). To investigate the biologic effect of MDM2 overexpression on TP53 status, we included tumor samples with high level MDM2 amplifications (estimated MDM2 copy number > 30). All cases were manually reviewed through visualization of bam files with the integrated genomic browser (IGV) and double check of tumor cell purity through review of microscopic slides and variant allelic fractions of the detected clonal mutations.
Immunohistochemistry
Representative areas from formalin-fixed, paraffin-embedded tissues were subjected to IHC, which was performed on 4-μm-tick sections using a Ventana auto-stainer and an ultra-View DAB Detection Kit (Ventana, Tucson, Arizona), according to the manufacturer’s instructions. Primary antibody for p53 (clone DO-7, catalog No.M7001, DAKO, Denmark, Glostrup, 1:1000, Mouse monoclonal) was used. The p53 IHC staining were evaluated by two pathologists (JK and YNS) and classified into 3 categories: overexpression (strong diffuse nuclear immunoreactivity in all tumor cells), null (complete absence of nuclear immunoreactivity in all tumor cells), and usual (neither overexpression nor null, variable nuclear immunoreactivity in tumor cells).
Statistical analysis
The R software (version 4.02 Vienna, Austria) was used to perform statistical analyses. Association between TP53 mutational status by NGS test and IHC staining was tested using the χ2 and/or the Fisher exact tests. P-values less than 0.05 were considered statistically significant.
Results
Malignant tumors of various organs were included in this study: anus in 2 (1.5%), brain in 7 (5.2%), breast in 6 (4.4%), large intestine in 107 (79.3%), lung in 2 (1.5%), ovary in 6 (4.4%), small intestine in 1 (0.7%), stomach in 2 (1.5%), and unknown in 2 (1.5%) cases (Table 1). Pathologic diagnosis of tumor was mostly adenocarcinoma (113 cases, 83.7%). Most cases were surgically resected specimens (125 cases, 92.6%) followed by forceps or needle biopsies (5 cases, 3.7%), and excisional biopsies (5 cases, 3.7%). Before obtaining tissue, neoadjuvant treatment was performed in 26 cases (19.3%).
Table 1.
Clinicopathologic characteristics of cases
| Characteristics | No of patients | % of patients |
|---|---|---|
| Sex | ||
| Male | 69 | 51.1 |
| Female | 66 | 48.9 |
| Age | ||
| < 60 | 55 | 40.7 |
| ≥ 60 | 80 | 59.3 |
| Primary site | ||
| Anus | 2 | 1.5 |
| Brain | 7 | 5.2 |
| Breast | 6 | 4.4 |
| Large intestine | 107 | 79.3 |
| Lung | 2 | 1.5 |
| Ovary | 6 | 4.4 |
| Small intestine | 1 | 0.7 |
| Stomach | 2 | 1.5 |
| Unknown | 2 | 1.5 |
| Diagnosis | ||
| Adenocarcinoma | 106 | 78.5 |
| Carcinoma | 2 | 1.5 |
| Endometroid adenocarcinioma | 1 | 0.7 |
| Gastrointestinal stromal tumor | 1 | 0.7 |
| Glioblastoma | 7 | 5.2 |
| Invasive ductal carcinoma | 6 | 4.4 |
| Malignant brenner tumor | 1 | 0.7 |
| Mucinous aadenocarcinoma | 6 | 4.4 |
| Papillary serous carcinoma | 4 | 3.0 |
| Squamous cell carcinoma | 1 | 0.7 |
| Procedure | ||
| Resection | 125 | 92.6 |
| Excision | 5 | 3.7 |
| Biopsy | 5 | 3.7 |
| Neoadjuvant treatment | ||
| Yes | 26 | 19.3 |
| No | 109 | 80.7 |
Through NGS analysis, TP53 mutation was observed in 104 out of 135 cases (77.0%). Of the 104 cases with the TP53 mutation, 72 types of TP53 mutations were observed (Supplementary data). Table 2 shows the association of TP53 mutational type and the IHC staining pattern. As a result of p53 IHC pattern, there were 86, 28 and 21 cases of overexpression, null, and usual pattern, respectively (Fig. 1). Most of the cases with p53 overexpression pattern except for 7 cases were accompanied by mutations (79/86, 91.9%) such as single nucleotide variation, In-frame indel, truncation, CN loss, and splice. Of the 7 cases without mutation, 4 cases received neo-adjuvant chemotherapy. Similarly, in the cases of p53 with null pattern, 25 out of 28 cases (89.3%) accompanied mutations. No mutations were observed in all cases (21/21, 100%) showing the p53 usual pattern.
