Abstract
Previously, we described a gene cluster of Enterococcus faecalis OG1RF that produced an antigenic polysaccharide when cloned in Escherichia coli. The polysaccharide antigen was not detectable in E. faecalis strains, however. Here, we show by reverse transcriptase-PCR that the 16 genes in this region are transcribed in OG1RF. Gene disruption of orfde4, encoding a putative glycosyl transferase, and orfde6, a putative dTDP-rhamnose biosynthesis gene, generated two OG1RF mutants. The mutants showed delayed killing and a higher 50% lethal dose in a mouse peritonitis model. In addition, two mucoid E. faecalis isolates from patients with chronic urinary tract infections were found to produce the polysaccharide antigen.
Polysaccharides (PS) of bacterial pathogens play important roles during infection. The capsule of Streptococcus pneumoniae is considered its most important virulence factor, since it enables the organism to persist in the host by conferring resistance to phagocytosis (6, 8). Capsular PS from Staphylococcus aureus types 5 and 8 were shown to induce cytokine release from human epithelial and endothelial cell lines (22). The serotype f PS (a rhamnose-glucose polymer) of Streptococcus mutans was found to stimulate the release of tumor necrosis factor α (23). Furthermore, being the dominant immunogens on the bacterial surface, some PS (e.g., the O antigens of gram-negative bacteria and capsules of streptococci and staphylococci) have been used as the basis for both vaccine development and serological typing of clinical isolates.
Enterococci are a leading cause of nosocomial infections and account for 5 to 15% of infective endocarditis in the United States (14), with most clinical isolates being Enterococcus faecalis. The development of multiple antibiotic resistance in enterococci in recent decades has posed a serious threat to effective therapy and raises awareness of the need for a better understanding of the pathogenicity of enterococci. Members of our group previously reported that a cloned gene cluster (epa, encoding proteins involved in the biosynthesis of an enterococcal PS antigen) of E. faecalis OG1RF produced a PS in Escherichia coli (27). The PS reacted with sera from four patients with enterococcal endocarditis. Sequence analysis showed that epa genes were similar to those involved in the biosynthesis and export of PS, including genes for rhamnose biosynthesis, glycosyl transferases, and ATP-binding cassette transporters. Insertion mutations in three of the genes, orfde4, orfde5, and orfde8, abolished immunoreactivity of the E. coli clone (27). However, we were not able to detect the PS antigen in several E. faecalis strains. Early studies of E. faecalis showed that some cell wall PS were antigenic and might be used for serological typing (1, 9, 19, 21). Neither the chemical compositions nor the genetic basis of these PS have been elucidated. In addition, Bottone et al. (2) recently described the first isolation of mucoid E. faecalis strains from patients with chronic urinary tract infections.
The biosynthesis of PS in many bacteria is regulated. The production of colanic acid in E. coli is elevated under certain growth conditions, such as in chemically defined media with high concentrations of phosphate and with high carbon-to-nitrogen ratios and temperatures below 25°C (24, 25). Alginate production in Pseudomonas aeruginosa is turned on in the lungs of cystic fibrosis patients (18). Since the synthesis of PS is a costly process, using ATP and sugars that would otherwise provide energy for cell activities, regulation is not surprising. Furthermore, since PS play specific roles in infection, it may also be advantageous to regulate their production during the stages of infection (25). Possibly the production of PS in E. faecalis is regulated.
In this study, we demonstrate that even though no antigen is detected, epa genes are transcribed in OG1RF with at least three transcriptional start sites. Furthermore, OG1RF mutants with disruptions in two of the genes showed a slightly higher 50% lethal dose (LD50) and a statistically significant delay in killing in a mouse peritonitis model. In addition, two recently reported mucoid E. faecalis strains showed positive reactions with specific antibodies against the PS produced by the E. coli clone.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. E. coli cells were grown in Luria-Bertani broth or on Luria-Bertani agar with appropriate antibiotics overnight at 37°C. Enterococci were grown in brain heart infusion (BHI) broth or on BHI agar (Difco) overnight at 37°C for routine purposes unless otherwise stated. The following antibiotics for selection of E. coli recombinants were used: chloramphenicol (CM) (25 μg/ml) and kanamycin (KM) (50 μg/ml). A concentration of 2,000 μg of KM per ml was used for selection of E. faecalis mutants. Serum samples were collected from patients with E. faecalis endocarditis infections and had high titers against E. faecalis strains (26). E1 and E2 are two mucoid E. faecalis isolates kindly provided by Edward J. Bottone at the Division of Infectious Diseases, the Mount Sinai Hospital, New York, N.Y. E1 is constitutively mucoid, while E2 is mucoid after incubation at room temperature but not at 37°C (E. J. Bottone, personal communication).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−φ80dlacZΔ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK−mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | |
| TX5159 | DH5α(pTEX5159) | 27 |
| E. faecalis | ||
| OG1RF | 13 | |
| TX5179 | OG1RF orfde4::Kmr | This study |
| TX5180 | OG1RF orfde6::Kmr | This study |
| E1 | Mucoid isolate from a patient with chronic urinary tract infection; constitutive mucoid phenotype | E. J. Bottone (2) |
| E2 | Mucoid isolate from a patient with chronic urinary tract infection; mucoid at room temperature | E. J. Bottone (2) |
| Plasmids and cosmids | ||
| pBeloBAC11 | Cosmid vector, F′ replicon, Cmr | H. Shizuya (26) |
| pTEX5159 | pBeloBAC11 with a 43-kb insert from OG1RF which contains a gene cluster for PS biosynthesis | 27 |
| pTEX4577 | pBluescript SK(−) with Ωkm2 inserted in the ScaI site, Kmr Aps | 15 |
| pTEX5177 | pTEX4577 containing an intragenic fragment of orfde4 inserted in the BamHI site | This study |
| pTEX5178 | pTEX4577 containing an intragenic fragment of orfde6 inserted in the BamHI site | This study |
DNA manipulations and transformation of E. coli.
DNA preparation, purification, restriction digestion, agarose gel electrophoresis, and ligation were performed using standard methods (17) or following the manufacturer's instructions, unless otherwise stated. Routine preparation of competent E. coli cells and transformation of DNA into E. coli were performed by a one-step procedure (5).
Preparation of total RNA.
The RNeasy Mini Kit (Qiagen, Santa Clarita, Calif.) was used to extract total RNA from E. faecalis, with slight modifications. All the solutions and utensils were treated with diethylpyrocarbonate (DEPC) to eliminate RNase. A single colony was inoculated into 5 ml of BHI broth and incubated at 37°C overnight with shaking. A fresh 5-ml culture was then inoculated with 50 μl of the overnight culture and incubated with shaking for 4 h. The cells were harvested and resuspended in 100 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.0]) containing 3 mg of lysozyme per ml. After incubation at room temperature for 20 min, 10 μl of 10% sodium dodecyl sulfate (SDS) and 350 μl of buffer RLT (Qiagen) were added and the mixture was vortexed vigorously. Subsequent steps were performed according to the instructions from the kit. After elution with DEPC-treated H2O, the sample was treated with 5 μl of RNase-free DNase (Promega) at 37°C for 30 min and then boiled for 10 min to inactivate the DNase. After phenol-chloroform extraction and ethanol precipitation, the sample was again resuspended in DEPC-treated H2O. The RNA concentration was measured in a spectrometer at 260 nm.