Table 2.
Immunohistochemical staining patterns and mutational analysis of TP53
| Mutation type | Immunohistochemical pattern of p53 | Total | ||
|---|---|---|---|---|
| Overexpression | Null type | Usual | ||
| SNV | 65 (75.6) | 0 (0) | 0 (0) | 65 |
| Frameshift indel | 0 (0) | 7 (25) | 0 (0) | 7 |
| In-frame indel | 4 (4.7) | 0 (0) | 0 (0) | 4 |
| Truncation | 1 (1.2) | 9 (32.1) | 0 (0) | 10 |
| Copy-number loss | 1 (1.2) | 0 (0) | 0 (0) | 1 |
| Splice | 2 (2.3) | 7 (25) | 0 (0) | 9 |
| SNV/Frameshift indel | 1 (1.2) | 1 (3.6) | 0 (0) | 2 |
| SNV/SNV | 5 (5.8) | 0 (0) | 0 (0) | 5 |
| Splice/Truncation | 0 (0) | 1 (3.6) | 0 (0) | 1 |
| Wild type | 7 (8.1) | 3 (10.7) | 21 (100) | 31 |
| Total | 86 | 28 | 21 | 135 |
The corresponding percentages are shown in parentheses
Fig. 1.
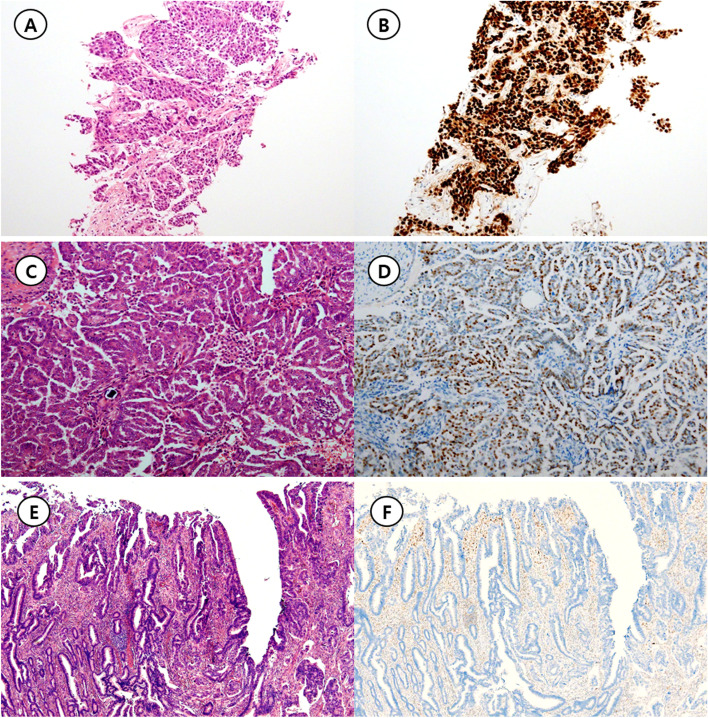
Representative figures for three p53 immunohistochemistry patterns. A Invasive ductal carcinoma of the breast harboring TP53 H193R mutation (Hematoxylin & Eosin, × 1.25 objective lens). B Diffuse strong immunoreactivity for p53, that is overexpression pattern (p53 immunohistochemistry, × 1.25 objective lens). C Low grade papillary serous carcinoma of the ovary without any oncogenic TP53 mutation (Hematoxylin & Eosin, × 10 objective lens) D Approximately a half of the tumor cells express p53 in the nuclei, that is usual pattern (p53 immunohistochemistry, × 10 objective lens). E Adenocarcinoma of the colon harboring TP53 H214Qfs*2 mutation (Hematoxylin & Eosin, × 4 objective lens). F Tumor cells are completely negative for p53 protein expression. Adjacent non-neoplastic cells showing p53 expression serves as an internal positive control. This pattern is classified as null pattern (p53 immunohistochemistry, × 4 objective lens)
The association of TP53 mutation type and p53 IHC pattern was better observed when the TP53 mutation were divided into IF-DBD and DIRUPTED according to the resulting sequence context (Table 3, p < 0.001). Because the in-frame SNVs or Indels are predicted to maintain most of the protein coding sequences intact while other alterations may disrupt amino acid sequences considerably, the p53 protein IHC pattern is predicted to be different between the two groups. Indeed, overexpression pattern was closely associated with the IF-DBD TP53 mutations (sensitivity 100%, specificity 81.7%, positive predictive value 87.2%, negative predictive value 100%), while null pattern was associated with the DIRUPTED TP53 mutations (sensitivity, 86.2%, specificity 97.2%, positive predictive value 89.3%, negative predictive value 96.3%). The usual p53 IHC pattern well predicted the absence of pathogenic TP53 mutations (sensitivity 67.7%, specificity 100%, positive predictive value 100%, negative predictive value 91.2%). Additionally, among 26 cases that received previous neoadjuvant treatment, there were 19, 6, and 1 cases of overexpression, null type, and usual pattern, respectively. And the association between TP53 mutation type and p53 IHC pattern was statistically significant (Table 4, p < 0.002).