RNA RT-PCR.
Murine leukemia virus (MuLV) reverse transcriptase (RT) (Perkin-Elmer) was used to synthesize cDNA for PCR. The Perkin-Elmer protocol for reverse transcription, with modifications, was used. A 20-μl reaction mixture contained 4 μl of MgCl2 (25 mM), 2 μl of 10× PCR buffer II (Perkin-Elmer), 2 μl of each dNTP (10 mM), 0.5 μl of RNase inhibitor (20 U/μl), 1 μl of reverse primer (20 μM), 0.5 μl of MuLV RT (50 U/μl), 25 ng of total RNA, and DEPC-treated H2O. For the DNA control, 5 ng of OG1RF genomic DNA was used instead of total RNA. When MuLV RT was not used, the volume of DEPC-treated H2O was increased correspondingly. The reaction was carried out at 42°C for 30 to 60 min, and then the mixture was heated at 95°C for 5 min in a Perkin-Elmer 9600 thermal cycler. The subsequent PCR was performed according to the Perkin-Elmer protocol. To each reverse transcription reaction mixture, 4 μl of 25 mM MgCl2, 8 μl of 10× PCR buffer II, 65.5 μl of H2O, 1 μl of each primer (20 μM), and 0.5 μl of AmpliTaq DNA polymerase (5 U/μl) were added. PCR cycling was done with the following conditions: 24 to 30 cycles of 96°C for 1 min; 94°C for 30 s, 50 to 55°C (based on the melting temperatures of the primers) for 30 s, and 72°C for 30 s, followed by 10 min at 72°C and holding at 4°C. The primers used in RT-PCR are listed in Table 2.
TABLE 2.
Primers used for RT-PCR
| Primera | Sequenceb | Position in source sequencec |
|---|---|---|
| GW301 | 5′ATGGATCCTATGAGCATGCAAGAAAT | 1 of orfde4, upper strandd |
| GW302 | 5′CATTGAATAACACTTGATTATGACC | (3′) 220 of orfde4, lower strand |
| GW303 | 5′ATGGATCCTATGAAAGGAATTATTTT | 1 of orfde6, upper strandd |
| GW304 | 5′CATCTGGGCTTTCTTGTACCGC | (3′) 232 of orfde6, lower strand |
| GW305 | 5′ATTGAGACAACGCAAAGTCAC | (5′) 49 of orfde2, upper strand |
| GW306 | 5′CACTCCAAAATGCTGCTAAAG | (3′) 302 of orfde2, lower strand |
| GW307 | 5′ATGGATCCTATGCCTACAGCAGGAG | 1 of orfde3, upper strandd |
| GW308 | 5′CAACGTCACAAAATCTATCCG | (3′) 223 of orfde3, lower strand |
| GW316 | 5′CGACAACTCATTAAACGACC | (5′) 258 of orfde8, upper strand |
| GW317 | 5′GAACCCACGCTTTGACTAAC | (3′) 504 of orfde8, lower strand |
| GW318 | 5′CAAAAGCTACTTTGCCCTGCC | (5′) 15 of orfde9, upper strand |
| GW319 | 5′ACGTAAACAAGTGTCGCCCC | (3′) 337 of orfde9, lower strand |
| GW320 | 5′GAGCGGGAAATCGGAAAGTG | (5′) 494 of orfde10, upper strand |
| GW321 | 5′CAATCAGAATTGCTGAGCCGAC | (3′) 682 of orfde10, lower strand |
| GW322 | 5′ACATTTTGCGGACGATTGC | (5′) 47 of orfde10, upper strand |
| GW323 | 5′ATGATTTTAAGGCCAAGTATGC | (5′) 77 of orfde11, upper primer |
| GW324 | 5′GAATGAAAAACAAGTGGGTG | (3′) 369 of orfde11, lower strand |
| GW325 | 5′TAGTCCAACCAAAAAATCTTACC | (5′) 90 of orfde12, upper strand |
| GW326 | 5′CTCTTCCATCTCTTCTCGGG | (3′) 362 of orfde12, lower strand |
| GW329 | 5′AATGGAAACAGACGAGAGTACC | (5′) 21 of orfde14, upper strand |
| GW330 | 5′CGGTTTGACTTTTGCTAAGG | (3′) 323 of orfde14, lower strand |
| GW331 | 5′AGCCTACACGTTAAACATTGA | (5′) 45 of orfde15, upper strand |
| GW332 | 5′TTCCGCTAAAACTGCCTCC | (3′) 231 of orfde15, lower strand |
| GW333 | 5′TACAATCACTAAAGAAAGCCC | (5′) 91 of orfde16, upper strand |
| GW334 | 5′GATGATTGAAAATGAAAAGGG | (3′) 262 of orfde16, lower strand |
| GW337 | 5′GAGTCAAAGATTAGCGGTAGTC | (5′) 24 of orfde5, upper strand |
| GW338 | 5′TCAAGGAGCAACAATAATTCAC | (3′) 269 of orfde5, lower strand |
| GW365 | 5′GAAGGCTACATTTTATCAGAAC | (5′) 295 of orfde7, upper strand |
| GW366 | 5′CGCTTCAAATTCTTTTAAGG | (3′) 524 of orfde7, lower strand |
| GW367 | 5′TTAAGAAGCCTGTTTGTGG | (5′) 218 of orfde13, upper strand |
| GW368 | 5′TTGTTTTGTAGACTAATGGG | (3′) 414 of orfde13, lower strand |
| GW371 | 5′TAAGAGGGATTGGTAACTTG | (3′) 244 of orfde5_6, lower strand |
Only the primers that yielded the best results for each ORF are listed.
Underlining indicates a BamHI site which is not present in the template sequence. Sequences run from the 5′ end to the 3′ end.
The position of a primer on the upper strand is the position of the 5′ end of the primer on the upper strand, and the position of a primer on the lower strand is the position of the 3′ end of the primer on the complementary upper strand.
These primers contain altered sequences (ATGGATCC) at the 5′ end.
Sequencing of RT-PCR products and sequence comparison.
The RT-PCR products were gel purified by using the Qiaquick Gel Extraction kit (Qiagen). After elution with 30 μl of H2O, 10 μl of each DNA solution was subjected to electrophoresis through an 0.8% agarose gel to check the bands. For sequencing, 3.5 or 7 μl of each DNA solution was used as the template and the upper primers were used as sequencing primers. The sequences were compared to each corresponding open reading frame (ORF) by using the Bestfit program of Genetics Computer Group (Madison, Wis.).
Cloning intragenic DNA fragments of orfde4 and orfde6 into plasmid pTEX4577.
Plasmid vector pTEX4577 was derived from pBluescript SK(−) containing a KM resistance marker selectable in both E. coli and E. faecalis (15). Intragenic DNA fragments of orfde4 and orfde6 were PCR amplified from chromosomal DNA of OG1RF with primers containing the recognition sequence for BamHI, as follows: GW345, GW346, GW347, and GW348 (Table 3). Each PCR mixture contained 2 μl of OG1RF chromosome DNA (5.6 ng/μl), 10 μl of 10× PCR buffer I (containing 15 mM MgCl2; Perkin-Elmer), 2 μl of each dNTP (100 mM; Perkin-Elmer), 1 μl of each primer (20 μM), 77 μl of H2O, and 1 μl of AmpliTaq DNA polymerase (5 U/μl; Perkin-Elmer). PCR cycling was performed in a Perkin-Elmer 9600 thermal cycler, as follows: 96°C for 1 min; 94°C for 20 s, 55°C for 20 s, and 72°C for 20 s (30 cycles); 72°C for 10 min; and holding at 4°C.
TABLE 3.
Primers used for cloning the intragenic fragments of orfde4 and orfde6
| Primer | Length | Sequencea | 5′ Nucleotide position in corresponding ORF |
|---|---|---|---|
| GW345 | 26-mer | ATAGGATCCATTTATTACTATCCATC | 165 of orfde4 |
| GW346 | 22-mer | ATGGGATCCACCGTTTCAAAAG | 671 of orfde4, complementary strand |
| GW347 | 24-mer | AGAGGATCCACCACGTTTTGAAAG | 165 of orfde6 |
| GW348 | 24-mer | TAAGGATCCGTGTGTGCCTGTATC | 667 of orfde6, complementary strand |
Underlined letters indicate BamHI site; bold letters indicate sequences from orfde4 or orfde6. Sequences run from the 5′ end to the 3′ end.
After amplification, a small aliquot of each reaction mixture was analyzed by agarose gel electrophoresis to determine the size of each product. The remaining reaction mixtures were purified using the Qiaquick PCR purification kit (Qiagen). After BamHI digestion of the purified PCR products and subsequent purification by the Qiaquick PCR purification kit to remove the BamHI restriction enzyme, each fragment was ligated to BamHI-digested pTEX4577 with T4 ligase (New England Biolabs). The ligation mixtures were transformed into DH5α and plated on LB agar-KM (50 μg/ml) with isopropyl-β-d-galactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Four KM-resistant white colonies were picked after overnight incubation at 37°C and analyzed by restriction enzyme digestions. A plasmid containing the intragenic fragment of orfde4 was designated pTEX5177, and one containing the orfde6 intragenic fragment was designated pTEX5178. Plasmids pTEX5177 and pTEX5178 were further subjected to DNA sequencing to verify the inserts.
Generation of OG1RF mutants with disruptions in orfde4 and orfde6.
DNA from plasmids pTEX5177 and pTEX5178 was prepared from 250 ml of overnight cultures using the standard alkaline-SDS method and purified in a CsCl-ethidium bromide gradient, followed by dialysis against 10 mM Tris, pH 8.0, and precipitation with ethanol (17). The final DNA concentration was adjusted to 2.5 μg/μl with sterile H2O. Preparation of OG1RF competent cells and electroporation of DNA into OG1RF were carried out as described previously (15), with slight modifications. For each electroporation, 5 μg of DNA and 50 μl of OG1RF competent cells were mixed and added to an ice-cold 0.1-cm gap electrode Gene Pulser/E. coli Pulser Cuvette (Bio-Rad). One pulse was given at 1.25 V with 400-Ω resistance and 25-μF capacity. Immediately after the pulse, 1 ml of Todd-Hewitt broth (THB)–0.25 M sucrose was added to the cuvette. After incubation at 37°C for 2 h, the cell mixture was centrifuged and the cell pellet was resuspended in 0.2 ml of THB–0.25 M sucrose and plated onto THB–S0.25 M sucrose agar containing 2,000 μg of KM per ml. Colonies appearing within 36 h of incubation were streaked onto BHI agar with 2,000 μg of KM per ml. Clones that grew up after overnight incubation at 37°C were further analyzed by colony PCR and Southern blot analysis.
Colony PCR of OG1RF mutants.
A single colony was picked with a 200-μl sterile pipette tip and resuspended in 100 μl of sterile H2O. GW378 and GW379 are oligonucleotides directing outward DNA synthesis from the ends of the KM resistance gene of pTEX4577. Their sequences are as follows: GW378, 5′ GTGATATTCTCATTTTAGCC; and GW379, 5′ GACTTACTGGGGATCAAGCC. GW378 and GW379, and primers to orfde4 and orfde6, GW301 and GW303 (Table 2), and GW345, GW346, GW347, and GW348 (Table 3) were used in different combinations for PCR. PCR mixtures contained 1 μl of the cell suspension, 4 μl of PCR buffer I (Perkin-Elmer), 1 μl of a mixture of the four dNTPs (2.5 mM [each]), 4 μl of each primer (1 μM), 25.5 μl of sterile H2O, and 0.5 μl of AmpliTaq DNA polymerase (5 U/μl; Perkin-Elmer). PCR cycling was performed in a Perkin-Elmer 9600 thermal cycler with the following conditions: 96°C for 1 min; 94°C for 20 s, 55°C for 20 s, and 72°C for 1 min (30 cycles); 72°C for 10 min; and holding at 4°C. One of each of the colonies showing the expected PCR products was designated TX5179 or TX5180 for interruptions in orfde4 and orfde6, respectively, and was subjected to Southern blot analysis for verification.
Southern blot analysis of OG1RF mutants.
Genomic DNA from OG1RF, TX5179, and TX5180 was prepared based on a procedure described for E. coli (7). Briefly, enterococcal strains were grown in 5 ml of BHI broth with appropriate antibiotics at 37°C overnight. The cells were harvested, washed once with 5 ml of J buffer (0.1 M Tris-Cl, 0.1 M EDTA, and 0.15 M NaCl [pH 8.0]) and resuspended in 0.16 ml of J buffer. The suspension was then treated with 20 μl of freshly made lysozyme solution (10 mg/ml in 0.25 M Tris-Cl, pH 8.0) at 37°C for 20 min, followed by 1 μl of RNase A (34 mg/ml; Sigma) at 37°C for 10 min and then 70°C for 3 min. After this, 16 μl of 30% Sarkosyl was added and the mixture was incubated at 70°C for 20 min and then at 37°C for 1 h. Then 0.4 mg of proteinase K was added, and the solution was incubated at 37°C for 2 to 4 h. Another 0.4 mg of proteinase K was added at the end of the incubation, and the solution was dialyzed against 0.01 M Tris (pH 8.0)–0.01 M EDTA (pH 8.0)–0.15 M NaCl at 37°C overnight. The DNA solution was extracted once with phenol-chloroform and once with chloroform, and dialyzed against TE for several hours at room temperature. The concentration of DNA was then determined in a fluorometer.
For Southern blot analysis, 3 μg of genomic DNA was digested with EcoRI and XbaI and subjected to agarose gel electrophoresis. Blotting was performed using Hybond-N+ nylon membrane (Amersham) and 0.4 N NaOH solution according to the manufacturer's instructions. DNA of plasmids pTEX5177 and pTEX5178 was digested with BamHI, and the insert fragments were purified using the Qiaquick Gel Extraction kit (Qiagen). The Random Primers DNA Labeling System (GIBCO BRL) was used to label the insert fragments with [α-32P]dCTP (Amersham). Hybridization was carried out at 65°C according to manufacturer's instructions.