Table 3.
Immunohistochemical staining patterns and predicted functional consequence of TP53 mutations
| Functional consequence | Immunohistochemical pattern of p53 | ||
|---|---|---|---|
| Overexpression | Null type | Usual | |
| IF-DBD | 75 (87.2) | 0 (0) | 0 (0) |
| DISRUPTED | 4 (4.7) | 25 (89.3) | 0 (0) |
| Wild type | 7 (8.1) | 3 (10.7) | 21 (100) |
| Total | 86 | 28 | 21 |
The corresponding percentages are shown in parentheses
Table 4.
Immunohistochemical staining patterns and predicted functional consequence of TP53 mutations within cases with previous neoadjuvant treatment
| Functional consequence | Immunohistochemical pattern of p53 | ||
|---|---|---|---|
| Overexpression | Null type | Usual | |
| IF-DBD | 14 (73.7) | 0 (0) | 0 (0) |
| DISRUPTED | 1 (5.3) | 4 (66.7) | 0 (0) |
| Wild type | 4 (2.1) | 2 (33.3) | 1 (100) |
| Total | 19 | 6 | 1 |
The corresponding percentages are shown in parentheses
Table 5 shows the p53 IHC pattern according to the MDM2 amplification status. The MDM2-amplified tumors showed more frequent usual p53 IHC pattern than MDM2-non-amplified tumors (p < 0.001, Fisher’s exact test). Null pattern was observed in only one case (1/12, 8.3%) in contrast to the assumption that p53 may be downregulated through MDM2 amplification. Instead, when we looked at the presence and the type of TP53 mutations in the MDM2-amplified tumors, the p53 IHC pattern just followed TP53 mutation status regardless of MDM2 amplification except for one case where p53 IHC overexpression pattern and wild type TP53 mutation was observed (Table 6).
Table 5.
Immunohisochemical staining pattern of p53 according to MDM2 amplification status
| MDM2 amplification status | ||
|---|---|---|
| No amplification | Amplification | |
| Null type | 27 (22.0) | 1 (8.3)a |
| Usual | 13 (10.6) | 8 (66.7) |
| Overexpression | 83 (67.5) | 3 (25.0) |
| Total | 123 | 12 |
The corresponding percentages are shown in parentheses
a This case harbored TP53 compound R337C and E204Gfs*43 mutations and classified as DISRUPTED in main analysis
Table 6.