Determination of growth curves and stability of single crossover disruptions in TX5179 and TX5180.
Overnight cultures of TX5179, TX5180, and OG1RF were diluted 100-fold with fresh BHI broth and incubated at 37°C with shaking. Klett units were measured every hour until the stationary phase was reached. Aliquots (100 μl) of each culture were taken every hour, and 100 μl of 10−4 to 10−8 dilutions of each sample were plated onto BHI agar plates. Colonies were counted after incubation at 37°C for 24 h.
TX5179 and TX5180 were grown in BHI broth without antibiotics at 37°C with shaking. Aliquots (100 μl) of each culture were taken after 24 h of growth, and 10-fold serial dilutions were made. Dilutions (10−5-, 10−6, and 10−7-fold; of each sample in 100-μl volumes) were plated on BHI and BHI-KM (2,000 μg/ml) agar plates and incubated at 37°C overnight. Colonies were counted, and the numbers from BHI agar plates and BHI-KM agar plates were compared.
Examination of OG1RF mutants in a mouse peritonitis model.
Testing of the mutants was performed as described previously (20). OG1RF and the mutants were incubated in BHI broth overnight at 37°C with shaking. The cells were harvested by centrifugation, washed with chilled 0.85% saline solution, and resuspended in 20 ml of ice-cold 0.85% saline to reach a density of ∼5 × 1010 CFU/ml. A series of twofold dilutions were then made with 50% sterile rat fecal extract to obtain four to five inocula for injection into mice. Meanwhile, 10-fold serial dilutions were made and aliquots of 10−6, 10−7, and 10−8 dilutions were plated onto BHI agar plates with proper antibiotics to obtain the actual titer. Outbred ICR female mice, four to six weeks old (Harlan Dawley, Houston, Tex.) were used. Each was injected intraperitoneally with 1 ml of cell suspension with a syringe with a 25-gauge needle. Observations were made every 3 h, and the number of surviving mice was recorded.
The LD50 was determined as described by Reed and Muench (16). Survival statistics were computed using the Kaplan-Meier method and compared by log rank using StatView (Abacus Concepts, Inc., Berkeley, Calif.). Spleens were recovered from dead mice injected with the highest inocula, homogenized, and suspended in 3 ml of chilled 0.85% saline. Serial dilutions of each spleen suspension were plated onto BHI agar plates and, for TX5179 and TX5180, also onto BHI agar plates containing 2 mg of KM per ml. Colonies were counted after incubation for 24 h at 37°C.
Elution of antibodies specific to the PS.
Antibodies were eluted as described previously (27). Briefly, cell lysates of TX5159 were treated with proteinase K, applied to SDS-polyacrylamide gel electrophoresis gels, transferred to nitrocellulose membranes, and incubated with one of the patient serum samples. Elution was carried out using 100 mM glycine (pH 2.5); the solution was neutralized with a 1/10 volume of 1 M Tris (pH 8.0) and stored at −20°C until used.
Immunoblot analysis of bacterial colonies.
Colonies were inoculated onto LB or BHI agar plates containing antibiotics and incubated at 37°C or room temperature for 24 h. NitroBind nitrocellulose transfer membranes (Micron Separations, Inc., Westborough, Mass.) were used to lift the colonies. Subsequent steps were performed as described previously (27).
RESULTS
Detection of mRNA transcripts from epa genes in OG1RF.
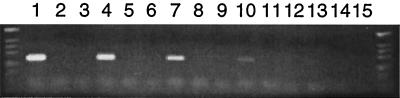
To determine if the ORFs in the epa region were transcribed, RT-PCR was performed. In the reactions using RNA as the template, only those reaction mixtures containing both RT and Taq polymerase yielded PCR products. Reaction mixtures containing only Taq polymerase and no RT (negative controls) did not yield PCR products (data not shown). This confirmed that the PCR products were derived from mRNA templates and not from contaminating DNA. In many cases a single RT-PCR band of the expected size was visualized after agarose gel electrophoresis (orfde3, orfde4, orfde5_6, orfde8, orfde9, orfde10, orfde12, orfde15, and orfde16). However, in some cases a band of the expected size as well as bands of other sizes were observed (orfde2, orfde5, orfde6, orfde7, orfde11, orfde13, and orfde14). Varying the RT-PCR conditions and designing new primers to different regions of the ORFs in the latter case improved the results with orfde5, orfde6, orfde11, and orfde13 but not with orfde2, orfde7, and orfde14. PCR controls using DNA instead of RNA (without RT) yielded only bands of the expected sizes.
To verify that the RT-PCR products observed on agarose gels were the expected products, each band of the correct size from orfde2 to orfde16 was gel purified and subjected to DNA sequencing. The DNA sequences were compared with the sequences of each corresponding ORF. The results showed that each product had the correct sequence (data not shown). This indicated that all 16 genes in the epa region are transcribed. To examine the nature of the bands of incorrect sizes, some of them were also gel purified and subjected to DNA sequencing. None of the sequences examined showed any homology to the corresponding ORFs, although sequences resembling those of the primers could be found at the ends. Thus, these bands do not represent RNAs derived from an epa gene.
Generation of OG1RF mutants.
Although PS could not be detected in E. faecalis, sera from patients with enterococcal infections contained anti-PS antibodies (27). If the PS was produced only during infection, it may be a virulence factor. To test this, mutations were made in orfde4 and orfde6, both necessary for PS production in E. coli. Intragenic fragments of orfde4 and orfde6 were cloned in pTEX4577, resulting in plasmids pTEX5177 and pTEX5178, respectively. In pTEX5177, the orfde4 intragenic fragment was in the same orientation as that of the T7 promoter, while in pTEX5178, the orfde6 fragment was in the orientation of the T3 promoter.
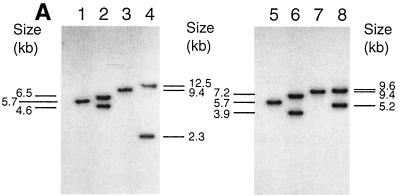
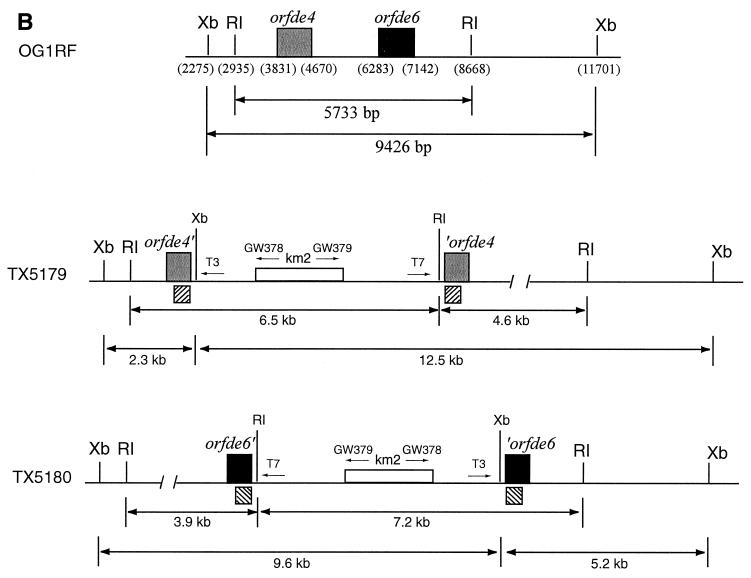
DNA of pTEX5177 and pTEX5178 was electroporated into OG1RF to generate single crossover mutants. Four resistant colonies for orfde4 and one for orfde6 were obtained. Colony PCR of these five clones produced bands of the expected sizes (data not shown). To verify the disruptions, one clone for orfde4 and one for orfde6, designated TX5179 and TX5180, respectively, were further analyzed by Southern blot hybridization (Fig. 1A). The results showed that the 5.7-kb EcoRI band in OG1RF was split into two bands of about 6.5 and 4.2 kb in TX5179 and into two bands of 7.2 and 3.9 kb in TX5180, while the 9.4-kb XbaI band was split into two bands of about 12.5 and 2.3 kb in TX5179 and into two bands of 9.6 and 5.2 kb in TX5180. These sizes are as expected from a single crossover at orfde4 or orfde6 (Fig. 1B).