Immunohistochemical staining patterns according to the TP53 mutation types in MDM2-amplified cases
| TP53 mutation types | Immunohistochemical pattern of p53 | ||
|---|---|---|---|
| Overexpression | Null type | Usual | |
| IF-DBD | 2 (66.7) | 0 (0) | 0 (0) |
| DISDRUPTED | 0 (0) | 1 (100) | 0 (0) |
| Wild type | 1 (33.3) | 0 (0) | 8 (100) |
| Total | 3 | 1 | 8 |
The corresponding percentages are shown in parentheses
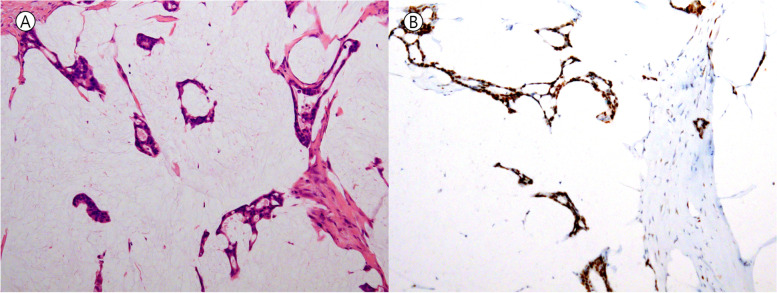
The p53 IHC pattern and TP53 mutation discordance was observed in 14 out of 135 cases (14/135, 10.4%) (Table 7), and could be divided into the following 2 groups; 1) Wild type TP53 gene showing either overexpression or null IHC pattern, and 2) DISRUPTED type TP53 mutation showing overexpression pattern. In group 1, 7 cases showed overexpression and 3 cases showed null p53 IHC pattern in the absence of TP53 mutations. Among the 7 cases showing p53 overexpression, 4 cases received neo-adjuvant chemoradiation therapy (Fig. 2). In addition, 2 out of 3 cases with p53 IHC null also received neo-adjuvant chemoradiation therapy. Since group 1 type discrepancies could also be seen due to failure of TP53 mutation detection, we re-checked tumor cell purity. However, the tumor cell purity was within acceptable range in all cases (Fig. 3). The other cases belonged to the group 2: DISRUPTED type TP53 mutation and overexpression p53 IHC pattern.
Table 7.
p53 immunohistochemical staining pattern and TP53 mutation discordant cases
| Primary Site | Diagnosis | P53 IHC | TP53 mutation type | Class | alteration | allele frquency | cbioportal | MDM2 amplification | Neoadjuvant treatment | |
|---|---|---|---|---|---|---|---|---|---|---|
| group 1 | Large intestine | Adenocarcinoma | NT | WT | WT | ND | negative | Not amplified | Done | |
| Large intestine | Adenocarcinoma | NT | WT | WT | ND | negative | Not amplified | Done | ||
| Large intestine | Adenocarcinoma | NT | WT | WT | ND | negative | Not amplified | |||
| Large intestine | Adenocarcinoma | OE | WT | WT | ND | negative | Not amplified | |||
| Large intestine | Mucinous adenocarcinoma | OE | WT | WT | ND | negative | Not amplified | Done | ||
| Large intestine | Adenocarcinoma | OE | WT | WT | ND | negative | Not amplified | |||
| Large intestine | Adenocarcinoma | OE | WT | WT | ND | negative | Not amplified | Done | ||
| Anus | Adenocarcinoma | OE | WT | WT | ND | negative | Not amplified | Done | ||
| Ovary | Papillary serous carcinoma | OE | WT | WT | ND | negative | Not amplified | Done | ||
| Brain | Glioblastoma | OE | WT | WT | ND | negative | Amplified | |||
| group 2 | Large intestine | Adenocarcinoma | OE | truncation | LOF | R342* | 0.23 | Likely oncogenic | Not amplified | |
| Large intestine | Adenocarcinoma | OE | splice site | LOF | X126_splice | 0.33 | Likely oncogenic | Not amplified | Done | |
| Large intestine | Adenocarcinoma | OE | CNV loss | LOF | loss | 0 | unknown | Not amplified | ||
| Large intestine | Adenocarcinoma | OE | splice site | LOF | X261_splice | 0.31 | Likely oncogenic | Not amplified |
ND Not detected, NT Null type, OE Overexpression, WT Wild type
Fig. 2.
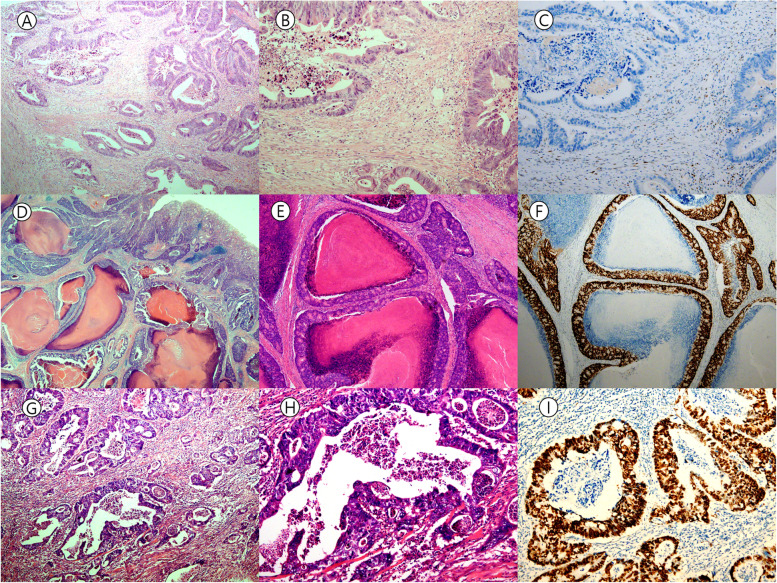
Mucinous adenocarcinoma of the colon that experienced neo-adjuvant chemoradiation therapy but did not show any TP53 mutation. A Several strips or clusters of tumor cells are floating in the mucin pool (Hematoxylin & Eosin, × 10 objective lens). B TP53 overexpression pattern is noted (p53 immunohistochemistry, × 10 objective lens)
Fig. 3.
Representative case with discrepancy between TP53 mutation and p53 IHC pattern. A&B Tumor area from which DNA was extracted for NGS analysis. Tumor cell purity is in acceptable range (Hematoxylin & Eosin, A: X4, B: X10 objective lens, respectively). C Null type pattern of p53 immunostaining is observed. No TP53 mutation was detected in this case (p53 immunohistochemistry, × 10 objective lens). D, E Adenocarcinoma of the cecum with comedo-type necrosis. Area from which DNA has been extracted is shown in E (Hematoxylin & Eosin, D: X1.25, and E: X4 objective lens, respectively). F Overexpression pattern of p53 immunostaining is noted in the absence of detectable TP53 mutation (p53 immunohistochemistry, × 4 objective lens). G, H Moderately differentiated adenocarcinoma of the rectum. DNA has been extracted from the area shown in G (Hematoxylin & Eosin, G: X4, and H: X10 objective lens, respectively). I Overexpression pattern of p53 immunostaining is noted in the absence of detectable TP53 mutation. This patient did not receive neoadjuvant chemoradiation therapy (p53 immunohistochemistry, × 10 objective lens)
Discussion
In this study, the presence of oncogenic or likely oncogenic TP53 mutations and their predicted functional consequences were well correlated with p53 IHC pattern in most cases. Sensitivity for p53 overexpression or null type pattern for prediction of IF-DBD type or DISRUPTED type TP53 mutations was 100% and 86.2%, respectively, and specificity for TP53 usual pattern for prediction of WT TP53 status was also high as 100%. MDM2 amplification did not significantly affect TP53 IHC pattern, in contrast to the expectation that MDM2 would act as a negative regulator of p53, resulting in loss of p53 protein expression. Taken together, the p53 IHC pattern may serve as a reasonable surrogate for TP53 mutations.
For the exceptions in those associations, some cases could be explained by predicted biological consequences. The 4 out of 7 cases with p53 overexpression in the absence of any TP53 mutation received neo-adjuvant chemoradiation therapy. As physiologic p53 accumulation is possible due to the neo-adjuvant chemoradiation therapy, p53 overexpression may be observed in the absence of TP53 gene mutations. However, the other occasions, such as p53 IHC null pattern in tumors harboring wild type TP53, p53 IHC overexpression pattern in tumors harboring DISRUPTED type TP53 mutations, or p53 overexpression in chemoradiation-naïve tumors with wild type TP53, are hard to explain. There might be additional factors affecting p53 IHC pattern other than the presence of the type of TP53 mutations that require further investigation.
As for the association between types of TP53 mutations and p53 IHC patterns, IF-DBD TP53 mutations were commonly associated with overexpression pattern while DISRUPTED TP53 mutations were associated with null pattern. Those associations have been explained by the accumulated biological experiment data that p53 protein encoded by IF-DBD TP53 mutations accumulate in the tumor cell nuclei through interfered MDM2-mediated ubiquitination, and DISRUPTED TP53 mutations result in non-sense mediated decay, or prematurely truncated mRNA, and thus null pattern of p53 IHC [22]. Of note, MDM2 amplification was not associated with p53 IHC null pattern, suggesting that TP53 mutation itself may have a greater influence on IHC pattern than the role of MDM2 as a p53 negative regulator.