FIG. 1.
(A) Southern blot analysis of OG1RF, TX5179, and TX5180. Genomic DNA of OG1RF, TX5179, and TX5180 was digested with EcoRI and XbaI, separated by electrophoresis in a 0.6% agarose gel and transferred onto a Hybond-N+ nylon membrane. The insert fragments of pTEX5177 and pTEX5178 were labeled with [32P]dCTP and used as probes. Lanes 1-4 were probed with the insert of pTEX5177, and lanes 5-8 were probed with that of pTEX5178. Hybridization was performed under high-stringency conditions at 65°C. Lane 1, OG1RF plus EcoRI; lane 2, TX5179 plus EcoRI; lane 3, OG1RF plus XbaI; lane 4, TX5179 plus XbaI; lane 5, OG1RF plus EcoRI; lane 6, TX5180 plus EcoRI; lane 7, OG1RF plus XbaI; lane 8, TX5180 plus XbaI. (B) Illustration of the local organizations of orfde4, orfde6, and the flanking regions in OG1RF, TX5179, and TX5180. RI, EcoRI; Xb, XbaI. The numbers in the parentheses indicate the nucleotide positions within the epa region in OG1RF. In TX5179, orfde4 was interrupted by pTEX5177, resulting in two partial copies, while in TX5180, orfde6 was interrupted by pTEX5178, resulting in two partial copies of orfde6. T3 and T7 are promoter regions on pTEX4577, and GW378 and GW379 are primers to the ends of the KM resistance gene. The arrows were not drawn to scale. The striped boxes underneath the disrupted ORFs indicate where the probes hybridized.
Growth curves of TX5179 and TX5180 and stability of single crossover disruptions.
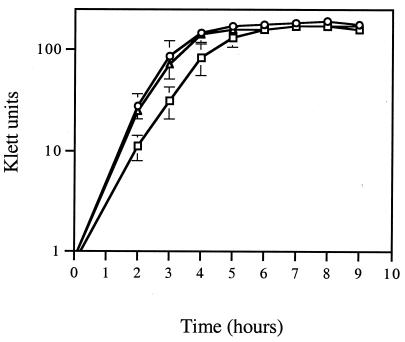
To determine if the growth of mutants TX5179 and TX5180 differed from that of wild-type OG1RF, the optical turbidity (in Klett units) and bacterial titers were measured (Fig. 2). TX5179 and TX5180 grew slightly slower than OG1RF and had slightly lower cell densities at stationary phase. The bacterial titer for OG1RF after growth for 24 h was (2.20 ± 0.28) × 109 CFU/ml, while the titers for TX5179 and TX5180 were (1.65 ± 0.13) × 109 CFU/ml and (1.80 ± 0.27) × 109 CFU/ml, respectively, similar to the turbidity of each culture (Fig. 2). Thus, the insertion mutations did not affect growth.
FIG. 2.
Growth curves of OG1RF, TX5179, and TX5180. Klett units were measured every hour until stationary phase. Circles, OG1RF; triangles, TX5180; boxes, TX5179.
The numbers of colonies of TX5179 and TX5180 on BHI agar plates were comparable to those on BHI-KM agar plates (1.24 × 109 versus 1.4 × 109 for TX5179 and 1.52 × 109 versus 1.45 × 109 for TX5180) after growth for 24 h in BHI medium, indicating that the insertion mutations in the chromosomes of TX5179 and TX5180 were stable.
Comparison of mRNA transcript levels in OG1RF and TX5180.
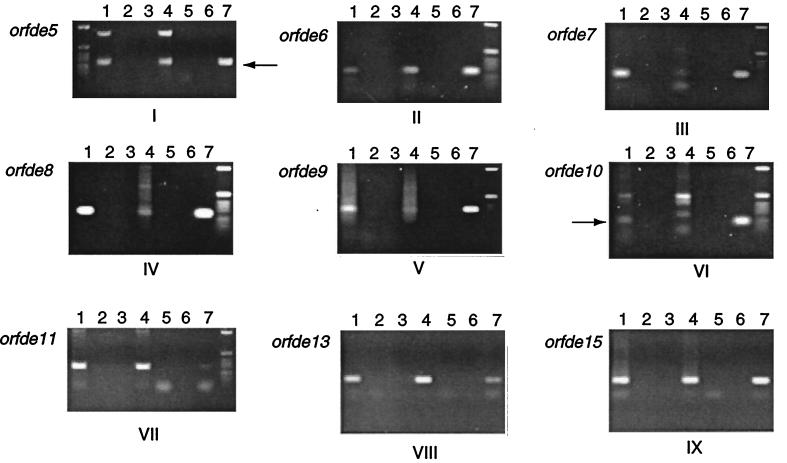
To determine the effect of gene disruption on the transcription of downstream genes, total RNA was extracted from OG1RF, TX5179, and TX5180 after 4 h of growth in BHI broth. Extraction from OG1RF and TX5180 yielded 300 to 400 ng of RNA per μl, whereas for unknown reasons, no RNA was obtained from TX5179 in two attempts. RT-PCR was performed on total RNA from OG1RF and TX5180, and transcription of a number of ORFs (orfde5 to orfde11, orfde13, and orfde15) was measured. After RT-PCR, the intensity of each band from TX5180 was compared to that of its corresponding band from OG1RF. The results indicated that the amount of mRNA transcripts of orfde7 to orfde10 in TX5180 was greatly reduced, while that of transcripts of orfde5, orfde6, orfde11, orfde13, and orfde15 was not (Fig. 3A). A band was observed for orfde6 because the region between the primers (GW303 and GW304) used in RT-PCR was not affected by the insertion. This suggests that there is a polar effect of the insertion on transcription of some epa genes.
FIG. 3.
(A). Comparison of the levels of mRNA transcripts of TX5180 and OG1RF. The target ORF is labeled next to each panel. The primers used for each ORF are as follows: panel I, GW337 and GW338; panel II, GW303 and GW304; panel III, GW365 and GW366; panel IV, GW316 and GW317; panel V, GW318 and GW319; panel VI, GW320 and GW321; panel VII, GW323 and GW324; panel VIII, GW367 and GW368; and panel IX, GW331 and GW332. The arrows next to the orfde5 and orfde10 panels indicate the specific bands (of expected sizes). The final concentration of each primer in the reaction was 400 nM for the reverse primers and 200 nM for the forward primers. A total of 25 ng of total RNA or 5 ng of chromosomal DNA was used as the template in each reaction. Lanes 1 to 3, OG1RF RNA; lanes 4 to 6, TX5180 RNA; and lanes 7, OG1RF chromosomal DNA. In addition, AmpliTaq DNA Polymerase (Taq) and/or MuLV RT, or neither, was used in each different reaction, as follows: lanes 1 and 4, RT and Taq; lanes 2, 5, and 7, Taq only; and lanes 3 and 6, neither. Reverse transcription was performed at 42°C for 40 min and then the mixture was heated at 95°C for 5 min. The PCR was carried out in a Perkin-Elmer 9600 thermal cycler using the following conditions: 94°C, 30 s; 55°C, 30 s; and 72°C, 30 s for 30 cycles. (B) RT-PCR of serial dilutions of OG1RF total RNA. Total RNA (25 ng/μl) was diluted 10-, 100-, 1,000-, and 10,000-fold with DEPC-treated H2O, and 1 μl of each dilution was used as the template for RT-PCR. GW301 and GW302 were used as primers. Lanes 1 to 3, undiluted RNA; lanes 4 to 6, 10-fold dilution; lanes 7 to 9, 100-fold dilution; lanes 10 to 12, 1,000-fold dilution; and lanes 13 to 15, 10,000-fold dilution. AmpliTaq DNA Polymerase (Taq) and/or MuLV RT, or neither, was used in each different reaction, as follows: lanes 1, 4, 7, 10, and 13, RT and Taq; lanes 2, 5, 8, 11, and 14, Taq only; and lanes 3, 6, 9, 12, and 15, neither. Reverse transcription and PCR were carried out using the same conditions as those described for panel A.
To examine the level of discrimination of RNA concentration in the RT-PCR system, total RNA (25 ng/μl) of OG1RF was diluted in a series of 10-fold dilutions and 1 μl of each dilution was subjected to RT-PCR with reaction and cycling conditions the same as those used for comparing RNAs of TX5180 and OG1RF. Primers GW301 and GW302 for orfde4 were used (Fig. 3B). The results showed that bands were observed with 25, 2.5, 0.25, and 0.025 ng of total RNA, but not with 2.5 pg of total RNA. The intensity of the bands decreased with the amount of RNA. Thus, under the conditions used, the system was able to discriminate a 10-fold difference in the amount of total RNA. Furthermore, the difference in the levels of intensity of bands in lanes 1 and 10 of Fig. 3B, indicative of a 1,000-fold difference in the RNA level, appeared to be of the same order of magnitude as the reduction in transcription of orfde7 to orfde9 in the mutant TX5180.
Examination of OG1RF mutants in a mouse peritonitis model.
To test if the disruptions in orfde4 and orfde6 had any effect on the virulence of OG1RF, TX5179, and TX5180 were examined in a mouse peritonitis model. In an initial experiment, a group of six mice was used for each of the four inocula made by twofold serial dilutions of overnight cultures with sterile rat fecal extract. Bacteria were recovered from the spleens of dead mice injected with the highest inocula of OG1RF, TX5179, or TX5180, and their titers on BHI agar plates and BHI-KM agar plates were determined. The titers for OG1RF, TX5179, and TX5180 on BHI plates were 3.6 × 108, 1.2 × 108, and 1.6 × 108 CFU/ml, respectively. The numbers of colonies from BHI plates were comparable to those from BHI-KM agar plates for both TX5179 and TX5180, indicating that the disruptions were stable in vivo.
The range of inocula used in this experiment did not allow the calculation of LD50, since the bacterial titers for OG1RF and the two mutants were not closely matched. However, when the data (not shown) were analyzed for the time to killing, TX5179 and TX5180 showed statistically significant delays in killing compared to that of OG1RF (Table 4, rows 1 and 2).
TABLE 4.
Statistical significance of the delayed killing of mice by TX5179 and TX5180 compared with OG1RF
| Strain | No. of CFU/mla
|
Pb | No. of CFU/mla
|
Pb | ||
|---|---|---|---|---|---|---|
| OG1RF | TX5179 | OG1RF | TX5180 | |||
| 1 | 2.3 × 108 (6) | 3.3 × 108 (6) | 0.0034 | 2.3 × 108 (6) | 3.2 × 108 (6) | 0.0178 |
| 2 | 2.3 × 108 (6) | 6.6 × 108 (6) | 0.0009 | |||
| 3 | 3.5 × 108 (6) | 4.0 × 108 (16) | <0.0001 | 3.5 × 108 (6) | 5.0 × 108 (16) | 0.0002 |
Size of the inoculum used for injection into the peritoneum of each mouse. Numbers in parentheses are the number of mice used for each inoculation. The time-to-death values for wild-type and mutants are shown in Fig. 4.
The P value was calculated in the StatView program using the log rank method. A P value of less than 0.05 is statistically significant.
The LD50 values calculated for the three strains in this experiment were as follows (in CFU per milliliter): for OG1RF, 2.4 × 108; for TX5179, 6.0 × 108; and for TX5180, 5.0 × 108.
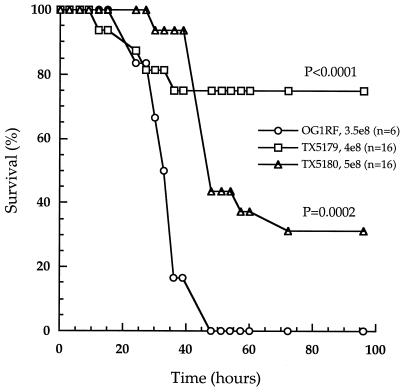
To further examine the effect of the mutations on the survival of mice, sixteen mice were used for each of the two mutants, with inoculum sizes of 4 × 108 CFU for TX5179 and 5 × 108 CFU for TX5180 (Table 4, row 3). Six mice were used for OG1RF at 3.5 × 108 CFU. The survival of the 16 mice injected with TX5179 and TX5180 was compared with that of the six mice injected with OG1RF. The percentage of surviving mice was plotted against time (Fig. 4). The two mutants resulted in a higher percentage of surviving mice and delayed killing compared with that of OG1RF. The P values were <0.0001 for TX5179 versus OG1RF and 0.0002 for TX5180 versus OG1RF, consistent with the results obtained in the first experiments.
FIG. 4.
Percentages of surviving mice injected with OG1RF, TX5179, or TX5180 over time. Circles, OG1RF; squares, TX5179; triangles, TX5180. The inoculum sizes and the number of mice used for each strain were as follows: OG1RF, 3.5 × 108 CFU (6 mice); TX5179, 4.0 × 108 CFU (16 mice): and TX5180, 5.0 × 108 CFU (16 mice). See Table 4 for statistical significance.