There have been several studies that attempted to correlate p53 IHC pattern with TP53 mutational type [22, 23, 29, 30]. In the study of Yemelyanova et al. [23], the results of p53 IHC were divided into 5 groups according to the percentage of positive tumor cells, and when positive tumor cells were more than 60% or 0%, the sensitivity to detect TP53 mutation was 94%. Additionally, in the study of Kobel et al. [22], the predictability of four types of p53 antibodies were evaluated by applying the same three-tier scoring system as in our study. Although it differed slightly depending on the type of antibody, the sensitivity and specificity for p53 IHC were 0.87 ~ 0.95 and 0.73 ~ 0.95 respectively. Singh et al. [29] also included cytoplasmic p53 IHC result as a mutant pattern, and the sensitivity and specificity for TP53 mutation were 97.7% and 88.89% respectively. These studies, together with our study, showed that p53 IHC is useful for the prediction of TP53 mutation type although not perfect.
Previous studies were mainly limited to one organ, but in this study, we could investigate p53 IHC patterns and TP53 mutations in various organs, including colon, brain, breast, ovary, lung, stomach and small intestine, by virtue of NGS. In addition, we also reviewed discrepant cases and the reasons for the discrepancies. Although some discrepancies could not be reasonably explained, we found that neoadjuvant chemoradiation may induce p53 overexpression in the absence of IF-DBD type TP53 mutations. Our study has some limitations. First, there may be selection bias due to availability of NGS data. In addition, the p53 IHC pattern was evaluated by only one type of monoclonal antibody, and detailed mechanistic study for discrepancies between p53 IHC pattern and TP53 mutation type could not be performed because of shortage of time and resources.
In conclusion, TP53 mutational status can be well predicted through three p53 IHC patterns (overexpression, null, and usual) in most cases: 1) overexpression for IF-DBD type TP53 mutations, 2) null for DISRUPTED type TP53 mutations, and 3) usual for the absence of TP53 mutations. In addition, MDM2 amplification does not seem to significantly affect the p53 IHC pattern. Finally, we propose that pattern-based approach (overexpression, null, and usual patterns) might be more informative in the interpretation of p53 IHC than traditional interpretation system based on positive tumor cell proportion.
Supplementary Information
Additional file 1: Supplementary data. Detailed p53 mutational features in all 135 cases.
Acknowledgements
Not applicable.
Abbreviations
- IHC
Immunohistochemical staining
- DISRUPTED
Truncations, frameshifts, splice site mutations, and deep deletions
- IF-DBD
In-frame mutations in the DNA binding domain
- NGS
Next-generation sequencing
Authors’ contributions
Y.-N.S., D.K, and J.K.conceptualized the research idea and conducted the research investigation. Y.-N.S. wrote the original draft. D.K and J.K. reviewed and edited the original draft. All of the authors have read the manuscript and confirmed its correctness and their contribution to its preparation. The author(s) read and approved the final manuscript.
Funding
This work is supported by a grant, 2017M3A9G5061816.
Declarations
Ethics approval and consent to participate
Informed consent was obtained from all patients. The study was approved by the ethnics board at Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea (2018–0976).