To compare the LD50 values of OG1RF and the mutants, the experiment was repeated using a slightly different range of inocula and six mice for each inoculum. The two mutants showed higher LD50 than OG1RF (Table 4). These results demonstrate that genes in the epa locus contribute to virulence in the mouse peritonitis model.
Reaction of two mucoid E. faecalis isolates with anti-PS antibody.
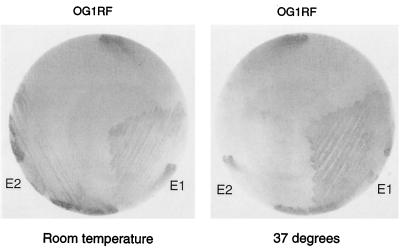
Two mucoid E. faecalis isolates from patients with chronic urinary tract infections (2), E1 and E2, were tested for reaction with anti-PS antibody. E1, E2, and OG1RF were streaked on BHI agar plates and incubated at room temperature or 37°C for 24 h. The colonies were lifted onto nitrocellulose membranes for immunoblotting with the antibody eluted from the E. coli clone expressing the PS antigen (Fig. 5). The results showed that E1 grown at both room temperature and 37°C reacted, E2 reacted only after incubation at room temperature, and, as found previously, OG1RF did not react at either temperature. The temperature dependence of the immunoreactivity paralleled that of mucoidy, suggesting that the PS being overproduced in the mucoid strains is in fact the antigen.
FIG. 5.
Immunoblot of two mucoid E. faecalis isolates with the eluted antibody. Strains were grown on BHI agar for 24 h at either room temperature or 37°C and then lifted onto nitrocellulose filters. Immunoblotting was carried out with the eluted antibody as described in Materials and Methods.
DISCUSSION
In a previous study, members of our group described a PS antigen biosynthesis gene cluster (epa) of E. faecalis (27). In this report we demonstrate that although the epa antigen is not detectable in laboratory-grown E. faecalis cells, the genes within epa are transcribed in E. faecalis and disruptions in two of the genes (orfde4 and orfde6) caused a statistically significant delay in killing in a mouse peritonitis model.
The E. coli clone containing the epa gene cluster, TX5159, was previously found to react with sera collected from four endocarditis patients infected with E. faecalis in different regions in the United States, suggesting that the PS was produced in all of these patients (27). However, we were able to detect the antigen neither in E. faecalis strain OG1RF under different growth conditions (including incubation in the peritonea of mice) nor in several other E. faecalis clinical isolates or laboratory strains. To determine if the genes within epa were transcribed, we performed RT-PCR analysis of the total RNA of OG1RF grown in BHI medium for 4 h. Each band of the correct size was confirmed by sequencing the purified PCR products. The results indicated that the orfde2 to orfde16 genes were transcribed. However, it is not clear whether the amount of mRNA represents a low basal level that must be induced for the production of the PS or whether this is sufficient mRNA for PS production.
To study the effects of mutations in this region, two mutants, TX5179 (in orfde4) and TX5180 (in orfde6), were generated. Members of our group previously showed that insertions in orfde4 abolished antigen production in the E. coli clone TX5159 (27). Transposon mutagenesis also indicated that one of the putative rhamnose biosynthesis genes, orfde8, was required for the immunoreactivity of TX5159, and orfde6 is predicted to encode the first enzyme in this rhamnose biosynthesis pathway.
Since the mutants were generated via single crossover events, resulting in two partial copies of orfde4 or orfde6, the stability of the crossovers was determined. The disruptions appeared to be stable, since very few cells became KM sensitive after growth in plain BHI broth for 24 h or incubation in the peritonea of mice for at least 12 h. The mutants grew slightly less well than OG1RF; however, the difference appeared to be minor.
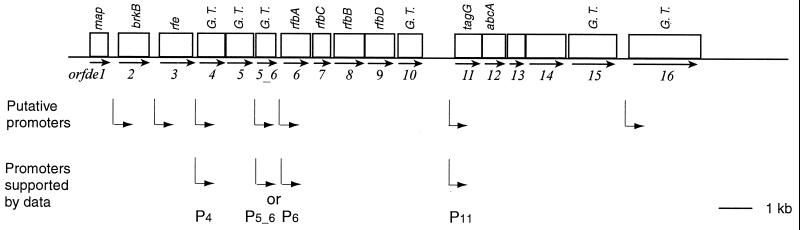
To examine if the disruptions had polar effects on transcription, RT-PCR was performed to measure the mRNA levels from several ORFs. The transcription of orfde7 to orfde10 in TX5180 was drastically reduced, while that of orfde11, orfde13, and orfde15 was not (Fig. 3A). This suggested that the orfde6 to orfde10 genes were in one transcriptional unit and that their transcription was independent of that of orfde11 and other downstream genes (Fig. 6). This is consistent with the results of DNA sequence analysis, which suggested that a promoter-like sequence was found upstream of orfde11 (27). In the study previously conducted by members of our group, analysis of Tn7 transposon mutants indicated that insertions in orfde3 or between orfde3 and orfde4 did not affect the immunoreactivity of TX5159, and an insertion in orfde4 was complemented by a plasmid, pTEX5175, that contains only orfde4 and orfde5. These results suggested that there should be transcriptional start sites upstream of orfde4 and orfde6 (27). Thus, epa should contain at least three transcriptional start sites (Fig. 6).
FIG. 6.
Hypothetical transcriptional organization of the epa region. Line 1, open reading frames in epa (open boxes) and their direction of transcription (arrows below the boxes). Line 2, putative promoters identified by homology to the E. coli −35 and −10 sequences recognized by ς70 (27); vertical lines indicate the position of each putative promoter, and arrows indicate their directions. Line 3, summary of possible promoters in epa that were supported by experimental data. P5_6 and P6 cannot be distinguished based on current data.
To test if the disruptions had any effect on the virulence of OG1RF, the two mutants were examined in a mouse peritonitis model. TX5179 and TX5180 appeared to be less virulent, as indicated by a high LD50 as well as to have delayed killing compared to that of OG1RF. The attenuation in the virulence of TX5179 and TX5180 in the mouse peritonitis model could reflect an involvement of this region in E. faecalis virulence. Considering the slightly slower growth of the mutants, the P values were calculated using higher initial inocula of the mutants versus OG1RF. In the case of TX5179, for example, a statistically significant P value (0.0034) was obtained when results from TX5179 with an inoculum of 6.6 × 108 CFU were compared with those from OG1RF with an inoculum of only 2.3 × 108 CFU. This suggested that the delayed killing was not due to the differences in growth rates.
The recent report of four mucoid encapsulated E. faecalis isolates from patients with chronic urinary tract infections by Bottone et al. (2) shed new light on this project. Two of the isolates, E1 and E2, were examined and showed positive reactions with the anti-PS antibody. E1 reacted after incubation at both room temperature and 37°C, while E2 reacted only after incubation at room temperature. These results suggested that the antigenic PS was overproduced in the mucoid strains.
The reasons that no antigenic PS was detected in wild-type E. faecalis remain to be determined. One possibility is that it is expressed only under certain conditions. In P. aeruginosa, the production of alginate is suppressed by genes in the muc locus until the organism invades the lungs of cystic fibrosis patients. The muc region was mapped to a separate locus from that of the biosynthesis genes (3, 4, 11, 12, 18). Therefore, it is possible that a negative regulatory region is also present in E. faecalis at a different location from epa and suppresses the PS production in E. faecalis. Under certain conditions, such as chronic infections or attachment to heart valves, the suppression could be turned off. The PS could then be produced, generating antibodies in the patients. By this model, in E. coli TX5159 carrying the cloned epa locus, the negative control region is absent or not functional and PS production is permitted. Since the orfde2 to orfde16 genes were transcribed in OG1RF grown in BHI medium, it is not clear if the level of transcription of these genes reflects a basal level produced under the negative control. Alternatively, the epa locus could be posttranscriptionally regulated or the target(s) of regulation could be outside of epa.
A second possibility is that some function(s) required for the synthesis or export of the PS is not present in the strains examined. For example, the capsulation gene cluster (cap) of Haemophilus influenzae type b is on a compound transposon flanked by two insertion element-like (IS1016) elements. The type b cap locus consists of a duplication of cap with an unduplicated small bridge region containing bexA between the two large repeats. Recombination between the large repeats can result in the deletion of bexA, leading to a capsule-deficient phenotype (10). If this were the case in E. faecalis, the lost function(s) must have been complemented in TX5159 by E. coli genes outside the rfb region (E. coli rfb was previously shown not to affect antigen production). A variation of this possibility is that there is a hot spot in this region for transposon or conjugative plasmid integration and/or excision. In support of this hypothesis, comparison of the OG1RF gene cluster with preliminary genome sequences of E. faecalis strain V583 recently released by the Institute of Genome Research showed that all epa genes are present in V583 but that the V583 orfde10 is interrupted by conjugative transposon-like sequences (unpublished data). We think this explanation is less likely, however, since the insertion mutations in the epa locus of OG1RF led to reduced virulence, suggesting that the PS functions during infections.
The possibility that the antigenic PS is produced in OG1RF but is different or modified compared to the antigen made in E. coli also cannot be excluded. The antibodies generated during infection may be directed against a degradation product of the native molecule. The partial PS may be the antigen made in E. coli. In the mucoid strains, the overproduction of the native PS may also provide enough of the partial molecule to be detected, whereas in the other E. faecalis strains tested, there would not be enough to detect.
In conclusion, we report here that genes orfde2 to orfde16 in the epa region were transcribed in OG1RF and that disruptions in orfde4 and orfde6 caused statistically significant delayed killing and a higher LD50 in a mouse peritonitis model. There are at least three transcriptional start sites hypothesized in epa and genes orfde6 to orfde10 are in one transcriptional unit. In view of the regulation of the production of many bacterial PS and the fact that E2 reacted only with the eluted antibody after incubation at room temperature, it is possible that E. faecalis also has adopted ways of manipulating the production or display of the antigenic PS. Comparisons of the genetic organizations of the epa region in OG1RF and in the mucoid E. faecalis strains and of levels of gene expression and isolation of the PS material and analysis of its chemical composition will yield information invaluable for addressing many of the issues discussed above. Further study on the involvement of epa in virulence should have important implications for prevention and treatment of enterococcal infections.
ACKNOWLEDGMENTS
We are grateful to Edward J. Bottone at the Division of Infectious Diseases, the Mount Sinai Hospital, for providing us with the mucoid strains, and to Robert Rakita and Bei Wang of the Division of Infectious Diseases, Department of Medicine, and Monjula Chidambaram of the Department of Microbiology and Molecular Genetics at the University of Texas Medical School at Houston for helpful advice and technical support.
This work was supported by grant AI 33516 from the NIH Division of Microbiology and Infectious Diseases to B.E.M.
REFERENCES
- 1.Bleiweis A S, Krause R M. The cell walls of group D streptococci. I. The immunochemistry of the type 1 carbohydrate. J Exp Med. 1965;122:237–249. doi: 10.1084/jem.122.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottone E J, Patel L, Patel P, Robin T. Mucoid encapsulated Enterococcus faecalis: an emerging morphotype isolated from patients with urinary tract infections. Diagn Microbiol Infect Dis. 1998;31:429–430. doi: 10.1016/s0732-8893(98)00027-3. [DOI] [PubMed] [Google Scholar]
- 3.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encoding homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher J C, Schurr M J, Yu H, Rowen D W, Deretic V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 1997;143:3473–3480. doi: 10.1099/00221287-143-11-3473. [DOI] [PubMed] [Google Scholar]
- 5.Chung C T, Niemela S Z, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVelasco E A, Verheul A F M, Verhoef J, Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebel-Tsipis J, Botstein D, Fox M S. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71:433–448. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- 8.Garcia E, Lopez R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 9.Jelinkova J, Rotta J. Identification and typing of enterococci. Methods Microbiol. 1978;12:199–220. [Google Scholar]
- 10.Kroll J S. The genetics of encapsulation in Haemophilus influenzae. J Infect Dis. 1992;165(Suppl.):93–96. doi: 10.1093/infdis/165-supplement_1-s93. [DOI] [PubMed] [Google Scholar]
- 11.Martin D W, Schurr M J, Mudd M H, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 12.Martin D W, Schurr M J, Mudd M H, Govan J R W, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin X, Singh K V, Xu Y, Weinstock G M, Murray B E. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob Agents Chemother. 1998;42:2882–2888. doi: 10.1128/aac.42.11.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Schurr M J, Martin D W, Mudd M H, Deretic V. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J Bacteriol. 1994;176:3375–3382. doi: 10.1128/jb.176.11.3375-3382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharpe M E, Shattock P M F. The serological typing of group D streptococci associated with outbreaks of neonatal diarrhoea. J Gen Microbiol. 1952;6:150–165. doi: 10.1099/00221287-6-1-2-150. [DOI] [PubMed] [Google Scholar]
- 20.Singh K V, Qin X, Weinstock G M, Murray B E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 21.Smyth J, Matthews H, Halpenny M K, Brandis H, Colman G. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. J Med Microbiol. 1987;23:45–54. doi: 10.1099/00222615-23-1-45. [DOI] [PubMed] [Google Scholar]
- 22.Soell M, Diab M, Haan-Archipoff G, Beretz A, Herbelin C, Poutrel B, Klein J. Capsular polysaccharide types 5 and 8 of Staphylococcus aureus bind specifically to human epithelial (KB) cells, endothelial cells and monocytes and induce release of cytokines. Infect Immun. 1995;63:1380–1386. doi: 10.1128/iai.63.4.1380-1386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soell M, Lett E, Holveck F, Scholler M, Wachsmann D, Klein J P. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen and mannan binding protein inhibits TNF-alpha release. J Immunol. 1995;154:851–860. [PubMed] [Google Scholar]
- 24.Stout V. Regulation of capsule synthesis includes interactions of the RcsC/RcsB regulatory pair. Res Microbiol. 1994;145:389–392. doi: 10.1016/0923-2508(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 25.Whitfield C, Valvano M A. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Jiang L, Murray B E, Weinstock G M. Enterococcus faecalis antigens in human infections. Infect Immun. 1997;65:4207–4215. doi: 10.1128/iai.65.10.4207-4215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Murray B E, Weinstock G M. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]