Competing interests
NO relevant conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18(2):89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35(6):672–688. doi: 10.1002/humu.22552. [DOI] [PubMed] [Google Scholar]
- 4.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 6.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 7.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 8.Ramos H, Soares MIL, Silva J, Raimundo L, Calheiros J, Gomes C, et al. A selective p53 activator and anticancer agent to improve colorectal cancer therapy. Cell Rep. 2021;35(2):108982. doi: 10.1016/j.celrep.2021.108982. [DOI] [PubMed] [Google Scholar]
- 9.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127(7):1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 11.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brachmann RK, Yu K, Eby Y, Pavletich NP, Boeke JD. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17(7):1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolova PV, Wong KB, DeDecker B, Henckel J, Fersht AR. Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations. EMBO J. 2000;19(3):370–378. doi: 10.1093/emboj/19.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hientz K, Mohr A, Bhakta-Guha D, Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2017;8(5):8921–8946. doi: 10.18632/oncotarget.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Liu N, Liu J, Liu Y, Zhang C, Long S, et al. Mutant p53 drives cancer chemotherapy resistance due to loss of function on activating transcription of PUMA. Cell Cycle. 2019;18(24):3442–3455. doi: 10.1080/15384101.2019.1688951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104(3):263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang H, Jiang AM, Qi CX. Aberrant nuclear p53 expression predicts hemizygous 17p (TP53) deletion in chronic lymphocytic leukemia. Am J Clin Pathol. 2010;133(1):70–74. doi: 10.1309/AJCPEPX1C7HHFELK. [DOI] [PubMed] [Google Scholar]
- 18.Hussain SP, Hofseth LJ, Harris CC. Tumor suppressor genes: at the crossroads of molecular carcinogenesis, molecular epidemiology and human risk assessment. Lung Cancer. 2001;34(Suppl 2):S7–15. doi: 10.1016/S0169-5002(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 19.Iggo R, Gatter K, Bartek J, Lane D, Harris AL. Increased expression of mutant forms of p53 oncogene in primary lung cancer. Lancet. 1990;335(8691):675–679. doi: 10.1016/0140-6736(90)90801-B. [DOI] [PubMed] [Google Scholar]
- 20.Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23(30):7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 21.Smith ND, Rubenstein JN, Eggener SE, Kozlowski JM. The p53 tumor suppressor gene and nuclear protein: basic science review and relevance in the management of bladder cancer. J Urol. 2003;169(4):1219–1228. doi: 10.1097/01.ju.0000056085.58221.80. [DOI] [PubMed] [Google Scholar]
- 22.Kobel M, Piskorz AM, Lee S, Lui S, LePage C, Marass F, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res. 2016;2(4):247–258. doi: 10.1002/cjp2.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih Ie M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24(9):1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 24.McCluggage WG, Soslow RA, Gilks CB. Patterns of p53 immunoreactivity in endometrial carcinomas: 'all or nothing' staining is of importance. Histopathology. 2011;59(4):786–788. doi: 10.1111/j.1365-2559.2011.03907.x. [DOI] [PubMed] [Google Scholar]
- 25.McManus DT, Yap EP, Maxwell P, Russell SE, Toner PG, McGee JO. p53 expression, mutation, and allelic deletion in ovarian cancer. J Pathol. 1994;174(3):159–168. doi: 10.1002/path.1711740304. [DOI] [PubMed] [Google Scholar]
- 26.Leitao MM, Soslow RA, Baergen RN, Olvera N, Arroyo C, Boyd J. Mutation and expression of the TP53 gene in early stage epithelial ovarian carcinoma. Gynecol Oncol. 2004;93(2):301–306. doi: 10.1016/j.ygyno.2004.01.043. [DOI] [PubMed] [Google Scholar]
- 27.Nenutil R, Smardova J, Pavlova S, Hanzelkova Z, Muller P, Fabian P, et al. Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J Pathol. 2005;207(3):251–259. doi: 10.1002/path.1838. [DOI] [PubMed] [Google Scholar]
- 28.Singer G, Stohr R, Cope L, Dehari R, Hartmann A, Cao DF, et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29(2):218–224. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 29.Singh N, Piskorz AM, Bosse T, Jimenez-Linan M, Rous B, Brenton JD, et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol. 2020;250(3):336–345. doi: 10.1002/path.5375. [DOI] [PubMed] [Google Scholar]
- 30.Sawada K, Momose S, Kawano R, Kohda M, Irie T, Mishima K, et al. Immunohistochemical staining patterns of p53 predict the mutational status of TP53 in oral epithelial dysplasia. Mod Pathol. 2022;35(2):177–185. doi: 10.1038/s41379-021-00893-9. [DOI] [PubMed] [Google Scholar]
- 31.Kim JE, Chun SM, Hong YS, Kim KP, Kim SY, Kim J, et al. Mutation burden and I index for detection of microsatellite instability in colorectal cancer by targeted next-generation sequencing. J Mol Diagn. 2019;21(2):241–250. doi: 10.1016/j.jmoldx.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary data. Detailed p53 mutational features in all 135 cases.